Abstract
The mouse Igκ gene locus has three known transcriptional enhancers: an intronic enhancer (Ei), a 3′ enhancer (E3′), and a further downstream enhancer (Ed). We previously discovered, using the chromosome conformation capture technique, that Ei and E3′ interact with a novel DNA sequence near the 3′ end of the Igκ locus, specifically in B cells. In the present investigation we examined the function of this far downstream element. The sequence is evolutionarily conserved and exhibits a plasmacytoma-cell-specific DNase I hypersensitive site in chromatin, henceforth termed HS10 in the locus. HS10 acts as a co-activator of E3′ in transient transfection assays. While HS10−/− mice exhibited normal patterns of B cell development, they were tested further along with E3′−/− and Ed−/− mice for their Igκ expression levels in plasma cells, and for both allelic and isotype exclusion in splenic B cells. HS10−/− and Ed−/− but not E3′−/− mice exhibited 2.5-fold lower levels of Igκ expression in antigenically challenged plasma cells. E3′−/− but not HS10−/− mice exhibited impaired IgL isotype and allelic exclusion in splenic B cells. We have suggestive results that Ed may also weakly participate in these processes. In addition, HS10−/− mice no longer exhibited regional chromosome interactions with E3′, and exhibited modestly reduced SHM in the Jκ-Cκ intronic region in germinal center B cells from Peyer’s patches. We conclude that the HS10, E3′ and Ed differentially regulate Igκ gene dynamics.
Keywords: B cells, gene regulation, knockout mice, enhancers, transcription, gene rearrangement, isotype exclusion, allelic exclusion
Introduction
The production of Ab molecules during B cell development occurs by the stage-specific and sequential rearrangement of Ab genes in the bone marrow. The IgH gene locus functionally rearranges first, by V(D)J joining events, leading to the pre-B cell stage of differentiation (1). Next, the Igκ locus is poised for rearrangement, and in response to the appropriate environmental cues, one of the 96 Vκ genes is semi-randomly selected for recombination to a Jκ region (2; for reviews, see refs. 3, 4). If Igκ V-J joining is productively unsuccessful because of out-of-reading frame recombination junctions, then the Igλ locus becomes activated for rearrangement and expression, which in wild type (WT) mice accounts for production of only approximately 5% of the total IgL chains. Interestingly, most λ-producing B cells possess aberrantly rearranged Igκ genes or ones deleted because of Vκ joining to a recombining sequence (RS) near the 3′-end of the locus (5–8).
Research in our laboratory has focused on the Igκ locus because it offers the opportunity to visualize chromatin remodeling events that are linked to gene activation, as well as to identify changes in chromatin structure that may precede gene rearrangement and transcriptional activation during B lymphocyte differentiation (9 and refs. within). Rearrangement of the Igκ locus results in its transcriptional activation because the process repositions a Vκ gene carrying its own promoter into a chromatin domain containing three powerful enhancers: an intronic enhancer (Ei) within the transcription unit (10), and two enhancers downstream of the transcription termination region (11), termed E3′ (12) and Ed (9). Experimental evidence supports the proposal that these enhancers mediate long-range interactions to trigger transcriptional activation via a regulated looping mechanism. In activated B cells, the three enhancers exhibit all possible pair-wise interactions with rearranged Vκ gene promoters and among themselves, with the looping out of the intervening DNA sequences (13). However, in unstimulated B cells, rearranged Vκ gene promoters only form complexes with either Ei or E3′, but not with Ed. This results in a transcriptionally poised state in the locus (14).
The functional significance of Ei, E3′ and Ed in Igκ gene dynamics in B lymphocytes has been previously studied by creating individual targeted deletions of each. The results of these experiments revealed that Ei and E3′ each play quantitative roles in gene rearrangement (15, 16), while E3′ and Ed each play quantitative roles in rearranged gene transcription (16, 17). Ed, the farthest downstream enhancer residing some 8 kb away from E3′, plays no role in gene rearrangement and maximally acts during the latest stages of B cell differentiation (17). However, double deletions of pairs of enhancers has severe consequences on the locus. Deletion of Ei and E3′ eliminates Igκ gene rearrangement (18), while deletion of E3′ and Ed abolishes Igκ gene transcription (19). These results reveal that these enhancers play redundant compensatory roles in this locus.
Previously we searched for new cis-acting elements that might be involved in Igκ gene regulation by using Ei or E3′ as anchor sites with chromosome conformation capture technology. We found that these enhancers exhibited B-cell-specific interactions spanning distances up to 40 kb with a DNA sequence near the 3′ end of the locus (13). In the current investigation we determine the function of this new far downstream DNA element, which proves to be evolutionarily conserved and forms a plasmacytoma-cell-specific DNase I hypersensitive site in chromatin (HS10). By creating a targeted homozygous deletion of HS10 and characterizing its resulting phenotype along with those resulting from homozygous deletions of either E3′ or Ed, we demonstrate that both HS10 and Ed, but not E3′, play pivotal roles in high-level transcription in plasma cells, whereas E3′ and Ed, but not HS10, significantly contribute to IgL isotype and Igκ allelic exclusion in splenic B cells.
Materials and Methods
Cell line culture
P815, EL-4, 3-1 and 38B9 cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum, 1% penicillin-streptomycin and 2 mM L-glutamine. S194 and A20 cells were cultured in Iscove's medium containing 5% fetal bovine serum, and MPC-11 cells were cultured in Dulbecco's modified Eagle's medium containing 20% horse serum. S194, A20, and MPC-11 cells were cultured at 37° with 10% CO2; all other lines were cultured at 37°C with 5% CO2.
Mapping DNase I hypersensitive sites
Cells were permeabilized with NP40 and treated with increasing concentrations of DNase I (Roche Applied Science, Indianapolis, IN) (0–32 µg/ml) for 3 min at room temperature (20). Cells were lysed and DNase I was quenched in lysis buffer containing Proteinase-K. Genomic DNA was purified by phenol:chloroform extraction and 10–15 µg were digested to completion with ScaI. Then DNA was transferred to Zeta-probe GT genomic membrane (Bio-Rad Laboratories, Richmond, CA) after electrophoretic separation on agarose gels. Prehybridization and hybridization were performed at 65°C with ExpressHyb hybridization buffer (Clonetech, Madison, WI). The membranes were hybridized with [α-32P] dCTP-labeled probe. Membranes were exposed to Eastman Kodak film with intensifying screens at −70°C or to PhosphorImager screens (Molecular Dynamics, Sunnyvale, CA). The probe was a 465 bp DNA fragment amplified by PCR, using primers that were Probe-f: 5′-TTCCCTTCTTCCTGCTAAGTCAA-3′ and Probe-r: 5′-GATGATGTGGGGTCTCAGATACC-3′. PCR reactions were performed for 35 cycles of 30 s at 94°C, 30 s at 56°C, and 30 s at 72°C.
Reporter gene constructions
Plasmids used in transient transfection luciferase assays were constructed as follows: A 1.3 kb fragment encompassing HS10 was amplified from mouse genomic DNA. MluI or XhoI sites were introduced into the PCR fragment by incorporating these restriction enzyme recognition sequences into the primers. PCR amplified HS10 fragments were inserted into the MluI or XhoI site of the E3′.κpLuc plasmid (21), designated here as HS10.E3′.κpLuc and E3′.HS10.κpLuc, respectively. The primer pairs for PCR amplification of HS10 fragments containing MluI or XhoI were, respectively: HS10-MluI-f: 5′-CGTGACGCGTTGCAAACAGCCATTCGCTAACA-3′ and HS10-MluI-r: 5′-CGTGACGCGTCATGCCTTTAATCACAGCCTAG-3′; HS10-XhoI-f: 5′-CGTGCTCGAGTGCAAACAGCCATTCGCTAACA-3′ and HS10-XhoI-r: 5′-CGTGCTCGAGCATGCCTTTAATCACAGCCTAG-3′. For HS10.κpLuc plasmid construction, a HS10 fragment was PCR amplified using primers HS10-MluI-f and HS10-XhoI-r, and then this fragment was cloned into MluI and XhoI sites of E3′.κpLuc to replace the E3′ enhancer. PCR reactions were performed for 35 cycles of 30 s at 94°C, 30 s at 56°C, and 1 min at 72°C.
Transient transfection luciferase assays
Cell lines were transiently transfected in triplicate using either ProFection DEAE-dextran (Promega, San Luis Obispo, CA) for S194 as previously described (9), or Lipofectamine 2000 (Invitrogen, Carlsbad, CA) for others. Typically, 106–107 cells and 1–2 µg of DNA were used per transfection, adjusted for construct sizes to provide equimolarity with respect to the κpLuc reporter, along with 30 ng of pRL-CMV Renilla luciferase reporter (Promega). Cells were harvested 24 h post-transfection and cell extracts were assayed for luciferase activity using the Dual-Luciferase™ reporter assay system (Promega) following the manufacturers' instructions. The Renilla luciferase activity was used to normalize transfection efficiencies.
Mouse strains
Mice possessing a 1.3 kb (1,327 bp) deletion of the HS10 element in the endogenous Igκ locus were generated by standard embryonic stem (ES) cell targeting technology; germline transmissible mice were bred with Cre recombinase expressing MORE mice to obtain neor deletion mice (22) (Supplemental Fig. S1). Mice bearing a human Cκ knocked-in gene were kindly provided by Michel C. Nussenzweig of Rockefeller University (23). E3′−/− mice, originally produced by Fred Alt’s laboratory (16), were kindly provided by Mark Schlissel. Ed−/− mice were previously established in our laboratory (17). All mice were extensively backcrossed onto a C57/BL6 genetic background. All mice strains were used in accordance with protocols approved by the UT Southwestern Medical Center Institutional Animal Care and Use Committee (IACUC).
