Abstract
S100 proteins are low molecular weight calcium binding proteins expressed in vertebrates. The family constitutes 21 known members that are expressed in several tissues and cell types and play a major role in various cellular functions. Uniquely, members of the S100 family have both intracellular and extracellular functions. Several members of the S100 family (S100A1, S100A2, S100A4, S1008, S100A9, S100A11, and S100B) have been identified in human articular cartilage, and their expression is upregulated in diseased tissue. These S100 proteins elicit a catabolic signaling pathway via receptor for advanced glycation end products (RAGE) in cartilage and may promote progression of arthritis. This review summarizes our current understanding of the role of S100 proteins in cartilage biology and in the development of arthritis.
1. S100 family of proteins
S100 protein was first discovered by Moore in brain tissue and named “S100-protein” because it was soluble in 100 percent saturated ammonium sulfate [1]. However, subsequent studies have shown that this fraction contained two related proteins, S100A and S100B [2]. Currently, the family of S100 proteins is comprised of 21 known members that are characterized by the presence of two EF-hand domains (calcium binding), one at each N-terminus and C- terminus separated by a hinge region. The carboxyl–terminal (c-terminus) EF-hand domain is referred to as the conical (higher affinity) calcium binding loop that encompasses 12 amino acids, whereas the low affinity N-terminal loop formed of 14 amino acids is known as the “pseudo” or S100-specific EF–hand domain [3, 4]. In humans, most S100 genes are clustered at chromosomal locus 1q21 except S100B, S100p and S100Z, which are mapped at chromosome 12q22, 4 and 5 [5–8]. Members of the S100 protein family have sequence homology between 22 to 57% with marked variance at the hinge region and C-terminus, which is thought to contribute to the diversity in their biological function [6]. S100 proteins are expressed in various tissues in a cell type-specific manner and with specific sub-cellular localization [9, 10].
In the cell, S100 proteins exist as dimers (homodimers or heterodimers) or multimers [11–13] wherein the monomeric units of S100 proteins are held together by non-covalent bonds [14–16]. The only exception to this rule is S100G, which always exists as a monomer [17, 18]. In addition, multimerization of S100 protein is also promoted by posttranslational modification of monomeric S100 proteins [19] and by binding of metal ions [11, 12, 20]. S100 proteins bind various divalent metal ions, including calcium (high affinity), zinc, and copper [21–27]. Binding of these ions to S100 protein also modulates their function by altering conformational changes. In the calcium-free state, the EF-hand in each monomer is in anti-parallel confirmation. Upon calcium binding, each S100 monomer opens up to accommodate a target; the S100 dimer can bind target proteins on opposite sides. Although S100 dimers are characterized by the same structural motif, differences in the primary sequence of individual helices, the hinge region, or the C-terminal region, as well as differences in the inter-helical angles in calcium-loaded dimers, may be important for the recognition of target proteins and specification of functional roles of individual S100 members.
2. Biological functions of S100 proteins
S100 proteins play a major role in a broad array of biological functions. They have both intracellular and extracellular functions. Their intracellular functions include modulation of enzyme activity, calcium homeostasis, cell growth and mobility, cell cycle regulation, cell differentiation and cell survival [28]. Extracellularly, S100 proteins interact with cell surface receptors, including receptor for advance glycation end products (RAGE) [28–30] and toll-like receptors (TLR) [31], [32] and participate in signal transduction.
S100 proteins lack intrinsic enzymatic activity, but they participate in biological function via protein-protein interaction and modulating the activity of their target molecule. Binding of S100B to nuclear Dbf2-related protein kinase (NDR kinase) directly blocks the recruitment of substrate on NDR kinase [33]. A similar mechanism has been proposed for S100A4-mediated regulation of MetAP2 activity [34]. On the other hand, binding of S100A1 to ryanodine receptor, a cellular mediator of calcium-induced calcium release, increases its affinity for ryanodine (its ligand) several fold, resulting in opening of the RyR channel and increased release of calcium from the sarcoplasmic reticulum in skeletal muscle [35], thus influencing intracellular calcium homeostasis.
Furthermore, studies have shown that multiple members of S100 family can interact with a specific target molecule and have differential effects. For example, S100A1, S100B, S100A4, and S100A11 are known to interact directly with cytoskeletal elements including microtubilin, microfilaments, intermediate filaments, actin, myosin, tropomyosin; regulating cytoskeletal dynamics; and playing a role in cell mobility and proliferation [28, 36–38]. Likewise, many S100 proteins, including S100A2, S100A4, and S100B, interact with tumor suppressor 53 (p53) and modulate its activity differentially. Binding of S100A4 and S100B to p53 negatively modulates its activity to cause cell growth arrest and apoptosis [39–41], whereas binding of S100A2 enhances p53's transcriptional activity [42]. Binding of multiple S100 proteins to a single target protein is not surprising given the significant homology in their primary sequence, and provides the mechanism for the similar functional role of the two different S100 proteins in different tissues. On the other hand, the differential affects of S100 proteins on the function of a single target protein also could explain functional diversity within a given tissue.
Some members of the S100 protein family are released into the extracellular environment, where they act as cytokines and are involved in cell proliferation and survival, cell migration, and inflammation [28, 29, 43–47]. These functions of S100 proteins are associated with numerous human pathologies, including cancer [48], neurodegenerative diseases such as Alzheimer's disease [49, 50], autoimmune diseases, and arthritis [51]. Thus, S100 proteins have garnered significant interest as potential therapeutic targets for various human disorders.
3. S100 proteins in cartilage
Cartilage is an avascular connective tissue present in the ear, nose, rib cage, intervertebral discs, and joints. Articular cartilage lines the end of bones in the joints. It provides smooth, frictionless movements of joints and helps distribute the load across the joints. Articular cartilage is made up of collagens (mainly collagen type II), aggrecan, large proteoglycans, and small biglycans. This matrix provides both tensile strength and flexibility to the tissue. Chondrocytes are the sole cellular component of the cartilage tissue responsible for cartilage homeostasis (matrix synthesis and repair). However, in arthritis (both rheumatoid arthritis [RA] and osteoarthritis [OA]), there is progressive loss of cartilage due to increased production of matrix-degrading enzymes (e.g. matrix metalloproteinases [MMPs] and aggrecanases) locally produced by chondrocytes in response to various catabolic stimuli. In RA, a large proportion of matrix-degrading enzymes come from the rheumatoid synovium, and chondrocytes also participate in the RA disease process at sites remote from the cartilage-pannus junction. Extracellular S100 proteins behave like cytokines and activate the RAGE signaling pathway, resulting in increased production of matrix-degrading enzymes, including MMPs and A Disintegrin And Metalloproteinase with Thrombospondin Motifs (ADAMTSs), which are involved in cartilage degradation and the development of arthritis.
S100 protein in chondrocytes was first identified by Stefansson et al [52] in human adult and fetal tissue using immunohistochemistry; they found S100 protein staining in both the cytoplasm and the nucleus. Subsequently, several groups have identified S100 protein in normal tissue and in tumors of cartilage and bone [53–56]. In earlier work, S100 protein was primarily studied as a marker for chondrocytic phenotype and/or chondrogenic origin [55, 57, 58]. However, additional studies have reported that the intensity of S100 protein increases in chondrocytes near cartilage lesions and suggested that S100 protein may be involved in the cartilage repair process.
