Abstract
Acute Promyelocytic Leukemia (APL) results from a reciprocal translocation that fuses the gene for the PML tumor suppressor to that encoding the retinoic acid receptor alpha (RARα). The resulting PML-RARα oncogene product interferes with multiple regulatory pathways associated with myeloid differentiation, including normal PML and RARα functions. The standard treatment for APL includes anthracycline-based chemotherapeutic agents plus the RARα agonist all-trans retinoic acid (ATRA). Relapse, which is often accompanied by ATRA resistance, occurs in an appreciable frequency of treated patients. One potential mechanism suggested by model experiments featuring the selection of ATRA resistant APL cell lines involves ATRA resistant versions of the PML-RARα oncogene, where the relevant mutations localize to the RARα ligand-binding domain (LBD). Such mutations may act by compromising agonist binding, but other mechanisms are possible. Here, we studied the molecular consequence of ATRA resistance by use of circular dichroism, protease resistance, and fluorescence anisotropy assays employing peptides derived from the NCOR nuclear co-repressor and the ACTR nuclear co-activator. The consequences of the mutations on global structure and co-factor interaction functions were assessed quantitatively, providing insights into the basis of agonist resistance. Attenuated co-factor switching and increased protease resistance represent features of the LBDs of ATRA-resistant PML-RARα, and these properties may be recapitulated in the full-length oncoproteins.
Keywords: all-trans retinoic acid, coactivator, corepressor, nuclear receptor, transcription regulation, fluorescence anisotropy
Acute promyelocytic leukemia (APL)1, a subtype of acute myeloid leukemia, is a rapid onset blood cancer caused by a massive proliferation of cells derived from the myelogenous lineage 1–3. APL results from a balanced chromosomal rearrangement between 15q22 and 17q21, producing aberrant gene fusions between the genes encoding for the promyelocytic leukemia (PML) and retinoic acid receptor α proteins 4–6. Studies with mouse transgenic models 7–9 indicate that the PML-RARα translocation is necessary and sufficient to confer most of the clinical manifestations of APL, making it an attractive target for molecular therapeutics 10.
The PML and RARα gene products that constitute the two halves of the PML-RARα oncoprotein both have significant roles in cellular growth control and differentiation. PML is a confirmed tumor suppressor gene that controls apoptosis, cell proliferation, and cellular senescence, organizing proteins in the nucleus to regulate their type and extent of post-translational modification 11 ,12. To carry out these functions, PML is organized into “nuclear bodies”, and these may serve to regulate apoptosis by sequestering important transcription factors, including p53 13. While the full scope of PML’s activities has yet to be established, its regulation of p53 has implications for the self-renewal properties of stem cells 14–16.
RARα is a member of the nuclear receptor superfamily, all of whose members are transcription factors possessing separate DNA and ligand binding domains 17. Nuclear receptors, including the RXR-RARα heterodimer complex, activate transcription in response to small molecule ligands 18. Activation depends on factor binding to characteristic sites (i.e. retinoic acid response elements, RAREs) located adjacent to Pol II –transcribed promoters 19. In addition to the requirement for heterodimer formation with RXR to bind DNA 20, RARα’s ability to modulate transcription is dependent on its interactions with nuclear co-activators (ACTR) and nuclear co-repressors (e.g., N-CoR and SMRT)21. These modulate transcription by virtue of their associated histone acetylase (HAT) and histone deacetylase (HDAC) activities, respectively 22,23. In the absence of ligand, RXR-RARα is complexed with nuclear co-repressors, including N-CoR and its tightly associated HDAC activities. These interactions promote the de-acetylation of chromatin and repression of Pol II transcription 24,25.
Owing to the ability of all trans retinoic acid (ATRA) to promote the differentiation of leukemic blasts during the early stages of treatment, APL has historically been considered to represent a model of a cancer treatable by differentiation therapy 26, 27. When patients are treated with ATRA as a single agent, the selection of ATRA-resistant blast cells can occur. While this occurs less frequently with patients treated with the more standard regimen of ATRA plus chemotherapeutic agents, relapse can occur, and is linked to ATRA resistance 28–30. While a host of different mechanisms for acquired ATRA resistance have been suggested, including increased ATRA catabolism and the up-regulation of cellular RA binding proteins, selection experiments employing the PML-related NB4 cell line suggest the potential importance of the development of mutations within the PML-RARα gene 28. In many of these cells, resistance appears to be linked to mutations in the RARα portion of the PML-RARα gene, but the relationship of genotype to phenotype appears to be complex 28. More recently, combination therapies have been devised for APL that feature the use of ATRA, arsenic trioxide (ATO), and anthracycline-based chemotherapy, and these have resulted in 5-year disease free remissions for approximately 90% of patients receiving the triple therapy 31,32. The molecular basis of this increased therapeutic efficacy remains to be fully characterized.
