Abstract
We have examined the subcellular localization of the KH-type splicing regulatory protein (KSRP). KSRP is a multidomain RNA-binding protein implicated in a variety of cellular processes, including splicing in the nucleus and mRNA localization in the cytoplasm. We find that KSRP is primarily nuclear with a localization pattern that most closely resembles that of polypyrimidine tract binding protein (PTB). Colocalization experiments of KSRP with PTB in a mouse neuroblastoma cell line determined that both proteins are present in the perinucleolar compartment (PNC), as well as in other nuclear enrichments. In contrast, HeLa cells do not show prominent KSRP staining in the PNC, even though PTB labeling identified the PNC in these cells. Because both PTB and KSRP interact with the c-src transcript to affect N1 exon splicing, we examined the localization of the c-src pre-mRNA by fluorescence in situ hybridization. The src transcript is present in specific foci within the nucleus that are presumably sites of src transcription but are not generally perinucleolar. In normally cultured neuroblastoma cells, these src RNA foci contain PTB, but little KSRP. However, upon induced neuronal differentiation of these cells, KSRP occurs in the same foci with src RNA. PTB localization remains unaffected. This differentiation-induced localization of KSRP with src RNA correlates with an increase in src exon N1 inclusion. These results indicate that PTB and KSRP do indeed interact with the c-src transcript in vivo, and that these associations change with the differentiated state of the cell.
INTRODUCTION
The small 18-nucleotide N1 exon is included in the c-src mRNA in neurons but is skipped in nonneuronal cells (Levy et al., 1987; Martinez et al., 1987). This regulation is mediated by regulatory sequences both flanking and within the N1 exon. A core regulatory element downstream of N1 is called the downstream control sequence, or DCS (Black, 1991, 1992; Chan and Black, 1995; Modafferi and Black, 1997, 1999). The DCS is comprised of both positive and negative regulatory elements (Chan and Black, 1995; Modafferi and Black, 1997). A CUCUCU element is bound by the polypyrimidine tract binding protein (PTB), which is required for splicing repression. In neuronal cells, this element is bound by the neuronal PTB homolog, nPTB (Chou et al., 2000; Markovtsov et al., 2000). Adjacent to the PTB binding site is the element UGCAUG that is crucial for enhancer activity. This element is bound by the KH-type splicing regulatory protein (KSRP) and other proteins (Min et al., 1997). KSRP cooperatively assembles onto a short DCS RNA with the proteins heterogeneous nuclear ribonucleoprotein (hnRNP) H and either nPTB or PTB in vitro (Min et al., 1997; Markovtsov et al., 2000). The DCS was originally characterized as a splicing enhancer, implying a positive role for the KSRP in splicing. However, PTB and its binding site in the DCS are required for N1 splicing repression in nonneural cells. Thus, roles for KSRP in splicing repression or derepression are also possible.
KSRP and its related proteins have a common domain structure, including conserved N- and C-terminal domains flanking a central region of four KH-type RNA-binding domains. These proteins have been implicated in a variety of nuclear and cytoplasmic processes. KSRP is a human homolog of the Drosophila splicing factor P-element somatic inhibitor (PSI). PSI acts as a splicing inhibitor by binding to a repressor element for the P-element third intron (Siebel et al., 1994). The C-terminal domain of PSI, which is similar to the C-terminal domain of KSRP, interacts with the U1 70K protein (Labourier et al., 2001). Mutation of this domain leads to splicing defects in transgenic flies (Labourier et al., 2001, 2002) In humans, there are at least two other proteins, in addition to KSRP, that are highly related to PSI across its multiple domains. One of these KSRP paralogs, the fuse-binding protein (FBP), is implicated in the transcriptional regulation of the c-myc gene (Davis-Smyth et al., 1996; Liu et al., 2000). KSRP (also called FBP2), and the other paralog FBP3 both show similar transcriptional activation properties to FBP (Duncan et al., 1996; Davis-Smyth et al., 1996). In other vertebrates, KSRP or related proteins have been implicated in mRNA localization. A rat KSRP homolog, MARTA1, binds to a dendritic targeting element of the microtubule-associated protein 2 mRNA (Rehbein et al., 2000, 2002). A chicken homolog, zipcode binding protein 2 (ZBP2), binds to a localization element within the β-actin mRNA (Gu et al., 2002). A Xenopus homolog, VgRBP71, is implicated in the localization of the VG1 mRNA (Kroll et al., 2002). KSRP was also copurified with the RNA editing complex for apolipoprotein B mRNA (Lellek et al., 2000). Finally, KSRP interacts with the AU-rich elements that destabilize fos mRNA and with the human exosome complex in vitro. Depletion/addback experiments in a cytoplasmic in vitro degradation system support a role for the protein in targeting mRNAs for degradation (Chen et al., 2001).
A number of nonmembrane-bound compartments within the mammalian cell nucleus can be identified by the presence of specific proteins and RNAs. These compartments include the nucleolus, nuclear speckles, Cajal bodies (coiled bodies or CBs), gems, Sam68 domains, promyelocytic leukemia (PML) bodies, paraspeckles, and perinucleolar compartments (PNCs) (Matera, 1999; Carmo-Fonseca, 2002; Lamond and Spector, 2003). PNCs are subnuclear compartments ranging in size from 0.5 to 1 μm that were originally defined by the presence of the Y small RNAs and other Pol III transcripts (Matera et al., 1995). PNCs are found at the periphery of the nucleolus and also contain the PTB protein (Ghetti et al., 1992; Huang et al., 1998). Like KSRP, PTB is a protein that is thought to be involved in both splicing regulation in the nucleus and additional functions in the cytoplasm (Valcarcel and Gebauer, 1997; Wagner and Garcia-Blanco, 2001). Recently, several additional factors have been identified in the PNC or similar perinucleolar structures, but the function of the PNC remains a mystery (Lee et al., 1996; Timchenko et al., 1996; Huang et al., 1998; Pinol-Roma et al., 1989; Huang, 2000; Huttelmaier et al., 2001; Wang et al., 2003).
Given the variety of nuclear and cytoplasmic functions ascribed to KSRP, we wanted to characterize its localization within cells relative to other proteins implicated in N1 exon regulation and relative to the src RNA transcript. We find that KSRP localizes strongly with PTB to the PNC in a neuroblastoma cell line, but this PNC-localized KSRP is much reduced in HeLa cells. The src pre-mRNA, although coincident with some of the nuclear PTB foci, is not in the PNC. Finally, the localization of KSRP with these src RNA foci requires differentiation of the neuroblastoma cells in culture.
MATERIALS AND METHODS
Plasmid Construction
All DNA constructs were made using standard cloning procedures. The green fluorescent protein (GFP)-KSRP fusion protein and GFP-KSRP deletion mutants were constructed by cloning full-length or fragments of KSRP, polymerase chain reaction (PCR)-amplified with Pfx Platinum DNA polymerase (Invitrogen, Carlsbad, CA) and specific primers, into the BglII and SalI sites of pEGFPC1 (BD Biosciences Clontech, Palo Alto, CA). All clones were confirmed by sequencing.
Cell Culture and Transfection
Human cervical carcinoma (HeLa) and mouse neuroblastoma (N1E-115) cells were cultured in DMEM or DMEM-F12, respectively, supplemented with 10% fetal calf serum (Invitrogen). For splicing assays, cells were grown on six-well dishes in monolayers. For localization experiments cells were grown either on two-well chamber slides (Nalge Nunc International, Naperville, IL) or on acid-washed coverslips in six-well dishes. Experiments done in the presence of RNA polymerase inhibitors contained 50 μg/μl α-amanitin and were incubated for 5 h before fixation (Kedinger et al., 1970; Lindell et al., 1970). For differentiation assays, N1E-115 cells were grown to confluence and then subcultured 24 h before the addition of 2% dimethyl sulfoxide (DMSO) directly to the culture medium (Kimhi et al., 1976). Media was changed daily, and cells were harvested after 120 h.
Transfections were performed using either Superfect (QIAGEN, Valencia, CA) or LipofectAMINE 2000 (Invitrogen). Briefly, N1E-115 cells were grown to 40-60% confluence and transiently transfected with Superfect as per the manufacturer's protocol. HeLa cells were grown to 80-90% confluence and transiently transfected with LipofectAMINE 2000 (Invitrogen) as per the manufacturer's protocol. Cells were harvested for analysis 36-48 h posttransfection.
Antibodies
A mouse mAb to KSRP (Ab5) was raised to the full-length protein and used at a dilution of 1:1000. A rabbit polyclonal antibody was raised against a peptide within the N terminus of PTB (PTB-NT) and used at a dilution of 1:1000. A rabbit polyclonal antibody against the GFP protein (1:1000) was a gift from Peter Tontonoz (University of California, Los Angeles). The Nova antibody was a gift from Robert Darnell (Rockefeller University). The anti-coilin and anti-PML antibodies (Santa Cruz Biotechnology, Santa Cruz, CA) were both used a dilution of 1:1000. The anti-fibrillarin antibody (Santa Cruz Biotechnology) was used at a dilution of 1:100. Horseradish peroxidase-conjugated goat-anti-mouse or goat anti-rabbit (Zymed Laboratories, South San Francisco, CA) secondary antibodies were used at a dilution of 1:2000.