Weiss and Dorfman [59] found that chondrocytes from different regions in cartilage lesions stained differently for S100 protein. Cells from degenerating and hypertrophic zones were stained strongly for S100 protein compared to proliferating columnar chondrocytes. Similarly, in osteoarthritic cartilage, S100 protein staining was seen in degenerative and osteophytic regions of cartilage compared to unaffected regions of cartilage [60], suggesting that S100 protein may be involved in cartilage calcification and mineralization. Wolff et al [61] and Leonardi et al [62] postulated that the presence of S100 protein in the cells near cartilage damage indicates that chondrocyte-like cells participate in early matrix repair. This notion of S100-positive cells in matrix repair was further supported by two independent studies. Itoi [63] reported that increased S100 protein staining in cultured cartilage cells related to production of type II collagen. In the second study, Sugimoto et al [64] showed that fewer cells stained positive for S100 protein in mild OA compared to moderate and severe OA. In mild OA, S100 protein staining was present in chondrocytes near the erosion of surface layers, whereas in severe OA, S100-positive cells were found in clusters. Based on these findings, the authors speculated that S100 protein could influence metabolic activity of cartilage matrix. These early studies suggested that S100 protein might be involved in cartilage matrix synthesis and repair. However, further studies are required to understand the molecular mechanism involved. When these earlier studies were performed, the S100 protein was thought to be a single protein. However, later studies have shown that the S100 protein fraction contains a number of structurally related proteins, which collectively form a family of S100 proteins.
Lately, there has been a renewed interest in S100 protein and its function in chondrocytes due to identification of multiple members of S100 proteins in the cartilage and synovial fluid of patients with OA and RA, and their ability to activate RAGE. RAGE is a transmembrane receptor of the immunoglobulin superfamily that has been implicated in several chronic diseases, including atherosclerosis, diabetes, Alzheimer's disease, and arthritis. RAGE was identified in cartilage and its expression is upregulated in diseased cartilage [65, 66]. Here, we summarize the function of various S100 proteins identified in cartilage to date.
S100B
S100B was one of the first members of the S100 family of proteins detected in cartilage [61]. However, its role in cartilage biology is not clearly understood. Recently, we have shown that extracellular S100B stimulates the RAGE signaling pathway in chondrocytes, resulting in activation of ERK and NF-κB signaling molecules and increased production of MMP-13 [30]. These data suggest that extracellular S100B is a pro-catabolic, pro-inflammatory factor that promotes cartilage degradation and has a role in the pathogenesis of OA and RA.
S100A2
S100A2 is a homodimeric protein that requires both calcium and zinc divalent metals for its activity. S100A2 was detected in both intracellular (chondrocytes) and extracellular (cartilage matrix) tissues. Extracellular cartilage matrix staining was confined to the calcifying areas of the epiphyseal cartilage [67], suggesting that S100A2 may be involved in calcification of cartilage.
S100A4
S100A4, also known as metastasin (Mts-1), fibroblast-specific protein (FSP1), p9ka, CAPL, and calvasculin, plays an important role in metastasis. We and others have shown that S100A4 is expressed in human articular chondrocytes, and its expression is upregulated both in RA and OA [30, 68]. S100A4 has both extracellular and intracellular functions. We have recently shown that chondrocytes secrete S100A4 in response to external stimuli [69] and that S100A4 secretion is independent of the Golgiendoplasmic reticulum secretory pathway [69]. Extracellular S100A4 stimulates chondrocytes by activating RAGE signaling, leading to increased matrix metalloproteinase-13 (MMP-13) [30]. In addition, we found that S100A4 translocated into the nucleus upon interleukin-1β (IL-1β) stimulation, and translocation required post-translational modification of S100A4 by the SUMO protein at K22 and K96. Nuclear S100A4 (nS100A4) was associated with the promoter region of MMP-13 in IL-1β-treated cells, suggesting that nS100A4 is a key factor in transcriptional regulation of the MMP-13 gene. Mutation of the SUMO moiety accepting lysine residues abolished the ability of S100A4 to be sumoylated and to translocate into the nucleus [70]. Blocking of sumoylation and nuclear transport of S100A4 inhibited the IL-1β-induced production of MMP-13. Taken together, these studies suggest that both extracellular and intracellular S100A4 protein is involved in cartilage degradation and in the pathophysiology of OA.
S100A8/S100A9
The alarmins S100A8 (MRP8) and S100A9 (MRP14) are abundantly expressed in myeloid cells and play a major role in inflammation. In joint tissues, S100A8 and A9 are expressed in synovium, bone, and cartilage [71]. In cartilage, S100A8 and S100A9 expression is restricted to non-proliferative hypertrophic chondrocytes [71] and thought to play a major role in calcification of the cartilage matrix. S100A8 expression is barely detectable in resting and unstimulated chondrocytes; however, pro-inflammatory cytokines such as TNFα, IL-1β, and IL-17 upregulate chondrocyte S100A8 expression [72]. Increased intracellular levels of S100A8 and S100A9 may lead to secretion of these proteins into extracellular environments. Secretion of S100A8 and A9 has been shown in macrophages, but it is not known if chondrocytes secrete these proteins.
Studies have shown high levels of S100A8 and A9 in the synovial fluid obtained from patients with OA or RA. However, levels of S100A8/A9 in synovial fluid from RA patients are 10-fold higher than in synovial fluid from OA patients [73]. It is not clear if chondrocytes contribute to the levels of S100A8/A9 in synovial fluid. However, activated macrophages and neutrophils in the inflamed synovium may contribute to increased levels of S100A8 and S100A9 proteins in synovial fluid from patients with OA and RA [74, 75]. The extracellular function of these alarmins is thought to occur via TLR-4 [32, 76]. Chondrocytes express both RAGE and TLR-4 receptors. Chondrocytes respond to extracellular S100A8/A9 stimulation by increasing the production of matrix-degrading enzymes, including matrix metalloproteinase (MMP-2,-3,-9 and -13) and ADAMTS-4 and -5 in RAGE- and TLR-4-independent pathways. These studies suggest that S100A8 and S100A9 play an important role in cartilage degradation via additional cell surface receptors.
Recently, Zreiqat et al [77] showed that S100A8 and S100A9 expression is upregulated in early OA but not late OA, suggesting that these alarmins may initiate initial cartilage degradation by upregulating MMPs and ADAMTSs. Deletion of S100A8 / S100A9 prevented cartilage degradation in an antigen-induced arthritis model [78], indicating that these proteins play a major role in cartilage degradation and development of inflammatory arthritis.
S100A11
S100A11, also known as S100C and calgizzarin, is a homodimeric protein that interacts with annexin I in a calcium-dependent manner and is a key protein in membrane organization [79, 80]. In cartilage, S100A11 expression is upregulated in OA and is thought to promote chondrocyte hypertrophy [65], characterized by a dysregulated matrix repair process, increased expression collagen type X, and matrix-degrading enzymes [65]. Chondrocytes releases S100A11 in response to CXCL8, IL-1β, and TNF-α stimulation [19, 65]. Extracellular S100A11 stimulates the RAGE-dependent signaling pathway, resulting in activation of p-38 MAP kinase and increased production of collagen type X, thereby promoting chondrocyte hypertrophy [65]. The extracellular function of S100A11 is attributed to its ability to form dimers. Cecil and Terkeltaub [19] reported that S100A11 monomers are covalently cross-linked by transamidation mediated by transglutaminase 2 (TG-2). Mutants of S100A11 that cannot undergo transamidation do not form dimers and fail to stimulate p38 MAP kinase signaling, but retain the capacity to interact with RAGE. These studies suggest that post-translational modification of S100A11 by TG2 is an important step in S100A11-mediated chondrocyte hypertrophy and thus differentiation and progression of OA.
4. S100 proteins and signal transduction
Extracellular functions of several S100 proteins are mediated by their interaction with RAGE, a member of the immunoglobulin superfamily expressed by diverse cell types, including chondrocytes [66]. Structurally, RAGE is composed of an extracellular domain (composed of two `c' type domains and one `v' type domain), a trans-membrane domain, and a short cytoplasmic domain [81]. Multiple isoforms of RAGE have been identified [82] that are generated due to alternate splicing of the RAGE gene [83]. The predominant forms of RAGE are full-length RAGE, a secreted form of RAGE (sRAGE), C-truncated RAGE (esRAGE), and an N-truncated RAGE. Chondrocytes express several forms of RAGE, including full-length RAGE, N-truncated RAGE and an unglycosylated form of RAGE [66], suggesting that multiple forms of RAGE may have specialized functions in cartilage tisue.
