Abstract
Sulfadoxine-pyrimethamine (SP) has been widely deployed in Africa for malaria control and molecular evidence of parasite drug-resistance is prevalent. However, the temporal effects on the selection of Plasmodium falciparum are not well understood. We conducted a retrospective serial cross-sectional study between 1997 and 2006 to investigate changes in drug-resistant malaria among pregnant women delivering at a single hospital in Blantyre, Malawi. P. falciparum parasites were genotyped for parasite clone multiplicity and drug-resistance mutations, and the strength of selection upon mutant genotypes was quantified. Five mutations in the dihydrofolate reductase and dihydropteroate synthase genes began at moderate frequencies and achieved fixation by 2005; the frequency of the highly-SP-resistant “quintuple mutant” haplotype increased from 19% to 100%. The selective advantage of alleles and haplotypes were quantified with selection coefficients: Selection was positive on all mutant alleles and haplotypes associated with SP resistance, and the relative fitness of the quintuple mutant haplotype was 0.139 (95% C.I. 0.067 – 0.211), indicating a substantial positive selective advantage. Mutations that confer higher levels of resistance to SP did not emerge. SP-resistant haplotypes were rapidly selected for and fixed in P. falciparum populations infecting pregnant women while SP was widely deployed in Malawi. These results underscore the pressing need for new preventive measures for pregnancy-associated malaria and provide a real-world model of the selection landscape malaria parasites.
Keywords: malaria, pregnancy-associated malaria, drug resistance, parasite evolution
1. Introduction
Pregnancy-associated malaria (PAM) may be responsible for up to 200,000 infant deaths every year (Steketee et al. 2001), and it is the most important preventable cause of poor birth outcomes in malaria-endemic areas in sub-Saharan Africa. The receipt of two to three doses of sulfadoxine-pyrimethamine (SP) as intermittent preventive therapy in pregnancy (IPTp-SP) decreases the risks of maternal malaria, maternal anemia, and low-birth weight (van Eijk et al. 2004). Although SP-resistant Plasmodium falciparum strains are prevalent across sub-Saharan Africa (Sridaran et al. 2010), SP has remained effective in most settings at preventing poor birth outcomes (ter Kuile et al. 2007).
Resistance to SP in P. falciparum is associated with the accumulation of single nucleotide polymorphisms (SNPs) in the parasite dihydrofolate reductase (dhfr) and dihydropteroate synthase (dhps) (Picot et al. 2009). These mutations are broadly distributed across sub-Saharan Africa (Sridaran et al. 2010), and parasite population genetic studies suggest that most derive not from de novo mutation but rather from the spread of resistant haplotypes from few origins (Roper et al. 2004; Maiga et al. 2007; Pearce et al. 2009). Few studies have explored temporal trends in these haplotypes (Raman et al. 2010; Abdel-Muhsin et al. 2004; Mockenhaupt et al. 2008; Nsanzabana et al. 2010), and none have comprehensively assessed mutants in SP-resistance in pregnant women. In Malawi, SP was adopted in 1993 as first-line treatment for uncomplicated malaria and for IPTp; first-line therapy was changed to an artemisinin-combination therapy in 2007, but SP continues to be employed for IPTp. Multiple reports document a high prevalence of SP-resistance mutations in adults and children with malaria (Plowe et al. 1997; Kublin et al. 2002; Bwijo et al. 2003; Nkhoma, et al. 2007), but no studies have explored the rate of the spread of this resistance.
Quantifying the rate of the spread of resistance can both inform understanding of the clinical durability of IPTp-SP and also assist predictive modeling of the rate of parasite evolution in response to current and future antimalarials. The QuEERPAM study (Queen Elizabeth Central Hospital Epidemiology of Resistance in Pregnancy-Associated Malaria) was a retrospective, serial cross-sectional molecular analysis of P. falciparum parasites infecting the peripheral blood of delivering women at a single hospital in Blantyre, Malawi. Herein, we describe temporal changes over 9 years in the frequency of drug-resistant genotypes and the multiplicity of parasite clones, and quantify the relative degree of selection on these haplotypes in the setting of intense drug pressure.
2. Materials and methods
2.1. Ethics statement
Ethics approval for this study was granted by the Research Ethics Committee of the College of Medicine, University of Malawi, and by the review boards of the Malawi Health Sciences Research Committee and the University of North Carolina at Chapel Hill.
2.2. Sample collection
Patient enrollment and sample collection have been described previously (Feng et al. 2010; Rogerson et al. 2000). Briefly, women delivering between 1997 and 2006 at Queen Elizabeth Central Hospital in Blantyre, Malawi, were invited to participate. Those who consented to participate were queried regarding demographic and clinical information, and peripheral blood was stored as red cell pellets at −40C or below prior to spotting onto filter paper for shipment to UNC. Thick blood smears were also prepared from peripheral blood as examined for malaria parasites (Feng et al. 2010); a random sample of approximately 25% of specimens between 1997 and 2005 which demonstrated Plasmodium falciparum parasites on microscopic examination were selected for genotyping.
2.3. Genotyping procedures
Three punches from each specimen were deposited in an individual well of a 96-well plastic plate. Genomic DNA (gDNA) was extracted with the invitrogen Purelink 96 kit (invitrogen, Foster City, CA) using a vacuum manifold.
The dhfr and dhps targets were amplified in separate reactions. The dhfr amplification included 300nM of primer 51-F (TGAGGTTTTTAATAACTACACATTTAGAGGTCT), 500nM of primer 164-R (TCGCTAACAGAAATAATTTGATACTCAT), 12.5uL of Qiagen HotStarTaq (Qiagen, Valencia, CA, USA), and 5uL of gDNA in a 25uL reaction. The dhps amplification included 900nM of primer 437-F (TGAAATGATAAATGAAGGTGCTAGTGT), 300nM of primer 613-R (GTTGTGTATTTATTACAACATTTTGATCATTC), 12.5uL of HotStarTaq, and 5uL of gDNA in a 25uL reaction. Cycling conditions for all amplifications were: 95C × 15m, (94C × 45s, 51C × 45s, 72C × 1m) × 45, then 72C × 10m. The pfmdr1 loci were genotyped by amplifying a segment at the 5-prime end of pfmdr1 in a nested reaction using 5uL of gDNA in a 25uL reactions (Vinayak et al. 2010). All reaction plates included appropriate negative and positive controls. Separate work areas were maintained for gDNA extraction, preparation of reaction plates, and analysis of amplified fragments, and filtered pipet tips were used for all procedures.
PCR products were electrophoresed for confirmation of amplification as indicated by a band at 500bp, 600bp, or 500bp, for the dhfr, dhps, and pfmdr1 fragments, respectively. PCR products were bidirectionally sequenced using ABI BigDye Terminator chemistry. Sequences were aligned with Sequencher v4.8 (Gene Codes, Ann Arbor, MI). Wildtype 3d7 sequences obtained from GenBank were used as referents for alignments: XM_001351443 for dhfr, Z30654 for dhps, and XM_001351751.1 for pfmdr1. Base calls at loci corresponding to codons 51, 59, 108 and 164 (for dhfr), codons 436, 437, 540, 581, and 613 (for dhps), and codons 86 and 184 (for pfmdr1) were confirmed by manual inspection. Aligned sequences were manually inspected both for mixed alleles (defined as peaks ≥10% of primary peak, above baseline) and for mutations at other loci; novel mutations were defined as single nucleotide polymorphisms evident in both sequences obtained from the same amplicon at a locus not included above.