Flow cytometry
Single-cell suspensions were prepared from bone marrow and spleens of 6–14 week old mice. Single-cell suspensions were stained with Abs and analyzed using a FACS Calibur with CellQuest software (BD Bioscience, San Diego, CA) or FlowJo software (Tree Star, Ashland, OR). For IgL isotype exclusion analysis, splenic cells were first stained with anti-FcγII/III (BD Bioscience, 553142, 1 µg/ml) at 4°C for 30 min. After washing, cells were stained with anti-mouse-Igκ-PE (BD Bioscience, 559940, 0.4 µg/ml), anti-mouse-Igλ1-3-FITC (BD Bioscience, 553434, 5 µg/ml) and anti-mouse-B220-APC (BD Bioscience, 553092, 0.5 µg/ml) at 4°C for 30 min. For Igκ allelic exclusion analysis, splenic cells were first stained with anti-FcγII/III at 4°C for 30 min. After washing, cells were stained with anti-mouse-Igκ-PE (0.4 µg/ml) at 4°C for 30 min. After extensive further washing, cells were then stained with anti-human-Igκ-APC (Southern Biotech, Birmingham, AL, 9230-11, 0.4 µg/ml) and anti-mouse-B220-FITC (BD Bioscience, 553038, 1 µg/ml) at 4°C for 30 min. Intracellular staining was preformed with a BD Cytofix/Cytoperm™ Kit (BD Bioscience), and Igκ levels were evaluated by mean fluorescence intensity (MFI) of anti-mouse-Igκ-PE. A PE-isotype Ab (BD Bioscience) was used as negative control for intracellular staining. Other reagents used were as follows: anti-IgM-APC (Southern Biotech); anti-CD43-PE (BD Bioscience); anti-CD25-biotin (BD Bioscience); and Streptavidin-PECy5.5 (BD Bioscience).
Real-time PCR for expression of Igκ and germline DNA levels
To examine Igκ gene expression, total RNA was extracted from 2–5×105 cells using Trizol reagent (Invitrogen). RNA samples were treated with DNase I (Invitrogen) to remove genomic DNA contamination and were reverse transcribed into cDNA using a SuperScript VILO cDNA Synthesis Kit (Invitrogen). Real-time PCR was performed to analyze Igκ expression levels with the following primers: sense 5′-GTATCCATCTTCCCACCATCCAGTGAGCAGTTAAC-3′ and antisense 5′-CTTGTGAGTGGCCTCACAGGTATAGCTGTTATGTC-3′. Transcript levels were calculated using the ΔCt method according to the manufacturer’s instructions and normalized to the cDNA levels of the mouse hypoxanthine-guanine phosphoribosyltransferase (HPRT) gene.
Germline retention of Igκ alleles were determined by real-time PCR assays, using forward and reverse primers that were complementary to sequences upstream (KGL-F) and downstream of Jκ1 (KGL-R), as described previously (19). Germline levels were normalized to the levels of an β-actin genomic region. The percentage of Igκ germline alleles was calculated by dividing the corresponding levels in WT or knockout mice B cells by those in embryonic stem (ES) cells.
Plasma cell purification
For isolating plasma cells from mice, splenic cells were first labeled with biotinylated anti-mouse-CD138 (BD Bioscience) Ab and then Streptavidin MicroBeads. CD138+ cells then were isolated with MACS columns. Isolated CD138+ cells were labeled with anti-mouse B220-FITC and anti-mouse-CD138-APC. Plasma cells (B220low and negative CD138high) were sorted on a MoFlo machine (Dako Cytomation, Carpinteria, CA).
Mouse immunization and ELISA analysis
WT, HS10−/−, E3′−/−, and Ed−/− mice, 10–16 weeks of age, were each immunized I.P. with 2 × 108 sheep red blood cells (SRBC) (Sigma-Aldrich, Carlsbad, CA) emulsified in complete Freund’s adjuvant (CFA) (Sigma-Aldrich). Spleens were harvested at the indicated times for analysis of Igκ expression in plasma cells. To induce a T-cell-dependent antibody response, WT, HS10−/− and Ed−/− mice were each immunized I.P. with 100 µg Keyhole limpet hemocyanin (KLH, Sigma-Aldrich) in emulsified CFA. To induce a T-cell-independent antibody response, mice were each immunized with 10 µg TNP-Ficoll (BioSearch Technologies, Novato, CA) I.P. in PBS. Sera were collected 7 days and 14 days later. Anti-KLH specific IgM or IgG Ab concentrations were analyzed with mouse Anti-KLH-IgM or Anti-KLH-IgG ELISA kits (Life Diagnostics, West Chester, PA). Anti-TNP specific IgM was analyzed by ELISA. Briefly, PVC microtiter plates (DYNEX technologies INC, Chantilly, VA) were coated with TNP-BSA (25 µg/ml) and incubated overnight at 4°C. After washing with PBS, diluted sera were added and incubated at 4°C overnight. Plates were subsequently washed three times and incubated with goat anti-mouse IgM-HRP (Southern Biotech) for 2 h. After washing three times, plates were developed with the ABTS substrate (Southern Biotech) following the manufacturer's instructions. Mouse anti-TNP antibody (BD Bioscience) was used to quantitate ELISA data.
Splenic cell culture
Single-cell suspensions of spleens from WT and HS10−/− mice were incubated with an optimal concentration of biotinylated-anti-CD43 (Ly-48, BD Bioscience) Ab and biotinylated-anti-Igλ1–3 (R26-24, BD Bioscience) Ab. Then MACS MS separation columns (Miltenyi Biotec, Auburn, CA) were used to deplete CD43+ and λ+ B cells. κ+ enriched B cells were cultured in RPMI 1640 medium supplemented with 10% FBS, penicillin/streptomycin, L-glutamine, and 50 mM β-mercaptoethanol at 106 cells/ml, and then were stimulated with 20 mg/ml LPS (Sigma-Aldrich), and 1.0 mg/ml anti-CD40 Ab (1C10, eBioscience, San Diego, CA) for 3 days at 37°.
Chromosome conformation capture
Chromosome conformation capture assays were performed as described using the previously reported PCR primers with minor modifications (13, 14). These previous studies established the validity of this technique in our hands by performing many controls, including the demonstration by genomic Southern assays that restriction enzyme digestion goes to completion at efficiencies ranging from 65 to 89% within cross-linked chromatin DNA for both transcriptionally active and inactive Igκ loci. 107 cells were cross-linked, lysed and nuclei were digested with 1000 IU of AseI at 37°C for 16 h. After ligation and subsequent DNA purification, the cross-linking frequencies between the anchor and test fragments, as measured by the amount of corresponding ligation product, were estimated by duplicate SYBR Green real-time PCR reactions relative to standards. The standards were used to normalize for the PCR primer efficiencies. BAC clone RP24-387E13, which contains all the relevant regions studied, was mixed at equal molar ratios with the PCR product spanning the house keeping gene, porphobilinogen deaminase (PBGD) AseI cutting site. The mixture was digested with AseI to completion and ligated at high concentration to generate eqimolar mixtures of all possible ligation products. The cross-linking and ligation efficiencies between different samples and different experiments were also normalized to the cross-linking frequencies of two adjacent AseI fragments of the PBGD gene. The relative cross-linking frequency was then calculated using the following formula: (PCR signal of a given Igκ gene ligation product from chromatin/PCR signal of that Igκ gene ligation product from naked DNA standard)/(PCR signal of a given PBGD gene ligation product from chromatin/PCR signal of that PBGD gene ligation product from naked DNA standard). The error bars presenting results represent the standard deviations from the means from at least three independent experiments.
SHM analyses
Single-cell suspensions prepared from Peyer’s patches were stained with PE-anti-B220 and FITC-PNA (Vector Laboratories, Burlingame, CA). B200+ PNAhigh GC cells were sorted on a MoFlo machine (Dako Cytomation). For the Jκ-Cκ intronic region SHM analysis, genomic DNA was purified from sorted WT and HS10−/− GC B cells. The Jκ-Cκ intronic regions from rearranged genes were PCR amplified by using the Phusion® Hot Start II High-Fidelity DNA Polymerase (Finnzymes; Thermo Scientific/Dharmacon, Lafayayette, CO) with a degenerate Vκ primer and a reverse primer located approximately 600 bp downstream of the Jκ5 as described elsewhere (17). Gel purified Vκ-Jκ5 PCR products were cloned into the StrataClone Blunt Vector Mix amp/kan (Agilent Technologies, La Jolla, CA). Vκ-Jκ5 clones were identified and sequenced by use of a T7 primer. Sequences were aligned with the mouse Jκ5 downstream sequence using the Vector NT I (Invitrogen) AlignX program and mismatches were scored as mutations in the 500 bp region downstream of Jκ5.
Results
Identification of HS10, a new DNase I hypersensitive site in the Igκ gene locus with E3′ co-activator activity
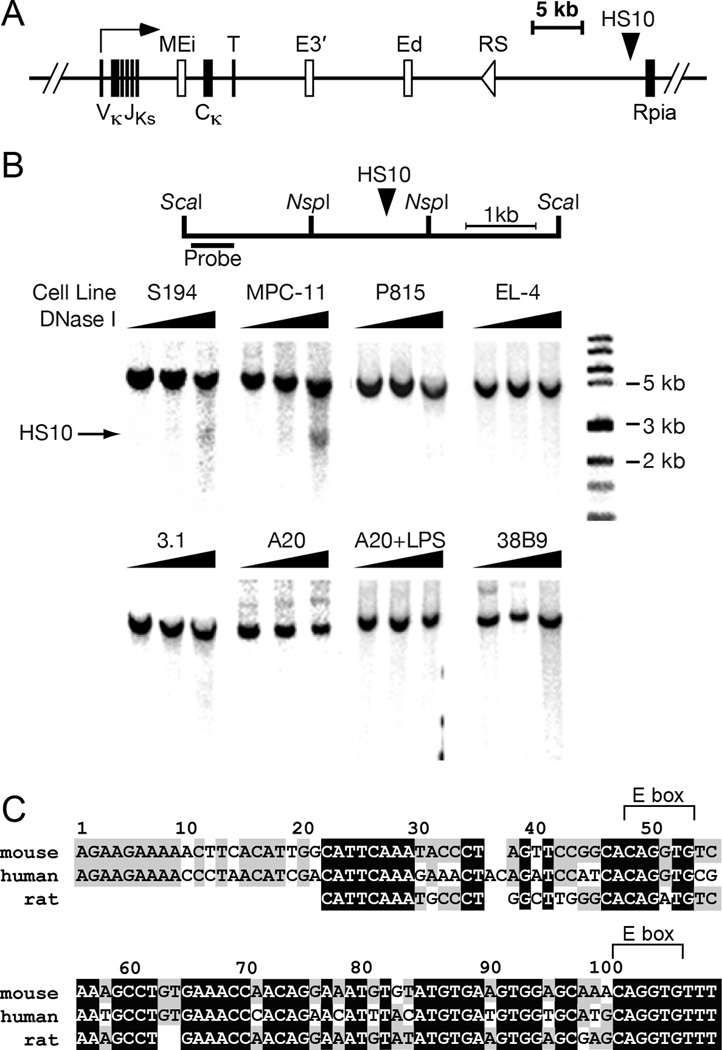
We identified a DNA sequence that exhibited B-cell specific, long range interactions with Ei and E3′ using chromosome conformation capture technology (13). This sequence resides some 40 kb downstream of Ei, within 2 kb of the neighboring housekeeping gene, ribose-5-phosphate isomerase (Rpia), at the 3′ end of the Igκ gene locus (Fig. 1A, HS10). Because functionally active cis-acting elements exhibit DNase I hypersensitive sites in chromatin (24), we first mapped the chromatin structure of the element as a function of B-cell differentiation using a panel of cell lines. The element formed a DNase I hypersensitive site, termed HS10, in the terminally differentiated plasmacytoma cell lines S194 and MPC-11, but not in other B-cell lines arrested at earlier developmental stages (3.1, A20, or 38B9), or non-B cells (P815 and EL-4) (Fig. 1B). Sequence analysis revealed that HS10 contains a core region 110 bp in length that is highly conserved among multiple species, which exhibits 76% identity with human and 87% identity with rat Igκ gene sequences (Fig. 1C). Two E-boxes are present in the core region, which are known to bind E2A proteins crucial for B cell development (31). Importantly, our previous studies have shown E-boxes with associated E2A proteins are present in Ei, E3′, and Ed in mouse B cells (13).