RAGE is activated by multiple ligands including S100 proteins, AGEs (advance glycation end products), amphoterin, amyloid-β, and lipopolysaccharide [81, 84–86]. The first step of RAGE activation involves increased production of reactive oxygen species (ROS) via activation of the NADPH oxidase complex [29, 87]. ROS then activates and recruits a cascade of signaling proteins, including RAS, Rac1 /Cdc 42, MAP kinases (ERK1/2, p-38 and JNK), phosphatidylinositol-3-kinase (PI3-K), AKT, Janus kinase (JAK), and NF-κB. The signaling pathway activated by RAGE depends on the kind of stimulus and cell type. Here, the focus is on the RAGE signaling pathways activated by S100 proteins.
Binding of S100B to RAGE has been shown to activate the Ras-MEK-ERK1/2-NF-κB pathway in neural cells, resulting in activation of small GTPases, Rac1 /Cdc 42 and neurite growth [88]. In addition, studies have shown that S100B also activates the phosphatidylinositol-3-kinase (PI3-K)-AKT pathway in a RAGE-dependent manner to increased cellular proliferation [89]. However, at micromolar concentrations, S100B protein activates RAGE and induces increased production of ROS and release of cytochrome-C from mitochondria, leading to apoptosis [88]. On the other hand, in microglia and monocytes, activation of RAGE by S100B results in activation of the Ras- Rac1 /Cdc 42-JNK-AP-1 pathway, leading to upregulation of the pro-inflammatory enzyme cyclo-oxygenase 2 (COX-2) [90, 91]. In addition to activation of MAP kinase-NF-κb pathways, stimulation of RAGE by S100B activates the JAK-STAT pathway. In vascular smooth muscle cells, binding of S100B protein to RAGE increases ROS production, which then recruits JAK2 and STAT 3, resulting in proliferation of vascular smooth muscle cells [92]. RAGE-mediated activation of the JAK-STAT pathway required activation of non-receptor Src tyrosine kinase, protein kinase C, and phospholipase D2 [92, 93].
In chondrocytes, activation of RAGE by S100A4 increased ROS production followed by the phosphorylation of Pyk-2, and activation of MAP kinases, ERK1/2, p-38 and JNK and the transcription factor NF-κB, resulting in increased production of MMP-13 [30]. Furthermore, stimulation of RAGE by S100A11 activated MAP kinases p-38, and promoted inflammation-associated chondrocyte hypertrophy [65]. These studies suggest that RAGE signaling plays an important role in the function of S100 proteins and their pathologies in cartilage. However, in a murine model of mild or severe joint instability, knocking down RAGE expression did not confer chondroprotection [94]. This result suggests that S100 proteins in cartilage may engage other receptors for their function, or that pathways activated by other receptors may modulate RAGE-S100 protein function in cartilage.
AGE-R1 (OST-48; oligosaccharyl transferase-4), a member of AGE receptor complex consisting of AGE-R2 (80 K-H phosphoprotein) and AGE-R3 (galectin-3), was recently shown to negatively regulate RAGE-mediated inflammatory response in mesangial cells [95]. The study showed that RAGE-AGE- mediated activation of MAP kinases (ERK1/2) and NF-κB was suppressed by overexpression of AGER1. On the other hand, knockdown of AGE-R1 enhanced AGE-mediated activation of MAP kinases (ERK1/2) and NF-κB more than twofold [95]. AGE-R1 is expressed in chondrocytes (R. Yammani and R. Loeser, unpublished observation), but its role in RAGE signaling and in chondrocyte biology is yet to be characterized. Interestingly, overexpression of CD36, a member of the class B scavenger receptor family that is involved in clearing AGEs from cells [96], was recently shown to suppress RAGE-S100A11-mediated catabolic functions in chondrocytes [94]. The study found that CD-36 expression was locally elevated at the site of cartilage lesions, and its expression was upregulated by IGF-1 and the activation of peroxisome proliferator activated receptor-γ (PPARγ). Both IGF-1 and PPAR promote cartilage anabolism, suggesting that CD36 expression may be chondroprotective and function opposite to RAGE. However, the role of CD 36 in cartilage and chondrocyte biology is not clearly understood.
CD36 is a multifunctional cell surface receptor expressed on many cells, including chondrocytes. It binds to multiple ligands such as oxidized phospholipoprotein, AGEs, and long-chain fatty acids and serves as an endocytic receptor [97–99]. It was first identified as a receptor for the matrix protein thrombospondin-1 [100, 101]. Binding of oxidized LDL (oxLDL) to CD 36 elicits a cascade of signaling pathways primarily involving the src family of non-receptor-bound tyrosine kinases (fyn and Lyn) and MAP kinase (ERK1/2, p38 and JNK), which then activates downstream pro-inflammatory genes [[102–105]]. Activation of CD 36 signaling pathways has been associated with numerous human diseases associated with chronic inflammation, such as atherosclerosis, thrombosis, Alzheimer's disease, and metabolic disease (type 2 diabetes) [106–109]. Interestingly, CD36 also serves as a co-receptor for the toll-like receptor (TLR) family of cell surface receptors [110], which promote cartilage degradation and development of both RA and OA. TLRs also share ligands (S100 and high-mobility group box [HMGB] proteins) with RAGE.
Thus, initial overexpression of CD36 may act to scavenge toxic proteins such as AGEs and S100 protein, and suppress RAGE signaling. However, increases in these ligands may then activate RAGE and TLRs, which then recruit CD36 into the mega-receptor complex, co-promote the catabolic pathway, and play a major role in arthritis. More studies are required to better understand the role of these receptors, and their interactions with each other and other proteins in cartilage health and disease.
5. Conclusions and future perspectives
Although multiple members of the S100 protein family are expressed in chondrocytes, very little is known about their biological function in cartilage, especially their intracellular functions. Current evidence suggests that S100 protein expression is upregulated in diseased cartilage, and that these proteins promote cartilage degeneration by activating RAGE signaling (Fig 1). However, studies in other cell types have suggested that RAGE is not a universal receptor for S100 proteins [78, 111–114], which may be true in chondrocytes as well. A recent study [94] showed that the absence of RAGE expression is not chondroprotective as expected, suggesting the role of an alternative S100 protein receptor. S100 proteins bind to other cell surface receptors, including TLR, CD36 [115], and carboxylated glycans [72, 115], all of which are expressed by chondrocytes. Thus, further studies are required to understand how S100 proteins are secreted by chondrocytes, which receptors are activated by S100 proteins, and the nature of their interactions to elucidate the role of S100 proteins in cartilage biology. Identification and characterization of these molecular events will provide new and novel targets for therapies.
Figure 1.