Samples positive in the dhfr and dhps reactions were genotyped at merozoite surface proteins 1 and 2 (msp1 and msp2) to determine the multiplicity of infection (MOI), following published protocols (Snounou et al. 1999; Kamwendo et al. 2002). Products were resolved on agarose gels, bands were counted and sized by two independent, masked observers, and differences were resolved by consensus.
2.4. Definitions and statistical analyses
Demographic and antenatal data were compared between years using one-way ANOVA, the Kruskal-Wallis rank test, or the chi-squared test. Numbers of bands among msp1 or msp2 family-specific amplifications were summed to generate a target-specific MOI.
Because the multiplicity of parasite clones that constitute a single infection affects the estimate of genotype frequencies, we employed MalHaploFreq in order to convert the prevalences of alleles and haplotypes into frequencies (Hastings and Smith 2008). In brief, the program incorporates a measure of the multiplicity of parasite clones and employs maximum likelihood analysis in order to estimate the frequency of molecular markers (genotype or haplotype) within a population. Subsequently, haplotypes for combined dhfr-dhps were assigned by multiplying the frequencies of individual dhfr and dhps haplotypes; though this approach may underestimate the frequency of the combined haplotype owing to linkage disequilibrium, it provides a reasonable estimate of frequency. Haplotypes for dhfr were based upon codons 51, 59, and 108 and classified as wildtype (no mutations), single mutants (single mutation), double mutants (two mutations), or triple mutants (all three mutants); those for dhps were based upon codons 437 and 540, and classified as wildtype (no mutations) or single (one mutation) or double (two mutations) mutants. Combined dhfr-dhps haplotypes followed similar convention.
Between years, MOI was compared with Kruskal-Wallis analysis of variance and haplotype frequencies with the likelihood ratio test.
In order to quantify the selective advantage or disadvantage of genotypes, selection coefficients were calculated to quantify the rate of change of alleles and haplotypes between subsequent generations of parasites; consistent with published analyses, we used an estimate of 6 parasite generations per year. Coefficients were computed by plotting the natural log of the ratio of the allele/haplotype frequency of interest to the frequency of alternate alleles/haplotypes, against parasite generations (assuming 6 P. falciparum generations per year) (Anderson and Roper 2005; Nsanzabana et al. 2010). To prevent division by zero, frequencies of 100% were included as 99.99%, and 0% as 0.01%. The coefficient from a linear regression line fitted to this plot was the selection coefficient; these coefficients were generated annually and for the overall 9-year period. Annual selection coefficients were calculated using regression lines fitted to data in the year before and after the year of interest. To explore the relationship between MOI and mutation frequency, correlations between the frequency of mutant alleles and haplotypes and mean annual MOI were calculated with Spearman's rank correlation.
All statistical analyses were performed using Stata/IC (v10, Stata Corp, College Station, TX) apart from those that employed MalHaploFreq (see above).
3. Results
3.1. Demographic and clinical data
Full data were available for 189 women from 1997–2004, ranging from 5 women in 1997 to 42 women in 2001 and 2004. The mean (SD) age of included women was 21.4 (4.3), and did not vary over the study period (Table 1). Primigravidae comprised 50.4% of the sample, and HIV-positive women 41.4%. SP use as IPTp changed significantly between 1997 and 2005: Beginning in 2001, no women denied receiving SP as IPTp. The timing of the last dose of SP was available from 1999 onwards, and varied significantly between years (p = 0.002): The proportion of women who received their last SP dose within 60 days prior to delivery increased from 55.6 to 81.8%.
Table 1.
Temporal trends in demographic and clinical data
| 1997 | 1998 | 1999 | 2000 | 2001 | 2002 | 2003 | 2004 | 2005 | p-valuea | |
|---|---|---|---|---|---|---|---|---|---|---|
| No. of women | 6 | 38 | 16 | 20 | 44 | 13 | 11 | 57 | 13 | -- |
|
| ||||||||||
| Age, y, mean | 19.3 | 20.7 | 20.5 | 20.9 | 21.2 | 22.7 | 22 | 22.3 | 22.7 | 0.500 |
|
| ||||||||||
| HIV pos, % | 40 | 39.5 | 29.4 | 47.4 | 29.3 | NA | 50 | 33.3 | NA | 0.827 |
|
| ||||||||||
| Primigravid, % | 83.3 | 52.6 | 41.2 | 40 | 64.4 | 53.9 | 36.4 | 56.4 | 53.4 | 0.808 |
|
| ||||||||||
| SP doses, % | ||||||||||
| 0 | 0 | 31.3 | 25 | 16.7 | 0 | 0 | 0 | 0 | 0 | |
| 1 | 75 | 46.9 | 50 | 72.2 | 44.4 | 25 | 30 | 21.2 | 15.4 | <0.001 |
| ≥2 | 25 | 21.8 | 25 | 11.2 | 55.6 | 75 | 70 | 78.8 | 84.6 | |
|
| ||||||||||
| SP doses, mean (SD) | 1.25 (0.5) | 1 (0.92) | 1 (0.73) | 1 (0.69) | 1.58 (0.55) | 1.92 (0.67) | 2 (0.94) | 2.21 (0.94) | 2.23 (0.83) | <0.001 |
|
| ||||||||||
| % last SP dose < 60 days prior to delivery b | NA | NA | 55.6 | 21.4 | 48.7 | 75 | 80 | 76.6 | 81.8 | 0.002 |
NA: not available; SD: standard deviation; HIV: human immunodeficiency virus; SP: sulfadoxine-pyrimethamine.
Calculated with oneway ANOVA for continuous data or the chi-squared test for categorical variables.
Of women who received SP.
3.2. Multiplicity of infection
Between years there were significant differences in mean number of msp1 (p = 0.025) and msp2 (p < 0.001) genotypes detected (Figure 1A). From 1997 to 2005, mean (SD) MOI decreased from 2.4 (1.1) to 1.5 (0.8) for msp1 and 3.8 (1.3) to 1.8 (1.2) for msp2. The proportion of women harboring more than one msp1 clone decreased from 80% to 43% (p = 0.116), and that for msp2 clones decreased from 100% to 45% (p = 0.016) (Figure 1B, 1C). There were no differences in MOI by gravidity or the receipt of SP within 60 days prior to delivery (data not shown).
Figure 1. Multiplicity of infection over time.
Mean (standard deviation) number of msp1 and msp2 variants harbored by included patients (A). Proportion of patients with numbers of distinct msp1 (B) and msp2 (C) variants per year.
Plotted points in A are offset within years to discriminate overlapping points.
