Abstract
Rab7 and Rab34 are implicated in regulation of lysosomal morphology and they share a common effector referred to as the RILP (Rab-interacting lysosomal protein). Two novel proteins related to RILP were identified and are tentatively referred to as RLP1 and RLP2 (for RILP-like protein 1 and 2, respectively). Overexpression of RILP caused enlarged lysosomes that are positioned more centrally in the cell. However, the morphology and distribution of lysosomes were not affected by overexpression of either RLP1 or RLP2. The molecular basis for the effect of RILP on lysosomes was investigated, leading to the demonstration that a 62-residue region (amino acids 272-333) of RILP is necessary for RILP's role in regulating lysosomal morphology. Remarkably, transferring this 62-residue region unique to RILP into corresponding sites in RLP1 rendered the chimeric protein capable of regulating lysosome morphology. A correlation between the interaction with GTP-bound form of both Rab proteins and the capability of regulating lysosomes was established. These results define a unique region in RILP responsible for its specific role in regulating lysosomal morphology as well as in its interaction with Rab7 and Rab34.
INTRODUCTION
As the terminal station of the endocytic/lysosomal pathway, the lysosome is responsible for degradation of “unwanted” proteins, glycans, lipids, and other molecules (Kornfeld and Mellman, 1989; Mellman, 1996; Gruenberg, 2001; Jerome and Yancey, 2003). Defects in lysosomal function have been shown to be associated with diverse human diseases, collectively referred to as lysosomal storage diseases (Neufeld, 1991; Walkley, 1998; Desnick and Schuchman, 2002; Germain, 2002; Ward et al., 2002). In normal cultured cells, lysosomes are distributed throughout in the cell with higher concentrations in the perinuclear region. In higher eukaryotic cells, specialized lysosomes with unique molecular properties are developed to carry out unique physiological processes, and they are collectively referred to as lysosome-related organelles (LROs; Dell'Angelica et al., 2000; Raposo and Marks, 2002; Shiflett et al., 2002). For example, melanosomes generated by melanocytes are important for pigmentation and protective role of the organism (Marks and Seabra, 2001). The class II compartments in antigen-presenting cells are LROs that are specifically involved in processing antigens into antigenic peptides for loading onto the class II major histocompatibility molecules, a key event in our immune defense (Pieters, 1997; Honey and Rudensky, 2003). Lytic granules in natural killer cells and cytotoxic T lymphocytes, platelet-dense granules in platelets, and granules in basophils and neutrophils are other examples of LROs (Dell'Angelica et al., 2000; Rendu and Brohard-Bohn, 2001; Djeu et al., 2002).
The morphology and distribution of lysosomes and/or LROs are intimately linked to their functions and the underlying molecular mechanisms are being actively investigated (Barbosa et al., 1996; Nagle et al., 1996; Oh et al., 1996, 2000; Gardner et al., 1997; Faigle et al., 1998; Dell'Angelica et al., 2000; Raposo et al., 2001; Shiflett et al., 2002; Ward et al., 2002). Several proteins have been recently shown to possess the property of clustering lysosomes in the perinuclear central region, including Vam6p, Rab7, Rab34, and RILP (Bucci et al., 2000; Caplan et al., 2001; Cantalupo et al., 2001; Jordens et al., 2001; Wang and Hong, 2002). Rab7 and Rab34 are both capable of interacting with RILP, and this interaction is confined to the wild-type and GTP-restricted mutant, but not the GDP-restricted mutant of these two Rabs (Cantalupo et al., 2001; Wang and Hong, 2002). The effect of Rab34 on lysosomal morphology and distribution is absolutely dependent on its interaction with RILP. Although the possibility that Rab7 could regulate lysosomes in a RILP-independent manner through other effectors has not been formally excluded, its ability to interact with RILP may be similarly important for Rab7 to regulate lysosomal morphology. In this case, the function of Rab7, Rab34, and RILP may converge on a common pathway. Vam6p seems to regulate lysosomes in a Rab7-independent manner, suggesting that it either functions downstream of or acts in parallel with Rab7 (Caplan et al., 2001).
In this report, we identified two novel proteins (RLP1 and RLP2) that are structurally related to RILP via the presence of two homologous regions. It was then functionally demonstrated that RILP, but not RLP1 and RLP2, has a specific role in regulating lysosomal morphology and distribution. A 62-residue region unique to the 401-residue RILP was revealed to be necessary for RILP's function to regulate lysosome as well as sufficient to confer a similar property, when transferred, to RLP1. A strong correlation was observed between the ability to regulate lysosomal morphology and interaction with Rab7 (or simultaneous interaction with both Rab7 and Rab34), suggesting this interaction could be the mechanism underlying RILP's ability to regulate lysosomes. This conclusion was further sustained by the observation that the 62-residue region of RILP can confer RLP1 the ability to interact efficiently with Rab7 and Rab34.
MATERIALS AND METHODS
Cells and Antibodies
Normal rat kidney (NRK) and Hela cells were grown in DMEM supplemented with 10% fetal bovine serum (Life Technologies, Ann Arbor, MI) in a 5% CO2 incubator at 37°C. The mAb (Mab) against myc tag, rat lamp1 and a Golgi SNARE GS28 were described earlier (Subramaniam et al., 1996; Wang and Hong, 2002). Mab against TGN38 was a gift from G. Banting (University of Bristol, UK; Luzio et al., 1990). Mab against transferrin receptor and EEA1 were from BD Transduction Laboratories (Lexington, KY). Polyclonal antibody against myc tag was from Upstate Biotechnology (Lake Placid, NY). FITC- and rhodamine-conjugated secondary antibodies were from Jackson ImmunoResearch (West Grove, PA). HRP-conjugated secondary antibodies were purchased from Pierce (Rockford, IL).
Examination of Expression of RILP Transcripts
Human multiple tissues cDNA panels (CLONTECH, Palo Alto, CA) were used for PCR-based analysis using the following oligonucleotides: primer 1 (5′ CAG TGC CGC TTC AGT CGG GAG GAG TTT 3′) and primer 2 (5′ GGC CTC TGG GGC GGC TGA GGC CCC 3′) were used to amplify a 489-base pair fragment coding for C-terminal region of RILP; primer 3 (5′ ATG GAC GTG TAC GAC ATC GCG TCG CTT 3′) and primer 4 (5′ TTC GAT CAG GGC TTT CCC CTG GGC CTC 3′) were used to amplify a 600-base pair fragment coding for N-terminal region of RLP1; primer 5 (5′ ATG GAG GAG CCC CCT GTG CGA GAA GAG 3′) and primer 6 (5′ GGT CTG TTT CCC CGA TCG AAA AAA GAA 3′) were used to amplify the entire coding region of RLP2; primer 7 (5′ TGA AGG TCG GAG TCA ACG GAT TTG GT 3′) primer 8 (5′ CAT GTG GGC CAT GAG GTC CAC CAC 3′) were used to amplify a control cDNA fragment of G3PDH. The PCR products were resolved by agarose gel electrophoresis.
Constructs for Expressing in Mammalian Cells
EST clones with open reading frames of human RILP (GenBank accession number AL568729, clone ID: CS0DE005YH19), mouse RLP1 (GenBank accession number AI789193, image: 1972593), and mouse RLP2 (GenBank accession number AW910174, image: 3156139) were obtained from ResGen (Invitrogen Corporation, Carlsbad, CA). For the expression construct of myc-tagged RILP, RLP1, and RLP2, the complete coding sequences of RILP, RLP1, and RLP2 were PCR-amplified from the corresponding EST clones and digested with EcoR1/NotI, then inserted into the EcoR1/NotI sites of pDMycneo vector (Seet and Hong, 2001) to generate myc-RILP, myc-RLP1 and myc-RLP2.