Western Blotting
Cells were lysed in low salt lysis buffer (1.0% NP-40, 50 mM Tris 8.0), and equal amounts of protein were loaded onto a 10% SDS-PAG. Proteins were transferred to nitrocellulose, blocked with 5% dry milk in 1× phosphate-buffered saline (PBS), pH 7.4, and probed with the appropriate primary antibody for 1 h at room temperature. Horseradish peroxidase-conjugated goat anti-mouse or goat anti-rabbit secondary antibodies (1:2000) were hybridized for 1 h at room temperature, and proteins were visualized using Supersignal West Pico chemiluminescent substrate (Pierce Chemical, Rockford, IL).
Reverse Transcription (RT)-PCR
Cytoplasmic RNA was isolated via the NP-40 lysis method followed by proteinase K treatment, acid-phenol extraction, and ethanol precipitation (Modafferi and Black, 1999). RT-PCR was initiated by annealing 1 μg of cytoplasmic RNA to 50 pmol random hexamers in 10-μl total volume at room temperature for 15 min in 1× first strand buffer (Invitrogen). These reactions were subjected to reverse transcription in 20-μl total volume at room temperature for 15 min and then at 42°C for 1 h. Two microliters of the resulting cDNA was used in a 50-μl PCR reaction with 50 pmol of each of the following primers: for endogenous src, a mixture of 1/10 5′-32P-labeled E-src-2 (5′-TGGATGGAGTCGGAGGGCGC-3′) with 9/10 cold E-src-2, and E-src-3 (5′-CTGTCCTTCAAGAAAG GGGAGC-3′) (Black, 1991); and for endogenous GAPDH, a mixture of 1/10 5′-32P-labeled GDH1 (5′-TCTTCACCACCATGGAGAAG) with 9/10 cold GDH1, and GDH2 (5′-ACCAAAGTTGTCATGGATGAC). PCR protocols included 25 or 30 cycles (94°C for 30 s, 48°C for 30 s, 72°C for 30 s) for src and 25 cycles (94°C for 30 s, 55°C for 30 s, 72°C for 30 s) for GAPDH. Then, 2.5 μl of the PCR reaction was run on a 6% denaturing PAG and exposed to a PhosphorImager screen. Bands were quantified using ImageQuant (Amersham Biosciences, Boston, MA) software, and percentage of exon inclusion was determined [(cpm exon included product/(cpm exon included product + exon skipped product)) × 100].
Preparation of Probes for Fluorescence In Situ Hybridization (FISH)
Src RNA probes to be used for fluorescence in situ hybridization were in vitro transcribed in the presence or absence of modified UTP. Briefly, template plasmid DNA was linearized with the appropriate restriction enzyme for 15 min at 37°C. Then, 200 ng of linearized template was added to the in vitro transcription reaction mix containing 2 μl of a 10 mM NTP mix, 3 μl of 10× transcription buffer (NEB), 3 μl of 0.1 M dithiothreitol, 0.5 μlofT7orSP6RNA polymerase (NEB), and distilled H2O up to 30 μl. RNAs to be labeled were transcribed in the presence of digoxigenin-UTP (DIG-UTP) (Roche Diagnostics, Indianapolis, IN) or fluorescein-UTP (Roche Diagnostics). All RNAs were run on 4% denaturing PAG, visualized by UV shadowing, excised from the gel, incubated overnight at 37 degrees in PCA buffer (20 mM Tris pH 7.5, 2 mM EDTA, 0.5 M ammonium acetate, 0.25% SDS), ethanol precipitated, and quantified before use. The MRP probe was a generous gift from Greg Matera (Case Western Reserve University).
Immunofluorescence, GFP-Fusions, and In Situ Hybridization
Cells were fixed 24 h postseeding for indirect immunofluorescence, or transfected with GFP fusion constructs 24 h postseeding, and then fixed 24 h posttransfection. All manipulations were performed at room temperature unless otherwise noted. All cells were fixed in freshly prepared 3% paraformaldehyde in 1× PBS, pH 7.4, for 10 min, and then washed three times in 1× PBS. Cells were permeabilized in 1× PBS + 0.5% Triton X-100 for 5 min, and then washed three times in 1× PBS. For immunofluorescence, blocking was performed for 30 min in 1× PBS + 3% bovine serum albumin (Sigma-Aldrich, St. Louis, MO). Cells were then incubated for 1 h with the appropriate primary antibody diluted in blocking buffer. Cells were rinsed three times in 1× PBS + 3% bovine serum albumin and incubated with either rhodaminered-X-conjugated or fluorescein isothiocyanate-conjugated donkey antimouse or anti-rabbit antibody (1:100; Jackson ImmunoResearch Laboratories, West Grove, PA) diluted in blocking buffer for 1 h. Cells were rinsed three times in blocking buffer and then mounted with Vectashield either with or without 4,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, Burlingame, CA), and sealed with clear nail polish. In the case of GFP-fusions without subsequent treatment, cells were fixed and mounted immediately for visualization.
Fluorescence in situ hybridization was performed by fixing and permeabilizing cells as described above. After rinsing twice with 1× PBS postpermeabilization, cells were rinsed once with 2× SSC. Then 20 μl of hybridization mix was added to a slide (200 ng of probe, 4 μl of deionized formamide, 8 μl of hybridization buffer (stock solution of 20 μl of 20× SSC, 40 μl of 50% dextran sulfate, 20 μl of a 10 mg/ml solution of Escherchia coli tRNA; Sigma-Aldrich), nH2O up to 20 μl). Coverslips were inverted onto the slide and sealed with rubber cement, and then incubated in a dark humid chamber overnight at 45°C. Rubber cement was removed and coverslips were floated in 2× SSC, rinsed once in 2× SSC for 15 min at 45°C, and twice in 2× SSC for 15 min at room temperature. Samples with fluorescein-UTP-labeled probes were then mounted in Vectashield. Samples with dig-labeled probes were inverted onto a drop containing rhodamine- or fluorescein-conjugated mouse anti-DIG antibody (1:200) (Roche Diagnostics) in 2× SSC for 1 h at room temperature. Fluorescence in situ hybridization to detect mitochondrial RNA processing (MRP) RNA was performed as described previously (Matera et al., 1995).
For colocalization experiments, coverslips were incubated with the appropriate primary antibody diluted in 2× SSC at room temperature for 1 h, and then washed three times in 2× SSC for 5 min each. Cells were then incubated with rhodamine-red-X-conjugated donkey anti-mouse or anti-rabbit (Jackson ImmunoResearch Laboratories) antibodies at room temperature for 1 h. They were then rinsed twice in 2× SSC, once in 1× SSC, once in 1× PBS, mounted on glass slides with Vectashield, and sealed with clear nail polish.
Microscopy
Phase-contrast images of live cells were photographed with a Nikon N6006 35-mm camera attached to a Nikon Diaphot-TMD phase contrast microscope with a 10× Plan lens. All fluorescence images were collected on a Leica DM IRBE inverted confocal laser-scanning microscope equipped with argon, krypton, and helium-neon lasers. DAPI stains were visualized using a Ti-Sapphire 2-photon laser. All confocal images were collected using a 63× Plan-Apo lens (Nikon) by using Leica confocal software. Stacks of 0.5-μm sections were collected for each sample and images were then selected which best represented the overall field. Intensity measurements were performed using Leica confocal software by drawing a line across the foci in question and measuring intensity across the line. Graphs were produced using the Leica confocal software. All confocal images are single sections.