3.3. Mutant genotypes
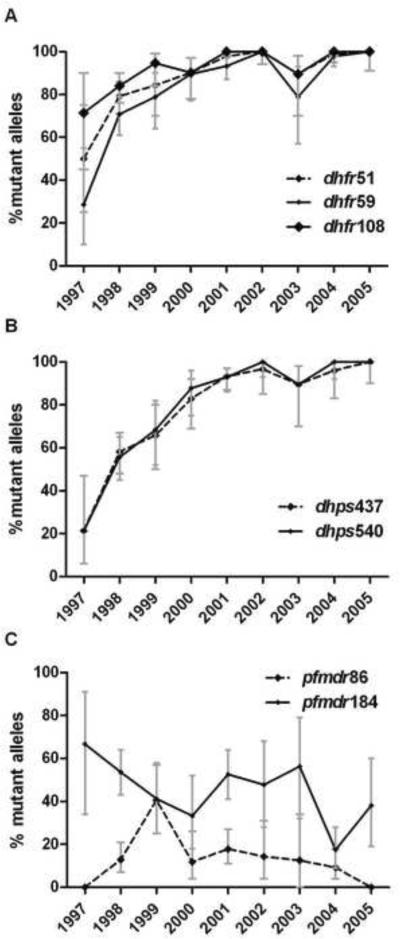
Mutations at codons 51, 59, and 108 of dhfr and codons 437 and 540 of dhps achieved 100% frequency by 2005 (Figure 2A, B). Mutant allele frequencies in 1997 ranged from 21% to 71%, and were generally lower in dhps than dhfr. Mutations in pfmdr1 decreased over time, but the trend was not significant (Figure 2C). There were no novel dhfr, dhps, or pfmdr1 mutations. There were no mutants at codon 164 of dhfr, and a mutation at codon 581 of dhps was detected in only 1 sample, in 2004.
Figure 2. Frequency of mutant dhfr (A), dhps (B), and pfmdr1 (C) alleles by year.
Points are frequencies, bars are 95% confidence intervals.
Allele frequencies were generated from allele prevalences using MalHaploFreq (Hastings and Smith 2008), which uses maximum likelihood analysis to estimate genotype frequencies based upon genotype prevalence and the multiplicity of parasite clones.
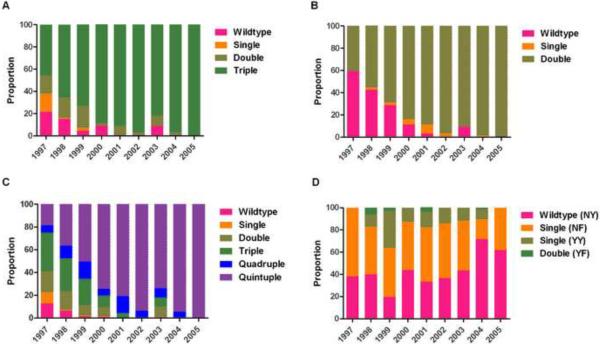
Haplotype frequencies for both dhfr (p < 0.0001) and dhps (p < 0.0001) differed significantly between years. The dhfr triple mutant haplotype increased from 45.6% (95% C.I. 25 – 68) to 100% (95% C.I. 91 – 100), and the dhps double mutant haplotype increased from 40.5% (95% C.I. 21 – 63) to 100% (95% C.I. 90 – 100); the combined quintuple mutant haplotype increased from 18.5% to 100% (Figure 3A, B, C). pfmdr1 haplotype frequencies did not vary significantly between years (Figure 3D).
Figure 3. Frequency of dhfr (A), dhps (B), dhfr-dhps (C), and pfmdr1 (D) haplotypes by year.
The frequencies of dhfr, dhps, and pfmdr1 haplotypes were calculated directly using MalHaploFreq (Hastings and Smith 2008), which uses maximum likelihood analysis to estimate genotype frequencies based upon genotype prevalence and multiplicity of parasite clones. The frequencies of dhfr-dhps haplotypes were calculated by multiplying frequencies of individual gene haplotypes, an approximation that cannot account for possible linkage disequilibrium between dhfr and dhps.
In stratified analyses, there were no differences in the frequency of mutations or haplotypes between primigravidae and multigravidae, nor between those who received SP late in pregnancy (within 60 days prior to delivery) and who received it early. Because no women denied the receipt of SP from 2001 onwards, a comparison of genotypes between SP-exposed and –unexposed was possible only for 1998–2000 (there were no non-exposed women in 1997); over these years, the frequency of dhfr haplotypes did not differ significantly in either SP-exposed women or SP-unexposed women (p > 0.05), though dhps haplotypes differed over these years in both groups (both p < 0.001).
3.4. Estimation of selection pressure on mutant genotypes
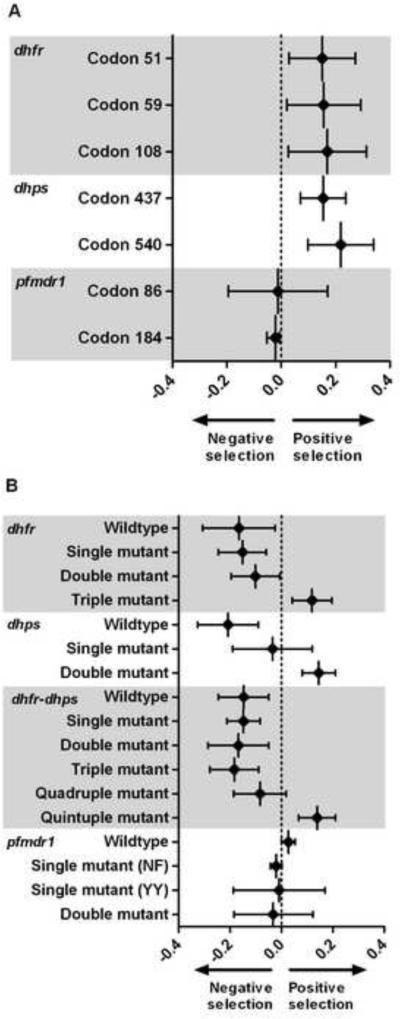
Selection coefficients were calculated to measure the relative fitness of alleles and haplotypes over time by quantifying the proportional survival of genotypes in subsequent parasite generations. Between 1997 and 2005, selection coefficients were significant for all dhfr and dhps mutations and nonsignificant on pfmdr1 alleles (Figure 4A). Over the study period, coefficients for mutations in codons 51, 59, and 108 in dhfr were 0.151 (95% C.I. 0.029 – 0.273), 0.156 (95% C.I. 0.020 – 0.293), and 0.170 (95% C.I. 0.026 – 0.314); those for mutations in codons 437 and 540 in dhps were 0.154 (95% C.I. 0.071 – 0.237) and 0.219 (95% C.I. 0.099 – 0.340). Selection on pfmdr1 mutations at codons 86 and 184 was close to null and statistically nonsignificant.
Figure 4. Selection coefficients on mutant alleles (A) and haplotypes (B).
Plotted points are point estimates, bars are 95% confidence intervals.
Plotted points are the slope of a regression line fitted to a plot of the ln(R/S) plotted against parasite generations (assumed 6/year), where R is the frequency of the mutant allele at the indicated codon (or haplotype), and S is the frequency of the wildtype allele (or other haplotype). Values greater than zero represent an allele/haplotype survival advantage in subsequent parasite generations; those less than zero indicate a survival disadvantage.