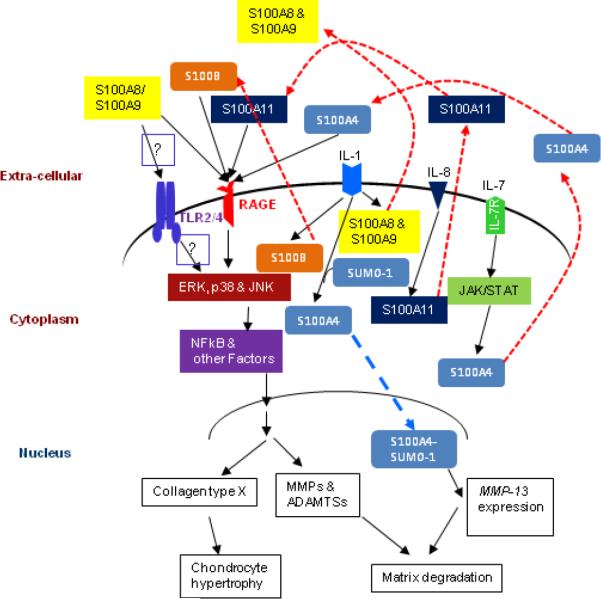
Biological roles of S100 proteins in cartilage. Increased expression of S100 proteins due to the activity of pro-inflammatory cytokines results in their secretion into the extracellular environment (red dashed lines). There, S100 proteins stimulate chondrocytes in an autocrine or paracrine manner via RAGE or TLR. Activation of RAGE or TLR signaling pathways causes chondrocyte hypertrophy and increased production of matrix-degrading enzymes (MMPs and ADAMTSs), which results in cartilage degradation and development of arthritis. Our recent study has shown that S100A4 translocates into the nucleus and promotes MMP-13 expression (blue dashed line). Question marks indicate still uncharacterized pathways in cartilage.
Highlight
S100 family of protein in cartilage.
Biological functions of S100 proteins in the cartilage.
Role of S100 proteins in the development of arthritis.
Acknowledgements
This work was supported by the National Institutes of Health (NIH) grant R03 AR061050. I thank Richard F. Loeser, M.D., for his critical review of the manuscript and Karen Klein, MA, ELS (Research Support Core, WFUHS) for her editorial contributions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Moore BW. A soluble protein characteristic of the nervous system. Biochemical and biophysical research communications. 1965;19:739–744. doi: 10.1016/0006-291x(65)90320-7. [DOI] [PubMed] [Google Scholar]
- [2].Donato R. Perspectives in S-100 protein biology. Review article. Cell calcium. 1991;12:713–726. doi: 10.1016/0143-4160(91)90040-l. [DOI] [PubMed] [Google Scholar]
- [3].Krebs J, Quadroni M, Van Eldik LJ. Dance of the dimers. Nature structural biology. 1995;2:711–714. doi: 10.1038/nsb0995-711. [DOI] [PubMed] [Google Scholar]
- [4].Fritz G, Botelho HM, Morozova-Roche LA, Gomes CM. Natural and amyloid self-assembly of S100 proteins: structural basis of functional diversity. The FEBS journal. 2010;277:4578–4590. doi: 10.1111/j.1742-4658.2010.07887.x. [DOI] [PubMed] [Google Scholar]
- [5].Schafer BW, Wicki R, Engelkamp D, Mattei MG, Heizmann CW. Isolation of a YAC clone covering a cluster of nine S100 genes on human chromosome 1q21: rationale for a new nomenclature of the S100 calcium-binding protein family. Genomics. 1995;25:638–643. doi: 10.1016/0888-7543(95)80005-7. [DOI] [PubMed] [Google Scholar]
- [6].Schafer BW, Heizmann CW. The S100 family of EF-hand calcium-binding proteins: functions and pathology. Trends in biochemical sciences. 1996;21:134–140. doi: 10.1016/s0968-0004(96)80167-8. [DOI] [PubMed] [Google Scholar]
- [7].Wicki R, Marenholz I, Mischke D, Schafer BW, Heizmann CW. Characterization of the human S100A12 (calgranulin C, p6, CAAF1, CGRP) gene, a new member of the S100 gene cluster on chromosome 1q21. Cell calcium. 1996;20:459–464. doi: 10.1016/s0143-4160(96)90087-1. [DOI] [PubMed] [Google Scholar]
- [8].Shang X, Cheng H, Zhou R. Chromosomal mapping, differential origin and evolution of the S100 gene family. Genet Sel Evol. 2008;40:449–464. doi: 10.1186/1297-9686-40-4-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Donato R. S100: a multigenic family of calcium-modulated proteins of the EF-hand type with intracellular and extracellular functional roles. The international journal of biochemistry & cell biology. 2001;33:637–668. doi: 10.1016/s1357-2725(01)00046-2. [DOI] [PubMed] [Google Scholar]
- [10].Mandinova A, Atar D, Schafer BW, Spiess M, Aebi U, Heizmann CW. Distinct subcellular localization of calcium binding S100 proteins in human smooth muscle cells and their relocation in response to rises in intracellular calcium. Journal of cell science. 1998;111(Pt 14):2043–2054. doi: 10.1242/jcs.111.14.2043. [DOI] [PubMed] [Google Scholar]
- [11].Moroz OV, Blagova EV, Wilkinson AJ, Wilson KS, Bronstein IB. The crystal structures of human S100A12 in apo form and in complex with zinc: new insights into S100A12 oligomerisation. Journal of molecular biology. 2009;391:536–551. doi: 10.1016/j.jmb.2009.06.004. [DOI] [PubMed] [Google Scholar]
- [12].Koch M, Bhattacharya S, Kehl T, Gimona M, Vasak M, Chazin W, Heizmann CW, Kroneck PM, Fritz G. Implications on zinc binding to S100A2. Biochimica et biophysica acta. 2007;1773:457–470. doi: 10.1016/j.bbamcr.2006.12.006. [DOI] [PubMed] [Google Scholar]
- [13].Ostendorp T, Leclerc E, Galichet A, Koch M, Demling N, Weigle B, Heizmann CW, Kroneck PM, Fritz G. Structural and functional insights into RAGE activation by multimeric S100B. The EMBO journal. 2007;26:3868–3878. doi: 10.1038/sj.emboj.7601805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Sastry M, Ketchem RR, Crescenzi O, Weber C, Lubienski MJ, Hidaka H, Chazin WJ. The three-dimensional structure of Ca(2+)-bound calcyclin: implications for Ca(2+)-signal transduction by S100 proteins. Structure. 1998;6:223–231. doi: 10.1016/s0969-2126(98)00023-9. [DOI] [PubMed] [Google Scholar]
- [15].Ishikawa K, Nakagawa A, Tanaka I, Suzuki M, Nishihira J. The structure of human MRP8, a member of the S100 calcium-binding protein family, by MAD phasing at 1.9 A resolution. Acta crystallographica. 2000;56:559–566. doi: 10.1107/s0907444900002833. [DOI] [PubMed] [Google Scholar]
- [16].Moroz OV, Antson AA, Dodson GG, Wilson KS, Skibshoj I, Lukanidin EM, Bronstein IB. Crystallization and preliminary X-ray diffraction analysis of human calcium-binding protein S100A12. Acta crystallographica. 2000;56:189–191. doi: 10.1107/s0907444999014936. [DOI] [PubMed] [Google Scholar]
- [17].Heizmann CW, Ackermann GE, Galichet A. Pathologies involving the S100 proteins and RAGE. Sub-cellular biochemistry. 2007;45:93–138. doi: 10.1007/978-1-4020-6191-2_5. [DOI] [PubMed] [Google Scholar]
- [18].van der Eerden BC, Hoenderop JG, de Vries TJ, Schoenmaker T, Buurman CJ, Uitterlinden AG, Pols HA, Bindels RJ, van Leeuwen JP. The epithelial Ca2+ channel TRPV5 is essential for proper osteoclastic bone resorption. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:17507–17512. doi: 10.1073/pnas.0505789102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Cecil DL, Terkeltaub R. Transamidation by transglutaminase 2 transforms S100A11 calgranulin into a procatabolic cytokine for chondrocytes. J Immunol. 2008;180:8378–8385. doi: 10.4049/jimmunol.180.12.8378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Streicher WW, Lopez MM, Makhatadze GI. Modulation of quaternary structure of S100 proteins by calcium ions. Biophysical chemistry. 2010;151:181–186. doi: 10.1016/j.bpc.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Pedrocchi M, Schafer BW, Durussel I, Cox JA, Heizmann CW. Purification and characterization of the recombinant human calcium-binding S100 proteins CAPL and CACY. Biochemistry. 1994;33:6732–6738. doi: 10.1021/bi00187a045. [DOI] [PubMed] [Google Scholar]