FIGURE 1.
Mapping and sequence analysis of DNase I hypersensitive site HS10. A, Schematic diagram depicting a rearranged Igκ locus. The coordinates of Ei, E3′, Ed, and HS10 in the NCDI37/mm9 mouse chromosome 6 sequence are respectively 70,675,570 to 70,676,084, 70,685,250 to 70,686,058, 70,693,704 to 70,694,944, and 70,713,758 to 70,715,084. B, upper: Schematic diagram of the positions of ScaI and NspI restriction sites, HS10 and the hybridization probe. lower: HS10 was mapped by indirect end-labeling after DNase I digestion of the various indicated permeabilized cell lines and ScaI digestion of the resulting purified DNA followed by Southern blotting. The solid arrow indicates the position of HS10, which is plasmacytoma-cell-specific. Cell lines correspond to plasmacytomas S194 and MPC-11 (25, 26), mastocytoma P815 (27), pro-B 38B9 (28), pre-B 3-1 (28), mature B A20 (29), and mature T EL-4 cells (30). C, HS10 sequences are conserved between mouse, human and rat. Blank spaces indicate no match, gray and black boxes indicate two or three matches between species, respectively. Two conserved E-boxes (CANNTG) are shown above the sequences.
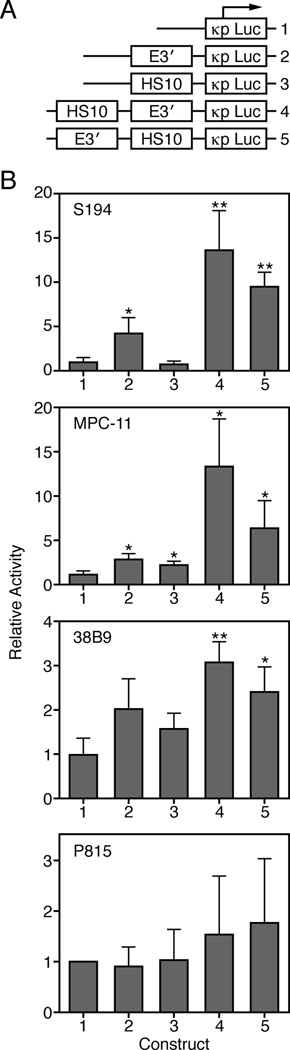
To determine if HS10 corresponded to a new transcriptional enhancer in the locus, we performed transient transfection assays with a luciferase reporter gene containing a Vκ gene promoter (Fig. 2A, construct 1). As a positive control, we found that when E3′ alone was inserted into this reporter it exhibited significant enhancer activities in both S194 and MPC-11 cells as expected, but not in 38B9 pro-B cells or P815 mastocytoma cells (Fig. 2A and B, construct 2). Insertion of HS10 alone resulted in only mild enhancer activity in MPC-11 cells (Fig. 2A and B, construct 3). By contrast, HS10 significantly stimulated transcription in constructs possessing E3′ in the plasmacytoma cell lines S194 and MPC-11, and also showed this trend but to a lesser extent in 38B9 pro-B cells, while HS10 was not active in P815 mastocytoma cells (Fig. 2A and B, constructs 4 and 5). We conclude that HS10 exhibits transcriptional synergism with E3′ in plasmacytoma cells, a result also observed earlier with Ei and Ed (9). However, HS10 does not stimulate transcription when combined with Ei in plasmacytoma cells (Supplemental Fig. S1A). We suspect that this is because Ei alone does not act as an enhancer transient transfection experiments with these cells (Supplemental Fig. S1A; ref. 9).
FIGURE 2.
DNA sequences encompassing HS10 specify transcriptional enhancer co-activator activity. A, Schematic diagram of lucifierase reporter gene constructs (labeled numbers 1–5, respectively). B, The luciferase activities of the above constructs was measured after transient transfection into the indicated cell lines. Data are representative of several independent experiments and are plotted as the activity of each construct relative to the activity of κpLuc after correction for the relative transfection efficiencies by monitoring the activity of a co-transfected pRL-CMV Renilla luciferase reporter gene. Data are presented as means ± SD. Statistical significance was determined by a paired Student’s t test. Double and single asterisks indicate p<0.01 and p<0.05, respectively, relative to the activity exhibited by the κpLuc construct.
HS10−/− mice exhibit normal surface Ig expression and B cell development in spleen and bone marrow
The observations that HS10 sequences are evolutionarily conserved, exhibit a plasmacytoma-specific DNase I hypersensitive site in chromatin, and possess enhancer co-activation activity, prompted us to examine the function of HS10 in the endogenous Igκ locus. We generated germline transmissible mice with a targeted 1.3 kb deletion of HS10, leaving only a single loxP site in its place, through standard embryonic stem cell targeting technology. The targeted and deleted loci were confirmed by Southern blotting and PCR assays (Supplemental Fig. S2). We found that HS10−/− mice exhibited no significant differences in bone marrow and spleen cell numbers or spleen weight compared to those of their WT littermates or age-matched WT mice controls [bone marrow cell numbers (2 femurs and 2 tibias): 50.9±4.2 vs 53.0±4.6 × 106, n=6, p=0.75 Student’s t test; spleen cell numbers: 82.2±15.8 vs 88.3±17.6 × 106, n=8, p=0.54 Student’s t test; or spleen weight: 89.4±13.6 vs 93.8±13.4 mg, n=6, p=0.57 Student’s t test]. Furthermore, HS10−/− mice exhibited similar levels of Igκ+ and Igλ+ B cells in spleen compared with WT mice (Fig. 3A). We further investigated the effect of HS10 deletion on the development of B cell subpopulations in bone marrow by FACS. The percentages of Igκ+ and Igλ+ cells in bone marrow were not significantly different between HS10−/− and WT mice (Fig. 3B). Finally, we evaluated early B cell compartments in bone marrow by FACS. We found no significant differences in the percentages of immature B cells (B220+IgM+) and circulating mature B cells (B220highIgM+) (Fig. 3C). No significant differences were also observed in the percentage of bone marrow B220+IgM− B cells, which include both pro-B and pre-B cells, from HS10−/− and WT mice (Fig. 3C). These B220+IgM− B cells were gated and further analyzed for their surface expression of CD43 and CD25. The results revealed that HS10−/− and WT mice have comparable levels of pro-B cells (B220+IgM−CD43+CD25−), large circling pre-B cells (B220+IgM−CD43+CD25+), and small pre-B cells (B220+IgM−CD43−CD25+) (Fig. 3C). In addition, we thoroughly analyzed FACS-sorted pre-B cells and found no differences in the levels of Igκ gene rearrangement nor Jκ region usage between HS10−/− and WT mice (Supplemental Fig. S3). Therefore, we conclude that deletion of HS10 causes no significant defects in B cell development or in cell surface Ig expression in spleen and bone marrow cells.
FIGURE 3.
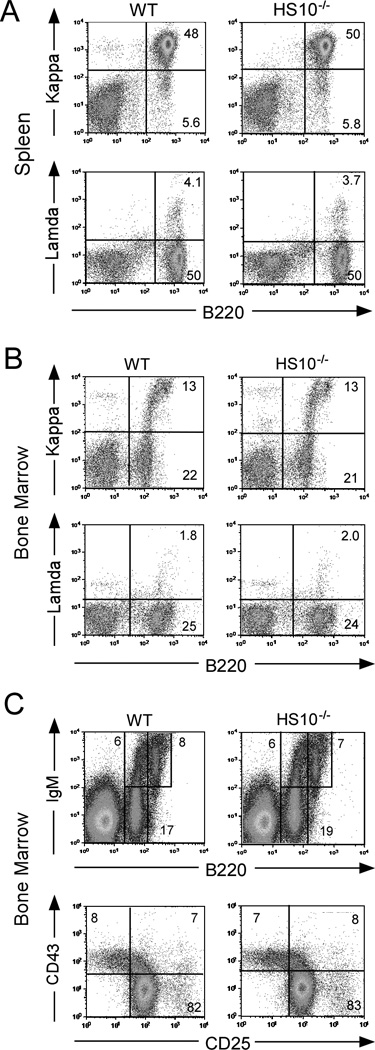
Flow-cytometric analysis of cell surface Ig expression and B cell development in spleen and bone marrow. A, FACS analysis of cell surface Ig expression in WT and HS10−/− mice. Single cell suspensions from spleen were simultaneously stained with anti-B220 and anti-Igκ Abs (upper); or anti-B220 and anti-Igλ Abs (lower). Stained cells were analyzed by FACS. Only cells residing in the lymphocyte gate were analyzed. Percentages of cells residing in various windows are shown in the figure sub-panels. Data are representative of independent FACS analyses from at least 6 mice of each genotype. The percentages of Igκ+ B cells between HS10−/− and WT were not significantly different, n=6, p=0.88 Student’s t test, nor were the percentages of Igλ+ B cells, n=6, p=0.54, Student’s t test. B, FACS analysis of cell surface Ig expression in bone marrow of WT and HS10−/− mice. Single cell suspensions from bone marrow were stained and analyzed as described in Fig. 3A. The percentages of Igκ+ B cells between HS10−/− and WT were not significantly different, n=6, p=0.73 Student’s t test, nor were the percentages of Igλ+ B cells, n=6, p=078, Student’s t test). C, Four color FACS analysis of B cell development in bone marrow of WT and HS10−/− mice. Single cell suspensions from bone marrow were stained with anti-B220 and anti-IgM Abs to assay for mature B (B220highIgM+) and immature B cells (B220+IgM+) (upper right subpanels). There were no significant differences between these cell populations, n=6, p=0.63, Student’s t test, n=6, p=0.95, Student’s t test, n=6, p=0.47, Student’s t test. B220+IgM− cells were gated and their expression of CD25 and CD43 were analyzed (lower panels). Data are representative of independent FACS analyses from at least 6 mice of each genotype. There were no significant differences between these cell populations, n=6, p=0.71, Student’s t test, n=6, p=0.87, Student’s t test, n=6, p=0.72, Student’s t test.