RESULTS
Cellular Localization of KSRP
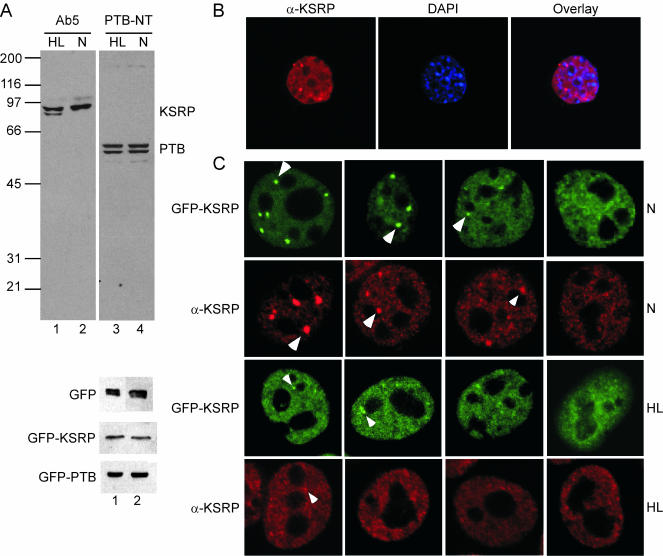
Given the numerous cellular processes in which KSRP has been implicated, we wanted to examine its intracellular localization and compare it with that of other splicing factors. We particularly wanted to look at cells showing the neuronal pattern of c-src splicing. We used N1E-115 mouse neuroblastoma cells, which include the N1 exon in ∼10% of their c-src mRNA. These were compared with HeLa (human cervical carcinoma) cells, which express relatively low levels of src mRNA, all skipping the N1 exon, and which are well characterized for splicing factor staining. The localization of KSRP was determined by immunofluorescence and by expression of a GFP-KSRP fusion protein. We used a mouse monoclonal antibody (mAb) (Ab5) raised to bacterially expressed full-length KSRP and a rabbit polyclonal antibody (PTB-NT) raised to the N-terminal 15-amino acid peptide of PTB. Western blot analysis of whole-cell extracts confirmed the specificity of the Ab5 and PTB-NT antibodies in both N1E-115 and HeLa cells (Figure 1A). The Ab5 antibody reacts with two KSRP bands in HeLa cell extracts (Figure 1A, lane 1) and one band in N1E-115 cells (Figure 1A, lane 2). Both of these HeLa bands are known to be KSRP from peptide analyses, but the nature of their difference is unknown (Min et al., 1997). The N1E-115 cells contain somewhat more KSRP than do HeLa cells. This is similar to the amount seen in WERI-1 retinoblastoma cells, which contain at least 4 × 104 KSRP molecules/cell. The PTB-NT antibody identifies the doublet of PTB splice variants in both N1E-115 and HeLa cells (Figure 1A, lanes 3 and 4). Transfection of the GFP, GFP-KSRP, and GFP-PTB fusion constructs, followed by Western blot analysis, verified the expression of the tagged proteins (Figure 1A). Immunolabeling with Ab5 showed KSRP strongly enriched in the nucleus (Figure 1B). This nuclear labeling was generally diffuse, with exclusion from nucleoli. The pattern of enrichment was identical for endogenous and overexpressed KSRP (Figure 1C), and similar to the localization observed for FBP (He et al., 2000). Colocalization of expressed GFP-KSRP with immunolabeled endogenous KSRP determined no difference in localization pattern (our unpublished data). Thus, overexpression of the protein did not alter its subcellular localization. In addition to the diffuse nuclear localization, KSRP was seen to be enriched in particular subnuclear structures (Figure 1C). These varied in number and size from cell to cell within the same preparation, ranging from zero to nine prominent inclusions in N1E-115 cells, with additional less intense nuclear enrichments. Interestingly, HeLa cells showed significantly fewer KSRP inclusions than N1E cells (up to four) that were much weaker in intensity. Figure 1C shows a panel of cells representing the range of staining patterns seen for the two cell types.
Figure 1.
KSRP localizes to the nucleus in HeLa and N1E-115 cells. (A) Top, Western blot of total cell lysate from HeLa (HL, lanes 1 and 3) or N1E-115 cells (N, lanes 2 and 4) probed with KSRP antibody (Ab5) or PTB antibody (PTB-NT). Bottom three panels, Western blot of HeLa (lane 1) or N1E-115 (lane 2) cell lysates after transient transfection with GFP (top), GFP-KSRP (middle), or GFP-PTB (bottom) expression constructs. Blots were probed with a GFP antibody. (B) KSRP is almost exclusively nuclear. HeLa cells were stained with the Ab5 antibody and with DAPI to visualize the nucleus. The left panel shows Ab5 staining, the middle panel DAPI, and the right panel shows the overlay of the two. (C) The range of staining patterns seen for GFP-KSRP (green) and for immunostained endogenous KSRP (α-KSRP, red). In N1E-115 cells (top two rows), KSRP is seen in more and brighter foci than in HeLa cells (bottom two rows). Arrowheads indicate subnuclear enrichments.
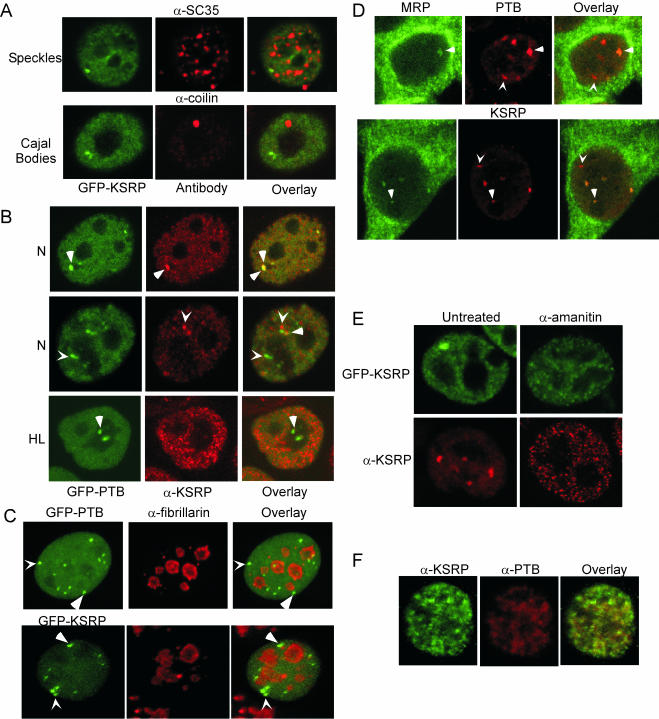
To identify these subnuclear compartments, experiments were performed with antibodies that identify various specific structures within the nucleus. Many, although not all, splicing factors are enriched in nuclear speckles (Lamond and Spector, 2003). However, an antibody to SC35, a protein present in nuclear speckles, showed that the KSRP enrichments do not coincide with nuclear speckles (Figure 2A) in N1E-115 cells. Similarly, KSRP is not present in CBs as seen by colocalization with an anti-coilin antibody (Figure 2A). Labeling with an antibody to the PML protein determined that KSRP is not present in PML bodies (our unpublished data).
Figure 2.
KSRP and PTB colocalize to the PNC. (A) Localization of GFP-KSRP compared with various endogenous proteins in N1E-115 cells. The left panels show GFP-KSRP images and the center panels show immunofluorescence images with anti-SC35 or anti-coilin to visualize the respective nuclear structures, and the right panels are overlays. (B) Colocalization of GFP-PTB with immunostained endogenous KSRP in N1E-115 cells (top two rows) and HeLa cells (bottom row). The left column is GFP-PTB, the middle column is KSRP, and the right column is the overlays. Arrowheads denote enrichments with both KSRP and PTB present. Chevrons denote either PTB-(green, left) or KSRP (red, middle)-specific enrichments. (C) PTB and KSRP enrichments are often but not always perinucleolar. Colocalization of GFP-KSRP or GFP-PTB with immunostained endogenous fibrillarin in N1E-115 cells. The left panel is either GFP-PTB or GFP-KSRP, the middle panel is fibrillarin, and the right panel is the overlay. Arrowheads denote enrichments associated with a nucleolus. Chevrons denote enrichments not associated with a nucleolus. (D) Some but not all PTB and KSRP enrichments contain MRP RNA. Colocalization of MRP RNA (green, left) with PTB (red, middle, top row) or KSRP (red, middle, bottom row). Arrowheads denote enrichments with both MRP RNA and either PTB or KSRP. Chevrons denote enrichments of either PTB or KSRP without MRP RNA. (E) PNC localization is dependent on RNA polymerase II transcription. Overexpressed (top row) or endogenous (bottom row) KSRP in N1E-115 cells either untreated (left column) or treated (right column) with 50 μg/ml α-amanitin before fixation. (F) KSRP and PTB show similar dispersion after inhibition of RNA polymerase II. N1E-115 cells were treated with 50 μg/ml α-amanitin before fixation, and then immunostained with both α-KSRP (green, left) and α-PTB (red, middle) antibodies. The right panel is the overlay.
KSRP's localization pattern most closely resembled that of PTB, with irregular enrichments frequently associated with the nucleolus (Figure 2B). In addition to its diffuse nuclear localization, PTB is enriched in the PNC. Colocalization experiments by using the Ab5 antibody and the GFP-PTB fusion protein showed that some of the subnuclear enrichments of KSRP coincide with the perinucleolar enrichments of PTB (Figure 2B, top row). Other patterns of labeling were also seen, including PTB enrichments that did not contain KSRP and KSRP enrichments that did not contain PTB (Figure 2B, middle row). Identical colocalization patterns were observed in the reverse experiment with the PTB-NT antibody and GFP-KSRP fusion, or using both KSRP and PTB antibodies rather than GFP-fusions (our unpublished data), indicating that the localization patterns are not due to problems of antibody specificity or the result of protein overexpression. Thus, KSRP is a component of the PNC in N1E-115 cells, but it is also seen in other structures that are not obviously juxtaposed to a nucleolus.