- [22].Heizmann CW, Cox JA. New perspectives on S100 proteins: a multi-functional Ca(2+)-, Zn(2+)- and Cu(2+)-binding protein family. Biometals. 1998;11:383–397. doi: 10.1023/a:1009212521172. [DOI] [PubMed] [Google Scholar]
- [23].Allen BG, Durussel I, Walsh MP, Cox JA. Characterization of the Ca2+-binding properties of calgizzarin (S100C) isolated from chicken gizzard smooth muscle. Biochemistry and cell biology = Biochimie et biologie cellulaire. 1996;74:687–694. doi: 10.1139/o96-075. [DOI] [PubMed] [Google Scholar]
- [24].Baudier J, Glasser N, Gerard D. Ions binding to S100 proteins. I. Calcium- and zinc-binding properties of bovine brain S100 alpha alpha, S100a (alpha beta), and S100b (beta beta) protein: Zn2+ regulates Ca2+ binding on S100b protein. The Journal of biological chemistry. 1986;261:8192–8203. [PubMed] [Google Scholar]
- [25].Franz C, Durussel I, Cox JA, Schafer BW, Heizmann CW. Binding of Ca2+ and Zn2+ to human nuclear S100A2 and mutant proteins. The Journal of biological chemistry. 1998;273:18826–18834. doi: 10.1074/jbc.273.30.18826. [DOI] [PubMed] [Google Scholar]
- [26].Filipek A, Heizmann CW, Kuznicki J. Calcyclin is a calcium and zinc binding protein. FEBS letters. 1990;264:263–266. doi: 10.1016/0014-5793(90)80263-i. [DOI] [PubMed] [Google Scholar]
- [27].Nishikawa T, Lee IS, Shiraishi N, Ishikawa T, Ohta Y, Nishikimi M. Identification of S100b protein as copper-binding protein and its suppression of copper-induced cell damage. The Journal of biological chemistry. 1997;272:23037–23041. doi: 10.1074/jbc.272.37.23037. [DOI] [PubMed] [Google Scholar]
- [28].Donato R. Intracellular and extracellular roles of S100 proteins. Microscopy research and technique. 2003;60:540–551. doi: 10.1002/jemt.10296. [DOI] [PubMed] [Google Scholar]
- [29].Donato R. RAGE: a single receptor for several ligands and different cellular responses: the case of certain S100 proteins. Current molecular medicine. 2007;7:711–724. doi: 10.2174/156652407783220688. [DOI] [PubMed] [Google Scholar]
- [30].Yammani RR, Carlson CS, Bresnick AR, Loeser RF. Increase in production of matrix metalloproteinase 13 by human articular chondrocytes due to stimulation with S100A4: Role of the receptor for advanced glycation end products. Arthritis and rheumatism. 2006;54:2901–2911. doi: 10.1002/art.22042. [DOI] [PubMed] [Google Scholar]
- [31].Loser K, Vogl T, Voskort M, Lueken A, Kupas V, Nacken W, Klenner L, Kuhn A, Foell D, Sorokin L, Luger TA, Roth J, Beissert S. The Toll-like receptor 4 ligands Mrp8 and Mrp14 are crucial in the development of autoreactive CD8+ T cells. Nature medicine. 2010;16:713–717. doi: 10.1038/nm.2150. [DOI] [PubMed] [Google Scholar]
- [32].Vogl T, Tenbrock K, Ludwig S, Leukert N, Ehrhardt C, van Zoelen MA, Nacken W, Foell D, van der Poll T, Sorg C, Roth J. Mrp8 and Mrp14 are endogenous activators of Toll-like receptor 4, promoting lethal, endotoxin-induced shock. Nature medicine. 2007;13:1042–1049. doi: 10.1038/nm1638. [DOI] [PubMed] [Google Scholar]
- [33].Bhattacharya S, Large E, Heizmann CW, Hemmings B, Chazin WJ. Structure of the Ca2+/S100B/NDR kinase peptide complex: insights into S100 target specificity and activation of the kinase. Biochemistry. 2003;42:14416–14426. doi: 10.1021/bi035089a. [DOI] [PubMed] [Google Scholar]
- [34].Endo H, Takenaga K, Kanno T, Satoh H, Mori S. Methionine aminopeptidase 2 is a new target for the metastasis-associated protein, S100A4. The Journal of biological chemistry. 2002;277:26396–26402. doi: 10.1074/jbc.M202244200. [DOI] [PubMed] [Google Scholar]
- [35].Treves S, Scutari E, Robert M, Groh S, Ottolia M, Prestipino G, Ronjat M, Zorzato F. Interaction of S100A1 with the Ca2+ release channel (ryanodine receptor) of skeletal muscle. Biochemistry. 1997;36:11496–11503. doi: 10.1021/bi970160w. [DOI] [PubMed] [Google Scholar]
- [36].Donato R. Functional roles of S100 proteins, calcium-binding proteins of the EF-hand type. Biochimica et biophysica acta. 1999;1450:191–231. doi: 10.1016/s0167-4889(99)00058-0. [DOI] [PubMed] [Google Scholar]
- [37].Sorci G, Agneletti AL, Bianchi R, Donato R. Association of S100B with intermediate filaments and microtubules in glial cells. Biochimica et biophysica acta. 1998;1448:277–289. doi: 10.1016/s0167-4889(98)00134-7. [DOI] [PubMed] [Google Scholar]
- [38].Bresnick AR. Molecular mechanisms of nonmuscle myosin-II regulation. Current opinion in cell biology. 1999;11:26–33. doi: 10.1016/s0955-0674(99)80004-0. [DOI] [PubMed] [Google Scholar]
- [39].Rustandi RR, Baldisseri DM, Weber DJ. Structure of the negative regulatory domain of p53 bound to S100B(betabeta) Nature structural biology. 2000;7:570–574. doi: 10.1038/76797. [DOI] [PubMed] [Google Scholar]
- [40].Chen H, Fernig DG, Rudland PS, Sparks A, Wilkinson MC, Barraclough R. Binding to intracellular targets of the metastasis-inducing protein, S100A4 (p9Ka) Biochemical and biophysical research communications. 2001;286:1212–1217. doi: 10.1006/bbrc.2001.5517. [DOI] [PubMed] [Google Scholar]
- [41].Grigorian M, Andresen S, Tulchinsky E, Kriajevska M, Carlberg C, Kruse C, Cohn M, Ambartsumian N, Christensen A, Selivanova G, Lukanidin E. Tumor suppressor p53 protein is a new target for the metastasis-associated Mts1/S100A4 protein: functional consequences of their interaction. The Journal of biological chemistry. 2001;276:22699–22708. doi: 10.1074/jbc.M010231200. [DOI] [PubMed] [Google Scholar]
- [42].Mueller A, Schafer BW, Ferrari S, Weibel M, Makek M, Hochli M, Heizmann CW. The calcium-binding protein S100A2 interacts with p53 and modulates its transcriptional activity. The Journal of biological chemistry. 2005;280:29186–29193. doi: 10.1074/jbc.M505000200. [DOI] [PubMed] [Google Scholar]
- [43].Reeves RH, Yao J, Crowley MR, Buck S, Zhang X, Yarowsky P, Gearhart JD, Hilt DC. Astrocytosis and axonal proliferation in the hippocampus of S100b transgenic mice. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:5359–5363. doi: 10.1073/pnas.91.12.5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Hu J, Castets F, Guevara JL, Van Eldik LJ. S100 beta stimulates inducible nitric oxide synthase activity and mRNA levels in rat cortical astrocytes. The Journal of biological chemistry. 1996;271:2543–2547. doi: 10.1074/jbc.271.5.2543. [DOI] [PubMed] [Google Scholar]