HS10 and Ed but not E3′ are required for maximal Igκ expression in plasma cells
Because HS10 forms a plasmacytoma cell-specific DNase I hypersensitive site, we decided to focus our further immediate attention on its potential function in splenic plasma cells before and after immunological challenge. We also evaluated the contributions of E3′ and Ed using knockout mice because their roles in plasma cells has not been previously addressed. Splenic cells from unimmunized mice were stained with anti-CD138 and anti-B220 Abs, and the percentage of plasma cells (B220low and negative CD138high) were evaluated by FACS. We found no significant differences in the percentages of plasma cells from HS10−/−, E3′−/− and Ed−/− mice compared to those of WT mice (Fig. 4A, upper panels) (HS10−/− vs WT, 0.25±0.08% vs. 0.26±0.10%, n=4, p=0.88, Student’s t test) (E3′−/− vs Ed−/− mice, 0.28±0.08% vs. 0.25±0.07%, n=4, p=0.59, Student’s t test) (E3′−/−- and Ed−/− vs HS10−/− and WT mice, n=4, p=0.94, one-way ANOVA). To trigger plasma cell expansion, we immunized WT, HS10−/−, E3′−/− and Ed−/− mice with sheep red blood cells in Freund’s complete adjuvant and monitored splenic plasma cell levels by FACS as before. We found that the plasma cell populations dramatically increased 5 days after immunization (Fig. 4A, lower panels), but that HS10−/−, E3′−/− and Ed−/− mice exhibited similar increases in the number of plasma cells compared to their WT littermates (WT vs HS10−/−, 2.01±0.27% vs. 2.05±0.3%, n=4, p=0.85, Student’s t test) (E3′−/− vs Ed−/− mice, 1.95±0.21% vs. 1.88±0.32%, n=4, p=0.72, Student’s t test).
FIGURE 4.
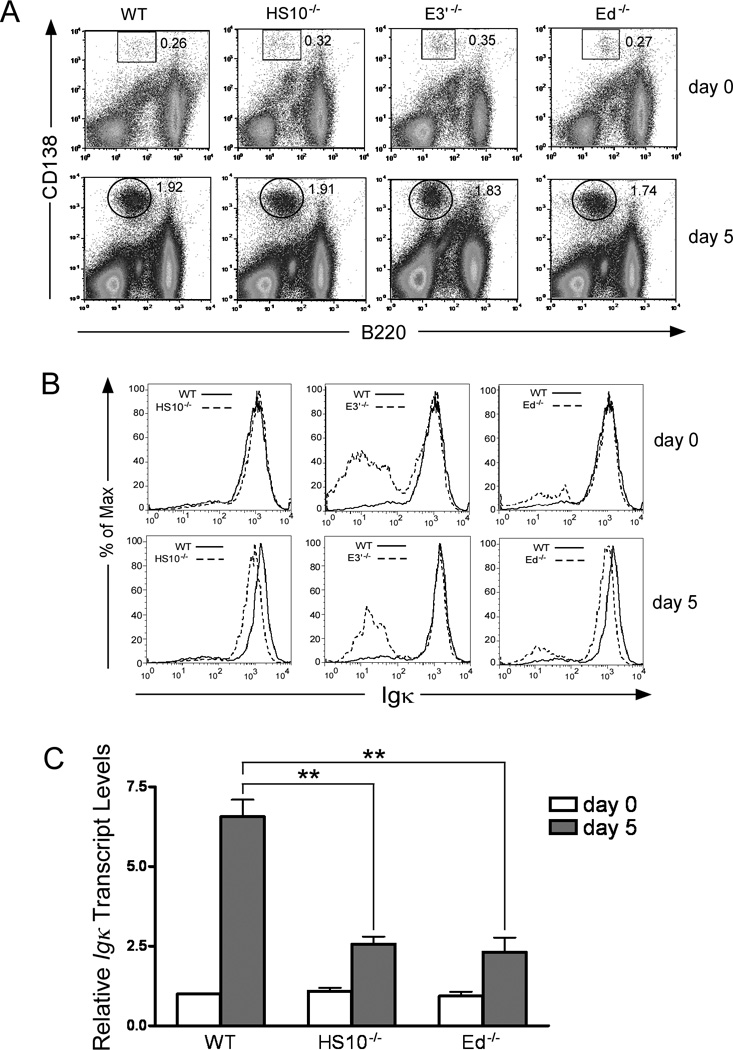
HS10 and Ed but not E3′ are required for maximal Igκ expression in plasma cells. A, FACS analysis of splenic plasma cell levels (B220low and negative CD138high, boxed or circled regions) from WT, HS10−/−, E3′−/− and Ed−/− mice, before (upper panels) and 5 days after immunization with sheep red blood cells (SRBC) (lower panels). Data are representative of independent FACS analyses from at least 6 mice of each genotype. B, FACS analysis of Igκ expression in permeabilized splenic plasma cells from WT, HS10−/−, E3′−/− and Ed−/− mice, before (upper panels) and 5 days after immunization with SCBC (lower panels). Data are representative of independent FACS analyses from at least 6 mice of each genotype. C, Realtime PCR assays of Igκ transcript levels in FACS sorted splenic plasma cells from WT, HS10−/−, E3′−/− and Ed−/− mice before and 5 days after SRBC immunization. Double asterisks indicate p< 0.01.
We also analyzed Igκ expression levels in plasma cells from WT, HS10−/−, E3′−/− and Ed−/− mice by intracellular Igκ staining, both before and after immunological challenge. FACS analysis revealed that before immunization, the plasma cells from WT, HS10−/−, E3′−/−, and Ed−/− mice expressed comparable levels of Igκ, which was indicated by the similar levels of Igκ mean fluorescent intensities (MFI): WT (1084±69), HS10−/− (1105±64), E3′−/− (1031±75), and Ed−/− (1071±63) (Fig. 4B, upper panels, n=4, p=0.48, One-way ANOVA). It is also to be noted that a significant fraction of plasma cells from E3′−/− mice are Igκ minus because they are Igλ+; this observation but to a lesser extent also occurs for Ed−/− mice (Fig. 4B; see also Fig. 7A). However, 5 days after immunization, the plasma cells from HS10−/− and Ed−/− mice, but not those from E3′−/− mice, expressed significantly lower levels of Igκ compared to those from WT cells (Fig. 4B, lower panels): MFIs (HS10−/− vs WT, 1140±72 vs. 1587±78, p=0.01, n=4, Student’s t test); Ed−/− vs E3′−/− vs WT (1124 ± 64 and 1598±71 and 1613±79, n=4, p<0.01; Ed−/− vs. WT or Ed−/− vs. E3′−/−, p<0.01, One-way ANOVA). These results indicate that both HS10 and Ed, but not E3′, are required for maximal Igκ expression in plasma cells.
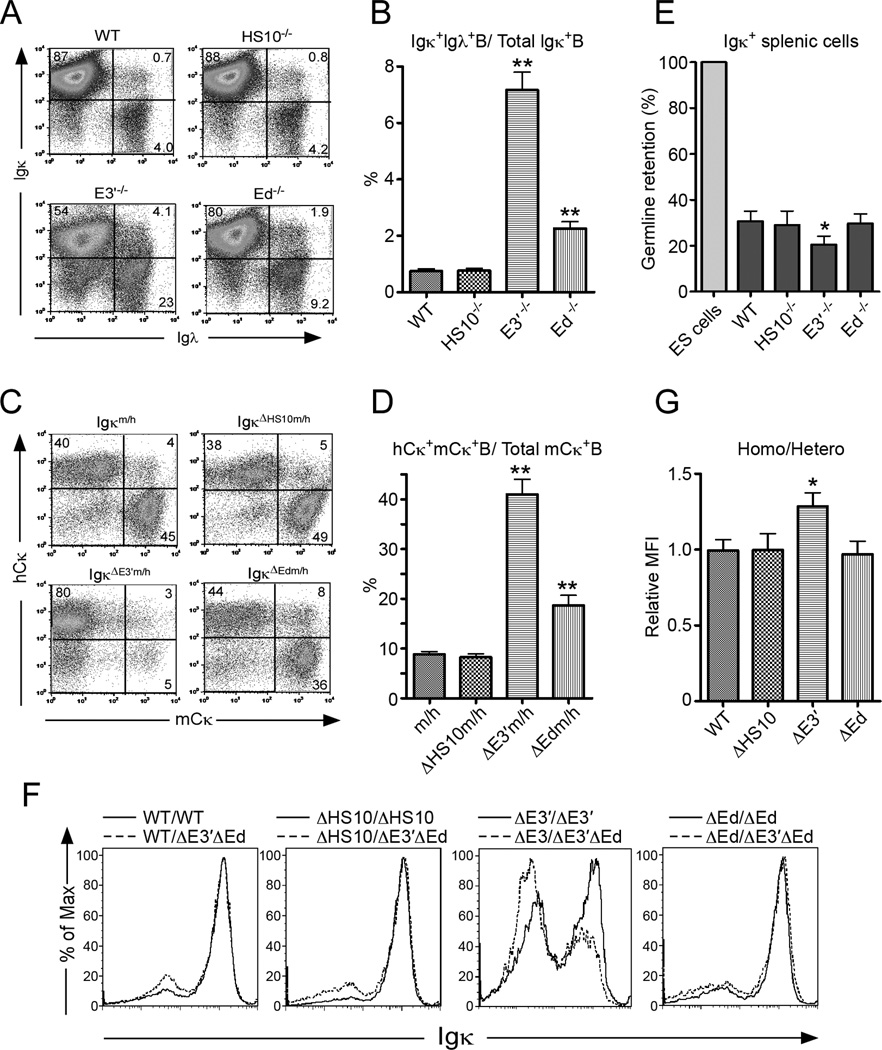
FIGURE 7.