We repeated selection analyses upon dhfr, dhps, and combined dhfr-dhps haplotypes (Figure 4B). The dhfr triple mutant haplotype (0.119; 95% C.I. 0.042 – 0.196), the dhps double mutant haplotype (0.146; 95% C.I. 0.081 – 0.211), and the combined quintuple mutant haplotype (0.139; 95% C.I. 0.067 – 0.211) were all under significant positive selective pressure. Most dhfr and dhps haplotypes containing wildtype alleles exhibited significant negative selective pressure as evidenced by selection coefficients less than zero. Coefficients for pfmdr1 haplotypes were clustered near null and statistically nonsignificant.
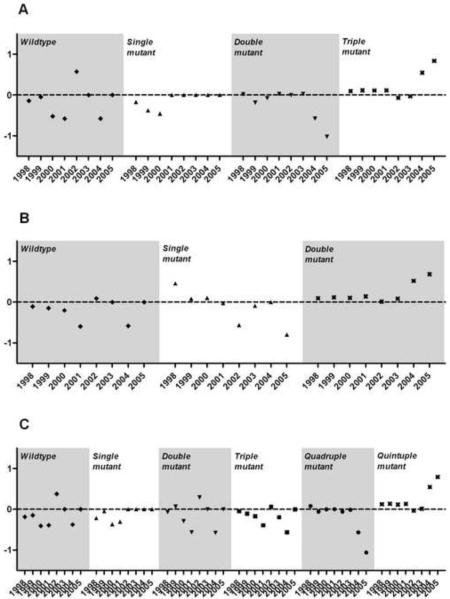
Annualized selection coefficients were calculated for all mutant alleles and haplotypes to examine temporal trends in the degree of selection (Figure 5). Nearly all coefficients were non-significant owing to small sample sizes that result from limiting the analyses to 3-year blocks. However, point estimates of selection increased in magnitude in the final 3 years of the study upon extant haplotypes: positively on the quintuple mutant, and negatively on partially-wildtype haplotypes.
Figure 5. Annual selection coefficients on dhfr (A), dhps (B) and combined dhfr-dhps (C) haplotypes.
Plotted points are point estimates.
Plotted points are the slope of a regression line fitted to a plot of the ln(R/S) plotted against parasite generations (assumed 6/year), where R is the frequency of the mutant haplotype indicated, and S is the frequency of other haplotypes. Annual selection coefficients were calculated as the slope of the line fitted to the ln(R/S) for the year of interest and the preceding and subsequent years. Values greater than zero represent a haplotype survival advantage in subsequent parasite generations; those less than zero indicate a survival disadvantage.
To examine for differences in selection by receipt of SP, coefficients for the year 1999 were stratified by SP exposure (the only year that was flanked by years in which women did not receive SP). Selection coefficients were nonsignificant for all alleles and haplotypes, except for the dhfr-dhps quintuple mutant haplotype: in SP-unexposed women, there was negative selection that was not significant (−0.091; 95% C.I. –10 – 10), but in SP-exposed women, there was significant positive selection (0.246; 95% C.I. 0.23 – 0.26).
3.5. Correlation of MOI with mutant genotypes
Annual mutant allele frequencies were significantly negatively correlated with mean MOI at msp2 for all dhfr and dhps loci and at msp1 for dhfr108 and dhps540 (Table 2). There was no significant correlation with pfmdr1 mutation frequency. Wildtype haplotypes of dhfr, dhps, and dhfr-dhps were significantly positively correlated with mean msp1 and msp2 MOI, and full-mutant haplotypes of these genes were negatively associated with mean msp1 and msp2 MOI.
Table 2.
Correlations between annual mean multiplicity of infection and the annual frequencies of mutant alleles and haplotypes
| Mean mspl MOI | p-value | Mean msp2 MOI | p-value | |
|---|---|---|---|---|
| Gene, mutant codon | ||||
| dhfr | ||||
| 51 | −0.60 | 0.086 | −0.80 | 0.010 |
| 59 | −0.59 | 0.096 | −0.76 | 0.016 |
| 108 | −0.71 | 0.034 | −0.76 | 0.018 |
| dhps | ||||
| 437 | −0.65 | 0.058 | −0.85 | 0.004 |
| 540 | −0.68 | 0.045 | −0.88 | 0.002 |
| pftndr1 | ||||
| 86 | −0.03 | 0.93 | 0.19 | 0.620 |
| 184 | 0.50 | 0.171 | 0.52 | 0.154 |
|
| ||||
| Gene, mutant haplotype | ||||
| dhfr | ||||
| Wildtype | 0.72 | 0.028 | 0.79 | 0.011 |
| Single | 0.73 | 0.025 | 0.84 | 0.004 |
| Double | 0.45 | 0.224 | 0.63 | 0.067 |
| Triple | −0.63 | 0.067 | −0.82 | 0.007 |
| dhps | ||||
| Wildtype | 0.68 | 0.045 | 0.82 | 0.002 |
| Single | −0.19 | 0.63 | 0.03 | 0.93 |
| Double | −0.68 | 0.042 | −0.90 | 0.001 |
| Combined dhfr-dhps | ||||
| Wildtype | 0.77 | 0.014 | 0.90 | 0.001 |
| Single | 0.75 | 0.020 | 0.91 | <0.001 |
| Double | 0.65 | 0.060 | 0.82 | 0.007 |
| Triple | 0.74 | 0.021 | 0.93 | <0.001 |
| Quadruple | 0.20 | 0.606 | 0.52 | 0.15 |
| Quintuple | −0.72 | 0.030 | −0.88 | 0.002 |
| pftndr1 | ||||
| Wildtype | −0.32 | 0.406 | −0.57 | 0.112 |
| Single (NF) | 0.42 | 0.265 | 0.52 | 0.154 |
| Single (YY) | −0.03 | 0.932 | 0.193 | 0.62 |
| Double (YF) | −0.20 | 0.604 | −0.09 | 0.815 |
Significance of correlations determined by Spearman's rank correlation; significant correlations highlighted in bold. MOI: multiplicity of infection; msp1: merozoite surface protein 1; msp2: merozoite surface protein 2.
4. Discussion
In this retrospective, serial cross-sectional study, we describe the selection and fixation of P. falciparum mutations associated with resistance to SP in parasites infecting pregnant women, and quantify the degree of selection on mutant alleles and haplotypes. While the quintuple mutant haplotype reached fixation, mutations associated with high-level SP-resistance (dhfr164 and dhps581) were absent or rare. Additionally, we demonstrate a clear decline in the clonal multiplicity of P. falciparum infections. The high prevalence of SP-resistance mutations in Malawian P. falciparum populations has been reported previously (Nkhoma et al. 2007), and the mutation frequencies in our study reflect both the general parasite population and pressure applied by increased SP use in pregnant women. Nevertheless, the QuEERPAM study is the first investigation of antimalarial drug resistance to quantify the rate of the expansion of resistance mutations from moderate frequencies until fixation, and the first large-scale, longitudinal investigation of resistance mutations in parasites infecting pregnant women.