- [45].Hu J, Ferreira A, Van Eldik LJ. S100beta induces neuronal cell death through nitric oxide release from astrocytes. Journal of neurochemistry. 1997;69:2294–2301. doi: 10.1046/j.1471-4159.1997.69062294.x. [DOI] [PubMed] [Google Scholar]
- [46].Perera C, McNeil HP, Geczy CL. S100 Calgranulins in inflammatory arthritis. Immunology and cell biology. 2010;88:41–49. doi: 10.1038/icb.2009.88. [DOI] [PubMed] [Google Scholar]
- [47].Hu SP, Harrison C, Xu K, Cornish CJ, Geczy CL. Induction of the chemotactic S100 protein, CP-10, in monocyte/macrophages by lipopolysaccharide. Blood. 1996;87:3919–3928. [PubMed] [Google Scholar]
- [48].Salama I, Malone PS, Mihaimeed F, Jones JL. A review of the S100 proteins in cancer. Eur J Surg Oncol. 2008;34:357–364. doi: 10.1016/j.ejso.2007.04.009. [DOI] [PubMed] [Google Scholar]
- [49].Marshak DR, Pena LA. Potential role of S100 beta in Alzheimer's disease: an hypothesis involving mitotic protein kinases. Progress in clinical and biological research. 1992;379:289–307. [PubMed] [Google Scholar]
- [50].Griffin WS. Inflammation and neurodegenerative diseases. The American journal of clinical nutrition. 2006;83:470S–474S. doi: 10.1093/ajcn/83.2.470S. [DOI] [PubMed] [Google Scholar]
- [51].Foell D, Roth J. Proinflammatory S100 proteins in arthritis and autoimmune disease. Arthritis and rheumatism. 2004;50:3762–3771. doi: 10.1002/art.20631. [DOI] [PubMed] [Google Scholar]
- [52].Stefansson K, Wollmann RL, Moore BW, Arnason BG. S-100 protein in human chondrocytes. Nature. 1982;295:63–64. doi: 10.1038/295063a0. [DOI] [PubMed] [Google Scholar]
- [53].Nakamura Y, Becker LE, Marks A. S-100 protein in tumors of cartilage and bone. An immunohistochemical study. Cancer. 1983;52:1820–1824. doi: 10.1002/1097-0142(19831115)52:10<1820::aid-cncr2820521010>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- [54].Okajima K, Honda I, Kitagawa T. Immunohistochemical distribution of S-100 protein in tumors and tumor-like lesions of bone and cartilage. Cancer. 1988;61:792–799. doi: 10.1002/1097-0142(19880215)61:4<792::aid-cncr2820610425>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- [55].Ushigome S, Takakuwa T, Shinagawa T, Takagi M, Kishimoto H, Mori N. Ultrastructure of cartilaginous tumors and S-100 protein in the tumors. With reference to the histogenesis of chondroblastoma, chondromyxoid fibroma and mesenchymal chondrosarcoma. Acta pathologica japonica. 1984;34:1285–1300. doi: 10.1111/j.1440-1827.1984.tb00555.x. [DOI] [PubMed] [Google Scholar]
- [56].Hasegawa T, Seki K, Yang P, Hirose T, Hizawa K, Wada T, Wakabayashi J. Differentiation and proliferative activity in benign and malignant cartilage tumors of bone. Human pathology. 1995;26:838–845. doi: 10.1016/0046-8177(95)90004-7. [DOI] [PubMed] [Google Scholar]
- [57].Li CL, Martinez V, He B, Lombet A, Perbal B. A role for CCN3 (NOV) in calcium signalling. Mol Pathol. 2002;55:250–261. doi: 10.1136/mp.55.4.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Chano T, Ishizawa M, Matsumoto K, Morimoto S, Hukuda S, Okabe H. The identity of proliferating cells in bone tumors with cartilaginous components: evaluation by double-immunohistochemical staining using proliferating cell nuclear antigen and S-100 protein. Eur J Histochem. 1995;39:21–30. [PubMed] [Google Scholar]
- [59].Weiss AP, Dorfman HD. S-100 protein in human cartilage lesions. J Bone Joint Surg Am. 1986;68:521–526. [PubMed] [Google Scholar]
- [60].Mohr W, Kuhn C, Pelster B, Wessinghage D. S-100 protein in normal, osteoarthrotic, and arthritic cartilage. Rheumatology international. 1985;5:273–277. doi: 10.1007/BF00541355. [DOI] [PubMed] [Google Scholar]
- [61].Wolff DA, Stevenson S, Goldberg VM. S-100 protein immunostaining identifies cells expressing a chondrocytic phenotype during articular cartilage repair. J Orthop Res. 1992;10:49–57. doi: 10.1002/jor.1100100106. [DOI] [PubMed] [Google Scholar]
- [62].Leonardi R, Villari L, Bernasconi G, Piacentini C, Baciliero U, Travali S. Cellular S-100 protein immunostaining in human dysfunctional temporomandibular joint discs. Archives of oral biology. 2000;45:411–418. doi: 10.1016/s0003-9969(99)00144-2. [DOI] [PubMed] [Google Scholar]
- [63].Itoi M. An Experimental study on the relationship between S-100 protein synthesis and differentiation of chondrocytes. Journal of Kyoto Prefectural university of Medicine. 1988;97:1395–1406. [Google Scholar]
- [64].Sugimoto S, Kusuzaki K, Takeshita H, Kuzuhara A, Tsuji Y, Yamashita F, Hirasawa Y, Ashihara T. The distribution of S-100 protein positive chondrocytes in the human articular cartilages under aging or diseased conditions. Nippon Seikeigeka Gakkai zasshi. 1991;65:902–908. [PubMed] [Google Scholar]
- [65].Cecil DL, Johnson K, Rediske J, Lotz M, Schmidt AM, Terkeltaub R. Inflammation-induced chondrocyte hypertrophy is driven by receptor for advanced glycation end products. J Immunol. 2005;175:8296–8302. doi: 10.4049/jimmunol.175.12.8296. [DOI] [PubMed] [Google Scholar]
- [66].Loeser RF, Yammani RR, Carlson CS, Chen H, Cole A, Im HJ, Bursch LS, Yan SD. Articular chondrocytes express the receptor for advanced glycation end products: Potential role in osteoarthritis. Arthritis and rheumatism. 2005;52:2376–2385. doi: 10.1002/art.21199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Balmain N, Moutahir F, Heizmann CW, Lieberherr M. Immunolocalization of S100A2 calcium-binding protein in cartilage and bone cells. Cellular and molecular biology (Noisy-le-Grand, France) 2003;49:485–486. [PubMed] [Google Scholar]
- [68].Hopwood B, Tsykin A, Findlay DM, Fazzalari NL. Microarray gene expression profiling of osteoarthritic bone suggests altered bone remodelling, WNT and transforming growth factor-beta/bone morphogenic protein signalling. Arthritis research & therapy. 2007;9:R100. doi: 10.1186/ar2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Yammani RR, Long D, Loeser RF. Interleukin-7 stimulates secretion of S100A4 by activating the JAK/STAT signaling pathway in human articular chondrocytes. Arthritis and rheumatism. 2009;60:792–800. doi: 10.1002/art.24295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Miranda KJ, Loeser RF, Yammani RR. Sumoylation and nuclear translocation of S100A4 regulate IL-1beta-mediated production of matrix metalloproteinase-13. The Journal of biological chemistry. 2010;285:31517–31524. doi: 10.1074/jbc.M110.125898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Zreiqat H, Howlett CR, Gronthos S, Hume D, Geczy CL. S100A8/S100A9 and their association with cartilage and bone. Journal of molecular histology. 2007;38:381–391. doi: 10.1007/s10735-007-9117-2. [DOI] [PubMed] [Google Scholar]