In contrast to the N1E-115 cells, HeLa cells showed less frequent and much weaker labeling of KSRP in perinucleolar structures. Although PTB labeling clearly identified the PNC in these cells, only a few of the PNCs seemed to contain KSRP (Figure 2B). To measure the difference between these two cells more carefully, we examined >500 of each and counted how frequently KSRP was found in the PTB-labeled PNCs. Analysis of this large sample of cells revealed that 81% of N1E-115 cells have KSRP colocalizing to the PTB-labeled foci. In contrast, only 30% of HeLa cells had KSRP protein present in the PNC (Table 1). Noticeable structural differences were also seen between the two cell types. In N1E-115 cells KSRP accumulates in larger, more intense foci. In HeLa cells the KSRP foci, if present at all, are smaller and fewer (Figure 2B).
Table 1.
Percentage of cells in which KSRP is found in PTB enrichments
| Cell type | Total cells | K&Pa | Percentage |
|---|---|---|---|
| N1E-115 | 553 | 449 | 81 |
| HeLa | 534 | 161 | 30 |
K&P, number of cells with coincident KSRP and PTB structures.
To more carefully examine the position of these enrichments relative to the nucleolus, we performed colabeling experiments with the GFP-KSRP and GFP-PTB fusion constructs and an antibody to the nucleolar protein fibrillarin (Figure 2C). Although many of the GFP-PTB foci were obviously juxtaposed to a nucleolus, some did not have adjacent fibrillarin labeling (Figure 2C, top row). Similarly, although many KSRP enrichments were present at the periphery of a nucleolus, some were not (Figure 2C, bottom row). In HeLa cells most of the PTB enrichments are PNCs (Huang et al., 1997). To confirm that some of the PTB and KSRP foci seen in N1E-115 cells were not PNCs, N1E-115 cells were subjected to fluorescence in situ hybridization with a probe for MRP RNA, another PNC marker, and then probed with either anti-PTB or anti-KSRP antibodies (Figure 2D). In both cases, some foci for the antibody-labeled proteins were also labeled with the MRP RNA probe, but some were not. Thus, in the N1E-115 cells, KSRP and PTB also exist in subnuclear structures that are not perinucleolar and do not contain MRP RNA. This is in contrast to HeLa cells, where the majority of PTB enrichments are perinucleolar compartments (Huang et al., 1997).
KSRP Localization Is Pol II Dependent
The localization of PTB to the PNC is dependent on multiple RNA recognition motifs, suggesting that PNC localization requires RNA binding (Huang et al., 1997). Moreover, inhibition of Pol II transcription causes PTB to partially relocate from the PNC to “rosettes” around nuclear speckles (Huang et al., 1998), further supporting an RNA-dependent localization of PTB. Because KSRP also binds to Pol II transcripts and is present in the PNC, we investigated whether Pol II inhibition affects KSRP's localization.
Cells were incubated in the presence of 50 μg/ml α-amanitin to selectively inhibit RNA polymerase II (Nguyen et al., 1996). As visualized either in cells expressing GFP-KSRP or in cells stained with the Ab5 antibody, PNC localization was lost upon inhibition of Pol II transcription (Figure 2E). Under these conditions, KSRP reorganized into many small, distinct foci within the nucleus. Although these foci seem similar to speckles, they are smaller and more concentrated. The pattern resembled that of PTB upon similar treatment with α-amanitin (Huang et al., 1998). Labeling with both anti-KSRP and anti-PTB antibodies showed that KSRP (Figure 2F, green, left) and PTB (Figure 2F, red, middle) were still present in some of the same enrichments upon inhibition of RNA polymerase II transcription.
The KH-RNA-Binding Domains Are Required for the PNC Localization of KSRP
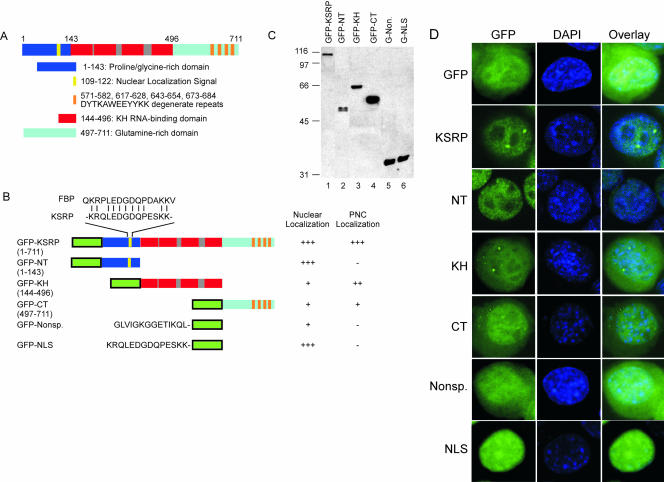
Localization of PTB to the PNC is dependent upon the presence of at least three of its four RRM RNA-binding domains (Huang et al., 1997). The domain structure of KSRP includes a proline-glycine-rich N terminus, four KH-type RNA binding domains, and a glutamine-rich C terminus. The C terminus contains four degenerate copies of the sequence DYTKAWEEYYKK thought to be involved in protein-protein interactions (Figure 3A) (Siebel et al., 1995; Duncan et al., 1996; Labourier et al., 2001). To delineate the sequence requirements for KSRP localization to both the nucleus and the PNC, we fused various portions of KSRP to GFP and transiently expressed each fusion in N1E-115 cells. Fusion constructs included the full-length protein (GFPKSRP), the N-terminal domain (GFP-NT, residues 1-143), the four KH-domains (GFP-KH, residues 144-496), and the C-terminal domain (GFP-CT, residues 497-711) (Figure 3B). Western blot analysis using a rabbit polyclonal anti-GFP antibody verified expression of these constructs in the N1E-115 cells (Figure 3C). Transient transfection of the empty GFP vector revealed GFP's typical diffuse localization in both the nucleus and cytoplasm (Figure 3D, top row). As seen previously, GFP-KSRP localized entirely to the nucleus with enrichment in PNCs (Figure 3D, KSRP). GFP-NT was also nuclear but lacked any observable enrichment in the PNC (Figure 3D, NT). GFP-KH localized to both the nucleus and the cytoplasm. Within the nuclear staining, there were small but clear PNC-like enrichments (Figure 3D, KH). These enrichments colocalized with PTB as seen with the full-length protein (our unpublished data). GFP-CT also localized to both nucleus and cytoplasm. However, this protein showed PNC staining that was at best much fainter and more diffuse than GFP-KH (Figure 3D, CT). Together, these data indicate that there is a nuclear localization signal present within the N terminus of KSRP but that the RNA-binding domains of KSRP are needed for clear PNC localization.
Figure 3.
KSRP deletion mutants identify regions important for nuclear and PNC localization. (A) The domain structure of KSRP. KSRP contains a proline/glycine-rich N terminus (dark blue) with a nuclear localization signal (NLS, yellow), a central domain with four KH-type RNA-binding domains (red), and an unusual C terminus (light blue) rich in glutamine with four degenerate copies of the repeat DYTKAWEEYYKK (orange). (B) Deletion mutants of KSRP. Mutants were fused to the C terminus of GFP (green), except in the cases of GFP-nonsp. and GFP-NLS, which were fused to the N terminus of GFP. Fusion constructs were rated on their localization to the nucleus and to the PNC. Three + signs indicate the highest amount of localization; a - sign indicates no localization. (C) Western blot analysis verifies expression of GFP-fusion constructs in N1E-115 cells. Lane 1 is GFP-KSRP, lane 2 is GFP-NT, lane 3 is GFP-KH, lane 4 is GFP-CT, lane 5 is GFP-nonspecific, and lane 6 is GFP-NLS. Size markers are noted to the left of the blot. Blot was probed with anti-GFP. For GFP alone see Figure 1A. (D) The KH-domains are sufficient for PNC localization. In the left column are GFP images of cells after transient transfection of each fusion construct. The middle column shows DAPI images, and the right column shows the overlays. GFP alone (top row) was compared with each of the fusion constructs indicated to the left of each row. DAPI staining identifies the nucleus in all panels.
Examination of the KSRP sequence identified a potential nuclear localization sequence within the N-terminal region similar to that of FBP (He et al., 2000) (Figure 3B). The KSRP sequence KRQLEDGDQPESKK also resembles a bipartite NLS comprised of two segments of basic amino acids separated by ∼10 nonbasic amino acid residues (Figure 3B). Fusion of this KSRP peptide sequence to the N terminus of GFP (pEGFPN1) and transient expression in N1E-115 cells confirmed that this sequence localized GFP entirely to the nucleus (Figure 3D, NLS versus Nonsp.). A nonspecific sequence behaved similarly to GFP alone and was distributed diffusely to both the nucleus and cytoplasm.
The src Transcript Is Enriched in Nuclear Foci
The PNC is thought to contain Pol II transcripts as well as its identified components (Huang et al., 1998). Because two proteins known to bind the src pre-mRNA, PTB and KSRP, were found in the PNC, we wanted to determine whether the src transcript itself was a PNC component. More generally, we wanted to determine whether the proteins we see binding the src pre-mRNA in vitro could be observed to colocalize with the RNA in vivo.