In the standard model for how the oncoprotein executes its transformative properties, PML-RARα interferes with a broad range of developmental and signaling pathways, which are themselves controlled by an array of diverse transcription factors. Until recently, interference with the differentiation and development programs of the PML and RARα parent genes was thought to be a critical mechanism by which PML-RARα blocks myeloid transformation 5,14,33. Significantly, the PML-RARα fusion protein requires substantially higher concentrations of retinoic acid (10−7 to 10−6 M, versus 10−9 M for the wild type) to dissociate co-repressors and activate transcription 34. In this view, PML-RARα’s effects would be rationalized by its ability to act as a dominant negative regulator of genes normally regulated by RARα. In the early stages of treatment with ATRA and chemotherapeutic drugs, the increased local concentration of ATRA would act to blunt many of these dominant negative effects.
Owing to the complexity of APL, the relative contributions of different potential mechanisms linking ATRA resistance to disease relapse are difficult to assess. Here, we chose to investigate one relatively straightforward mechanism that can be observed in NB-4 cells (derived from APL) in culture, namely the accumulation of mutations that encode substitutions in the RARα ligand-binding domain of PML-RARα 35. Biochemical characterization of these mutants suggests that they have lost the ability to bind ATRA, and/or release co-repressor, even at concentrations of ATRA that are much higher than what can be achieved pharmacologically. Here, a variety of different approaches were employed to characterize the molecular basis of ATRA resistance in the context of a RARα ligand binding domain model. These studies indicate that attenuated co-factor resistance and altered resistance to proteolytic degradation are both characteristics associated with ATRA resistance, consistent with evolving models to explain the molecular basis of APL.
MATERIALS AND METHODS
Materials
All buffers and solutions were prepared from reagent grade chemicals and deionized glass distilled water. Unless otherwise noted, all chemicals and biochemicals were purchased from Fisher or Sigma. Enzymes for molecular biology were obtained from New England Biolabs. Ni-NTA agarose was purchased from Qiagen. ATRA was obtained from Sigma, and then prepared in 80:20 ethanol:dimethylsulfoxide and then stored at −80 °C. In titration experiments with purified receptor, the volume of ATRA added was small relative to the total volume, such that the concentration of ethanol:dimethylsulfoxide never exceeded 2%. Rhodamine Red™ –C2 – maleimide was purchased from Invitrogen.
Preparation of the ligand binding domain of RARα
The wild type human ligand binding domain of the retinoic acid receptor α (herein, RARα LBD) was amplified from the pSG-5 derivative plasmid (a gift of Wilson Miller, Lady Davis Institute for Medical Research) by use of the polymerase chain reaction. The 738-nucleotide fragment was cloned into plasmid pQE-30 (Qiagen) to produce pMF-1, a bacterial expression plasmid in which a His6 affinity tag is appended to the N-terminal end of the inserted open reading frame. The human RXRα LBD, comprising residues 201–462, was amplified from cDNA (Origene) and then cloned into expression plasmid pET-3a (Novagen), which does not encode an affinity tag. Substitutions corresponding to ATRA resistant mutations (M297L, L398P, I410T and M413T) were introduced into the RARα open reading frame of pMF-1 by use of the QuickChange™ oligonucleotide directed mutagenesis procedure (Stratagene), employing 30–39 nucleotide double stranded mutagenic primers. The RARα LBD protein was expressed in E. coli cells grown at 37° C to an OD600 of 0.6, followed by a four hour induction with 0.5 mM IPTG at 20° C. The LBD of RXRα was similarly expressed in E. coli and induced at 37° C. The recovery of stable and soluble RARα was enhanced by purification in the presence of RXR. Cell pellets derived from over-expression of the LBDs from RXRα and RARα were combined and purified by nickel-NTA affinity chromatography. The pellets were sonicated in Buffer A (500 mM NaCl, 20 mM Tris [pH 8.1], 10 mM β-mercaptoethanol, 5 mM imidazole), and then loaded and washed on a Ni-NTA column with Buffer B (Buffer A plus 20 mM imidazole). Bound RARα LBD was eluted by use of a 18–250 mM gradient in imidazole. Fractions containing LBD were identified by SDS-PAGE, pooled, dialyzed into storage buffer (Buffer A with no imidazole), and then concentrated. The Bradford method was used to determine protein concentrations of the final LBD preparations. Owing to the presence of only a single tryptophan, protein concentrations of the RARα LBD cannot be determined from the absorbance at 280 nm. The homogeneity of the RARα LBD in the final preparations was confirmed by N-terminal protein sequencing, performed by the University of Texas Medical Branch, Biomolecular Resource Facility, and MALDI-TOF mass spectrometry.