- [72].van Lent PL, Grevers LC, Blom AB, Arntz OJ, van de Loo FA, van der Kraan P, Abdollahi-Roodsaz S, Srikrishna G, Freeze H, Sloetjes A, Nacken W, Vogl T, Roth J, van den Berg WB. Stimulation of chondrocyte-mediated cartilage destruction by S100A8 in experimental murine arthritis. Arthritis and rheumatism. 2008;58:3776–3787. doi: 10.1002/art.24074. [DOI] [PubMed] [Google Scholar]
- [73].Baillet A, Trocme C, Berthier S, Arlotto M, Grange L, Chenau J, Quetant S, Seve M, Berger F, Juvin R, Morel F, Gaudin P. Synovial fluid proteomic fingerprint: S100A8, S100A9 and S100A12 proteins discriminate rheumatoid arthritis from other inflammatory joint diseases. Rheumatology (Oxford, England) 2010;49:671–682. doi: 10.1093/rheumatology/kep452. [DOI] [PubMed] [Google Scholar]
- [74].Sellam J, Berenbaum F. The role of synovitis in pathophysiology and clinical symptoms of osteoarthritis. Nature reviews. 2010;6:625–635. doi: 10.1038/nrrheum.2010.159. [DOI] [PubMed] [Google Scholar]
- [75].Youssef P, Roth J, Frosch M, Costello P, Fitzgerald O, Sorg C, Bresnihan B. Expression of myeloid related proteins (MRP) 8 and 14 and the MRP8/14 heterodimer in rheumatoid arthritis synovial membrane. The Journal of rheumatology. 1999;26:2523–2528. [PubMed] [Google Scholar]
- [76].van Lent PL, Grevers LC, Schelbergen R, Blom A, Geurts J, Sloetjes A, Vogl T, Roth J, van den Berg WB. S100A8 causes a shift toward expression of activatory Fcgamma receptors on macrophages via toll-like receptor 4 and regulates Fcgamma receptor expression in synovium during chronic experimental arthritis. Arthritis and rheumatism. 2010;62:3353–3364. doi: 10.1002/art.27654. [DOI] [PubMed] [Google Scholar]
- [77].Zreiqat H, Belluoccio D, Smith MM, Wilson R, Rowley LA, Jones K, Ramaswamy Y, Vogl T, Roth J, Bateman JF, Little CB. S100A8 and S100A9 in experimental osteoarthritis. Arthritis research & therapy. 2010;12:R16. doi: 10.1186/ar2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].van Lent PL, Grevers L, Blom AB, Sloetjes A, Mort JS, Vogl T, Nacken W, van den Berg WB, Roth J. Myeloid-related proteins S100A8/S100A9 regulate joint inflammation and cartilage destruction during antigen-induced arthritis. Annals of the rheumatic diseases. 2008;67:1750–1758. doi: 10.1136/ard.2007.077800. [DOI] [PubMed] [Google Scholar]
- [79].Pan J, Rintala-Dempsey AC, Li Y, Shaw GS, Konermann L. Folding kinetics of the S100A11 protein dimer studied by time-resolved electrospray mass spectrometry and pulsed hydrogen-deuterium exchange. Biochemistry. 2006;45:3005–3013. doi: 10.1021/bi052349a. [DOI] [PubMed] [Google Scholar]
- [80].Rety S, Osterloh D, Arie JP, Tabaries S, Seeman J, Russo-Marie F, Gerke V, Lewit-Bentley A. Structural basis of the Ca(2+)-dependent association between S100C (S100A11) and its target, the N-terminal part of annexin I. Structure. 2000;8:175–184. doi: 10.1016/s0969-2126(00)00093-9. [DOI] [PubMed] [Google Scholar]
- [81].Neeper M, Schmidt AM, Brett J, Yan SD, Wang F, Pan YC, Elliston K, Stern D, Shaw A. Cloning and expression of a cell surface receptor for advanced glycosylation end products of proteins. The Journal of biological chemistry. 1992;267:14998–15004. [PubMed] [Google Scholar]
- [82].Ding Q, Keller JN. Evaluation of rage isoforms, ligands, and signaling in the brain. Biochimica et biophysica acta. 2005;1746:18–27. doi: 10.1016/j.bbamcr.2005.08.006. [DOI] [PubMed] [Google Scholar]
- [83].Ding Q, Keller JN. Splice variants of the receptor for advanced glycosylation end products (RAGE) in human brain. Neuroscience letters. 2005;373:67–72. doi: 10.1016/j.neulet.2004.09.059. [DOI] [PubMed] [Google Scholar]
- [84].Yan SD, Chen X, Fu J, Chen M, Zhu H, Roher A, Slattery T, Zhao L, Nagashima M, Morser J, Migheli A, Nawroth P, Stern D, Schmidt AM. RAGE and amyloid-beta peptide neurotoxicity in Alzheimer's disease. Nature. 1996;382:685–691. doi: 10.1038/382685a0. [DOI] [PubMed] [Google Scholar]
- [85].Schmidt AM, Yan SD, Yan SF, Stern DM. The biology of the receptor for advanced glycation end products and its ligands. Biochimica et biophysica acta. 2000;1498:99–111. doi: 10.1016/s0167-4889(00)00087-2. [DOI] [PubMed] [Google Scholar]
- [86].Hori O, Brett J, Slattery T, Cao R, Zhang J, Chen JX, Nagashima M, Lundh ER, Vijay S, Nitecki D, et al. The receptor for advanced glycation end products (RAGE) is a cellular binding site for amphoterin. Mediation of neurite outgrowth and co-expression of rage and amphoterin in the developing nervous system. The Journal of biological chemistry. 1995;270:25752–25761. doi: 10.1074/jbc.270.43.25752. [DOI] [PubMed] [Google Scholar]
- [87].Basta G, Lazzerini G, Del Turco S, Ratto GM, Schmidt AM, De Caterina R. At least 2 distinct pathways generating reactive oxygen species mediate vascular cell adhesion molecule-1 induction by advanced glycation end products. Arteriosclerosis, thrombosis, and vascular biology. 2005;25:1401–1407. doi: 10.1161/01.ATV.0000167522.48370.5e. [DOI] [PubMed] [Google Scholar]
- [88].Huttunen HJ, Kuja-Panula J, Sorci G, Agneletti AL, Donato R, Rauvala H. Coregulation of neurite outgrowth and cell survival by amphoterin and S100 proteins through receptor for advanced glycation end products (RAGE) activation. The Journal of biological chemistry. 2000;275:40096–40105. doi: 10.1074/jbc.M006993200. [DOI] [PubMed] [Google Scholar]
- [89].Leclerc E, Fritz G, Weibel M, Heizmann CW, Galichet A. S100B and S100A6 differentially modulate cell survival by interacting with distinct RAGE (receptor for advanced glycation end products) immunoglobulin domains. The Journal of biological chemistry. 2007;282:31317–31331. doi: 10.1074/jbc.M703951200. [DOI] [PubMed] [Google Scholar]
- [90].Bianchi R, Adami C, Giambanco I, Donato R. S100B binding to RAGE in microglia stimulates COX-2 expression. Journal of leukocyte biology. 2007;81:108–118. doi: 10.1189/jlb.0306198. [DOI] [PubMed] [Google Scholar]
- [91].Bianchi R, Giambanco I, Donato R. S100B/RAGE-dependent activation of microglia via NF-kappaB and AP-1 Co-regulation of COX-2 expression by S100B, IL-1beta and TNF-alpha. Neurobiology of aging. 2010;31:665–677. doi: 10.1016/j.neurobiolaging.2008.05.017. [DOI] [PubMed] [Google Scholar]
- [92].Shaw SS, Schmidt AM, Banes AK, Wang X, Stern DM, Marrero MB. S100B-RAGE-mediated augmentation of angiotensin II-induced activation of JAK2 in vascular smooth muscle cells is dependent on PLD2. Diabetes. 2003;52:2381–2388. doi: 10.2337/diabetes.52.9.2381. [DOI] [PubMed] [Google Scholar]
- [93].Reddy MA, Li SL, Sahar S, Kim YS, Xu ZG, Lanting L, Natarajan R. Key role of Src kinase in S100B-induced activation of the receptor for advanced glycation end products in vascular smooth muscle cells. The Journal of biological chemistry. 2006;281:13685–13693. doi: 10.1074/jbc.M511425200. [DOI] [PubMed] [Google Scholar]