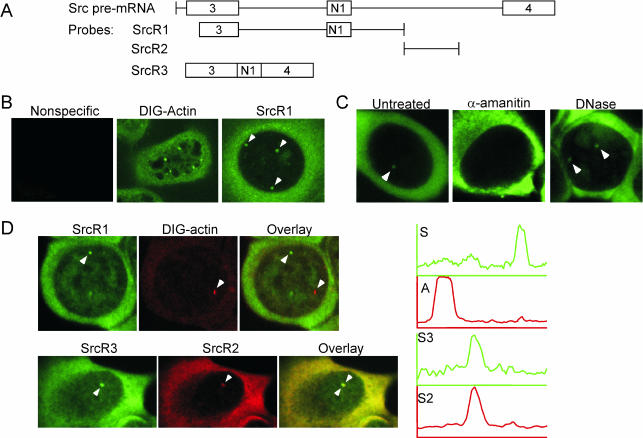
To observe the src transcript, FISH was performed on N1E-115 cells. RNA probes labeled with either fluorescein-UTP or digoxigenin-UTP, and covering various regions of the src transcript were transcribed in vitro and hybridized to fixed, permeabilized cells (Figure 4A). FISH studies of other Pol II transcripts generally reveal one or more distinct foci of staining corresponding to RNA synthesis at the gene loci as well as a background of more diffuse staining (Misteli and Spector, 1999; Smith and Lawrence, 2000; Muhlemann et al., 2001; Shopland et al., 2001, 2002; Levsky et al., 2002; Femino et al., 2003; Oleynikov and Singer, 2003). Probes specific for src RNA localized diffusely in the cytoplasm and, as expected, were enriched in the nucleus in distinct foci (Figure 4B). A digoxigenin-labeled actin probe was used as a positive control, and gave the expected cytoplasmic stain with enriched foci in the nucleus (Figure 4B) (Oleynikov and Singer, 2003). A nonspecific adenovirus probe did not have any discernible fluorescence (Figure 4B). All src RNA probes tested revealed similar localization patterns (our unpublished data). N1E-115 cells are highly aneuploid (192 chromosomes; http://www.atcc.org/SearchCatalogs/longview.cfm?view=ce,1961028,CRL-2263&text=N1E%2D115&max=20), and as expected, each nucleus had multiple foci for sites of both actin and src RNA. To verify that these enrichments were RNA, cells were incubated with 50 μg/ml α-amanitin to inhibit Pol II transcription before fixation and hybridization with SrcR1. This treatment eliminated all nuclear enrichments (Figure 4C). In contrast, DNase treatment had no effect on the enrichments (Figure 4C). Thus, these foci are likely to be RNA at actively transcribed loci.
Figure 4.
src pre-mRNA is enriched in specific, RNA polymerase II-dependent foci within the nucleus. (A) Diagrams of the RNA probes used for FISH to the src transcript. (B) src RNA is enriched in nuclear foci. A nonspecific adenovirus probe (left), a digoxigenin-labeled actin probe (middle), and a src probe (SrcR1, right). Arrowheads indicate foci of src RNA. (C) src RNA enrichments are RNA polymerase II-dependent but unaffected by DNase. Cells were either untreated (left), treated with 50 μg/ml α-amanitin in the cell culture medium (middle), or treated with DNase (right) in the FISH hybridization mixture. All cells were probed with SrcR1. Arrowheads indicate src RNA enrichments. (D) src RNA enrichments are specific. Top row, cells were cohybridized with a digoxigenin-labeled actin probe and fluorescein-labeled SrcR1 (top row). The left panel is src RNA, the middle panel is actin RNA, and the right panel is the overlay. Graphs to the right indicate areas of intensity as measured across a line connecting the foci labeled with arrowheads. Green is src RNA intensity and red is actin RNA intensity. Bottom row, cells were cohybridized with two different src RNA probes, SrcR2 and SrcR3, that cover nonoverlapping regions of the gene. The left panel is fluorescein-labeled SrcR3, the middle panel is DIG-labeled SrcR2, and the right panel is the overlay. Graphs indicate intensity across the foci labeled with arrowheads. Green is SrcR3 and red is SrcR2.
The src RNA nuclear enrichments were compared with each other and with those labeled with the actin probe. Colocalizations were performed with the DIG-labeled actin RNA probe and fluorescein-labeled SrcR1. When both the actin probe and the src probe were simultaneously hybridized to N1E-115 cells, separate enrichments occurred in the nucleus as expected for separate gene loci. Intensity measurements revealed separate intensity peaks for src RNA and for actin RNA (Figure 4D). In contrast, simultaneous hybridization using DIG-labeled SrcR2 and fluorescein-labeled SrcR3 (Figure 4A), which cover nonoverlapping regions of the src transcript, showed src RNA nuclear enrichments that coincide (Figure 4D). Intensity measurements revealed coincident peaks of intensity for each src RNA probe (Figure 4D). Thus, these enriched foci are specifically the src locus and are not due to cross-hybridization of the probe to some other locus.
To examine whether the src RNA was present in the PNC, FISH-labeled cells were colabeled with the anti-fibrillarin antibody. Although some foci of src RNA were observed near a nucleolus, there was no consistent juxtaposition of the src RNA foci and nucleoli (our unpublished data). Thus, the src RNA is not a likely component of the PNC.
Colocalization of KSRP and PTB with the src Transcript
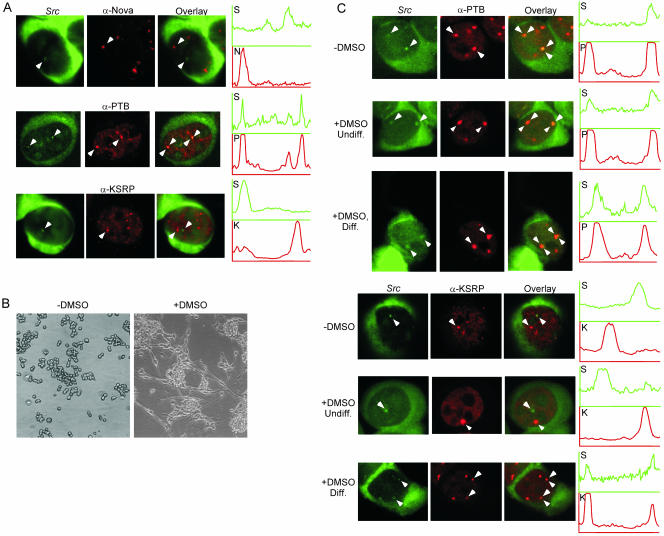
We next wanted to determine whether PTB and KSRP are present within the src transcript enrichments. Colocalizations were performed using SrcR1 and either PTB-NT or Ab5. Immunostaining with the PTB-NT antibody after FISH revealed that, in normally cultured N1E-115 cells, PTB is present in the src RNA foci (Figure 5A). As expected, there were additional PTB enrichments that did not contain src RNA. Staining with the KSRP antibody after FISH did not show a large enrichment of KSRP in the src RNA foci, although lower levels of KSRP were sometimes present (Figure 5A). The brightest foci of KSRP are distinct from the src RNA enrichments within the nucleus. The Nova RNA-binding protein has been implicated in several neuronal alternative splicing events, but not src (Jensen et al., 2000). As expected, the enrichments of Nova staining in N1E-115 cells do not coincide with the src loci (Figure 5A).
Figure 5.
Differentiation of N1E-115 cells causes KSRP to relocalize to the src RNA foci. (A) Undifferentiated cells subjected to FISH with SrcR1 before immunolocalization to visualize PTB or KSRP. The left panels show src RNA staining; the middle panels show Nova, PTB, or KSRP immunostaining as labeled; and the right panels are overlays. Arrowheads denote subnuclear enrichments. Graphs indicate fluorescence intensity across the foci labeled with arrowheads. Green is the src RNA signal (S) and red is either the Nova (top row, N), PTB (middle row, P), or KSRP (bottom row, K) immunostain signal. (B) Phase images of N1E-115 cells either undifferentiated (left) or differentiated with 2% DMSO in the culture medium for 5 d (right). (C) Differentiation induces colocalization of KSRP with src. Cells were either untreated (rows 1 and 4, -DMSO) or treated with DMSO for 5 d (rows 2, 3, 5, and 6). Undiff. (rows 2 and 5) indicates cells in the DMSO-treated cultures that maintained their round shape and failed to extend neurites. Diff. (rows 3 and 6) indicates cells in the DMSO-treated cultures that exhibit neurite extension. The left image of each row shows the src RNA image in green. The middle image of each row shows the immunofluorescence for PTB or KSRP. The overlays are shown at the right. Graphs indicate intensity plots across the foci labeled with arrowheads. Green is src RNA intensity (S), red is either PTB (P) or KSRP (K).