For the chimeric protein expression constructs between RILP (R) and RLP1 (R1), the chimeric sequences R(1-198)R1(257-406), R(1-300)R1(350-406), R(1-333)R1(350-406), R1(1-256)R(199-401), R1(1-325)R(272-401), R1(1-338)R(284-401), and R1(1-325)R(272-333)R1(350-406) were generated by PCR and then subcloned into the pDmycneo vector as described above. All the constructs were confirmed by sequencing.
Rab7Q67L and Rab34Q111L mutants were generated through standard PCR mutagenesis as described previously (Wang and Hong, 2002). For the GST-fusion protein expression constructs, the PCR products of Rab7Q67L and Rab34Q111L were cloned into pGEX-4T-1 vector to generate GST-Rab7Q67L and GST-Rab34Q111L. The GST-Rab33bQ92L construct was prepared using a similar approach.
Transfection and Immunofluorescence Microscopy
Normal rat kidney (NRK) and Hela cells were grown in DMEM supplemented with 10% fetal bovine serum (Life Technologies, Ann Arbor, MI) in a 5% CO2 incubator at 37°C. Transfection was performed using Lipofectamine (Life Technologies, Gaithersburg, MD) as described previously (Wong et al., 1999; Wang and Hong, 2002). The transfected cells were washed with PBSCM (PBS containing 1 mM CaCl2 and 1 mM MgCl2) and then fixed with 3% paraformaldehyde in PBSCM at 4°C. After sequential washing with PBSCM supplemented with 50 mM NH4Cl and the PBSCM, cells were permeabilized with 0.1% saponin (Sigma, St. Louis, MO) in PBSCM for 15 min at room temperature. The permeabilized cells were incubated with primary antibodies in fluorescence dilution buffer (PBSCM containing 5% normal goat serum, 5% fetal bovine serum, and 2% bovine serum albumin) for 1 h at room temperature. After three washes, cells were incubated with FITC- or rhodamine-conjugated secondary antibodies in fluorescence dilution buffer for 1 h at room temperature, then washed six times, and mounted with Vectashield (Vector Laboratories, Burlingame, CA). Confocal microscopy was performed with a Carl Zeiss Axioplan II microscope (Thornwood, NY) equipped with confocal scanning laser. For brefeldin A or nocodazole treatment, cells were incubated with 10 μg/ml brefeldin A (Epicentre Technologies, Madison, WI) or Nocodazole (Sigma) for 1 h at 37°C before fixation.
GST Pull-down Experiment and Immunoblot Analysis
The expression and production of recombinant GST fusion proteins were performed as described (Wang and Hong, 2002). For GST-pull-down experiments, HeLa cells were transfected with myc-tagged constructs as indicated in the text. After 18 h, cells were harvested and lysed in the binding buffer (containing 20 mM HEPES, pH 7.4, 100 mM NaCl, 5 mM MgCl2, 1% TX-100, and EDTA-free proteinase inhibitor cocktail from Roche, Nutley, NJ) for 1 h at 4°C. The lysates were spun down using TLA-100 rotor at 55,000 rpm for 30 min. The supernatants were incubated with 50 μg GST, GST-Rab7Q67L, Rab33bQ92L, or GST-Rab34Q111L bound to the GST-sepharose 4B resin and loaded with 100 μM GTP-γ-S. After overnight incubation, the resin was sequentially washed with buffer 1 (20 mM HEPES, pH 7.4, 500 mM NaCl, 5 mM MgCl2, and 1% TX-100), buffer 2 (20 mM HEPES, pH 7.4, 100 mM NaCl, 5 mM MgCl2, and 0.5% TX-100), and buffer 3 (20 mM HEPES, pH 7.4, 100 mM NaCl, and 5 mM MgCl2). The samples were then resolved by SDS-PAGE. For immunoblotting, proteins resolved by SDS-PAGE were transferred to nitro-cellulose filter. The filter was blocked with 5% milk in PBS and then incubated with primary antibody against myc tag for 30 min at room temperature. The filter was washed and incubated with HRP-conjugated secondary antibody for 30 min at room temperature. The blots were detected by the use of ECL system (Pierce).
Biochemical Subcellular Fractionation
Hela cells expressed myc-RILP, RLP1, and RLP2 (via transient transfection) were broken with Ball-homogenizer in homogenization buffer (20 mM HEPES, pH 7.3, 100 mM NaCl, 5 mM MgCl2, and EDTA-free proteinase inhibitor cocktail). The homogenate was centrifuged at 2000 × g for 5 min to remove nuclei. The postnuclear supernatant was then spun at 85,000 rpm for 30 min using a TLA-100 rotor. The supernatant was kept as cytosol fraction. The pellet was dissolved in homogenization buffer containing 1% Triton-100 for 1 h on ice. The extracted proteins were kept as membrane fraction after a 14,000 rpm spin (30 min) to remove insoluble materials. The protein concentrations of both fractions were quantified, and ∼15 μg of cytosolic and membrane proteins was analyzed by SDS-PAGE and processed for Western blotting to detect myc-RILP, RLP1, and RLP2, syntaxin5 (serving as a control for membrane proteins) (Subramaniam et al., 1997), or RhoGDI1 (serving as a control for cytosolic proteins; Wong et al., 1999).
RESULTS
Identification of Two Proteins (RLP1 and RLP2) Related to RILP
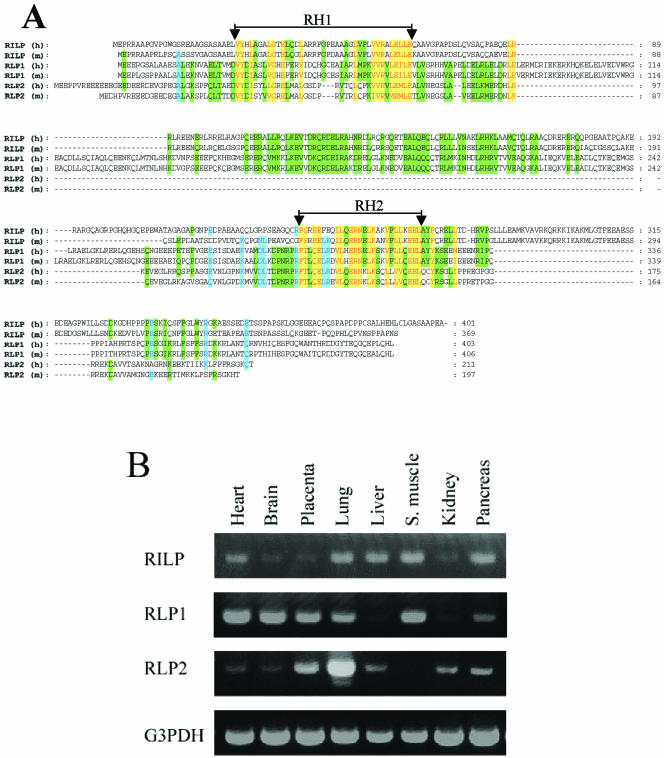
In views of the recent illustration that lysosome-associated Rab7 as well as Golgi-enriched Rab34 may share RILP as a common effector (Cantalupo et al., 2001; Wang and Hong, 2002), we searched databases for RILP related proteins. Two novel proteins bearing significant amino acid similarities (in two small regions) to RILP were identified and were tentatively named RLP1 (RILP-like protein 1) and RLP2 (RILP-like protein 2). Several EST clones for human (GenBank accession numbers BI755032, BQ892034, and BM698721) and mouse (GenBank accession numbers NM021430 and AB041584) RLP1 were identified. The coding region for the full-length human RLP1 were assembled from the sequences of BI755032, BQ892034, and BM698721, whereas both NM021430 and AB041584 contain the entire coding region sequence for mouse RLP1. Human and mouse RLP1 have 403 and 406 amino acids, respectively, and are 92% identical to each other. The complete coding region of RLP2 is represented by several human EST clones (GenBank accession numbers BC013042, AK056934, and XM055952) and mouse EST clones (GenBank accession numbers BC003324 and NM030259). Human and mouse RLP2 consist of 211 and 197 residues, respectively, and are 72% identical to each other. Because human (NP113618 and CAC33443) and mouse (assembled from EST clones BF784741, BY724781, and AV090951) RILP are also ∼72% identical, it seems that RLP1 is more conserved between human and mouse than do RILP and RLP2. RLP1 and RLP2 are ∼32% identical and share ∼22 and 18% amino acid identities, respectively, with RILP. The alignment of human and mouse RILP, RLP1, and RLP2 is shown in Figure 1A. Although the overall amino acid similarities among RILP, RLP1, and RLP2 are not very high, homologies among RILP, RLP1, and RLP2 in two confined small regions (residues 28-66 and residues 243-269) (RH1 and RH2 for RILP homology 1, and 2 of 39 and 27 amino acids, respectively) are obvious (Figure 1A). Proteins homologous to RILP and/or RLPs were found in several multicellular organisms such as Drosophila melanogaster (GenBank accession number NP-569910), Anopheles gambiae (GenBank accession number EAA09406), and Caenorhabditis elegans (GenBank accession number NP741113). Again, the amino acid homologies among RILP and/or RLPs of different species are similarly confined to the RH1 and RH2 regions, indicating that RH1 and RH2 are signatures of this protein family. However, unicellular organisms such as Saccharomyces cerevisiae and Schizosaccharomyces pombe do not seem to contain any structural homologues of RILP or RLPs, suggesting that RILP and RLPs may have evolved to accommodate functions that are more relevant to higher eukaryotes or that the functional counterparts in these unicellular organisms do not share obvious amino acid similarities.