The selection landscape of P. falciparum has generated much interest as a means to identify and modify factors that facilitate the origination and spread of resistance in parasite populations (Mackinnon and Marsh 2010). In this observational study, we cannot establish causality between SP exposure and the expansion of dhfr and dhps mutants alleles and haplotypes; however, their expansion, coupled with the lack of expansion of pfmdr1 mutations that have not been associated with SP resistance, suggests that SP use provided a major selective pressure. Because of the heterogeneity of proposed mediators of the spread of resistance – among them malaria transmission intensity, antimalarial drug use, acquired and innate host immunity, the nature of parasite genomic correlates, and the clinical epidemiology of malaria – population-level longitudinal field studies are limited in number, and much knowledge of selection and spread is inferred from modeling studies in vitro (Lozovsky et al. 2009) and in silico (Antao and Hastings 2011; Mackinnon 2005) or from microsatellite analyses of contemporary parasites derived from cross-sectional surveys (Roper et al. 2004; Pearce et al. 2009). Pregnancy-associated malaria provides a unique model by providing a proxy for acquired immunity (gravidity), standardizing exposure to antimalarials (IPTp with SP), and allowing for an approximation of clinical drug effectiveness (placental malaria and birth outcomes). Additionally, our study benefits from consistent enrollment over many years, which limits sources of bias in allele frequencies.
In contrast to previous field studies in which resistance stabilized at moderate frequencies despite onoing SP use (Pearce et al. 2003; Plowe et al. 2004), those in our study were driven to fixation. This may reflect the study of parasites that were subject to drug pressure at both the population and individual levels. On a population level, Malawi adopted SP as first-line malaria treatment SP in 1993, and fully-mutant dhps and dhfr haplotypes were prevalent in children in the early 2000s (Sridaran et al. 2010). On an individual level, most women had received SP as IPTp, and thus these parasitemias may represent drug-exposed, highly-resistant parasite populations. Though the receipt of SP late in pregnancy may be hypothesized to promote more within-host selection (Menendez et al. 2011; Harrington et al. 2009), there were no differences in selection on genotypes between women who received early versus late SP, and the partial malaria immunity of pregnant women may attenuate the phenomenon of in-host selection (Rogerson et al. 2010; Cravo et al. 2001). Taken together, these data suggest that the the deployment of SP for malaria control can result in rapid fixation of a range of SP-resistance alleles and haplotypes.
The degree of positive selection is directly correlated with the rapidity of the spread of resistance mutations in parasite populations (Hastings 2011), and was relatively consistent between SP-resistance genotypes, with coefficients between 15% and 22% on mutant alleles and 12% to 15% on mutant haplotypes. These coefficients exceed those reported in other epidemiologic settings: in a low-transmission area in South Africa, selection upon the dhfr triple mutant haplotype was estimated at 4.8% in the setting of widespread SP use (Anderson and Roper 2005), while in a moderate-transmission area of PNG over 12 years the selection was estimated at 2.2%, though this varied with the use of SP (Nsanzabana et al. 2010). Several factors may account for this disparity. Because we investigated parasites in women who had largely already received SP as IPTp, these parasites likely represent at least partial treatment failures as opposed to only incident, antimalarial-naïve parasitemias (see above), potentially inflating selection coefficients. Additionally, our population was semi-immune adults, as opposed to children, in whom clone selection and susceptibility to drug may be altered (O'Meara et al. 2006). Finally, our study population had a much higher – though declining – MOI that reflects a greater intensity of P. falciparum transmission, which is a postulated mediator of the spread of resistance (Hastings and Watkins 2005).
Along this line, the temporal trends in MOI and mutant genotype frequencies were significantly negatively correlated over the full course of the study. The hypothesized relationship between transmission intensity (which may be approximated by MOI) and the selection of resistance mutations is the subject of much debate (Ariey and Robert 2003; Hastings 2003; Hastings and Watkins 2005; Talisuna et al. 2007). Convincing field data are scant, but some ecological analyses have suggested that the spread of resistance may be fastest at the extremes of transmission intensity, owing to counterbalancing effects -- within the populations of parasites that constitute individual infections -- of opportunities for parasite “out-crossing” between genetically distinct parasite clones and the effect of competition between resistant and susceptible strains (Talisuna et al. 2003; Bell et al. 2006). In our study, yearly selection coefficients – despite skewing in the final years owing to allele frequencies which approached fixation – were fairly consistent upon fully-mutant dhfr and dhps haplotypes despite a significant coincident fall in MOI. Though causality cannot be inferred from observational data, this consistency suggests that MOI did not substantially mediate the rate of mutant selection in our cohort.
Why did mutations at codon 164 of dhfr and codon 581 of dhps fail to emerge? Both mutations were detected as early as 2001 in Malawi (Alker et al. 2005; Juliano et al. 2008) and the mutations are prevalent in other African settings (Harrington et al. 2009), but despite conditions which clearly favored the fixation of other SP-resistance mutations, only 1 isolate possessed dhps581 and none harbored the dhfr164 mutation. Importantly, both mutations have been convincingly associated with reduced in vitro (Brooks et al. 1994; Sirawaraporn et al. 1997) and in vivo (Krudsood et al. 2005; Gesase et al. 2009) efficacy of SP, and their emergence has been associated with the loss of efficacy of IPTp-SP (Harrington et al. 2011). It has been hypothesized that African P. falciparum populations lack undescribed genetic traits that compensate for a relative loss of enzyme fitness and therefore allow for the propagation of these mutations (Nzila et al. 2005); however, in vitro modeling of evolutionary trajectories of the P. falciparum dhfr have suggested that the dhfr164 mutation is itself compensatory and serves to restore fitness that was compromised by other dhfr mutations (Brown et al. 2010). Alternatively, their absence may also derive from population genetic dynamics. Notably, all mutations that were ultimately driven to fixation began the study with frequencies in excess of 20%, suggesting that their selection to fixation may be evidence of positive frequency-dependent selection upon P. falciparum drug-resistance genotypes (Weedall and Conway 2010). Under this scenario, the directional selection exerted by drug pressure abrogates countervailing pressure to maintain genotypic diversity within P. falciparum populations, limiting the spread of low-level drug-resistance mutants if their fitness advantage is marginal relative to that of the prevailing drug-resistant haplotypes. As an alternative, the spread of mutant genotypes may be facilitated by their acquisition of some threshold absolute frequency, at which selection pressures operate more efficiently to increase gene frequency (Mackinnon 2005). Finally, parasite population size may mediate the selection of mutant genotypes by favoring fewer mutations with relatively larger effects on phenotype (Anderson et al. 2011). Longitudinal studies in other epidemiologic settings can help to understand these phenomena of the P. falciparum selection landscape.
This investigation has several potential limitations. The maximum-likelihood approach to estimating gene and haplotype frequency could have biased our outcomes owing to the underestimation of infection multiplicity provided by traditional msp1/msp2 genotyping (Juliano et al. 2010), but this approach did not substantially alter the point estimates, though it did narrow the confidence intervals. Additionally, the use of linear regression to estimate selection coefficients does not adequately model non-linear changes in mutation and haplotype frequencies over the entire sample (Hastings et al. 2002); nevertheless, yearly selection coefficients (Figure 5) did not demonstrate clear patterns in temporal selection of haplotypes on a finer scale. Secondly, in some years, the number of included women was low, though we did not identify associations between frequencies and population size, our analyses incorporate measures of precision that respond to these differences, and the high MOI in these years resulted in a larger number of parasite strains being genotyped. Finally, we studied only parasites in peripheral blood at delivery, and could not analyze placental parasite populations that may differ from those in the periphery (Kamwendo et al. 2002).