Circular dichroism
Circular dichroism (CD) was performed on a Jasco 815 spectropolarimeter equipped with a Peltier stage. CD spectra were acquired in a 1 mm quartz cell using a 4 nm bandwidth and a scan rate of 0.5 nm/sec, over the wavelength rage from 300 to 190 nm. The spectra were collected in the presence and absence of 1 mM AM580, employing a protein concentration of 50–100 µM and a buffer of 500 mM NaCl, 20 mM Tris [pH 8.1], 10 mM β-mercaptoethanol. The data were converted to molar ellipticity (deg cm2 dmol−1) using the equation [θ]222 = (100 qobs)/cl, where qobs is the ellipticity measured in millidegrees, c is the concentration of the LBD (M), and l is the optical path length (cm).
Protease Sensitivity Assays
RARα LBD (5 µL of 1.88 mg/mL stock) and ATRA (0.5 µL of a 10 µM stock) were incubated at 22 °C for 30 min in the dark. Trypsin was added in 0.5 µL increments to a final concentrations of 0 – 2.1 µg/mL in 20 mM Tris, pH 8.1, 0.5 M NaCl. The reactions were incubated 15 minutes at 22° C, and then terminated by mixing with an equal volume of SDS/β-mercaptoethanol/bromophenol blue loading dye and heating to 22° C for five minutes. The terminated reactions were run on 12% SDS-PAGE gels.
Peptide synthesis
The peptides synthesized included the following: N-CoR, RID2 (NCoR1_HUMAN, NCBI accession NP_006302), H2N-CGHSFADPASNGLEDIIRKALMGSF-COOH; scrNCoR (scrambled NCoR sequence), H2N-CASSFRIENDLPKFMSHALIGGGDA-COOH; ACTR (SRC-3, NR box3 NCoA3_HUMAN, NCBI accession Q9Y6Q9), H2N-CKKENNALLRYLLDRDDPSDAL-COOH; scrACTR (scrambled ACTR sequence), H2N-CLELYNARDSDKNKPLDRLDLA-COOH. The peptides were synthesized by the Protein Core Facility at the University of Vermont on a Protein Technologies Inc. Symphony™ multiple peptide synthesizer via Fmoc chemistry, utilizing preloaded Wang resins purchased from Synbiosci Corporation (Livermore, CA). Double coupling using standard O-benzotriazole-N,N,N’,N’-tetramethyl-uronium-hexafluoro-phosphate (HBTU) activation was employed for peptide elongation. Cleavage of peptides from the resin was accomplished by treatment of the dried resin with 96:2:2 trifluoroacetic acid (TFA)/ triisopropylsilane (TIS)/ H2O for two hours. Following filtration of the resin, the cleavage supernatant was evaporated to one-fifth its original volume in a stream of nitrogen, and precipitated by addition of cold anhydrous diethyl ether. Each peptide was synthesized with a cysteine residue at its N-terminus, allowing conjugation of fluorophores by use of standard maleimide chemistry.