Figure 1.
(A) Identification of two novel proteins (RLP1 and RLP2) related to RILP. The amino acid sequences of human (h) and mouse (m) RILP, RLP1, and RLP2 were aligned. The two regions with highest homology were indicated as RH1 (RILP Homology 1) and RH2, respectively. The residues identical or conserved in all proteins were shown in red with a yellow background. Residues identical or conserved in five proteins were shown in blue with a light blue background, while residues identical or conserved in four proteins were shown in black with a green background. (B) RILP, RLP1, and RLP2 are expressed in diverse tissues. The tissue distributions of RILPs were examined by PCR approach using CLONTECH multiple tissue cDNA panels (the PCR product of G3PDH was used for normalization).
The expression profile of RILP and RLPs transcripts in various tissues was compared by a PCR-based approach using the Clontech multiple tissues cDNA panels. As shown in Figure 1B, RILP, RLP1, and RLP2 are expressed in diverse tissues examined at varying levels. The levels of RLP1 in the liver and kidney seem to be lower than those in other tissues. The highest level of RLP2 was detected in the lung, whereas skeletal muscle does not seem to have detectable levels of RLP2. These observations indicate that RILP, RLP1, and RLP2 are likely to participate in nonoverlapping functions in different tissues.
RILP, but not RLP1 or RLP2, Regulates Lysosomal Morphology and Distribution
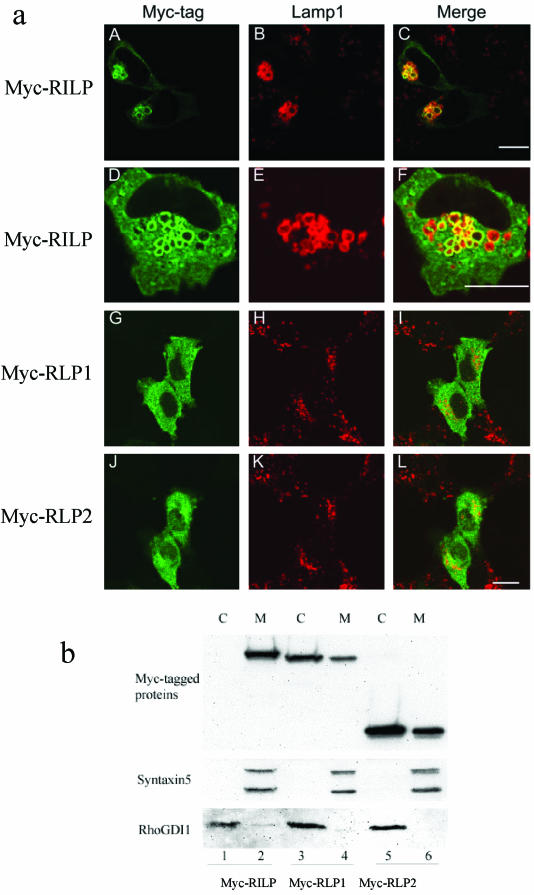
When RILP was overexpressed in transfected NRK cells, two noticeable phenotypes were observed for lysosomes marked by Lamp1 (Figure 2a). First, lysosomes are clustered in the peri-nuclear region (A-C) and this phenotype is similar to that observed when RILP was overexpressed in HeLa cells as reported earlier (Cantalupo et al., 2001; Jordens et al., 2001). Another interesting and prominent phenotype is that fewer but enlarged lysosomes are detected (A-F), which was less apparent in HeLa cells (Cantalupo et al., 2001). Furthermore, RILP was clearly enriched in the enlarged lysosomes (A and D). Because RLP1 and RLP2 are related to RILP, we have investigated whether they also possess a similar property of affecting lysosomes. When RLP1 and RLP2 were each overexpressed in NRK or other cells, however, the morphology, number and appearance of lysosomes were essentially unaffected (H and K), and the majority of RLP1 (G) and RLP2 (J) appears to be cytosolic. The subcellular distribution of RILP, RLP1, and RLP2 was also investigated by biochemical fractionation (Figure 2b). Transfected cells were fractionated into total cytosolic and membrane fractions. Equal amounts of cytosolic and membrane proteins were resolved by SDS-PAGE and analyzed by immunoblot. As shown, RILP was mainly detected in the membrane fraction, in the same way as syntaxin5 was confined to the membrane fraction (lane 2; Subramaniam et al., 1997), whereas the majority RhoGDI1 was detected in the cytosolic fraction (lane 1; Wong et al., 1999). Under similar conditions, RLP1 (lane 3) and RLP2 (lane 5) were mainly detected in the cytosolic fraction. These observations suggest that RILP is able to associate with lysosomes and to affect their morphology and distribution, whereas RLP1 and RLP2 are mainly cytosolic and does not possess the ability to regulate lysosomes.
Figure 2.
(a) RILP but not RLP1 or RLP2 causes enlarged lysosomes positioned centrally in the cell. NRK cells were transfected with constructs for the expression of myc-RILP (A-F), myc-RLP1 (G-I) and myc-RLP2 (J-L). The transfected cells were fixed and double-labeled with polyclonal antibody against the myc tag to reveal the expressed RILP and RLPs (A, D, G, and J) and mAb against rat Lamp1 to reveal lysosomes (B, E, H, and K). RILP colocalizes with lysosomal Lamp1 and causes enlarged lysosomes (A-F), but RLP1 and RLP2 are cytosolic and have no effects on lysosomal morphology (G-L). Bar, 10 μm. (b) RILP is mainly confined to the membrane, while RLP1 and RLP2 are distributed predominantly in the cytosol. Cells expressing Myc-RILP (lanes 1 and 2), Myc-RLP1 (lanes 3 and 4), Myc-RLP2 (lane 5-6) were fractionated into cytosolic (C; lanes 1, 3, and 5) and membrane (M; lanes 2, 4, and 6) fractions, respectively. The fractions were resolved by SDS-PAGE and analyzed by immunoblot to detected the Myc-tagged proteins (top panel) as well as syntaxin5 (serving as membrane protein control) (middle panel) and RhoGDI1 (serving as cytosolic protein control; bottom panel).