E3′ and Ed, but not HS10, are involved in IgL isotype and Igκ allelic exclusion. A, Analysis of Igκ and Igλ isotype exclusion. Single cell suspensions from spleen were simultaneously stained with anti-B220, anti-Igκ, and anti-Igλ Abs. The expression of Igκ and Igλ in WT, HS−/−, E3′−/−, and Ed−/− mice splenic B220+ cells were analyzed by FACS. Data are representative of independent FACS analyses from at least 4 mice of each genotype. B, Percent of Igκ+ cells that also express Igλ. Data are presented as means ± SD. Double and single asterisks indicate p< 0.01 and p< 0.05, respectively. C, Analysis of Igκ allelic exclusion. Single cell suspensions from spleen of Igκm/h, IgκΔHS10m/h, IgκΔE3’m/h, and IgκΔEdm/h mice were simultaneously stained with anti-B220, anti-mouse-Igκ (mCκ), and anti-human-Igκ (hCκ). The expression of mCκ and hCκ were analyzed by FACS. Percentages of cells residing in various windows are shown in the figure sub-panels. Data are representative of independent FACS analyses from at least 4 mice of each genotype. D, Percent of mCκ+ cells that also express hCκ. Data are presented as means ± SD. Double asterisks indicate p<0.01. E, Results of a real-time PCR assay used to detect germline retention of Igκ sequences in splenic B220+ Igκ+(hi) B cells from the indicated lines of mice. The percentages of germline retention were calculated by dividing the germline sequence levels of B cells by those of ES cells. Data are presented as means ± SD (n=3). The single asterisk indicates p<0.05. F, The levels of Igκ gene expression in B220+Igκ+ splenic B cells between the indicated pairs of homo- and heterozygotic genetic lines was assayed for by determining Mean Florescent Intensities (MFI) of Igκ in B220+ gated cells. G, Bi-allelic expression was evaluated by measuring the relative MFI between homozygotes and the heterozygotes from each pair of groups. The single asterisk indicates p<0.05.
Finally, to confirm the above results we performed real-time PCR assays to quantitate Igκ gene transcript levels in plasma cell populations before and 5 days after immunization. In agreement with the above results, approximately 2.5-fold lower Igκ transcript levels were observed in isolated plasma cells from HS10−/− and Ed−/− mice splenic cells after immunization as compared to WT mice levels (Fig. 4C) (p<0.01, n=4, Student’s t test). We conclude that HS10 and Ed, but not E3′, are required for maximal Igκ expression in plasma cells under conditions of strong Ag-induced immune response challenge.
HS10 and Ed are required for maximal T-cell dependent, but not T-cell independent Igκ Ab responses
Because the above results revealed that HS10 and Ed are important elements in maximizing Ab production in plasma cells, we decided to evaluate their roles further in responding to either T-cell dependent or T-cell independent Ag challenges. For this purpose we immunized WT, HS10−/− and Ed−/− mice with the T-cell-dependent Ag, KLH, as well as the T-cell-independent Ag, TNP-Ficoll, and assayed the resulting Ab titers in sera by ELISA. As shown in Fig. 5A, 7 days after immunization, high levels of anti-KLH (IgM) were detected and remained at similar levels after 2 weeks. However, we found that the levels of anti-KLH-IgM in HS10−/− and Ed−/− mice sera were significantly lower than those of WT mice. In addition, although the serum levels of anti-KLH (IgG) 7 days after immunization were quite low, they dramatically increased after 2 weeks. HS10−/− and Ed−/− mice produced significantly less anti-KLH-IgG 2 weeks after immunization compared to WT mice (Fig. 5B, right panel). By contrast, no significant differences in anti-TNP-IgM were observed among WT, HS10−/− and Ed−/− mice 7 or 14 days after immunization (Fig. 5C). We conclude that HS10 and Ed are important elements in maximizing the T-cell dependent immune response, presumably through signaling by CD40, but not in the T-cell independent process.
FIGURE 5.
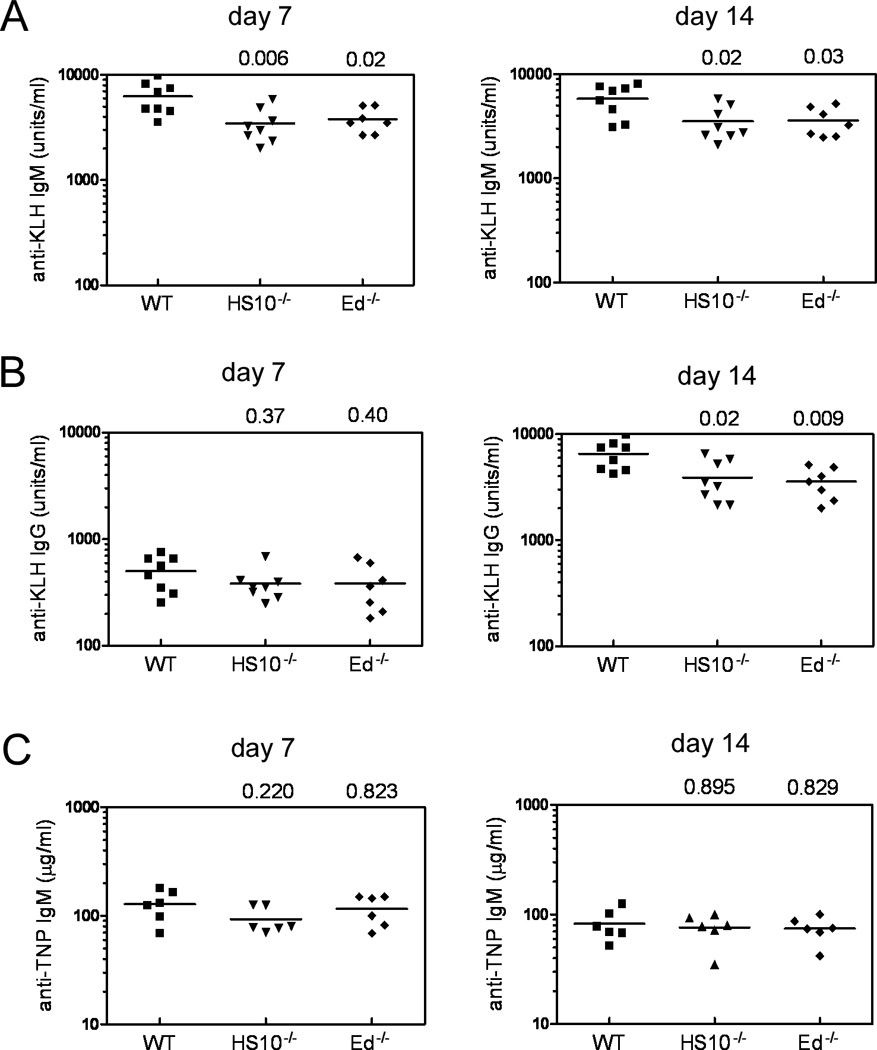
Both HS10 and Ed are required for maximal Igκ expression during a T-cell-dependent Ab response. A, B, and C, production of anti-KLH and anti-TNP Abs after immunization of WT, HS10−/− and Ed−/− mice. The results show sera titers measured by ELISA assays 7 and 14 days after immunization. Sera titers were only at background levels at day 0 (± 0.2). P values compared to WT levels that were determined by one-way ANOVA using SPSS 11.5 software are shown at the top of panels A, B and C.
Effect of HS10 on chromosome conformation
In order to approach understanding the mechanism of HS10 function, we utilized the chromosome conformation capture technique to address the consequences of deleting HS10 on various pair wise interactions between the enhancers and a DNA restriction fragment that would bear HS10 in WT mice (Fig. 6). Unfortunately, because this analysis requires biochemical amounts of cells, plasma cells are in too small amounts for this experiment to be practical or feasible. Therefore, as an alternative we generated biochemical amounts of activated splenic cells for this analysis, by culturing splenic cells for 3 days with anti-CD40 Abs and LPS to trigger them ex vivo to differentiate towards the plasma cell phenotype. As a negative control we used samples from CD3ε+ T cells. The results of the chromosome conformation capture assays revealed that B-cell-specific pair wise interactions between Ei and E3′ or Ei and Ed were similar between WT and HS10−/− mice samples (Fig. 6B and C). It is noteworthy, however, that the interactions between Ei and HS10 previously seen in plasmacytoma cells (13) were not observed in stimulated splenic cells from WT mice (Fig. 6D). However, these culturing conditions do not achieve the level of Igκ gene expression exhibited by in vivo antigen-stimulated CD138+ plasma cells. Nevertheless, upon HS10 deletion, regional interactions with E3′ were significantly diminished (Fig. 6E). We conclude that HS10 itself is largely responsible for the previously observed chromosome conformation capture results.
FIGURE 6.