In this investigation of the molecular epidemiology of pregnancy-associated malaria, we employ a serial study design and a population genetics approach to quantify the degree and tempo of selection for drug-resistance mutations. The fixation of mutations associated with resistance to SP serves as both a note of caution regarding the clinical durability of SP to prevent pregnancy-associated malaria and a real-world model of the genomic adaptive landscape of P. falciparum.
Highlights
Plasmodium falciparum mutations conferring drug resistance are widespread
We quantified the evolution of drug-resistant haplotypes in Malawi over 9 years
Mutations achieved fixation at 5 loci, but failed to appear at two loci associated with high-level drug-resistance
Mutant haplotypes expanded at a rate of 13% over each subsequent parasite generation
Longitudinal surveillance of malaria drug resistance can assist predictive modeling
Acknowledgements
We thank RJ Nemeyer and Mike Mistarz (each with the University of North Carolina, Chapel Hill) for their laboratory work and Andrew Read (Pennsylvania State University) for thoughtful discussion of the project. Additionally, Alfredo Mayor (Universitat de Barcelona), Julie Gutman (Centers for Disease Control and Prevention), Brian Greenwood (London School of Hygiene and Tropical Medicine) and our anonymous reviewers provided helpful manuscript suggestions. The staff of Queen Elizabeth Central Hospital in Blantyre provided excellent care of the patients, and ultimately, we are indebted to these study participants.
Funding This work was supported by the Malaria in Pregnancy Consortium (MiP), which is funded through a grant from the Bill & Melinda Gates Foundation to the Liverpool School of Tropical Medicine (to F.O.tK.). Sample collection was funded by a Career Development Fellowship and a Senior Overseas Biomedical Research Fellowship awarded by the Wellcome Trust (to S.J.R.) and by grants from NIH (#AI 49084), NIH-FIC (#5 D43 TW00908), and the Center for AIDS Research at the University North Carolina (to S.R.M.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Competing interestsAll authors declare that they have no commercial or other associations that may constitute conflicts of interest.
References
- Abdel-Muhsin AM, Mackinnon MJ, Ali E, Nassir el KA, Suleiman S, Ahmed S, Walliker D, Babiker HA. Evolution of drug-resistance genes in Plasmodium falciparum in an area of seasonal malaria transmission in Eastern Sudan. J Infect Dis. 2004;189:1239–1244. doi: 10.1086/382509. [DOI] [PubMed] [Google Scholar]
- Alker AP, Mwapasa V, Purfield A, Rogerson SJ, Molyneux ME, Kamwendo DD, Tadesse E, Chaluluka E, Meshnick SR. Mutations associated with sulfadoxine-pyrimethamine and chlorproguanil resistance in Plasmodium falciparum isolates from Blantyre, Malawi. Antimicrob Agents Chemother. 2005;49:3919–3921. doi: 10.1128/AAC.49.9.3919-3921.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson T, Nkhoma S, Ecker A, Fidock D. How can we identify parasite genes that underlie antimalarial drug resistance? Pharmacogenomics. 2011;12:59–85. doi: 10.2217/pgs.10.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson TJ, Roper C. The origins and spread of antimalarial drug resistance: lessons for policy makers. Acta Trop. 2005;94:269–280. doi: 10.1016/j.actatropica.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Antao T, Hastings IM. Environmental, pharmacological and genetic influences on the spread of drug-resistant malaria. Proc Biol Sci. 2011;278:1705–1712. doi: 10.1098/rspb.2010.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariey F, Robert V. The puzzling links between malaria transmission and drug resistance. Trends Parasitol. 2003;19:158–160. doi: 10.1016/s1471-4922(03)00054-0. author reply 160-151. [DOI] [PubMed] [Google Scholar]
- Bell AS, de Roode JC, Sim D, Read AF. Within-host competition in genetically diverse malaria infections: parasite virulence and competitive success. Evolution. 2006;60:1358–1371. [PubMed] [Google Scholar]
- Brooks DR, Wang P, Read M, Watkins WM, Sims PF, Hyde JE. Sequence variation of the hydroxymethyldihydropterin pyrophosphokinase: dihydropteroate synthase gene in lines of the human malaria parasite, Plasmodium falciparum, with differing resistance to sulfadoxine. Eur J Biochem. 1994;224:397–405. doi: 10.1111/j.1432-1033.1994.00397.x. [DOI] [PubMed] [Google Scholar]
- Brown KM, Costanzo MS, Xu W, Roy S, Lozovsky ER, Hartl DL. Compensatory mutations restore fitness during the evolution of dihydrofolate reductase. Mol Biol Evol. 2010;27:2682–2690. doi: 10.1093/molbev/msq160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bwijo B, Kaneko A, Takechi M, Zungu IL, Moriyama Y, Lum JK, Tsukahara T, Mita T, Takahashi N, Bergqvist Y, Bjorkman A, Kobayakawa T. High prevalence of quintuple mutant dhps/dhfr genes in Plasmodium falciparum infections seven years after introduction of sulfadoxine and pyrimethamine as first line treatment in Malawi. Acta Trop. 2003;85:363–373. doi: 10.1016/s0001-706x(02)00264-4. [DOI] [PubMed] [Google Scholar]
- Cravo P, Culleton R, Hunt P, Walliker D, Mackinnon MJ. Antimalarial drugs clear resistant parasites from partially immune hosts. Antimicrob Agents Chemother. 2001;45:2897–2901. doi: 10.1128/AAC.45.10.2897-2901.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G, Simpson JA, Chaluluka E, Molyneux ME, Rogerson SJ. Decreasing burden of malaria in pregnancy in Malawian women and its relationship to use of intermittent preventive therapy or bed nets. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesase S, Gosling RD, Hashim R, Ord R, Naidoo I, Madebe R, Mosha JF, Joho A, Mandia V, Mrema H, Mapunda E, Savael Z, Lemnge M, Mosha FW, Greenwood B, Roper C, Chandramohan D. High resistance of Plasmodium falciparum to sulphadoxine/pyrimethamine in northern Tanzania and the emergence of dhps resistance mutation at Codon 581. PLoS One. 2009;4:e4569. doi: 10.1371/journal.pone.0004569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington WE, Mutabingwa TK, Kabyemela E, Fried M, Duffy PE. Intermittent treatment to prevent pregnancy malaria does not confer benefit in an area of widespread drug resistance. Clin Infect Dis. 2011;53:224–230. doi: 10.1093/cid/cir376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington WE, Mutabingwa TK, Muehlenbachs A, Sorensen B, Bolla MC, Fried M, Duffy PE. Competitive facilitation of drug-resistant Plasmodium falciparum malaria parasites in pregnant women who receive preventive treatment. Proc Natl Acad Sci U S A. 2009;106:9027–9032. doi: 10.1073/pnas.0901415106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings I. How artemisinin-containing combination therapies slow the spread of antimalarial drug resistance. Trends Parasitol. 2011;27:67–72. doi: 10.1016/j.pt.2010.09.005. [DOI] [PubMed] [Google Scholar]