Numerical analysis of normally cultured N1E-115 cells revealed that all of the src RNA enrichments contain PTB (Table 2). In contrast, only 11% of the src RNA enrichments coincide with a KSRP enrichment (Table 3). Most cells show no foci containing both src RNA and KSRP. Many KSRP foci are perinucleolar and costain with PTB (Figure 2). However, the src RNA is not in these structures, again consistent with its absence from the PNC.
Table 2.
Colocalization of PTB and src RNA in N1E-115 cells
| Cells |
Foci |
||||||
|---|---|---|---|---|---|---|---|
| Total cells | P&Sa | Percentage | PTB foci | Src RNA foci | No. of foci with PTB & Src | Percentage of Src foci with PTB | |
| −DMSO | 8 | 8 | 100 | 43 | 22 | 22 | 100 |
| +DMSO | 13 | 13 | 100 | 104 | 71 | 66 | 93 |
P&S, number of cells with coincident PTB and src structures.
Table 3.
Colocalization of KSRP and src RNA in N1E-115 cells
| Cells |
Foci |
||||||
|---|---|---|---|---|---|---|---|
| Total cells | K&Sa | Percentage | KSRP foci | Src RNA foci | No. of foci with KSRP & Src | Percentage of Src foci with KSRP | |
| −DMSO | 11 | 3 | 27 | 60 | 28 | 3 | 11 |
| +DMSO | 21 | 21 | 100 | 172 | 103 | 97 | 94 |
K&S, number of cells with coincident KSRP and src structures.
The above-mentioned experiments were done with N1E-115 cells that were actively dividing in nonconfluent culture. We found that cells in denser culture seemed to have a higher coincidence of KSRP and src RNA in subnuclear foci (our unpublished data). N1E-115 cells grown to high density begin to exhibit differentiated characteristics, such as exit from the cell cycle and neurite outgrowth. To investigate whether differentiation affects the localization of the src transcript with the PTB and KSRP proteins, N1E-115 cells were treated with 2% DMSO for 5 d before fixation for FISH and immunofluorescence (Kimhi et al., 1976). After DMSO treatment, the culture contains a mixture of cells. Some cells are clearly differentiated and exhibit long neurite extension. Other cells in the same culture remain round without extended neurites, similar to untreated N1E-115 cells. Phase contrast images of the cells before and after addition of DMSO reveal that after 5 d of treatment ∼30% of the cells have extended neurites (Figure 5B). Differentiated cells were defined morphologically in the following experiments as cells showing neurites extended at least 30 μm, or approximately three times the length of the nucleus. As observed previously, untreated cells have PTB present in nearly all of the src RNA foci and KSRP was present in only a minor fraction of the src RNA foci (Figure 5C). The cells treated with DMSO but not exhibiting neurite extension looked similar to untreated cells. PTB and src RNA still colocalized as measured visually and by intensity plots (Figure 5C). Also like the untreated cells, in DMSO-treated cells without neurite outgrowth, KSRP and the src RNA were clearly present in separate foci and separate intensity peaks (Figure 5C). After neurite extension, PTB was still clearly in the src RNA foci, and the number of foci per cell increased (Figure 5C; Table 2). Most interestingly, cells with extended neurites exhibited a very different pattern of KSRP localization. In these morphologically differentiated cells, the number of src RNA foci that contained KSRP increased significantly from the 11% seen in undifferentiated cells to 94% (Figure 5C; Table 3). Again, the total number of KSRP foci per cell also increased upon N1E-115 cell differentiation (Table 3). Thus, neuronal differentiation of N1E-115 cells (induced by DMSO and measured morphologically by neurite extension) caused a relocalization of KSRP to the src RNA foci.
Differentiation of N1E-115 Cells Increases Exon N1 Inclusion
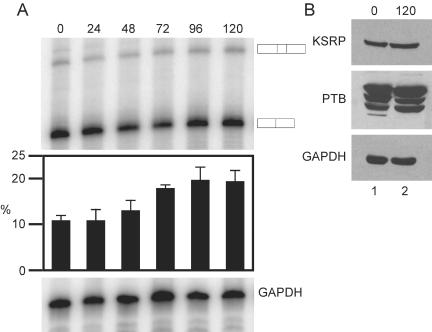
The relocalization of the c-src splicing regulatory factors led us to examine whether src transcription or N1 exon splicing were also changing upon N1E-115 differentiation. N1E-115 cells were harvested immediately before and every 24 h after addition of 2% DMSO to the culture medium for 5 d and cytoplasmic RNA was isolated. RT-PCR measurements of c-src and GAPDH mRNA revealed only small changes in overall levels of the two mRNAs over the time course of differentiation (Figure 6A). However, over the same time course, the level of N1 inclusion increased from ∼10% to a peak of 21% at 96 h of DMSO treatment (Figure 6A). Given that only ∼30% of the cells differentiated in the presence of DMSO, it is likely that exon N1 inclusion levels are higher in the differentiated cells, but that the effect is diluted by the lower inclusion in the remaining undifferentiated cells. Thus, the induced colocalization of KSRP with src RNA coincides with a change in N1 exon splicing.
Figure 6.
Differentiation of N1E-115 cells increases N1 inclusion. (A) RT-PCR analysis of cytoplasmic RNA isolated immediately before and every 24 h after the addition of 2% DMSO. The products for N1 exon inclusion and exclusion are noted at the right. The bar graph below illustrates exon inclusion levels with error bars, indicating the SD for n = 3. GAPDH was used as a control for the total RNA level. (B) Protein expression levels remain unaffected by differentiation of N1E-115 cells with DMSO. Western blot analysis of whole-cell extracts either immediately before (lane 1) or 5 d after (lane 2) addition of DMSO to the culture medium. The blot was probed with Ab5 (top row), PTB-NT (middle row), or GAPDH (bottom row) as a loading control.
Western blot analysis of undifferentiated and differentiated cells revealed no changes in the level of the PTB and KSRP proteins (Figure 6B). Thus, changes in the expression of these two proteins do not seem to be the cause of the change in protein localization or in N1 exon inclusion.
DISCUSSION
KSRP and closely related proteins have been implicated in a variety of processes, including transcription, splicing, RNA editing, RNA localization, and RNA degradation. Several of these roles require that a fraction of the protein be cytoplasmic. In our experiments, we see very little KSRP in the cytoplasm, although a small percentage of the total protein could reside there. Interestingly, ZBP2, a chicken homolog of KSRP involved in cytoplasmic localization of the β-actin mRNA shows more significant cytoplasmic localization in addition to its predominant nuclear staining (Gu et al., 2002). This protein is nearly identical to KSRP in the KH and C-terminal domains but has novel insertions in its N terminus. These insertions immediately flank the nuclear localization sequence that we identify here. Thus, it will be interesting to examine how these sequence changes might modify the protein's activity.
We examined the subcellular localization of KSRP in relation to two nuclear molecules that interact with it in vitro, PTB and c-src pre-mRNA. We find that in N1E-115 mouse neuroblastoma cells, KSRP is seen in enriched foci above a background of diffuse nuclear staining. These enrichments can be distinguished by their other components and by the cell growth conditions under which they are observed. In undifferentiated N1E-115 cells, some of the KSRP foci are in perinucleolar compartments, where they costain with PTB. In differentiated N1E-115 cells, some of the KSRP foci are at sites of c-src RNA synthesis.
The Association of KSRP with the PNC Varies with Cell Type
In normal undifferentiated N1E-115 cells, many KSRP foci are perinucleolar compartments, defined by the presence of the PTB protein and MRP RNA and by proximity to the nucleolus. Similar to PTB, KSRP localization to the PNC requires active transcription and its RNA binding domains. Thus, the recruitment of KSRP to the PNC likely involves its interaction with RNA. Significantly, although HeLa cells have clear PNC structures containing PTB, these structures contain little KSRP. The absence of KSRP from HeLa PNCs may reflect the absence of the appropriate RNA target in these cells. Because the PNCs in N1E cells do not apparently contain c-src pre-mRNA, this missing RNA target is likely not the c-src transcript (although this transcript is low in HeLa cells). Furthermore, because the src RNA foci are not obviously perinucleolar, the PNC localization of KSRP may not be related to its role in splicing.