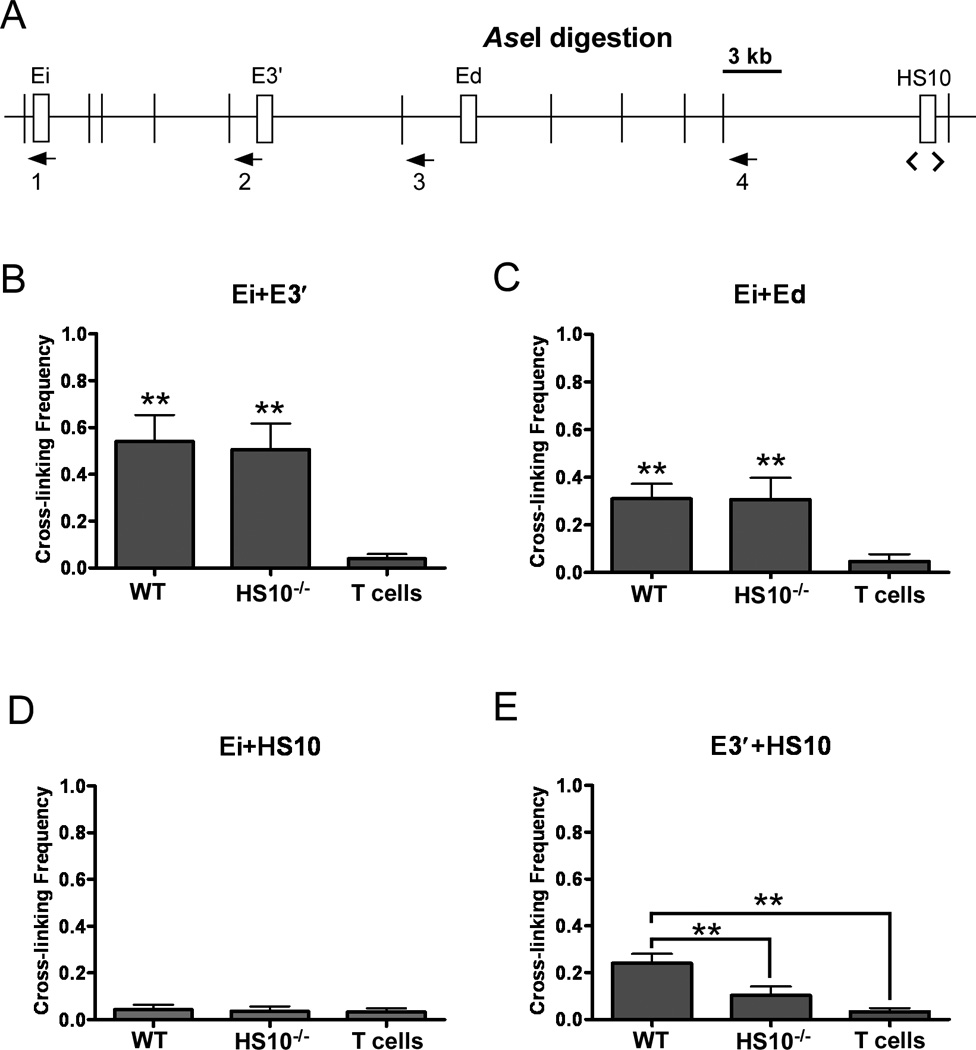
Importance of HS10 in pair wise interactions between selected cis-acting sequences in the Igκ locus in stimulated splenic B cells from WT and HS10−/− mice, or in T cells from WT mice. A, Map of the 3′ region of the mouse Igκ locus. Cis-acting sequences are indicated by rectangles. Numbered arrowheads indicate the primers used in selected chromosome conformation capture assays. Narrow vertical lines depict AseI enzyme cut sites. The region of the HS10 deletion is demarcated by the underlying brackets. B, Interactions between Ei and E3′ were assayed using PCR primers 1 and 2 (n=3, WT versus T, p<0.01; WT versus HS10−/−, p=0.73). C, Interactions between Ei and Ed were assayed using PCR primers 1 and 3 (n=3, WT versus T, p<0.01; WT versus HS10−/−, p=0.96). D, Interactions between Ei and HS10 were assayed using PCR primers 1 and 4 (n=3, WT versus T, p=0.54; WT versus HS10−/−, p=0.71; HS10−/− versus T, p=0.83). E, Interactions between E3′ and HS10 were assayed using PCR primers 2 and 4 (n≥3, WT versus T, p<0.01; WT versus HS10−/−, p<0.01; HS10−/− versus T, p=0.09). Data are presented as means ±SD and p values were determined by one-way ANOVA using SPSS 11.5 software. Double asterisks indicate p<0.01 with respect to a comparison with T cell samples, except for panel E that includes the additional indicated comparison.
E3′ and Ed, but not HS10, are involved in IgL isotype and Igκ allelic exclusion
We next assayed for the role of HS10 in specifying IgL isotype and Igκ allelic exclusion. We also evaluated how E3′−/− and Ed−/− mice would behave in these assays, because the functions of these enhancers in these processes have not been previously directly addressed. We first examined the population of Igκ+Igλ+ cells in the spleen from WT, HS10−/−, E3′−/−, and Ed−/− mice by FACS analysis. As shown in Fig. 7A, like WT mice, a normal very low level of isotype co-expressing B cells (B220+Igκ+Igλ+) was observed in HS10−/− mice (WT, 0.68±0.05%) (HS10−/−, 0.73±0.04%). Surprisingly, relative to WT mice levels, a dramatic increase in isotype co-expressing cells was detected in E3′−/− mice (4.46±0.35%), and a less striking but significant approximately two-fold increase was also observed in Ed−/− mice (1.92±0.20%). Because the number of splenic Igκ+ B cells in E3′−/− and Ed−/− mice were less than WT mice, we evaluated isotype exclusion by comparing the ratios of the number of double producers (B220+Igκ+Igλ+) to the total number of B cells that express Igκ (B220+Igκ+Igλ+ plus B220+Igκ+Igλ−). As shown in Fig. 7B, isotype co-expression was increased significantly in both E3′−/− and Ed−/− mice (n=4, p<0.01, WT vs. E3′−/− or WT vs. Ed−/−, One-way ANOVA), but not in HS10−/− mice.
As one approach to assay for allelic exclusion, we bred our mouse lines with human Cκ knockin mice (23) to generate heterozygotes (Igκm/h). As shown in Fig 7C, HS10 deficiency apparently did not alter the pattern of allelic exclusion, because like WT m/h heterozygotes, most cells producing molecules with mouse Cκ exons from ΔHS10 alleles did not produce counterparts also possessing human Cκ exons. In marked contrast, because of competition between ΔE3′ mouse Cκ alleles and WT knockin human Cκ alleles, ΔE3′ alleles were only able to contribute to surface Igκ expression in around 10% of the total splenic B cells. Although much less striking, heterozygotes possessing ΔEd alleles also exhibited more human Cκ+ B cells than mouse Cκ+ B cells, indicating that Ed may also playing a role in allelic choice. In the same way as we quantitated isotype exclusion, we evaluated bi-allelic expression by comparing the ratios of the number of double producers (B220+mCκ+hCκ+) to the number of total B cells that had expressed mouse Igκ (B220+mCκ+hCκ+ plus B220+mCκ+hCκ−). These results suggest that there is a severe impairment of allelic exclusion in heterozygotes possessing ΔE3′ alleles compared to those possessing WT or ΔHS10 alleles, and that heterozygotes possessing ΔEd alleles may also exhibit a modest but significant increase in bi-allelic expression (Fig. 7D, p<0.01, n=4, WT vs. E3′−/− or WT vs. Ed−/−, One-way ANOVA).
As another approach to assay for allelic exclusion, we measured the extent of rearrangement of the locus in Igκ+ splenic B cells using real-time PCR to quantitate the level of germline Igκ sequences from HS10−/−, E3′−/−, Ed−/−, and WT mice. As shown in Fig. 7E, the levels of such germline sequences in HS10−/−, Ed−/−, and WT mice samples were each about 30% of those of ES cell controls, while these levels were only about 20% from E3′−/− mice, a value statistically significantly lower than the other controls (p<0.05). This result further supports increased bi-allelic expression E3′−/− mice.
As a final approach to assay for bi-allelic expression, we bred our mouse lines with E3′−/−Ed−/− mice, which are inactive for Igκ gene expression (19), to generate heterozygotes. We then compared the level of Igκ gene expression between these heterozygotic lines and their homozygous counterparts in B220+Igκ+ splenic cells by measuring their respective Mean Florescent Intensities (MFI) of Igκ. We reasoned that if significant bi-allelic expression occurs in the homozygotes, then it should be detectably higher than the level of monoallelic expression in the heterozygotes. As shown in Fig 7E, HS10 or Ed deficiency did not alter the levels of Igκ, because HS10−E3′+Ed+/HS10−E3′+Ed+ homozygotes and HS10−E3+Ed+/HS10+E3−Ed− heterozygotes, or HS10+E3′+Ed−/HS10+E3′+Ed− homozygotes and HS10+E3′+Ed−/HS10+E3′−Ed− heterozygotes, expressed comparable levels of Igκ, just like WT control mice after being crossed with E3′−/−Ed−/− mice. In marked contrast, HS10+E3′−Ed+/HS10+E3′−Ed+ homozygotes expressed significantly higher levels of Igκ compared to HS10+E3′−Ed+/HS10+E3′−Ed− heterozygotes, consistent with significant bi-allelic expression. We quantitated these results by measuring the relative increase in MFI of Igκ in B220+ Igκ+ B cells between homozygotes and the heterozygotes for each pair of groups, an approach that revealed a statistically significant difference for the ΔE3′ pair (Fig. 7G; p<0.05, n=4, WT vs. E3′, One-way ANOVA). In conclusion, each of these three different approaches yield results that are strongly suggestive that allelic exclusion is hampered in mice deficient in E3′.
SHM is only modestly decreased in the Jκ-Cκ intronic region of HS10−/− GC B cells
The E box motif has been reported to enhance SHM (32, 33). In addition, the Ig transcription efficiency has also been correlated with levels of SHM (34). Because HS10 contains two E box sites and we also observed reduced Igκ expression in HS10−/− stimulated plasma B cells along a reduced T-cell-dependant antibody response, we investigated the possibility that SHM might be decreased in HS10−/− GC B cells. We purified B220+PNAhigh GC B cells from Peyer’s patches of HS10−/− and WT mice by flow cytometry. Genomic DNA was isolated from these GC B cells and VκJκ rearrangements were amplified using high fidelity PCR. VκJκ rearrangement products were gel purified, cloned and sequenced. We found that the mutation frequency within a 500 bp intronic region downstream of Jκ5 was 14.5 × 10−3 mutations per base in WT GC B cells. In HS10−/− GC B cells the mutation frequency in the corresponding region was 11.6 × 10−3 mutations per base (Fig. 8). Thus, the overall mutation frequency in HS10−/− GC B cells was reduced by only 20% compared with that of WT GC B cells, which although small was statistically significant (p<0.01). Among clones bearing mutations, the mutation frequency was 20.1 × 10−3 mutations per base in WT GC B cells, and 17.1 × 10−3 mutations per base in HS10−/− GC B cells, a 15% reduction (Fig. 8). Control experiments revealed that error frequency of the high fidelity PCR was only about 0.2 mutations per kb. Furthermore, we found that the percentage of clones with no mutations was 33% in HS10−/− GC B cells, compared to 25% in WT GC B cells (Fig. 8). In summary, these data indicate that Igκ SHM is modestly decreased in HS10−/− GC B cells from Peyer’s patches.
FIGURE 8.