- Hastings IM. Malaria control and the evolution of drug resistance: an intriguing link. Trends Parasitol. 2003;19:70–73. doi: 10.1016/s1471-4922(02)00017-x. [DOI] [PubMed] [Google Scholar]
- Hastings IM, Smith TA. MalHaploFreq: a computer programme for estimating malaria haplotype frequencies from blood samples. Malar J. 2008;7:130. doi: 10.1186/1475-2875-7-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings IM, Watkins WM. Intensity of malaria transmission and the evolution of drug resistance. Acta Trop. 2005;94:218–229. doi: 10.1016/j.actatropica.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Hastings IM, Watkins WM, White NJ. The evolution of drug-resistant malaria: the role of drug elimination half-life. Philos Trans R Soc Lond B Biol Sci. 2002;357:505–519. doi: 10.1098/rstb.2001.1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliano JJ, Porter K, Mwapasa V, Sem R, Rogers WO, Ariey F, Wongsrichanalai C, Read A, Meshnick SR. Exposing malaria in-host diversity and estimating population diversity by capture-recapture using massively parallel pyrosequencing. Proc Natl Acad Sci U S A. 2010;107:20138–20143. doi: 10.1073/pnas.1007068107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliano JJ, Trottman P, Mwapasa V, Meshnick SR. Detection of the dihydrofolate reductase-164L mutation in Plasmodium falciparum infections from Malawi by heteroduplex tracking assay. Am J Trop Med Hyg. 2008;78:892–894. [PMC free article] [PubMed] [Google Scholar]
- Kamwendo DD, Dzinjalamala FK, Snounou G, Kanjala MC, Mhango CG, Molyneux ME, Rogerson SJ. Plasmodium falciparum: PCR detection and genotyping of isolates from peripheral, placental, and cord blood of pregnant Malawian women and their infants. Trans R Soc Trop Med Hyg. 2002;96:145–149. doi: 10.1016/s0035-9203(02)90284-1. [DOI] [PubMed] [Google Scholar]
- Krudsood S, Imwong M, Wilairatana P, Pukrittayakamee S, Nonprasert A, Snounou G, White NJ, Looareesuwan S. Artesunate-dapsone-proguanil treatment of falciparum malaria: genotypic determinants of therapeutic response. Trans R Soc Trop Med Hyg. 2005;99:142–149. doi: 10.1016/j.trstmh.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Kublin JG, Dzinjalamala FK, Kamwendo DD, Malkin EM, Cortese JF, Martino LM, Mukadam RA, Rogerson SJ, Lescano AG, Molyneux ME, Winstanley PA, Chimpeni P, Taylor TE, Plowe CV. Molecular markers for failure of sulfadoxine-pyrimethamine and chlorproguanil-dapsone treatment of Plasmodium falciparum malaria. J Infect Dis. 2002;185:380–388. doi: 10.1086/338566. [DOI] [PubMed] [Google Scholar]
- Lozovsky ER, Chookajorn T, Brown KM, Imwong M, Shaw PJ, Kamchonwongpaisan S, Neafsey DE, Weinreich DM, Hartl DL. Stepwise acquisition of pyrimethamine resistance in the malaria parasite. Proc Natl Acad Sci U S A. 2009;106:12025–12030. doi: 10.1073/pnas.0905922106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackinnon MJ. Drug resistance models for malaria. Acta Trop. 2005;94:207–217. doi: 10.1016/j.actatropica.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Mackinnon MJ, Marsh K. The selection landscape of malaria parasites. Science. 2010;328:866–871. doi: 10.1126/science.1185410. [DOI] [PubMed] [Google Scholar]
- Maiga O, Djimde AA, Hubert V, Renard E, Aubouy A, Kironde F, Nsimba B, Koram K, Doumbo OK, Le Bras J, Clain J. A shared Asian origin of the triple-mutant dhfr allele in Plasmodium falciparum from sites across Africa. J Infect Dis. 2007;196:165–172. doi: 10.1086/518512. [DOI] [PubMed] [Google Scholar]
- Menendez C, Serra-Casas E, Scahill MD, Sanz S, Nhabomba A, Bardaji A, Sigauque B, Cistero P, Mandomando I, Dobano C, Alonso PL, Mayor A. HIV and placental infection modulate the appearance of drug-resistant Plasmodium falciparum in pregnant women who receive intermittent preventive treatment. Clin Infect Dis. 2011;52:41–48. doi: 10.1093/cid/ciq049. [DOI] [PubMed] [Google Scholar]
- Mockenhaupt FP, Bedu-Addo G, Eggelte TA, Hommerich L, Holmberg V, von Oertzen C, Bienzle U. Rapid increase in the prevalence of sulfadoxinepyrimethamine resistance among Plasmodium falciparum isolated from pregnant women in Ghana. J Infect Dis. 2008;198:1545–1549. doi: 10.1086/592455. [DOI] [PubMed] [Google Scholar]
- Nkhoma S, Molyneux M, Ward S. Molecular surveillance for drug-resistant Plasmodiumfalciparum malaria in Malawi. Acta Trop. 2007;102:138–142. doi: 10.1016/j.actatropica.2007.04.006. [DOI] [PubMed] [Google Scholar]
- Nsanzabana C, Hastings IM, Marfurt J, Muller I, Baea K, Rare L, Schapira A, Felger I, Betschart B, Smith TA, Beck HP, Genton B. Quantifying the evolution and impact of antimalarial drug resistance: drug use, spread of resistance, and drug failure over a 12-year period in Papua New Guinea. J Infect Dis. 2010;201:435–443. doi: 10.1086/649784. [DOI] [PubMed] [Google Scholar]
- Nzila A, Ochong E, Nduati E, Gilbert K, Winstanley P, Ward S, Marsh K. Why has the dihydrofolate reductase 164 mutation not consistently been found in Africa yet? Transactions of the Royal Society of Tropical Medicine and Hygiene. 2005;99:341–346. doi: 10.1016/j.trstmh.2004.07.002. [DOI] [PubMed] [Google Scholar]