Conjugation of fluorophore to cofactor peptides
The peptides were conjugated with Rhodamine Red™-C2-maleimide (Invitrogen) by incubation for 30 minutes at a ratio of 2:1 (peptide:dye) in methanol at 22° C. Dye-conjugated peptides were then purified from unconjugated peptides by chromatography on a Waters SymmetryPrep™ C18 7 µm 1900 × 150 mm preparatory column attached to a Shimadzu preparatory HPLC system. The peptides were loaded on the column pre-equilibrated with 0.1% trifluoroacetic acid (TFA)/H2O, and then eluted by a gradient of 0.1% TFA/ acetonitrile (ACN), up to 50% ACN, developed over 50 minutes. Elution profiles were detected at 214 nm and 254 nm. As determined by mass spectrometry performed by the University of Vermont Protein Core Facility, a final peptide:dye binding stoichiometry of 1:1 was routinely obtained for all conjugated peptides. The dye-conjugated peptides were reconstituted in 25% methanol to 300 µM, and then diluted to 10 µM in 500 mM NaCl, 20 mM Tris (pH 8.1) immediately before use.
Fluorescence anisotropy assay
Quantitative measurements of RARα LBD binding to cofactor-derived peptides were performed using fluorescence anisotropy. These assays employed a fixed concentration of each rhodamine red-conjugated cofactor peptide, the intrinsic anisotropies of which were measured as described below. At constant temperature and viscosity, the rotational relaxation time of the fluorophore-labeled peptide increases proportional to its molecular volume, indicating the binding by a larger protein 36–38. Varying amounts of RARα LBD (0 – 40 µM) in a buffer of 500 mM NaCl, 20 mM Tris (pH 8.1), 10 mM β-mercaptoethanol were incubated with the fixed concentration (50 nM) of peptide for 30 minutes at 22°C before measuring the anisotropy. Control binding reactions employed rhodamine red-conjugated peptides based on scrambled versions of the N-CoR and ACTR sequences. Steady-state anisotropies were measured using excitation and emission wavelengths at 570 nm and 590 nm, respectively, at 20°C, employing a Photon Technology International QM-6 fluorometer equipped with a 75-watt xenon arc lamp as an excitation source, and slit widths of 5 nm. ATRA was added to samples incrementally to final concentrations of 10–521 nM, and then steady-state measurements were repeated. All measurements were carried out at least twice, using different protein preparations.
Steady-state anisotropy data were collected using the time-based function for 10 seconds, with a one second integration time, and the data were averaged. Total fluorescence emission was simultaneously measured for each reaction, and any observed changes in fluorescence quantum yields were included in the data analysis as described in 37. Fraction bound was calculated as fB=(r−rF)/[(r−rF)+R(rB−rF)]; where r is the measured anisotropy at a given [RARα LBD]; rF and rB are the free anisotropy and maximum bound anisotropy, respectively; R is the change in fluorescence intensity with increasing [RARα LBD], defined as R=IB/IF, where IF and IB are the free and maximum bound intensities, respectively. Intensity indicates total polarized intensity, I(parallel) + 2I(perpendicular). As labeled peptide concentrations were substantially below the dissociation constant, the dissociation constants could be obtained from plots of fraction bound versus the concentration of RARα LBD, fitting the data by non-linear regression to the equation: q = [R]/(Kd+[R]); where θ = fraction bound, and [R] = the concentration of added (and thus free) RARα LBD.