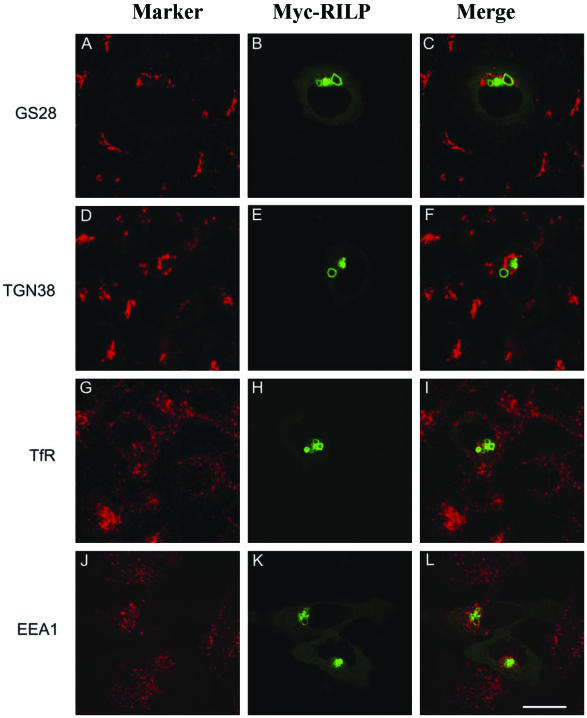
RILP Specifically Affects Lysosomes but not Other Organelles
The effect of overexpressed RILP on other organelles was also compared in this experimental setting (Figure 3). Again, overexpressed RILP was detected in enlarged lysosomal structures located centrally in the cells (B, E, H, and K). For Golgi stack marked by GS28 (Subramanian et al., 1996; A), the trans-Golgi network (TGN) marked by TGN38 (Luzio et al., 1990; D), the sorting/early and recycling endosomes marked by the transferrin receptor (TfR; Marsh et al., 1995; G), and the sorting/early endosomes marked by EEA1 (Mu et al., 1995; J), RILP did not exhibit any significant effects. In addition, structures of early secretory pathway such as ER exit sites marked by COPII, transport intermediates and cis-Golgi marked by Bet1 and the KEDL receptor were not affected by overexpressed RILP (unpublished observations). These results, taken together, suggest that RILP has a specific role in regulating lysosomal morphology and distribution. Similarly, these organelles were not affected by overexpression of either RLP1 or RLP2 (our unpublished observations).
Figure 3.
RILP regulates specifically lysosomes but not Golgi apparatus, early and recycling endosomes. NRK cells (A-I) or Hela cells (J-L) were transfected with myc-RILP expressing construct. The transfected cells were processed for double labeling using antibodies against myc tag and GS28 (A-C), myc tag and TGN38 (D-F), myc tag and transferrin receptor (G-I), or myc tag and EEA1 (J-L). As shown, RILP has no effects on Golgi apparatus, early and recycling endosomes. Note that enlarged lysosomes induced by RILP were not so obvious in HeLa cells, in which lysosomes are collapsed into compact structures. Bar, 10 μm.
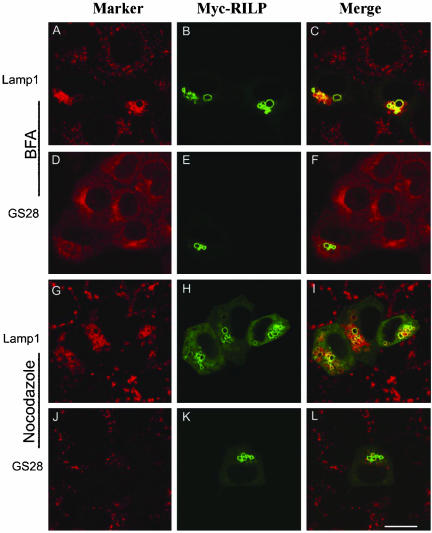
The Effect of RILP on Lysosomes Was Not Affected by Brefeldin A or Nocodazole
Many intracellular membrane trafficking processes are dependent on ARF family GTPases and their guanine nucleotide exchange factors (GEFs) as well as the microtubular network. We were interested in examining whether the observed effect of RILP could be abolished by the treatment of brefeldin A, which inhibits ARFs function by inhibiting the GEFs (Chavrier and Goud, 1999; Klausner et al., 1992; Sata et al., 1998, 1999); and/or nocodazole, which disrupts the microtubule network (Lippincott-Schwartz, 1998; Thyberg and Moskalewski, 1999). As shown in Figure 4, after brefeldin A treatment, GS28 was mainly redistributed into the ER (D), consistent with previous results (Subramaniam et al., 1995). However, both RILP and Lamp1 were still observed in fewer but enlarged lysosomes in cells treated with brefedin A (A, B, and E). The distribution of lysosomes in cells not overexpressing RILP was not affected by brefeldin A treatment (A and C), in agreement with previous results (Klausner et al., 1992). These observations suggest that RILP affects lysosomes through a pathway independent of ARFs and their GEFs. When cells were treated with nocodazole, GS28 was dispersed into many smaller mini-Golgi structures distributed throughout the cell (J), whereas lysosomes in nontransfected cells were not significantly affected, although their sizes do appear a little larger (G and I). The distribution of overexpressed RILP as well as the affected lysosomes marked by Lamp1 is not significantly affected by nocodazole treatment (G, H, and K).
Figure 4.
The effect of RILP on lysosomes is not affected by treatment with brefeldin A (BFA) or nocodazole. NRK cells transfected with myc-RILP expressing construct were treated with BFA (A-F) or nocodazole (G-L) for 1 h at 37°C (both at 10 μg/ml). The cells were then processed for double-labeling using antibodies against myc tag and Lamp1 (A-C and G-I) or myc tag and GS28 (D-F and J-L). As shown, the Golgi apparatus (marked by GS28) was redistributed into ER-like labeling by BFA (D-F) or fragmented by nocodazole treatment (J-L), but the effect of RILP on lysosomes is not affected by either treatments (A-C and G-I). Bar, 10 μm
Identification of a 62-residue Region Unique to RILP That Is Essential for Regulating Lysosomes
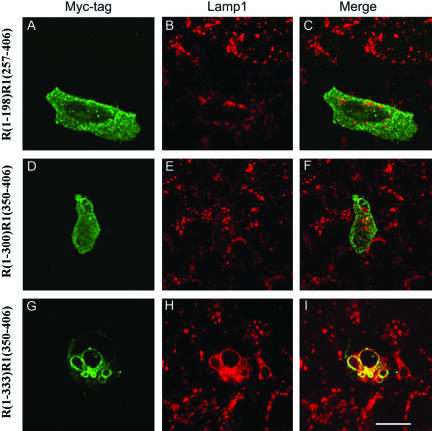
The specific regulation of lysosomes by RILP but not by its related RLP1 or RLP2 raises an intriguing question as to the structural basis that distinguishes it from its related proteins in this specific biological function. To address this fundamentally important issue, we have taken the approach of constructing chimeric proteins between RILP and RLP1 because they are more closely related among the three RILPs and have similar sizes. We first created chimeric proteins consisting of N-terminal region of RILP (R) of different lengths fused to the C-terminal region of RLP1 (R1) by swapping them at corresponding sites (Figure 5). A chimeric protein, R(1-198)R1(257-406), consisting of N-terminal 198-residue (amino acid 1-198) region of RILP fused to the C-terminal region (residue 257-406) of RLP1 did not significantly affect the morphology and distribution of lysosomes (A-C). Similarly, lysosomes were essentially not affected by the expression of R(1-300)R1(350-406) (D-F), another chimeric protein consisting of N-terminal region (residues 1-300) of RILP and the C-terminal region (residues 350-406) of RLP1. These results suggest that the N-terminal 300-residue region of RILP, when fused to the C-terminal region of RLP1, does not possess the ability to change lysosomal morphology and that residues after position 300 of RILP are important. With this in mind, another chimeric protein, R(1-333)R1(350-406), was created and found to be able to cause fewer but enlarged endosomes (G-I). This result suggests that the key region responsible for regulating lysosomal morphology lies in the N-terminal 333 residues of RILP. These results defined the C-terminal boundary (residue 333) of the region in RILP that is involved in lysosomal regulation.