- O'Meara WP, Smith DL, McKenzie FE. Potential impact of intermittent preventive treatment (IPT) on spread of drug-resistant malaria. PLoS Med. 2006;3:e141. doi: 10.1371/journal.pmed.0030141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce RJ, Drakeley C, Chandramohan D, Mosha F, Roper C. Molecular determination of point mutation haplotypes in the dihydrofolate reductase and dihydropteroate synthase of Plasmodium falciparum in three districts of northern Tanzania. Antimicrob Agents Chemother. 2003;47:1347–1354. doi: 10.1128/AAC.47.4.1347-1354.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce RJ, Pota H, Evehe MS, Ba el H, Mombo-Ngoma G, Malisa AL, Ord R, Inojosa W, Matondo A, Diallo DA, Mbacham W, van den Broek IV, Swarthout TD, Getachew A, Dejene S, Grobusch MP, Njie F, Dunyo S, Kweku M, Owusu-Agyei S, Chandramohan D, Bonnet M, Guthmann JP, Clarke S, Barnes KI, Streat E, Katokele ST, Uusiku P, Agboghoroma CO, Elegba OY, Cisse B, IE AE, Giha HA, Kachur SP, Lynch C, Rwakimari JB, Chanda P, Hawela M, Sharp B, Naidoo I, Roper C. Multiple origins and regional dispersal of resistant dhps in African Plasmodium falciparum malaria. PLoS Med. 2009;6:e1000055. doi: 10.1371/journal.pmed.1000055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picot S, Olliaro P, de Monbrison F, Bienvenu AL, Price RN, Ringwald P. A systematic review and meta-analysis of evidence for correlation between molecular markers of parasite resistance and treatment outcome in falciparum malaria. Malar J. 2009;8:89. doi: 10.1186/1475-2875-8-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowe CV, Cortese JF, Djimde A, Nwanyanwu OC, Watkins WM, Winstanley PA, Estrada-Franco JG, Mollinedo RE, Avila JC, Cespedes JL, Carter D, Doumbo OK. Mutations in Plasmodium falciparum dihydrofolate reductase and dihydropteroate synthase and epidemiologic patterns of pyrimethamine-sulfadoxine use and resistance. The Journal of infectious diseases. 1997;176:1590–1596. doi: 10.1086/514159. [DOI] [PubMed] [Google Scholar]
- Plowe CV, Kublin JG, Dzinjalamala FK, Kamwendo DS, Mukadam RA, Chimpeni P, Molyneux ME, Taylor TE. Sustained clinical efficacy of sulfadoxinepyrimethamine for uncomplicated falciparum malaria in Malawi after 10 years as first line treatment: five year prospective study. BMJ. 2004;328:545. doi: 10.1136/bmj.37977.653750.EE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman J, Little F, Roper C, Kleinschmidt I, Cassam Y, Maharaj R, Barnes KI. Five years of large-scale dhfr and dhps mutation surveillance following the phased implementation of artesunate plus sulfadoxine-pyrimethamine in Maputo Province, Southern Mozambique. Am J Trop Med Hyg. 2010;82:788–794. doi: 10.4269/ajtmh.2010.09-0401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogerson SJ, Chaluluka E, Kanjala M, Mkundika P, Mhango C, Molyneux ME. Intermittent sulfadoxine-pyrimethamine in pregnancy: effectiveness against malaria morbidity in Blantyre, Malawi, in 1997-99. Trans R Soc Trop Med Hyg. 2000;94:549–553. doi: 10.1016/s0035-9203(00)90083-x. [DOI] [PubMed] [Google Scholar]
- Rogerson SJ, Wijesinghe RS, Meshnick SR. Host immunity as a determinant of treatment outcome in Plasmodium falciparum malaria. Lancet Infect Dis. 2010;10:51–59. doi: 10.1016/S1473-3099(09)70322-6. [DOI] [PubMed] [Google Scholar]
- Roper C, Pearce R, Nair S, Sharp B, Nosten F, Anderson T. Intercontinental spread of pyrimethamine-resistant malaria. Science. 2004;305:1124. doi: 10.1126/science.1098876. [DOI] [PubMed] [Google Scholar]
- Sirawaraporn W, Sathitkul T, Sirawaraporn R, Yuthavong Y, Santi DV. Antifolate-resistant mutants of Plasmodium falciparum dihydrofolate reductase. Proc Natl Acad Sci U S A. 1997;94:1124–1129. doi: 10.1073/pnas.94.4.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snounou G, Zhu X, Siripoon N, Jarra W, Thaithong S, Brown KN, Viriyakosol S. Biased distribution of msp1 and msp2 allelic variants in Plasmodium falciparum populations in Thailand. Trans R Soc Trop Med Hyg. 1999;93:369–374. doi: 10.1016/s0035-9203(99)90120-7. [DOI] [PubMed] [Google Scholar]
- Sridaran S, McClintock SK, Syphard LM, Herman KM, Barnwell JW, Udhayakumar V. Anti-folate drug resistance in Africa: meta-analysis of reported dihydrofolate reductase (dhfr) and dihydropteroate synthase (dhps) mutant genotype frequencies in African Plasmodium falciparum parasite populations. Malar J. 2010;9:247. doi: 10.1186/1475-2875-9-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steketee RW, Nahlen BL, Parise ME, Menendez C. The burden of malaria in pregnancy in malaria-endemic areas. Am J Trop Med Hyg. 2001;64:28–35. doi: 10.4269/ajtmh.2001.64.28. [DOI] [PubMed] [Google Scholar]
- Talisuna AO, Langi P, Mutabingwa TK, Van Marck E, Speybroeck N, Egwang TG, Watkins WW, Hastings IM, D'Alessandro U. Intensity of transmission and spread of gene mutations linked to chloroquine and sulphadoxine-pyrimethamine resistance in falciparum malaria. Int J Parasitol. 2003;33:1051–1058. doi: 10.1016/s0020-7519(03)00156-5. [DOI] [PubMed] [Google Scholar]
- Talisuna AO, Okello PE, Erhart A, Coosemans M, D'Alessandro U. Intensity of malaria transmission and the spread of Plasmodium falciparum resistant malaria: a review of epidemiologic field evidence. Am J Trop Med Hyg. 2007;77:170–180. [PubMed] [Google Scholar]
- ter Kuile FO, van Eijk AM, Filler SJ. Effect of sulfadoxine-pyrimethamine resistance on the efficacy of intermittent preventive therapy for malaria control during pregnancy: a systematic review. JAMA : the journal of the American Medical Association. 2007;297:2603–2616. doi: 10.1001/jama.297.23.2603. [DOI] [PubMed] [Google Scholar]
- van Eijk AM, Ayisi JG, ter Kuile FO, Otieno JA, Misore AO, Odondi JO, Rosen DH, Kager PA, Steketee RW, Nahlen BL. Effectiveness of intermittent preventive treatment with sulphadoxine-pyrimethamine for control of malaria in pregnancy in western Kenya: a hospital-based study. Trop Med Int Health. 2004;9:351–360. doi: 10.1111/j.1365-3156.2004.01196.x. [DOI] [PubMed] [Google Scholar]
- Vinayak S, Alam MT, Sem R, Shah NK, Susanti AI, Lim P, Muth S, Maguire JD, Rogers WO, Fandeur T, Barnwell JW, Escalante AA, Wongsrichanalai C, Ariey F, Meshnick SR, Udhayakumar V. Multiple genetic backgrounds of the amplified Plasmodium falciparum multidrug resistance (pfmdr1) gene and selective sweep of 184F mutation in Cambodia. J Infect Dis. 2010;201:1551–1560. doi: 10.1086/651949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weedall GD, Conway DJ. Detecting signatures of balancing selection to identify targets of anti-parasite immunity. Trends Parasitol. 2010;26:363–369. doi: 10.1016/j.pt.2010.04.002. [DOI] [PubMed] [Google Scholar]