RESULTS
Selection of ATRA resistance mutations for functional characterization
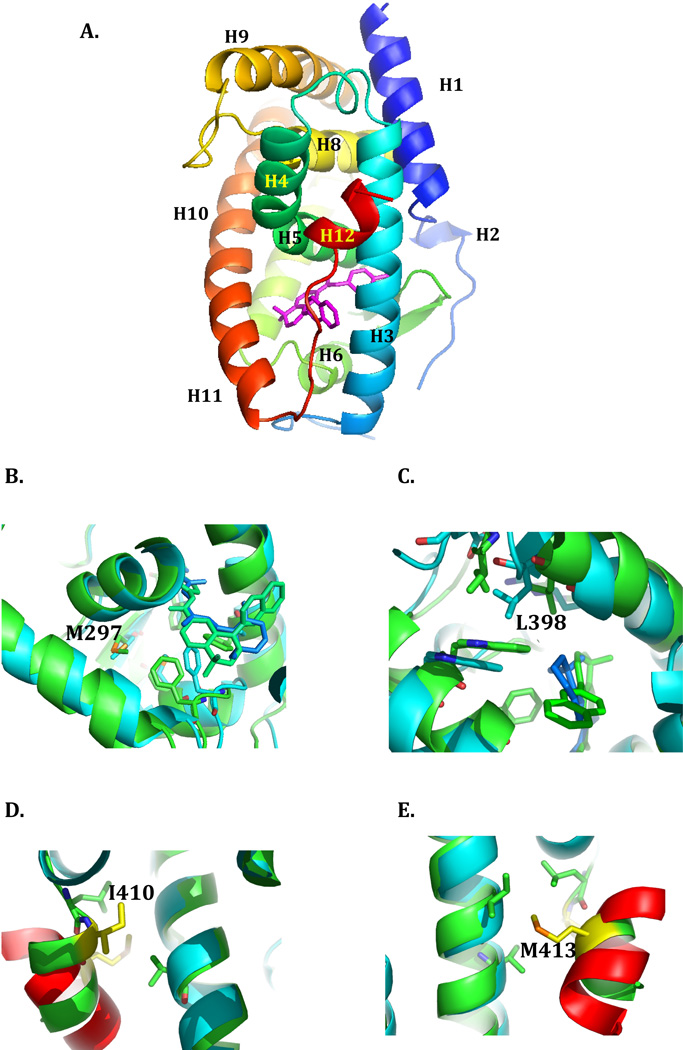
The interactions of RARα with nuclear co-regulators are mediated by the ligand-binding domain (LBD, also referred to as domain E). LBDs of this class of receptor share a common all-α protein fold consists of twelve α-helices packed into a three-layer sandwich, with a short β hairpin motif 39. ATRA binding causes repositioning of H11 and H12 C-terminal helices of the RARα LBD, remodeling the receptor to allow co-repressor release and co-activator recruitment 40. This exchange of the co-repressor for the co-activator is necessary for the subsequent stimulation of Pol II transcription 41–43. ATRA resistant mutants of PML-RARα that localized to the RARα LBD included variants derived from ATRA-resistant isolates of the PML-RARα cell line NB-4, and from ATRA-treated and relapsed patients 35,44–46. Four mutants that encompass different regions of the RARα LBD were selected for this study, consisting of L398P, M297L, I410T, and M413T. Their locations in the RARα LBD structure are indicated in Figure 1.
Figure 1. The ligand binding domain of RARα.
The structure of the ligand binding domain of RARα in its antagonist bound form (PDB ID:1DKF) is depicted in ribbon representation, with the N- to C-terminus in rainbow coloring. The individual helices are labeled, and the positions of mutations described in the text indicated. The antagonist ligand BMS600 is stick rendered in magenta. Figure 1 was generated using PyMOL 68. The sources of the mutants (summarized in 35) are as follows: M297L, referred to as APL Case 2, was first described in 45; L398P is referred to in the literature as NB4-R4 44; I410T is referred to as NB4-A1 49; M413T is referred to APL Case 9 46. The LBDs of RARα in the antagonist bound state (PDB:1DKF) and RARβ in the agonist bound state (PDB:1XDK) were superimposed using the MagicFit option of Swiss PDB Viewer. The RARα (antagonist bound) structure is colored green, and the RARβ (agonist bound) is colored cyan. The fragment of the TRAP220 co-activator bound to the RARβ is colored red. A, the region around Met297. B, the region around Leu398. C, the region around Ile410. D, the region around Met 413.
Expression and purification of the RARα LBD
As described in “Materials and Methods”, the mutations described above were introduced into a plasmid construct encoding the RARα LBD. In this over-expression construct, a His6 affinity tag is encoded at the NH2 end of the LBD to aid in purification. Initial efforts to express and purify the RARα LBD from cells grown at 37 °C resulted in unstable, aggregation prone protein, so the purification protocol was modified by lowering the growth temperature to 20 °C, and by carrying out the purification of the RARα LBD in the presence of the RXR LBD. The latter receptor domain did not possess an N-terminal affinity tag, so its co-purification with the RAR LBD would not be expected. The absence of RXR LBD in our RARα LBD preparations was confirmed by N-terminal sequencing, which provided no evidence of the sequence MASMTGGQQM, which would indicate the presence of RXR LBD (Supplementary Figure S1). The potential contamination of the RAR LBD preparation with RXR was thus below the limits of detection, and not deemed to present a concern in our characterization of RARα LBD. Receptor preparations obtained by His6 affinity chromatography were typically > 90% pure (Supplementary Figure S2). Attempts to increase purity by additional chromatographic steps, including size exclusion chromatography, were not successful, owing to a high propensity of both wild type and mutant receptors to undergo aggregation.