Figure 5.
Defining the C-terminal boundary of the RILP region that is responsible for regulating lysosomes. NRK cells were transfected with constructs for expressing myc tagged chimeric proteins consisting of the N-terminal part of RILP (R) and C-terminal part of RLP1 (R1). The transfected cells were then processed for double labeling using antibodies against myc tag and Lamp1. The chimera consisting of N-terminal half of RILP, R(1-198)R1(257-406), or 300-residue N-terminal part of RILP, R(1-300)R1(350-406), had no effects on lysosomes (A-F), but the chimera composed of 333-residue of N-terminal part of RILP, R(1-333)R1(350-406), does (G and H). This defines residue 333 as the C-terminal boundary. Bar, 10 μm
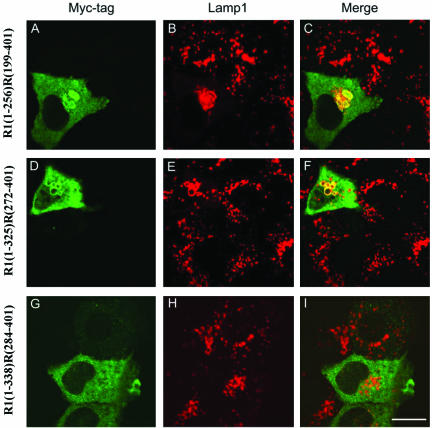
Another set of chimeric proteins were created in which the N-terminal portion of RLP1 (R1) of varying lengths was fused to the C-terminal region of RILP (R) at the respective sites. As shown in Figure 6, chimeric proteins R1(1-256)R(199-401) as well as R1(1-325)R(272-401) possess the similar property of being able to induce fewer but enlarged lysosomes that are positioned centrally (A-F), suggesting that the N-terminal 271-residue region of RILP could be functionally replaced by the corresponding portion of RLP1 (residues 1-325). However, another chimeric protein R1(1-338)R(284-401) failed to associate with lysosomes and to affect lysosomal morphology, suggesting that the RILP residues 272-284 could not be functionally replaced by that of RLP1. These results, taken together, defined the N-terminal boundary (residue 272) of the region in RILP that is important for its unique ability to regulate lysosomes. In conjunction with results shown in Figure 5, these observations establish a 62-residue region consisting of amino acids 272-333 of RILP that is essential for its unique property in regulating lysosomes, while the 271-residue N-terminal region and the 68-residue C-terminal region could each be replaced by the corresponding regions of RLP1.
Figure 6.
Defining the N-terminal boundary of the RILP region that is responsible for regulating lysosomes. NRK cells were transfected with constructs for expressing myc tagged chimeric proteins consisting of N-terminal part of RLP1 (R1) and C-terminal part of RILP (R). The cells were then processed for double labeling using antibodies against myc tag and Lamp1. Chimeric proteins containing C-terminal half of RILP, R1(1-256)R(199-401), (A-C), and C-terminal residues 272-401 of RILP, R1(1-325)R0(272-401), (D-F), affected lysosomes. The C-terminal part of RILP consisting of residues 284-401 lost the ability to affect lysosomes (G-I). This defines residue 272 as the N-terminal boundary of RILP. Bar, 10 μm
The 62-residue RILP Unique Region Is Sufficient to Render RLP1 Capable of Regulating Lysosomes
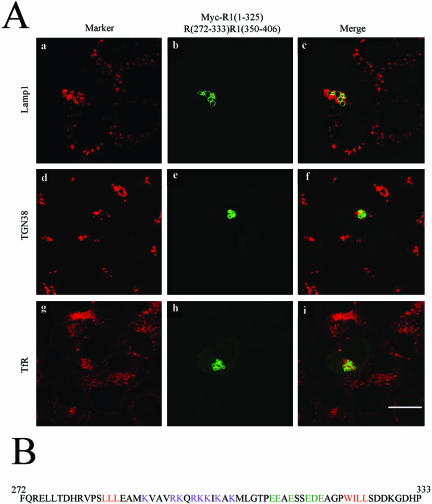
As this 62-residue region is necessary for RILP's ability to regulate lysosomes, it was of great interest to examine whether this region itself is sufficient to confer the ability to RLP1 when transferred to the corresponding site. In this consideration, a chimeric protein R1(1-325)R(272-333)R1(350-406) was constructed. It consists of the 325-residue N-terminal region of RLP1, followed by this 62-residue region of RILP, and terminates with the C-terminal (residues 350-406) region of RLP1. When expressed in transfected NRK cells, this chimeric protein was capable of clustering lysosomes into the central region and inducing the appearance of fewer but much-enlarged lysosomes (Figure 7A, a-c). The TGN marked by TGN38 (panels e and f), and early and recycling endosomes marked by TfR (g-i) were essentially not affected by this chimeric protein, establishing that this 62-residue region is sufficient to confer RLP1 the ability to associate with and regulate lysosomes.
Figure 7.
(A) A unique 62-residue (amino acids 272-333) region of RILP (R1) is sufficient to render RLP1 (R2) capable of regulating lysosomes. NRK cells were transfected with a construct for expressing myc tagged chimera, R1(1-325)R(272-333)R1(350-406). The cells were then processed for double-labeling using antibodies against myc tag and Lamp1 (a-c), myc tag and GS28 (d--f) or myc tag and transferrin receptor (g-i). As shown, residues 272-333 are sufficient for conferring RILP's property to RLP1 in regulating lysosomes. (B) The amino acid sequence of residues 272-333 of RILP. The positive and negative charged residues in the central part were shown in dark pink and dark green, respectively, with the embracing hydrophobic residues marked in red.