- [94].Cecil DL, Appleton CT, Polewski MD, Mort JS, Schmidt AM, Bendele A, Beier F, Terkeltaub R. The pattern recognition receptor CD36 is a chondrocyte hypertrophy marker associated with suppression of catabolic responses and promotion of repair responses to inflammatory stimuli. J Immunol. 2009;182:5024–5031. doi: 10.4049/jimmunol.0803603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Lu C, He JC, Cai W, Liu H, Zhu L, Vlassara H. Advanced glycation endproduct (AGE) receptor 1 is a negative regulator of the inflammatory response to AGE in mesangial cells. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:11767–11772. doi: 10.1073/pnas.0401588101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Ohgami N, Nagai R, Ikemoto M, Arai H, Kuniyasu A, Horiuchi S, Nakayama H. Cd36, a member of the class b scavenger receptor family, as a receptor for advanced glycation end products. The Journal of biological chemistry. 2001;276:3195–3202. doi: 10.1074/jbc.M006545200. [DOI] [PubMed] [Google Scholar]
- [97].Ohgami N, Nagai R, Ikemoto M, Arai H, Miyazaki A, Hakamata H, Horiuchi S, Nakayama H. CD36, serves as a receptor for advanced glycation endproducts (AGE) Journal of diabetes and its complications. 2002;16:56–59. doi: 10.1016/s1056-8727(01)00208-2. [DOI] [PubMed] [Google Scholar]
- [98].Endemann G, Stanton LW, Madden KS, Bryant CM, White RT, Protter AA. CD36 is a receptor for oxidized low density lipoprotein. The Journal of biological chemistry. 1993;268:11811–11816. [PubMed] [Google Scholar]
- [99].Abumrad NA, el-Maghrabi MR, Amri EZ, Lopez E, Grimaldi PA. Cloning of a rat adipocyte membrane protein implicated in binding or transport of long-chain fatty acids that is induced during preadipocyte differentiation. Homology with human CD36. The Journal of biological chemistry. 1993;268:17665–17668. [PubMed] [Google Scholar]
- [100].Dawson DW, Pearce SF, Zhong R, Silverstein RL, Frazier WA, Bouck NP. CD36 mediates the In vitro inhibitory effects of thrombospondin-1 on endothelial cells. The Journal of cell biology. 1997;138:707–717. doi: 10.1083/jcb.138.3.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Asch AS, Silbiger S, Heimer E, Nachman RL. Thrombospondin sequence motif (CSVTCG) is responsible for CD36 binding. Biochemical and biophysical research communications. 1992;182:1208–1217. doi: 10.1016/0006-291x(92)91860-s. [DOI] [PubMed] [Google Scholar]
- [102].Huang MM, Bolen JB, Barnwell JW, Shattil SJ, Brugge JS. Membrane glycoprotein IV (CD36) is physically associated with the Fyn, Lyn, and Yes protein-tyrosine kinases in human platelets. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:7844–7848. doi: 10.1073/pnas.88.17.7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Chen K, Febbraio M, Li W, Silverstein RL. A specific CD36-dependent signaling pathway is required for platelet activation by oxidized low-density lipoprotein. Circulation research. 2008;102:1512–1519. doi: 10.1161/CIRCRESAHA.108.172064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Bull HA, Brickell PM, Dowd PM. Src-related protein tyrosine kinases are physically associated with the surface antigen CD36 in human dermal microvascular endothelial cells. FEBS letters. 1994;351:41–44. doi: 10.1016/0014-5793(94)00814-0. [DOI] [PubMed] [Google Scholar]
- [105].Moore KJ, El Khoury J, Medeiros LA, Terada K, Geula C, Luster AD, Freeman MW. A CD36-initiated signaling cascade mediates inflammatory effects of beta-amyloid. The Journal of biological chemistry. 2002;277:47373–47379. doi: 10.1074/jbc.M208788200. [DOI] [PubMed] [Google Scholar]
- [106].Silverstein RL, Febbraio M. CD36, a scavenger receptor involved in immunity, metabolism, angiogenesis, and behavior. Science signaling. 2009;2:re3. doi: 10.1126/scisignal.272re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Silverstein RL, Febbraio M. CD36 and atherosclerosis. Current opinion in lipidology. 2000;11:483–491. doi: 10.1097/00041433-200010000-00006. [DOI] [PubMed] [Google Scholar]
- [108].Silverstein RL. Inflammation, atherosclerosis, and arterial thrombosis: role of the scavenger receptor CD36. Cleveland Clinic journal of medicine. 2009;76(Suppl 2):S27–30. doi: 10.3949/ccjm.76.s2.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Park L, Wang G, Zhou P, Zhou J, Pitstick R, Previti ML, Younkin L, Younkin SG, Van Nostrand WE, Cho S, Anrather J, Carlson GA, Iadecola C. Scavenger receptor CD36 is essential for the cerebrovascular oxidative stress and neurovascular dysfunction induced by amyloid-beta. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:5063–5068. doi: 10.1073/pnas.1015413108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Stewart CR, Stuart LM, Wilkinson K, van Gils JM, Deng J, Halle A, Rayner KJ, Boyer L, Zhong R, Frazier WA, Lacy-Hulbert A, El Khoury J, Golenbock DT, Moore KJ. CD36 ligands promote sterile inflammation through assembly of a Toll-like receptor 4 and 6 heterodimer. Nature immunology. 2010;11:155–161. doi: 10.1038/ni.1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Kiryushko D, Novitskaya V, Soroka V, Klingelhofer J, Lukanidin E, Berezin V, Bock E. Molecular mechanisms of Ca(2+) signaling in neurons induced by the S100A4 protein. Molecular and cellular biology. 2006;26:3625–3638. doi: 10.1128/MCB.26.9.3625-3638.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Foell D, Wittkowski H, Vogl T, Roth J. S100 proteins expressed in phagocytes: a novel group of damage-associated molecular pattern molecules. Journal of leukocyte biology. 2007;81:28–37. doi: 10.1189/jlb.0306170. [DOI] [PubMed] [Google Scholar]
- [113].Sorci G, Riuzzi F, Agneletti AL, Marchetti C, Donato R. S100B inhibits myogenic differentiation and myotube formation in a RAGE-independent manner. Molecular and cellular biology. 2003;23:4870–4881. doi: 10.1128/MCB.23.14.4870-4881.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Riuzzi F, Sorci G, Donato R. S100B stimulates myoblast proliferation and inhibits myoblast differentiation by independently stimulating ERK1/2 and inhibiting p38 MAPK. Journal of cellular physiology. 2006;207:461–470. doi: 10.1002/jcp.20580. [DOI] [PubMed] [Google Scholar]
- [115].Kerkhoff C, Sorg C, Tandon NN, Nacken W. Interaction of S100A8/S100A9-arachidonic acid complexes with the scavenger receptor CD36 may facilitate fatty acid uptake by endothelial cells. Biochemistry. 2001;40:241–248. doi: 10.1021/bi001791k. [DOI] [PubMed] [Google Scholar]