In order to better understand the aggregation phenomenon, samples of both wild type and selected mutant receptors were submitted to the HHMI/W.M. Keck Foundation Biotechnology Resource Laboratory at Yale University, which performed a coupled size exclusion chromatography-light scattering analysis. These experiments indicated that while typical receptor preparations quickly proceed to an aggregated state (~300 kD) in the absence of ATRA, these dissociate to monomers (~30 kD) in the presence of ATRA (Supplementary Figure S3).
Circular Dichroism Studies
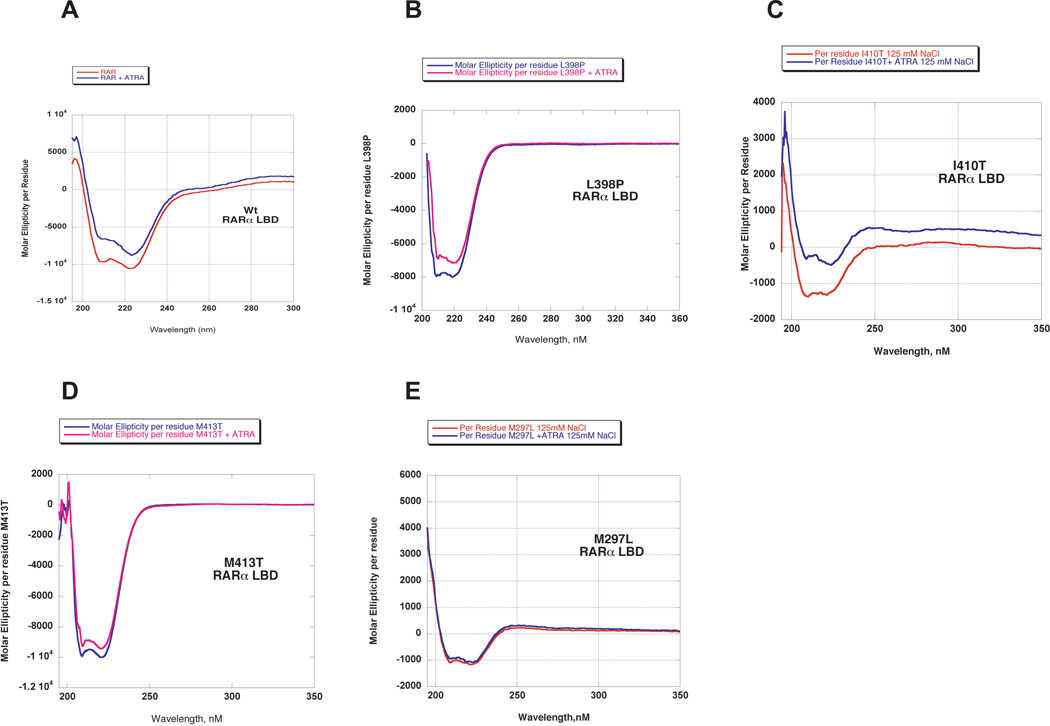
As a first step in assessing whether or not the ATRA resistance mutations confer significant changes in protein secondary structure, circular dichroism experiments were performed. Representative spectra for purified wild type and mutant LBD at concentrations between 10– 50 µM are shown in Figure 2. As indicated by the presence of significant negative ellipticity at 222 and 208 nm, both the wild type and the mutant LBD retain a significant proportion of α-helical structure. Deconvolution of the spectra indicates an approximate helical content in the range of 26–30%, in good agreement with similar measurements reported for the LBD of RARγ 47. Truncation of the data below 190 nm precluded accurate estimation of the content of β sheets and turns. Measurements were also performed in the presence of the ligand ATRA. These experiments showed that the presence of the ligand only had a minor effect on helical content, imparting a decrease of less than 5%. Interestingly, the mutant M297L showed the least degree of change in helical content in the presence of ATRA.
Figure 2. Secondary structure analysis of wild type and mutant RARα ligand binding domains.
CD spectra are plotted as the molar ellipticity [θ] (in units of deg-cm2-decimole−1) versus wavelength. A, wild type RARα LBD; B, L398P RARα LBD; C, I410T RARα LBD; D, M413T RARα LBD; E, M297L RARα LBD.