Correlation between the Ability to Regulate Lysosomes and That to Interact with Rab7 and Rab34
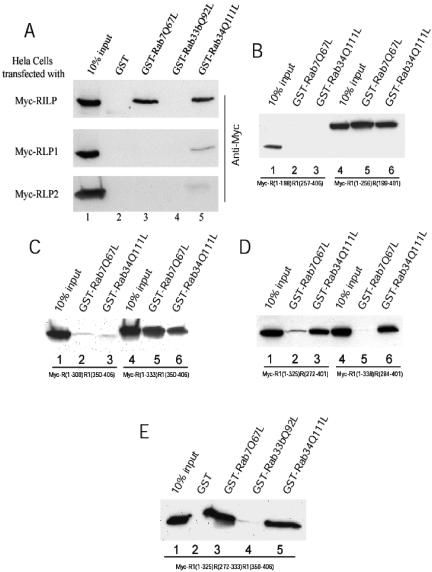
Although RILP overexpression alone could effectively affect lysosomal distribution and morphology, the fact that it interacts with Rab7 and Rab34 has prompted us to investigate whether there is any correlation between the ability to affect lysosomes with that to interact with Rab7 and/or Rab34. RILP, RLP1, RLP2, and various chimeric proteins were transiently expressed in transfected cells and the resulting cell lysates were incubated with immobilized GST, GST-Rab7Q67L, GST-Rab33bQ92L, and GST-Rab34Q111L. The amounts of proteins retained by the respective beads were determined by immunoblot analysis (Figure 8). As shown in Figure 8A, RILP was efficiently retained by immobilized GST-Rab7Q67L and GST-ab34Q111L, but not by GST or Rab33bQ92L. Much less RLP1 was retained by immobilized GST-Rab34Q111L, whereas RLP1 was not retained by GST-Rab7Q67L, GST, or GST-Rab33bQ92L. RLP2 did not show any interaction with Rab7 or Rab34. Among the three chimeric proteins that failed to affect lysosomes, R(1-198)R1(257-406) and R(1-300)R1(350-406) did not interact with either Rab7Q67L or Rab34Q111L (B, lanes 1-3; and C, lanes 1-3, respectively), whereas R1(1-338)R(284-401) interacted only with Rab34Q111L (D, lanes 4-6). Among the three chimeric proteins that have the ability to affect lysosomes, R(1-333)R1(350-406) (C, lanes 4-6), R1(1-256)R(199-401) (B, lanes 4-6), and R1(1-325)R(272-401) (D, lanes 1-3), all interacted with both Rab7Q67L and Rab34Q111L, although the interaction between R2(1-325)R1(272-401) and Rab7Q67L is not as strong as the other two. Importantly, the 62-residue unique RILP region could confer RLP1, in the chimeric protein R1(1-325)R(272-333)R1(350-406), the ability to interact efficiently with Rab7 as well as to enhance the interaction with Rab34 (E). These results suggest that, among RILP, RLP1, RLP2, and various chimeric proteins, there is a strong correlation between the ability to regulate lysosomes and the capability to interact with Rab7 (or simultaneously with both Rab7 and Rab34). Because RLP1 and R1(1-338)R(284-401), which interacts with Rab34 but not Rab7, failed to affect lysosomes, it seems that the ability to interact with Rab34 alone is not sufficient for this function. Because we have failed to create chimeric proteins that interact only with Rab7 but not Rab34, we can only conclude that interaction with Rab7 (or simultaneous interaction with Rab7 and Rab34) could be important for RILP's specific role to regulate lysosomal morphology and distribution. Furthermore, the 62-residue RILP region seems to play a dominant and transferable role in both interaction with Rabs and regulation of lysosomal morphology and distribution.
Figure 8.
Correlation between the ability to interact with Rab proteins and regulation of lysosomal morphology. Hela cells were transfected with myc tagged constructs as indicated. The cell lysates were incubated with GST, GST-Rab7Q67L, GST-Rab33bQ92L or GST-Rab34Q111L coupled to GST-Sepharose 4B resin in the presence of 100 μM GTP-γ-S. After extensive washing, the bound proteins were resolved by SDS-PAGE and detected by immunoblotting using antibody against Myc tag. (A) RILP interacts with both Rab7 and Rab34, whereas RLP1 interacts only with Rab34 weakly. The top panel shows that RILP can specifically bind to GST-Rab7Q67L or GST-Rab34Q111L but not GST or GST-Rab33bQ92L. The middle panel indicates RLP1 interacts with Rab34Q111L weakly but not with GST, Rab7Q67L, or Rab33bQ92L. RLP2 did not interact with any Rab protein (bottom panel). (B) C-terminal half of RILP(R1) is responsible for the interaction with Rab7 and Rab34. GST-Rab7Q67L and Rab34Q111L can pull down the chimeric protein R1(1-256)R(199-401) (lanes 4-6) but cannot bind to R(1-198)R1(257-406) (lanes 1-3). (C) Defining C-terminal boundary of the region of RILP (R) responsible for interaction with Rab7 and Rab34. GST-Rab7Q67L and Rab34Q111L can pull down the chimeric protein R(1-333)R1(350-406) (lanes 4-6) but cannot bind to R(1-300)R1(350-406) (lanes 1-3). (D) Defining the N-terminal boundary of the region of RILP (R) responsible for interaction with Rab7 and Rab34. GST-Rab7Q67L and Rab34Q111L can pull down the chimeric protein R1(1-325)R(272-401) (lanes 1-3), whereas R1(1-338)R(284-401) can be pulled down by Rab34Q111L but not Rab7Q67L (lanes 4-6). (E) The 62-residue (amino acids 272-333) region of RILP (R) is sufficient to confer the ability to RLP1(R1) for interaction with Rab7 and Rab34. As shown, efficient retention of Myc-R1(1-325)R(272-333)R1(350-406) by immobilized GST-Rab7Q67L (lane 3) and GST-Rab34Q111L (lane 5) but not GST (lane 2) or GST-Rab33b(Q92L) (lane 4) was observed.
DISCUSSION
In our present study, we have uncovered two proteins related to RILP and these three proteins may represent a small family of proteins sharing limited homology. Interestingly, the homology among them as well as among RILP and/or RLPs from various multicellular organisms is confined mainly to two small regions referred to as the RH1 and RH2 region of 39 and 27-residue, respectively. Because RH1 and RH2 regions are detected only in RILP and RLPs but not in other proteins, they are likely to contribute to some structural framework and/or interacting module(s) for other proteins. Further experiments are needed to establish the specific function of RH1 and RH2 regions. It is also of interest to note that no structural counterparts were detected in fully sequenced and well-annotated genome and proteome of unicellular organisms such as S. cerevisia and S. pombe, implying that RILP and RLPs may participate in some functions that are more relevant to the developmental and physiological processes of multicellular organisms. It is also possible that these unicellular organisms have functional homologues that are not structurally related to RILP or RLPs. In view of the fact that these unicellular organisms have only few but large lysosomes (vacuoles), they may not need RILP, which apparently serves to induce fewer but larger lysosomes.
The functional aspects of RILP, RLP1, and RLP2 were examined by investigating their potential effect on morphology and distribution of lysosomes and other organelles. Remarkably, RILP, but not RLP1 or RLP2, has a specific role in regulating lysosomal morphology and distribution. RILP is associated with lysosomes, whereas RLP1 and RLP2 are mainly distributed in the cytosol. Other organelles such as TGN, Golgi stack, early and recycling endosomes, and structures of the early secretory pathway were not affected by RILP (or RLP1 or RLP2), suggesting a very specific role of RILP in regulating only lysosomes. The regulatory role of RILP on lysosomes is not affected by treatment with brefeldin A. Our earlier demonstration that RILP-dependent regulation of lysosomes by Rab34 is insensitive to brefeldin A is consistent with this current observation (Wang and Hong, 2002). It is also consistent with our knowledge that brefeldin A treatment has no significant effect on lysosomes under normal culture conditions (Klausner et al., 1992). These results suggest that unlike the secretory pathway and early parts of the endocytic pathway, the lysosome and its regulation by RILP is likely not regulated by ARFs and their GEFs, which are the direct targets of brefeldin A. Disruption of microtubular network by the treatment of nocodazole caused fragmentation of the Golgi apparatus marked by GS28, whereas the effect of RILP in inducing fewer and centrally-localized and enlarged lysosomes was not affected. Our earlier study suggests that Rab34 regulates lysosomal distribution in a microtubule-dependent manner, although it is mediated by interaction with RILP (Wang and Hong, 2002).
How do we explain this apparent difference in microtubule-dependence between Rab34 and RILP? One key difference is that Rab34 seems to act via a long-range effect primarily from the Golgi apparatus, whereas RILP acts in cis directly on lysosomes. It is likely that microtubule-dependent effect of Rab34 could be due to a possibility that effective and productive “signaling” from Rab34 to RILP is dependent on intact microtubule. Another difference between Rab34 and RILP is that RILP is more potent than Rab34 in inducing the formation of fewer and enlarged lysosomes. Because of their sizes, these enlarged lysosomes will naturally be clustered in the more spacious central region, regardless whether the microtubular network is intact or not. The less spacious peripheral region of cultured cells will not be favorable for these enlarged lysosomes. Because of the smaller-sized lysosomes in Rab34-expressing cells, the microtubule may play a role in actively clustering these lysosomes in the perinuclear region and disruption of microtubules by nocodazole resulted in free diffusion of these lysosomes to the peripheral region. The combination of these two possibilities, together with other factors, could be the basis for the difference in microtubule requirement in Rab34- and RILP-regulated lysosomal distribution.