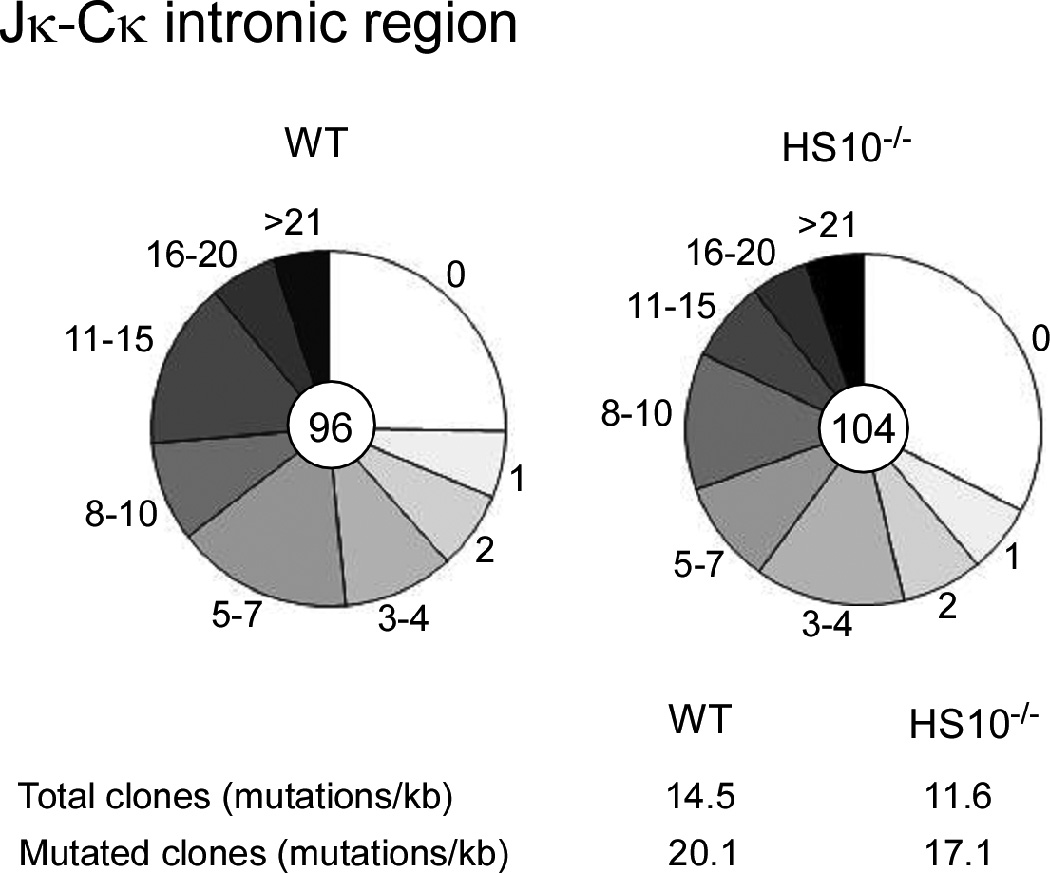
Analysis of Igκ SHM in GC B cells. B220+PNAhigh GC B cells were sorted from 4–6-month-old WT (left) and HS10−/− (right) mice, and SHM in a 500 bp Jκ-Cκ region that is immediately 3' of Vκ-Jκ5 recombination products was analyzed. Each pie slice represents the proportions of sequences having the indicated numbers of mutations. The numbers of plasmid clones that were sequenced are shown in pie centers. The mutation frequencies from the total or mutated clones are shown at the bottom.
Discussion
The DNA sequence corresponding to HS10 was first identified through exploratory experiments utilizing chromosome conformation capture assays to search for sequences that interacted with the Igκ gene enhancers (13). In the present investigation, we found that HS10 is a plasmacytoma-cell-specific DNase I hypersensitive site in chromatin and that its underlying sequence is evolutionarily conserved. These observations provided the impetus for uncovering the function of HS10, because every B-cell-specific hypersensitive site that has been identified previously in the mouse Igκ locus has proven to have a significant function in B cells. HS10−/− mice exhibited normal B cell development, surface Igκ expression, and Vκ-Jκ rearrangement (Fig. 3 and Supplemental Fig. S3). In plasma cells, instead, we found that HS10 was required for maximal Igκ expression but only after immunological challenges, which were T-cell-dependent. This was also true for Ed but not for E3′. In the case of T-cell-dependent responses, B cells receive not only a primary stimulation that is transduced through BCR cross linking, but also co-stimulatory signaling from CD40 or TLR ligands (35–38). Our results thus imply that HS10 and Ed may be activated by co-stimulatory signaling and mediate a special gene conformation that facilitates maximal Igκ gene expression during T-cell-dependent Ab responses.
It is not without president that the order of sequences along a multi-gene family correlates with the developmental order of their sequence usage (39). Interestingly, the locations of the cis-acting sequences 5′ to 3′ along the Igκ locus correlates with the stages of their functional usage during B cell development: Ei and E3′ are required for Vκ-Jκ joining (18), E3′ and Ed are essential for rearranged gene transcription (19), and Ed and HS10 are necessary for maximal Igκ expression in differentiating plasma cells (Figs. 4 and 5). Our previous chromosome conformation capture experiments reveal that these cis-acting elements form pair wise interactions with each other (13, 14), and work by others and us had revealed that strong phenotypes only result when pairs of these cis-acting sequences are deleted together (18, 19). These observations suggest the Ed and HS10 may have overlapping functions in plasma cells. This view is consistent with the observation that HS10 exhibits pair wise interactions with Ei and E3′, but not with Ed, in plasmacytoma cells (13). Our current chromosome conformation capture experiments reveal that HS10 is not required for Ei-E3′ or Ei-Ed interactions but is required for maximal regional interactions with E3′ in stimulated splenic cells.
Our results also revealed that E3′, but not HS10, contribute to IgL isotype and Igκ allelic exclusion. We have suggestive results that Ed may also weakly participate in these processes. In these cases the level of surface Igκ is detectably lower than that observed in WT cells, which would lead to less tonic BCR signaling and a less stringent suppression of continued RAG expression (40–43), which presumably accounts for the observed increases in isotype co-expression and bi-allelic expression due to continued gene rearrangements.
One of our experimental designs used to assay for allelic exclusion put two Igκ alleles in competition in the same cells, an allele carrying entirely WT cis-acting sequences marked by a human Cκ exon, and another allele with or without selected deletions in either HS10, E3′ or Ed, possessing a mouse Cκ exon. This approach not only revealed the percentage of cells that are double Igκ producers, but also allowed measurement of the allelic preferences for expression. The most striking skewed result in expression was observed in IgκΔE3’m/h heterozygotes, where less than 10% of the splenic B cells expressed the E3′-deleted allele. This test also revealed that the HS10 deletion had no effect whatsoever on splenic cell Igκ expression, in agreement with our earlier results of FACS analyses of HS10−/− cells.
Previous studies of SHM in mice possessing targeted Igκ gene enhancer deletions in the endogenous locus have revealed that E3' and Ed are each necessary to achieve WT levels of SHM in the 5' region of the Igκ gene’s J-C intron (17, 44). They further showed that Ei in the native locus plays no role in determining the level of SHM (44). We have found that HS10−/− mice exhibited a modest 20% reduction in SHM over WT levels. We conclude that HS10 plays only a very minor role in contributing to SHM under the conditions studied, where like the previous studies (17, 44), there was no specific Ag challenge. Notably, both E3' and Ed are enhancers that contribute maximally to Igκ gene transcription levels and each play a more significant role in SHM (17, 19, 44).
HS10 resides near the 3′ end of the Igκ locus, about 2 kb away from the downstream Rpia1 housekeeping gene. Rpia is expressed at the same low level in B cells as in fibroblasts (45), indicating that this housekeeping gene’s expression is totally buffered from the extraordinarily high-level of expression exhibited by the Igκ gene in B cells. Moreover, we have found that HS10−/− mice still express the Rpia gene at the same low constitutive level in B cells at different developmental stages and in non-B cells (Supplemental Fig. S4). Furthermore, the results of transient transfection and stable integration experiments proved that HS10 lacked silencer or insulator activities (Fig. 2 and Supplemental Fig. S1), but revealed that HS10 is a transcriptional co-activator of E3′ but not of Ei (Fig. 2 and Supplemental Fig. S1A).
In conclusion, the results of the present study together with our previously published studies on the mouse Igκ locus have revealed multiple levels of communication between distal cis-acting regulatory sequences through long range interactions (9, 13, 14, 17, 19). In the future it will be germane to determine how these elements contribute to the three-dimensional organization of the locus in B cell nuclei and to the determine what other genes share transcription factories and common regulatory circuits with the Igκ locus transcriptional interactome.
Supplementary Material
Acknowledgements
We thank Mark Schlissel for providing mice and Jose Cabrera for graphic illustrations. We are indebted to Nicolai van Oers for his insightful comments and suggestions.
Abbreviations used in this paper
- CFA
complete Freund’s adjuvant
- Ei
intronic enhancer
- E3′
3′ enhancer
- Ed
downstream enhancer
- ES
embryonic stem
- HS10
DNase I hypersensitive site number 10 in the region downstream of Vκ21-1
- MEi
matrix association region and Ei
- RS
recombination sequence
- SRBC
sheep red blood cells
Footnotes
This investigation was supported by Grants GM29935 and AI067906 from the National Institutes of Health and Grant I-0823 from the Robert A. Welch Foundation to WTG.
None of the authors have any financial interest related to this work.