The known components of the PNC are an unusual collection of molecules. Stable Pol III RNAs (such as the Y and RNase P RNAs) are found in the PNC, but without the proteins that usually assemble with them (such as the Ro and Th proteins). Also present in the PNC are hnRNP-like proteins (such as PTB and KSRP) whose known functions involve binding Pol II transcripts, although PTB has also been shown to bind some of the Y RNAs (Huang, 2000; Fabini et al., 2001; Wang et al., 2003). This composition makes speculation as to the function of the PNC difficult. One proposal is a role in the maturation or assembly of the Pol III RNAs into functional RNPs (Matera et al., 1995). The absence from the PNC of proteins known to be in the functional Pol III RNPs might indicate that the RNAs are at an early stage of their biogenesis (Matera et al., 1995; Wang et al., 2003). This is still a possible model. However, the change in KSRP association with the PNC that we observe between HeLa and N1E-115 cells indicates that PNC structure is variable, depending on cell type and presumably gene expression profile. PNCs are more frequently seen in highly transformed cells, but it was not known that the individual components of the structure could vary with cell type (Huang et al., 1997). This change in PNC composition would seem inconsistent with certain possible housekeeping functions of the structure (such as RNP assembly) and imply a function affecting specific transcripts in specific cells. Another possible function for the PNC is as a trafficking point for molecules destined for the cytoplasm (Matera et al., 1995). This function could clearly change with the gene expression profile of cells. An interesting example is Raver-1, a PTB-interacting protein that is found in both the PNC and the cytoplasm. On myotube differentiation, this protein is seen to redistribute from the nucleus to microfilament attachment sites at the cell surface (Huttelmaier et al., 2001). It is interesting to speculate that the PNC fraction of KSRP may be destined to take part in the protein's cytoplasmic functions.
The Association of KSRP with the c-src RNA Foci Varies with Neuronal Differentiation
Fluorescence in situ hybridization to src transcripts identifies discrete foci of nuclear localization, presumably corresponding to sites of gene transcription, as seen with other transcripts (Misteli and Spector, 1999; Smith and Lawrence, 2000; Muhlemann et al., 2001; Shopland et al., 2001, 2002; Levsky et al., 2002; Femino et al., 2003; Oleynikov and Singer, 2003). These src RNA foci are, for the most part, not perinucleolar and are not apparently related to the PNC. In addition to the PNC, both PTB and KSRP showed subnuclear enrichments that were not perinucleolar, and some of these PTB foci coincide with the sites of the src transcript. In contrast, KSRP was generally not found in these src RNA enrichments in undifferentiated N1E cells. KSRP is not necessarily completely absent from the src RNA foci (because there is always a diffuse background of KSRP within the nucleus), but it is not enriched in the manner seen with PTB. Neuronal differentiation of the N1E-115 cells changes the pattern of KSRP staining significantly, leading to a large increase in the number of cells showing a coincidence of KSRP with src RNA foci. These results provide an in vivo confirmation of the interactions of PTB and KSRP with src pre-mRNA observed in vitro (Markovstov et al., 2000).
PTB is a repressor of N1 exon splicing that binds in multiple copies to the c-src transcript (Chou et al., 2000). Biochemical analyses indicate that PTB binds to the src transcript under conditions of both exon repression and splicing. Thus, it is not surprising that PTB is associated with the src RNA foci under both growth conditions. Moreover, because the N1 exon is only partially included in these cells, the observed association of PTB with the src transcript presumably includes conditions both of exon inclusion and exon exclusion.
The observed recruitment of KSRP to src RNA foci upon differentiation demonstrates the utility of combining immunofluorescence with FISH in examining the changing interactions of RNA-binding proteins with specific transcripts in vivo. The mechanism of KSRP recruitment to the src RNA foci is an interesting question. The simplest models are that either new factors are expressed or existing factors are modified upon neuronal differentiation that alters the interaction of KSRP with the RNA. The induced recruitment of KSRP also has interesting implications for the role of this protein in src splicing.
Although drastically changing the interaction of KSRP with the src RNA loci (from 11 to 94% of src RNA loci containing KSRP), DMSO-induced differentiation of N1E-115 cells also led to a modest increase in src exon N1 splicing (from 10 to 21% exon inclusion). Because only ∼30% of the N1E-115 cells differentiate in the presence of DMSO, it is possible that N1 inclusion in differentiated cells is higher than what is observed in the whole population. Thus, it is tempting to relate the movement of KSRP to the src locus to the change in src splicing. However, to look at this more carefully we need to examine the KSRP and c-src RNA localization in primary cells or cell lines that undergo more efficient differentiation, or where we can enrich the differentiated population of cells. So far this has proven difficult. Moreover, by this method, we cannot assess where on the src transcript the protein is binding and whether it is assembling on the DCS in vivo as seen in vitro. Answering these questions are directions for future experiments.
Acknowledgments
We thank Robert Darnell and Peter Tontonoz for providing antibodies, Greg Matera for the MRP probe, Jason Underwood for the full-length KSRP clone, Julia Nikolic and Martin Hoang Luu for excellent technical assistance generating the Ab5 antibody, Matthew Schibler for assistance with the confocal microscope, Sandra Wolin for helpful comments on the manuscript, and finally members of the Black laboratory for helpful discussion. This work was supported by National Institutes of Health grant GM RO1 49662 (to D.L.B) and by U.S. Public Health Service National Research Service Award GM07185 (to M.P.H.). D.L.B. is an investigator of the Howard Hughes Medical Institute.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-09-0692. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-09-0692.
References
- Black, D.L. (1991). Does steric interference between splice sites block the splicing of a short c-src neuron-specific exon in non-neuronal cells? Genes Dev. 5, 389-402. [DOI] [PubMed] [Google Scholar]
- Black, D.L. (1992). Activation of c-src neuron-specific splicing by an unusual RNA element in vivo and in vitro. Cell 69, 795-807. [DOI] [PubMed] [Google Scholar]
- Carmo-Fonseca, M. (2002). The contribution of nuclear compartmentalization to gene regulation. Cell 108, 513-521. [DOI] [PubMed] [Google Scholar]
- Chan, R.C., and Black, D.L. (1995). Conserved intron elements repress splicing of a neuron-specific c-src exon in vitro [published erratum appears in Mol. Cell. Biol. 1997 May;17(5): 2970]. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mol. Cell. Biol. 15, 6377-6385. [DOI] [PMC free article] [PubMed]
- Chen, C., Gherzi, R., Ong, S., Chan, E.L., Raijmakers, R., Pruijn, G.J.M., Stoeklin, G., Moroni, C., Mann, M., and Karin, M. (2001). AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107, 451-464. [DOI] [PubMed] [Google Scholar]
- Chou, M.Y., Underwood, J.G., Nikolic, J., Luu, M.H., and Black, D.L. (2000). Multisite RNA binding and release of polypyrimidine tract binding protein during the regulation of c-src neural-specific splicing. Mol. Cell 5, 949-957. [DOI] [PubMed] [Google Scholar]
- Davis-Smyth, T., Duncan, R.C., Zheng, T., Michelotti, G., and Levens, D. (1996). The far upstream element-binding proteins comprise an ancient family of single-strand DNA-binding transactivators. J. Biol. Chem. 271, 31679-31687. [DOI] [PubMed] [Google Scholar]
- Duncan, R., Collins, I., Tomonaga, T., Zhang, T., and Levens, D. (1996). A unique transactivation sequence motif is found in the carboxyl-terminal domain of the single-strand-binding protein FBP. Mol. Cell. Biol. 16, 2274-2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabini, G., Raijmakers, R., Hayer, S., Fouraux, M.A., Pruijn, G.J., and Steiner, G. (2001). The heterogeneous nuclear ribonucleoproteins I and K interact with a subset of the ro ribonucleoprotein-associated Y RNAs in vitro and in vivo. J. Biol. Chem. 276, 20711-20718. [DOI] [PubMed] [Google Scholar]
- Femino, A.M., Fogarty, K., Lifshitz, L.M., Carrington, W., and Singer, R.H. (2003). Visualization of single molecules of mRNA in situ. Methods Enzymol. 361, 245-304. [DOI] [PubMed] [Google Scholar]
- Ghetti, A., Pinol-Roma, S., Michael, W.M., Morandi, C., and Dreyfuss, G. (1992). hnRNP I, the polypyrimidine tract-binding protein: distinct nuclear localization and association with hnRNAs. Nucleic Acids Res. 20, 3671-3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, W., Pan, F., Zhang, H., Bassell, G.J., and Singer, R.H. (2002). A predominantly nuclear protein affecting cytoplasmic localization of β-actin mRNA in fibroblasts and neurons. J. Cell Biol. 156, 41-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, L., Weber, A., and Levens, D. (2000). Nuclear targeting determinants of the far upstream element binding protein, a c-myc transcription factor. Nucleic Acids Res. 28, 4558-4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, S. (2000). Review: perinucleolar structures. J. Struct. Biol. 129, 233-240. [DOI] [PubMed] [Google Scholar]