The molecular basis responsible for the RILP-specific regulation of lysosomes was then investigated by constructing chimeric proteins between RILP and RLP1 and testing their effect on lysosomal morphology. These efforts have led to the finding that the N-terminal 271-residue region as well as the C-terminal 68-residue region of RILP could each be replaced by the corresponding region of RLP1, without affecting the ability to regulate lysosomal morphology. A 62-residue region (amino acid 272-333) of RILP is apparently necessary for this unique biological property. This conclusion was further extended by our demonstration that this region, when transferred to RLP1, could sufficiently confer the chimeric protein the ability to effectively regulate lysosomes. Visual examination of the amino acid sequence of this 62-residue region revealed that the central 41-residue subregion (residues 285-325) consists of an N-terminal half that is highly negatively charged and C-terminal half that is highly positively charged embraced at both ends by hydrophobic residues (Figure 7B).
The mechanistic aspects underlying the specific role of RILP was then investigated by assessing the ability of RILP, RLP1, and RLP2 as well as various chimeric proteins to interact with GTP-bound form of Rab7 and Rab34 (as well as GST and Rab33b as negative controls) because of the fact that RILP is an interacting protein for both Rab7 and Rab34 (Cantalupo et al., 2001; Jordens et al., 2001; Wang and Hong 2002). Interestingly, all proteins, including RLP1, RLP2, R(1-198)R1(257-406), R(1-300)R1(350-406), and R1(1-338)R(284-401), that failed to regulate lysosomes do not possess the ability to interact with Rab7Q67L. Among these four proteins, RLP1 and R1(1-338)R(284-401) are capable of interacting with Rab34Q111L, suggesting that interaction with Rab34 alone is not sufficient for these proteins to affect lysosome but a correlation between interaction with Rab7 and ability to regulate lysosome was noticed. On the other hand, all proteins, including RILP, R(1-333)R1(350-406), R1(1-256)R(199-401), R1(1-325)R(272-401), as well as R1(1-325)R(272-333)R1(350-406), that are capable of regulating lysosomes possess the ability to interact with both Rab7 and Rab34. These results provide a strong correlation between the ability to interact with Rab7 (or interaction with both Rab7 and Rab34) and the property to regulate lysosomes. Because chimeric proteins capable of interacting with Rab7 but not Rab34 were not observed, we could not distinguish whether interaction with Rab7 alone is sufficient to affect lysosomes or simultaneous interaction with both Rab7 and Rab34 is important. The most intriguing observation is that transferring the 62-residue RILP unique region to RLP1 not only render the chimeric protein capable of regulating lysosomes but also confer the chimeric protein to interact specifically and efficiently with Rab7 and Rab34. Taken together, it is reasonable to propose that interaction with Rab7 (or simultaneous interaction with Rab7 and Rab34) is likely the underlying mechanism for RILP and some chimeric proteins to regulate lysosomes. Although an earlier study showed that RILP's function in regulating lysosomes is not inhibited by dominant-negative mutant of Rab7, although indicating a Rab7-independent process for RILP action (Cantalupo et al., 2001), this apparent discrepancy is actually not contradictory to our conclusion. This is because that the dominant-negative Rab7 mutant does not possess the ability to interact with RILP and the exogenous RILP remains capable of interacting with endogenous wild-type Rab7 to mediate this Rab7(and Rab34)-dependent regulation on lysosomes.
The effect of RILP on lysosomal morphology and distribution is physiologically relevant. The observed phenotypes are very similar to those observed in Chediak-Higashi syndrome cells with defect in beige/Lyst (Barbosa et al., 1996; Shiflett et al., 2002; Ward et al., 2002). As overexpression of Beige/lyst resulted in many smaller lysosomes that are positioned (more peripherally) throughout the cell (phenotypes opposite to RILP overexpression (Perou et al., 1997) and overexpression of dominant-negative mutants of Beige results in larger lysosomes (Ward et al., 2003), it seems logical to propose that the morphology and distribution of lysosomes are subjected to two apparent opposite regulatory machineries: RILP and other proteins such as Rab7, Rab34, and Vam6p on one end, whereas the other end of the balance is beige/lyst and its interacting and/or regulatory proteins. These two opposite forces could be linked/integrated by some unknown mechanisms, which serve to strike a proper balance depending on the needs of cell and tissue physiology.
Acknowledgments
We thank Drs. Bor Luen Tang, Jill Tham, and Koh Pang Lim for their careful reading of the manuscript. W.H. is also a faculty member of the Department of Biochemistry, National University of Singapore. This work was supported by A*Star (Agency for Science, Technology and Research; to W.H.).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-06-0413. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-06-0413.
References
- Barbosa, M.D. et al. (1996). Identification of the homologous beige and Chediak-Higashi syndrome genes. Nature 382, 262-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucci, C., P. Thomsen, P. Nicoziani, J. McCarthy, and van Deurs, B. (2000). Rab 7, a key to lysosome biogenesis. Mol. Biol. Cell 11, 467-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantalupo, G., Alifano, P., Roberti, V., Bruni, C.B., and Bucci, C. (2001). Rab-interacting lysosomal protein (RILP): the Rab7 effector required for transport to lysosomes. EMBO J. 20, 683-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan, S., Hartnell, L.M., Aguilar, R.C., Naslavsky, N., and Bonifacino, J.S. (2001). Human Vam6p promotes lysosome clustering and fusion in vivo. J. Cell Biol. 154, 109-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavrier, P., and Goud, B. (1999). The role of ARF and Rab GTPases in membrane transport. Curr. Opin. Cell Biol. 11, 466-475. [DOI] [PubMed] [Google Scholar]
- Dell'Angelica, E.C., Mullins, C., Caplan, S., and Bonifacino, J.S. (2000). Lysosome-related organelles. FASEB J. 14, 1265-1278. [DOI] [PubMed] [Google Scholar]
- Desnick, R.J., and Schuchman, E.H. (2002). Enzyme replacement and enhancement therapies: lessons from lysosomal disorders. Nat. Rev. Genet. 3, 954-966. [DOI] [PubMed] [Google Scholar]
- Djeu, J.Y., Jiang, K., and Wei, S. (2002). A view to a kill: signals triggering cytotoxicity. Clin. Cancer Res. 8, 636-640. [PubMed] [Google Scholar]
- Faigle, W., Raposo, G., Tenza, D., Pinet, V., Vogt, A.B., Kropshofer, H., Fischer, A., de Saint-Basile, G., and Amigorena, S. (1998). Deficient peptide loading and MHC class II endosomal sorting in a human genetic immunodeficiency disease: the Chediak-Higashi syndrome. J. Cell Biol. 141, 1121-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner, J.M. et al. (1997). The mouse pale ear (ep) mutation is the homologue of human Hermansky-Pudlak syndrome. Proc. Natl. Acad. Sci. USA. 94, 9238-9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain, D.P. (2002). Lysosomes and lysosomal storage diseases. J. Soc. Biol. 196, 127-134. [PubMed] [Google Scholar]