Limited proteolysis experiments
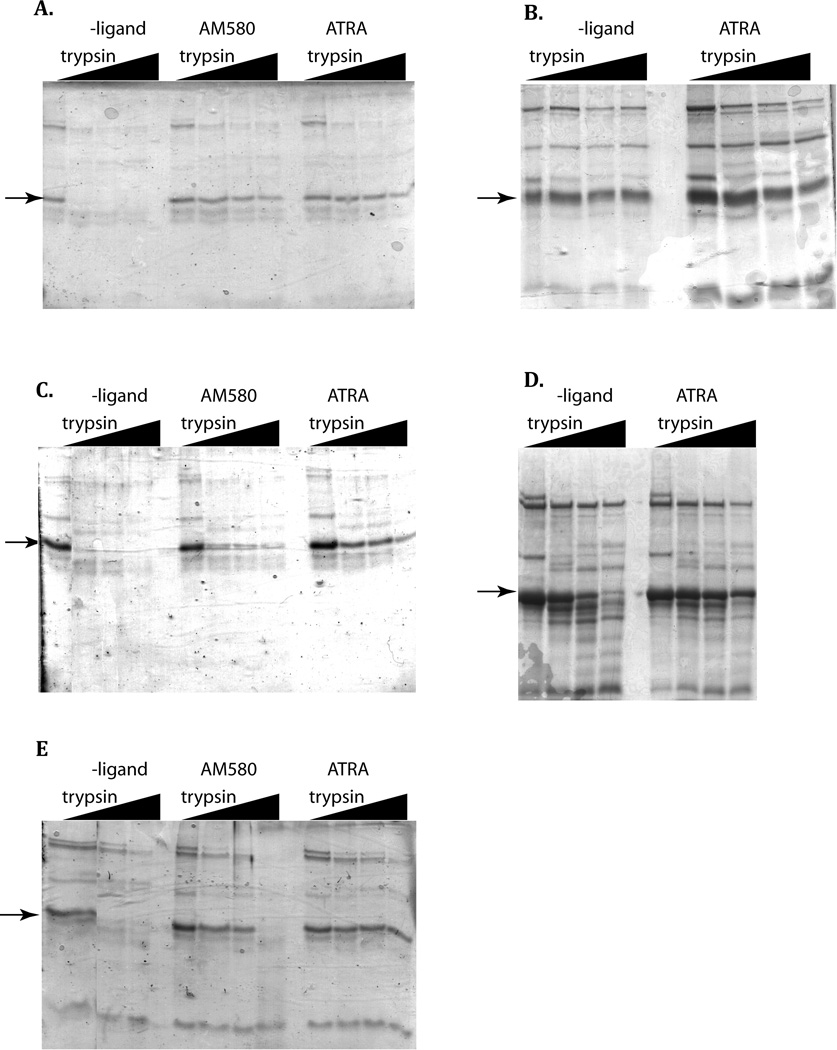
One mechanism by which ATRA promotes differentiation of APL cells is by induction of proteolysis of PML-RARα48. Accordingly, at least part of the ATRA resistant phenotype could be due to increased resistance of the mutants to proteolysis. In order to test this hypothesis, and gain insights into potential structural changes associated with the mutants, limited proteolysis experiments were performed using trypsin. Previously, similar experiments using PML-RARα fusion proteins produced by coupled transcription/translation in a reticulocyte lysate indicated that ATRA resistant mutants alter resistance of the fusion protein to protease 35,49. The experiments performed here differ in that only the LBD was examined. As shown in Figure 3A, incubation of the wild type LBD with trypsin in the absence of ATRA led to the rapid disappearance of the 25 kD band corresponding to the LBD. In the presence of either agonist ligand ATRA or AM580, a significant fraction of full-length receptor remained at the end of the 15 min incubation time. Strong protection by the ligands was also observed for the I410T (Figure 3C) and M297L mutants (Figure 3E). The remaining two mutants (L398P and M413T) exhibited a ladder of proteolytic products in the absence and presence of the ATRA ligands, but substantial amounts of near full-length product remained even when ATRA was absent. For these mutants, it appears that the mutation significantly alters the resistance of the LBD to trypsinolysis, such that any potential protective effect of the ATRA ligand is masked by other, mutationally-linked structural effects. Thus, among the four mutants, there are at least two capable of interacting with the ligand (I410T and M297L), and two (L398P and M413T) for which interactions with the ligand cannot be reliably assessed by this method.