- Huang, S., Deernick, T.J., Ellisman, M.H., and Spector, D.L. (1997). The dynamic organization of the perinucleolar compartment in the cell nucleus. J. Cell Biol. 137, 965-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, S., Deernick, T.J., Ellisman, M.H., and Spector, D.L. (1998). The perinucleolar compartment and transcription. J. Cell Biol. 143, 35-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttelmaier, S., Illenberger, S., Grosheva, I., Rudiger, M., Singer, R.H., and Jockusch, B.M. (2001). Raver 1, a dual compartment protein, is a ligand for PTB/hnRNPI and microfilament attachment proteins. J. Cell Biol. 155, 775-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, K.B., Dredge, B.K., Stefani, G., Zhong, R., Buckanovich, R.J., Okano, H.J., Yang, Y.Y., and Darnell, R.B. (2000). Nova-1 regulates neuron-specific alternative splicing and is essential for neuronal viability. Neuron 25, 359-371. [DOI] [PubMed] [Google Scholar]
- Kedinger, C., Gniazdowski, J.L., Mandel, J.R, Jr., Gissinger, F., and Chambon, P. (1970). α-Amanitin: a specific inhibitor of one of two DNA-dependent TNA polymerase activities from calf thymus. Biochem. Biophys. Res. Commun. 38, 165-171. [DOI] [PubMed] [Google Scholar]
- Kimhi, Y., Palfrey, C., Spector, I., Barak, Y., and Littauer, U.Z. (1976). Maturation of neuroblastoma cells in the presence of dimethylsulfoxide. Proc. Natl. Acad. Sci. USA 73, 462-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroll, T.T., Zhao, W., Jiang, C., and Huber, P.W. (2002). A homolog of FBP2/KSRP binds to localized mRNAs in Xenopus oocytes. Development 129, 5609-5619. [DOI] [PubMed] [Google Scholar]
- Labourier, E., Adams, M.D., and Rio, D.C. (2001). Modulation of P-element pre-mRNA splicing by a direct interaction between PSI and U1 snRNP 70K protein. Mol. Cell 8, 363-373. [DOI] [PubMed] [Google Scholar]
- Labourier, E., Blanchette, M., Feiger, J.W., Adams, M.D., and Rio, D.C. (2002). The KH-Type RNA-binding protein PSI is required for Drosophila viability, male fertility, and cellular mRNA processing. Genes Dev. 16, 72-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond, A.I., and Spector, D.L. (2003). Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell. Biol. 4, 605-612. [DOI] [PubMed] [Google Scholar]
- Lee, B., Matera, A.G., Ward, D.C., and Craft, J. (1996). Association of RNase mitochondrial RNA processing enzyme with ribonuclease P in higher ordered structures in the nucleolus: a possible coordinate role in ribosome biogenesis. Proc. Natl. Acad. Sci. USA 93, 11471-11476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lellek, H., Kirsten, R., Diehl, I., Apostel, F., Buck, F., and Greeve, J. (2000). Purification and molecular cloning of a novel essential component of the apolipoprotein B mRNA editing enzyme-complex. J. Biol. Chem. 275, 19848-19856. [DOI] [PubMed] [Google Scholar]
- Levsky, J.M., Shenoy, S.M., Pezo, R.C., and Singer, R.H. (2002). Single-cell gene expression profiling. Science 297, 836-840. [DOI] [PubMed] [Google Scholar]
- Levy, J.B., Dorai, T., Wang, L.H., and Brugge, J.S. (1987). The structurally distinct form of pp60c-src detected in neuronal cells is encoded by a unique c-src mRNA. Mol. Cell. Biol. 7, 4142-4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindell, T.J., Weinberg, F., Morries, P.W., Roeder, R.G., and Rutter, W.J. (1970). Specific inhibition of nuclear RNA polymerase II by α-amanitin. Science 170, 447-449. [DOI] [PubMed] [Google Scholar]
- Liu, J., Collins, I., Ge, H., Libutti, D., Li, J., Egly, J.M., and Levens, D. (2000). The FBP interacting repressor targets TFIIH to inhibit activated transcription. Mol. Cell 5, 331-341. [DOI] [PubMed] [Google Scholar]
- Markovtsov, V., Nikolic, J.M., Goldman, J.A., Turck, C.W., Chou, M.Y., and Black, D.L. (2000). Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol. Cell. Biol. 20, 7463-7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez, R., Mathey, P.B., Bernards, A., and Baltimore, D. (1987). Neuronal pp60c-src contains a six-amino acid insertion relative to its non-neuronal counterpart. Science 237, 411-415. [DOI] [PubMed] [Google Scholar]
- Matera, A.G. (1999). Nuclear bodies: multifaceted subdomains of the interchromatin space. Trends Cell Biol. 9, 302-309. [DOI] [PubMed] [Google Scholar]
- Matera, A.G., Frey, M.R., Margelot, K., and Wolin, S.L. (1995). A perinucleolar compartment contains several RNA polymerase III transcripts as well as the polypyrimidine tract binding protein, hnRNP I. J. Cell Biol. 129, 1181-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min, H., Turck, C.W., Nikolic, J.M., and Black, D.L. (1997). A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes Dev. 11, 1023-1036. [DOI] [PubMed] [Google Scholar]
- Misteli, T., and Spector, D.L. (1999). RNA polymerase II targets pre-mRNA splicing factors to transcription sites in vivo. Mol. Cell 3, 697-705. [DOI] [PubMed] [Google Scholar]
- Modafferi, E.F., and Black, D.L. (1997). A complex intronic splicing enhancer from the c-src pre-mRNA activates inclusion of a heterologous exon. Mol. Cell. Biol. 17, 6537-6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modafferi, E.F., and Black, D.L. (1999). Combinatorial control of a neuronspecific exon. RNA 5, 687-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlemann, O., Mock-Casagrande, C.S., Wang, J., Li, S., Custodio, N., Carmo-Fonseca, M., Wilkinson, M.F., and Moore, M.J. (2001). Precursor RNAs harboring nonsense codons accumulate near the site of transcription. Mol. Cell 8, 33-43. [DOI] [PubMed] [Google Scholar]
- Nguyen, V.T., Giannoni, F., Dubois, M.F., Seo, S.J., Vigneron, M., Kedinger, C., and Bensaude, O. (1996). In vivo degradation of RNA polymerase II largest subunit triggered by alpha-amanitin. Nucleic Acids Res. 24, 2924-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleynikov, Y., and Singer, R.H. (2003). Real-time visualization of ZBP1 association with beta-actin mRNA during transcription and localization. Curr. Biol. 13, 199-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinol-Roma, S., Swanson, M.S., Gall, J.G., and Dreyfuss, G. (1989). A novel heterogeneous nuclear RNP protein with a unique distribution on nascent transcripts. J. Cell Biol. 109, 2575-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehbein, M., Kindler, S., Horke, S., and Richter, D. (2000). Two trans-acting rat-brain proteins, MARTA1 and MARTA2, interact specifically with the dendritic targeting element in MAP2 mRNAs. Brain Res. Mol. Brain Res. 79, 192-201. [DOI] [PubMed] [Google Scholar]
- Rehbein, M., Wege, K., Buck, F., Schweizer, M., Richter, D., and Kindler, S. (2002). Molecular characterization of MARTA1, a protein interacting with the dendritic targeting element of MAP2 mRNAs. J. Neurochem. 82, 1039-1046. [DOI] [PubMed] [Google Scholar]
- Shopland, L.S., Byron, M., Stein, J.L., Lian, J.B., Stein, G.S., and Lawrence, J.B. (2001). Replication-dependent histone gene expression is related to Cajal body (CB) association but does not require sustained CB contact. Mol. Biol. Cell 12, 565-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shopland, L.S., Johnson, C.V., and Lawrence, J.B. (2002). Evidence that all SC-35 domains contain mRNAs and that transcripts can be structurally constrained within these domains. J. Struct. Biol. 140, 131-139. [DOI] [PubMed] [Google Scholar]
- Siebel, C.W., Admon, A., and Rio, D.C. (1995). Soma-specific expression and cloning of PSI, a negative regulator of P element pre-mRNA splicing. Genes Dev. 9, 269-283. [DOI] [PubMed] [Google Scholar]
- Siebel, C.W., Kanaar, R., and Rio, D.C. (1994). Regulation of tissue-specific P-element pre-mRNA splicing requires the RNA-binding protein PSI. Genes Dev. 8, 1713-1725. [DOI] [PubMed] [Google Scholar]
- Smith, K.P., and Lawrence, J.B. (2000). Interactions of U2 gene loci and their nuclear transcripts with Cajal (coiled) bodies: evidence for PreU2 within Cajal bodies. Mol. Biol. Cell 11, 2987-2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timchenko, L.T., Miller, J.W., Timchenko, D.R., DeVore, D.R., Datar, K.V., Lin, L., and Roberts, R. (1996). Identification of a (CUG)n triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nucleic Acids Res. 24, 4407-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valcarcel, J., and Gebauer, F. (1997). Post-transcriptional regulation: the dawn of PTB. Curr. Biol. 7, R705-R708. [DOI] [PubMed] [Google Scholar]
- Wagner, E.J., and Garcia-Blanco, M.A. (2001). Polypyrimidine tract binding protein antagonizes exon definition. Mol. Cell. Biol. 21, 3281-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C., Politz, J.C., Pederson, T., and Huang, S. (2003). RNA polymerase III transcripts and the PTB protein are essential for the integrity of the perinucleolar compartment. Mol. Biol. Cell 14, 2425-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]