- Gruenberg, J. (2001). The endocytic pathway: a mosaic of domains. Nat. Rev. Mol. Cell Biol. 2, 721-730. [DOI] [PubMed] [Google Scholar]
- Honey, K., and Rudensky, A.Y. (2003). Lysosomal cysteine proteases regulate antigen presentation. Nat. Rev. Immunol. 3, 472-482. [DOI] [PubMed] [Google Scholar]
- Jerome, W.G., and Yancey, P.G. (2003). The role of microscopy in understanding atherosclerotic lysosomal lipid metabolism. Microsc. Microanal. 9, 54-67. [DOI] [PubMed] [Google Scholar]
- Jordens, I., Fernandez-Borja, M., Marsman, M., Dusseljee, S., Janssen, L., Calafat, J., Janssen, H., Wubbolts, R., and Neefjes, J. (2001). The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr. Biol. 11, 1680-1685. [DOI] [PubMed] [Google Scholar]
- Klausner, R.D., Donaldson, J.G., and Lippincott-Schwartz, J. (1992). Brefeldin A: insights into the control of membrane traffic and organelle structure. J. Cell Biol. 116, 1071-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornfeld, S., and Mellman, I. (1989). The biogenesis of lysosomes. Annu. Rev. Cell Biol. 5, 483-525. [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz, J. (1998). Cytoskeletal proteins and Golgi dynamics. Curr. Opin. Cell Biol. 10, 52-59. [DOI] [PubMed] [Google Scholar]
- Luzio, J.P., Brake, B., Banting, G., Howell, K.E., Braghetta, P., and Stanley, K.K. (1990). Identification, sequencing and expression of an integral membrane protein of the trans-Golgi network (TGN38). Biochem. J. 270, 97-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks, M.S., and Seabra, M.C. (2001). The melanosome: membrane dynamics in black and white. Nat. Rev. Mol. Cell Biol. 2, 738-748. [DOI] [PubMed] [Google Scholar]
- Marsh, E.W., Leopold, P.L., Jones, N.L., and Maxfield, F.R. (1995). Oligomerized transferrin receptors are selectively retained by a lumenal sorting signal in a long-lived endocytic recycling compartment. J. Cell Biol. 129, 1509-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellman, I. (1996). Endocytosis and molecular sorting. Annu. Rev. Cell Dev. Biol. 12, 575-625. [DOI] [PubMed] [Google Scholar]
- Mu, F.T. et al. (1995). EEA1, an early endosome-associated protein. EEA1 is a conserved alpha-helical peripheral membrane protein flanked by cysteine “fingers” and contains a calmodulin-binding IQ motif. J. Biol. Chem. 270, 13503-13511. [DOI] [PubMed] [Google Scholar]
- Nagle, D.L. et al. (1996). Identification and mutation analysis of the complete gene for Chediak-Higashi syndrome. Nat. Genet. 14, 307-311. [DOI] [PubMed] [Google Scholar]
- Neufeld, E.F. (1991). Lysosomal storage diseases. Annu. Rev. Biochem. 60, 257-280. [DOI] [PubMed] [Google Scholar]
- Oh, J., Bailin, T., Fukai, K., Feng, G.H., Ho, L., Mao, J.I., Frenk, E., Tamura, N., and Spritz, R.A. (1996). Positional cloning of a gene for Hermansky-Pudlak syndrome, a disorder of cytoplasmic organelles. Nat. Genet. 14, 300-306. [DOI] [PubMed] [Google Scholar]
- Oh, J., Lui, Z.-X., Feng, G.H., Raposo, G., and Spritz, R.A. (2000). The Hermansky-Pudlak syndrome (HPS) protein is part of a high molecular weight complex involved in biogenesis of early melanosomes. Hum. Mol. Genet. 9, 375-385. [DOI] [PubMed] [Google Scholar]
- Perou, C.M., Leslie, J.D., Green, W., Li, L., Ward, D.M., and Kaplan, J. (1997). The Beige/Chediak-Higashi syndrome gene encodes a widely expressed cytosolic protein. J. Biol. Chem. 272, 29790-29794. [DOI] [PubMed] [Google Scholar]
- Pieters, J. (1997). MHC class II compartments: specialized organelles of the endocytic pathway in antigen presenting cells. Biol. Chem. 378, 751-758. [PubMed] [Google Scholar]
- Raposo, G., and Marks, M.S. (2002). The dark side of lysosome-related organelles: specialization of the endocytic pathway for melanosome biogenesis. Traffic. 3, 237-248. [DOI] [PubMed] [Google Scholar]
- Raposo, G., Tenza, D., Murphy, D.M., Berson, J.F., and Marks, M.S. (2001). Distinct protein sorting and localization to premelanosomes, melanosomes, and lysosomes in pigmented melanocytic cells. J. Cell Biol. 152, 809-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendu, F., and Brohard-Bohn, B. (2001). The platelet release reaction: granules'constituents, secretion and functions. Platelets 12, 261-273. [DOI] [PubMed] [Google Scholar]
- Sata, M., Donaldson, J.G., Moss, J., and Vaughan, M. (1998). Brefeldin A-inhibited guanine nucleotide-exchange activity of Sec7 domain from yeast Sec7 with yeast and mammalian ADP ribosylation factors. Proc. Natl. Acad. Sci. USA 95, 4204-4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sata, M., Moss, J., and Vaughan, M. (1999). Structural basis for the inhibitory effect of brefeldin A on guanine nucleotide-exchange proteins for ADP-ribosylation factors. Proc. Natl. Acad. Sci. USA 96, 2752-2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seet, L.F., and Hong, W. (2001). Endofin, an endosomal FYVE domain protein. J. Biol. Chem. 276, 42445-42454. [DOI] [PubMed] [Google Scholar]
- Shiflett, S.L., Kaplan, J., and Ward, D.M. (2002). Chediak-Higashi Syndrome: a rare disorder of lysosomes and lysosome related organelles. Pigment Cell Res. 15, 251-257. [DOI] [PubMed] [Google Scholar]
- Subramaniam, V.N., Krijnse-Locker, J., Tang, B.L., Ericsson, M., Yusoff, A.R., Griffiths, G., and Hong, W. (1995). Monoclonal antibody HFD9 identifies a novel 28 kDa integral membrane protein on the cis-Golgi. J. Cell Sci. 108, 2405-2414. [DOI] [PubMed] [Google Scholar]
- Subramaniam, V.N., Peter, F., Philp, R., Wong, S.H., and Hong, W. (1996). GS28, a 28-kilodalton Golgi SNARE that participates in ER-Golgi transport. Science 272, 1161-1163. [DOI] [PubMed] [Google Scholar]
- Subramaniam, V.N., Loh, E., and Hong, W. (1997). N-Ethylmaleimide-sensitive factor (NSF) and alpha-soluble NSF attachment proteins (SNAP) mediate dissociation of GS28-syntaxin 5 Golgi SNAP receptors (SNARE) complex. J. Biol. Chem. 272, 25441-25444. [DOI] [PubMed] [Google Scholar]
- Thyberg, J., and Moskalewski, S. (1999). Role of microtubules in the organization of the Golgi complex. Exp. Cell Res. 246, 263-279. [DOI] [PubMed] [Google Scholar]
- Walkley, S.U. (1998). Related cellular pathology of lysosomal storage disorders. Brain Pathol. 8, 175-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, T., and Hong, W. (2002). Interorganellar regulation of lysosome positioning by the Golgi apparatus through Rab34 interaction with Rab-interacting lysosomal protein. Mol. Biol. Cell 13, 4317-4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, D.M., Shiflett, S.L., and Kaplan, J. (2002). Chediak-Higashi syndrome: a clinical and molecular view of a rare lysosomal storage disorder. Curr. Mol. Med. 2, 469-477. [DOI] [PubMed] [Google Scholar]
- Ward, D.M., Shiflett, S.L., Huynh, D., Vaughn, M.B., Prestwich, G., and Kaplan, J. (2003). Use of expression constructs to dissect the functional domains of the CHS/Beige protein: identification of multiple phenotypes. Traffic 4, 403-415. [DOI] [PubMed] [Google Scholar]
- Wong, S.H., Xu, Y., Zhang, T., Griffiths, G., Lowe, S.L., Subramaniam, V.N., Seow, K.T., and Hong, W. (1999). GS32, a novel Golgi SNARE of 32 kDa, interacts preferentially with syntaxin 6. Mol. Biol. Cell 10, 119-134. [DOI] [PMC free article] [PubMed] [Google Scholar]