References
- 1.Yancopoulos GD, Alt FW. Developmental controlled and tissue-specific expression of unrearranged VH gene segments. Cell. 1985;40:271–281. [PubMed] [Google Scholar]
- 2.Brekke KM, Garrard WT. Assembly and analysis of the mouse immunoglobulin kappa gene sequence. Immunogenetics. 2004;56:490–505. doi: 10.1007/s00251-004-0659-0. [DOI] [PubMed] [Google Scholar]
- 3.Schatz DG, Ji Y. Recombination centres and the orchestration of V(D)J recombination. Nature Rev. Immunol. 2011;11:251–263. doi: 10.1038/nri2941. [DOI] [PubMed] [Google Scholar]
- 4.Schlissel MS. Regulation of activation and recombination of the murine Igκ locus. Immunol. Rev. 2004;200:215–223. doi: 10.1111/j.0105-2896.2004.00157.x. [DOI] [PubMed] [Google Scholar]
- 5.Durdik JM, Moore MW, Selsing E. Novel κ light chain rearrangements in mouse λ light chain producing B lymphocytes. Nature. 1984;307:749–752. doi: 10.1038/307749a0. [DOI] [PubMed] [Google Scholar]
- 6.Nadel B, Cazenave PA, Sanchez P. Murine lambda gene rearrangements: the stochastic model prevails over the ordered model. EMBO J. 1990;9:435–440. doi: 10.1002/j.1460-2075.1990.tb08128.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shimizu T, Iwasato T, Yamagishi H. Deletions of immunoglobulin Cκ region characterized by circular excision products in mouse splenocytes. J. Exp. Med. 1991;173:1065–1072. doi: 10.1084/jem.173.5.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vela JL, Aït-Azzouzene D, Duong BH, Ota T, Nemazee D. Rearrangement of mouse immunoglobulin kappa deleting element recombining sequence promotes immune tolerance and lambda cell production. Cell. 2008;28:161–170. doi: 10.1016/j.immuni.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu ZM, George-Raizen JB, Li S, Meyers KC, Chang MY, Garrard WT. Chromatin structural analyses of the mouse Igκ gene locus reveal new hypersensitive sites specifying a transcriptional silencer and enhancer. J. Biol. Chem. 2002;277:32640–32649. doi: 10.1074/jbc.M204065200. [DOI] [PubMed] [Google Scholar]
- 10.Queen C, Baltimore D. Immunoglobulin gene transcription is activated by downstream sequence elements. Cell. 1983;33:741–748. doi: 10.1016/0092-8674(83)90016-8. [DOI] [PubMed] [Google Scholar]
- 11.Xu M, Barnard MB, Rose SM, Cockerill PN, Huang S-Y, Garrard WT. Transcription termination and chromatin structure of the active immunoglobulin κ gene locus. J. Biol. Chem. 1986;261:3838–3845. [PubMed] [Google Scholar]
- 12.Meyer KB, Neuberger MS. The immunoglobulin κ locus contains a second, stronger B cell specific enhancer which is located downstream of the constant region. EMBO J. 1989;8:1959–1964. doi: 10.1002/j.1460-2075.1989.tb03601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Z, Garrard WT. Long range interactions between three transcriptional enhancers, active Vκ gene promoters and a 3′ boundary sequence spanning 46 kb. Mol. Cell. Biol. 2005;25:3220–3231. doi: 10.1128/MCB.25.8.3220-3231.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Z, Ma Z, Terada LS, Garrard WT. Divergent roles of RelA and c-Rel in establishing chromosomal loops upon activation of the Igκ gene. J. Immunol. 2009;183:3819–3830. doi: 10.4049/jimmunol.0901781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu Y, Davidson L, Alt FW, Baltimore D. Deletion of the Igκ light chain intronic enhancer/matrix attachment region impairs but does not abolish Vκ-Jκ rearrangement. Immunity. 1996;4:377–385. doi: 10.1016/s1074-7613(00)80251-4. [DOI] [PubMed] [Google Scholar]
- 16.Gorman JR, Van der Stoep N, Monroe R, Cogne M, Davidson L, Alt FW. The Igκ 3′ enhancer influences the ratio of Ig(kappa) versus Ig(lambda) B lymphocytes. Immunity. 1996;5:241–252. doi: 10.1016/s1074-7613(00)80319-2. [DOI] [PubMed] [Google Scholar]
- 17.Xiang Y, Garrard WT. The downstream transcriptional enhancer, Ed, positively regulates mouse Igκ gene expression and somatic hypermutation, J. Immunol. 2008;180:6725–6732. doi: 10.4049/jimmunol.180.10.6725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Inlay M, Alt FW, Baltimore D, Xu Y. Essential roles of the κ light chain intronic enhancer and 3' enhancer in κ rearrangement and demethylation. Nat. Immunol. 2002;3:463–468. doi: 10.1038/ni790. [DOI] [PubMed] [Google Scholar]
- 19.Zhou X, Xiang Y, Garrard, WT WT. TheIgκ enhancers, E3′ and Ed, are essential for triggering transcription. J. Immunol. 2010;185:7544–7552. doi: 10.4049/jimmunol.1002665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aaslabd R, Stewart AF. Analysis of DNase I hypersensitive sites in chromatin by cleavage in permeabilized cells. Methods Mol. Biol. 1999;119:355–362. doi: 10.1385/1-59259-681-9:355. [DOI] [PubMed] [Google Scholar]
- 21.Fulton R, Van Ness BG. Nucleic Acids Res. 1993;21:4941–4947. doi: 10.1093/nar/21.21.4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tallquist MD, Soriano P. Epiblast-restricted Cre expression in MORE mice: A tool to distinguish embryonic vs. extra-embryonic gene function. Genesis. 2000;26:113–115. doi: 10.1002/(sici)1526-968x(200002)26:2<113::aid-gene3>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 23.Casellas R, Shih T-AY, Kleinewietfeld M, Jankovic M, Nemazee D, Rajewsky K, Nussenzweig MC. Contribution of receptor editing to the antibody repertoire. Science. 2001;291:1541–1544. doi: 10.1126/science.1056600. [DOI] [PubMed] [Google Scholar]
- 24.Gross DS, Garrard WT. Nuclease hypersensitive sites in chromatin. Ann. Rev. Biochem. 1988;57:159–197. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]
- 25.Horibata K, Harris AW. Mouse myelomas and lymphomas in culture. Exp. Cell Res. 1970;60:61–77. doi: 10.1016/0014-4827(70)90489-1. [DOI] [PubMed] [Google Scholar]
- 26.Seidman JG, Leder P. A mutant immunoglobulin light chain is formed by aberrant DNA-and RNA-splicing events. Nature. 1980;286:779–783. doi: 10.1038/286779a0. [DOI] [PubMed] [Google Scholar]
- 27.Lundak RL, Raidt DJ. Cellular immune response against tumor cells. Cell Immunol. 1973;9:60–66. doi: 10.1016/0008-8749(73)90167-6. [DOI] [PubMed] [Google Scholar]
- 28.Lieber MR, Hesse JE, Mizuuchi K, Gellert M. Developmental stage specificity of the lymphoid V(D)J recombination activity. Genes Dev. 1987;1:751–761. doi: 10.1101/gad.1.8.751. [DOI] [PubMed] [Google Scholar]
- 29.Kim KJ, Kanellopoulos-Langevin C, Merwin RM, Sach DH, Asofsky R. Establishment and characterization of BALB/c lymphoma line with B cell properties. J. Immunol. 1979;122:549–554. [PubMed] [Google Scholar]
- 30.Shevach EM, Stobo JD, Green I. Immunoglobulin and θ-bearing murine leukemias and lymphomas. J. Immunol. 1972;108:1146–1151. [PubMed] [Google Scholar]
- 31.Quong MW, Romanow WJ, Murre C. E protein function in lymphocyte development. Ann. Rev. Immunol. 2002;20:301–322. doi: 10.1146/annurev.immunol.20.092501.162048. [DOI] [PubMed] [Google Scholar]
- 32.Michael N, Shen HM, Longerich S, Kim N, Longacre A, Storb U. The E box motif CAGGTG enhances somatic hypermutation without enhancing transcription. Immunity. 2003;19:235–242. doi: 10.1016/s1074-7613(03)00204-8. [DOI] [PubMed] [Google Scholar]
- 33.Schoetz U, Cervelli M, Wang YD, Fiedler P, Buerstedde JM. E2A expression stimulates Ig hypermutation. J. Immunol. 2006;177:395–400. doi: 10.4049/jimmunol.177.1.395. [DOI] [PubMed] [Google Scholar]
- 34.Odegard VH, Schatz DG. Targeting of somatic hypermutation. Nat. Rev. Immunol. 2006;6:573–583. doi: 10.1038/nri1896. [DOI] [PubMed] [Google Scholar]
- 35.Haxhinasto SA, Bishop GA. Synergistic B cell activation by CD40 and the B cell antigen receptor: role of B lymphocyte antigen receptor-mediated kinase activation and tumor necrosis factor receptor-associated factor regulation. J. Biol Chem. 2004;279:2575–2582. doi: 10.1074/jbc.M310628200. [DOI] [PubMed] [Google Scholar]
- 36.Meyer-Bahlburg A, Khim S, Rawlings DJ. B cell intrinsic TLR signals amplify but are not required for humoral immunity. J. Exp. Med. 2007;204:3095–3101. doi: 10.1084/jem.20071250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rubtsov AV, Swanson CL, Troy S, Strauch P, Pelanda R, Torres RM. TLR agonists promote marginal zone B cell activation and facilitate T-dependent IgM responses. J. Immunol. 2008;180:3882–3888. doi: 10.4049/jimmunol.180.6.3882. [DOI] [PubMed] [Google Scholar]
- 38.Jain S, Chodisetti SB, Agrewala JN. CD40 signaling synergizes with TL-2 in the BCR independent activation of resting B cells. PLoS One. 2011;6:e20651. doi: 10.1371/journal.pone.0020651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pearson CJ, Lemons D, McGinnis W. Modulating Hox gene functions during animal body pattering. Nature Rev. Genet. 2005;6:893–904. doi: 10.1038/nrg1726. [DOI] [PubMed] [Google Scholar]
- 40.Monroe JG. ITAM-mediated tonic signalling through pre-BCR and BCR complexes. Nature Rev. Immunol. 2006;6:283–294. doi: 10.1038/nri1808. [DOI] [PubMed] [Google Scholar]
- 41.Tze LE, Schram BR, Lam KP, Hogquist KA, Hippen KL, Liu J, Shinton SA, Otipoby KL, Rodine PR, Vegoe AL, Kraus M, Hardy RR, Schlissel MS, Rajewsky K, Behrens TW. Basal immunoglobulin signaling actively maintains developmental stage in immature B cells. PLoS Biol. 2005;3:e82. doi: 10.1371/journal.pbio.0030082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keren Z, Diamant E, Ostrovsky O, Bengal E, Melamed D. Modification of ligand-independent B cell receptor tonic signals activates receptor editing in immature B lymphocytes. J. Biol. Chem. 2004;279:13418–13424. doi: 10.1074/jbc.M311970200. [DOI] [PubMed] [Google Scholar]
- 43.Verkoczy L, Duong B, Skog P, Ait-Azzouzene D, Puri K, Vela JL, Nemazee D. Basal B cell receptor-directed phosphatidylinositol 3-kinase signaling turns off RAGs and promotes B cell-positive selection. J. Immunol. 2007;178:6332–6341. doi: 10.4049/jimmunol.178.10.6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Inlay MA, Gao HH, Odegard VH, Lin T, Schatz DG, Xu Y. Roles of the Ig kappa light chain intronic and 3' enhancers in Igκ somatic hypermutation. J. Immunol. 2006;177:1146–1151. doi: 10.4049/jimmunol.177.2.1146. [DOI] [PubMed] [Google Scholar]
- 45.Apel TW, Scherer A, Adchi T, Auch D, Ayane, M M, Reth M. The ribose-5-phosphate-isomerase-encoding gene is located immediately downstream from that encoding murine immunoglobulin κ. Gene. 1995;156:191–197. doi: 10.1016/0378-1119(94)00901-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.