Figure 3. Wild type and mutant RARα LBDs exhibit altered protease resistance.
Limited trypsin digestion was performed for 15 minutes at 22 °C in the dark, as described in “Materials and Methods”. The arrows indicate the intact RARα LBD, which has Mr of 28,637. A, wild type; B, L398P; C, I410T; D., M413T; E, M297L.
Measurement of dissociation constants of co-repressor peptide models by fluorescence anisotropy
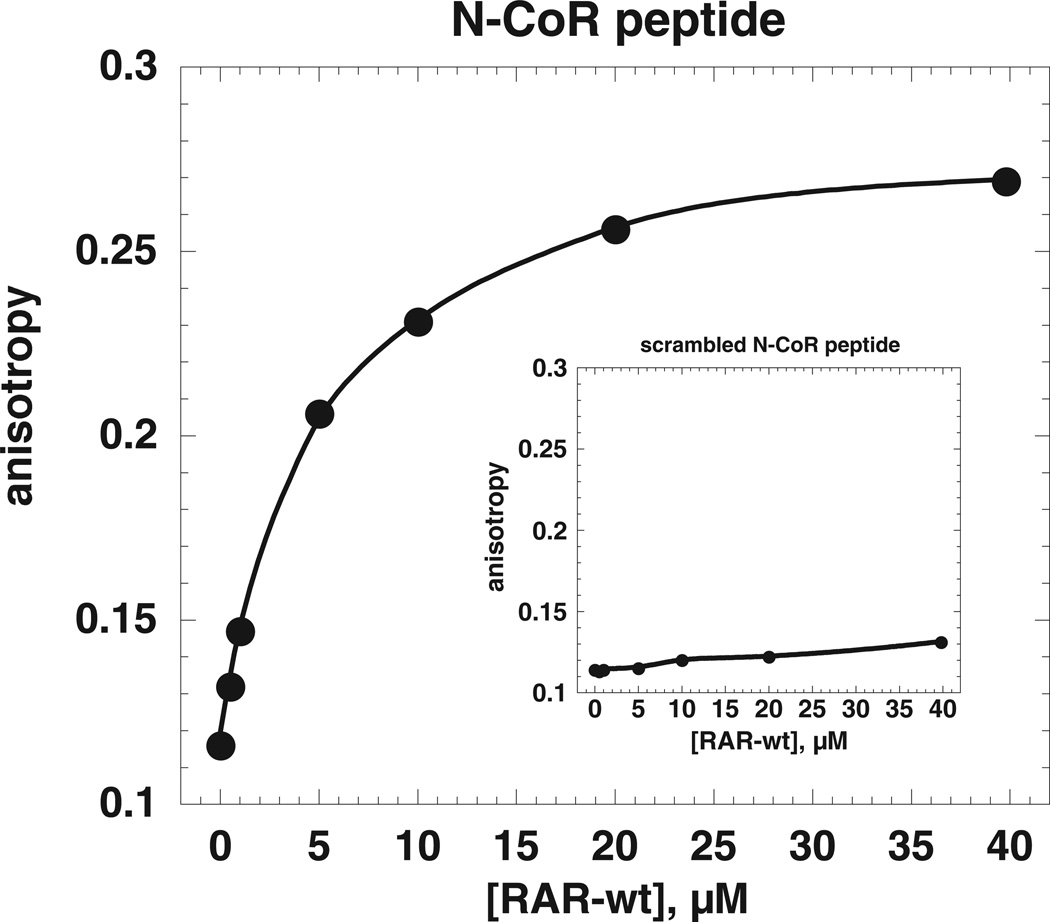
Prior work showed that ATRA resistant versions of PML-RARα are altered in their response to agonist, which in the non-mutated fusion protein causes a decreased interaction with nuclear co-repressor and a strengthened interaction with nuclear co-activator 35,50. To investigate this phenomenon in a more clearly defined model system where quantitative measurements about the relative strengths of different interactions can be performed, a fluorescence anisotropy assay was developed employing rhodamine conjugated peptides based on the RID II motif of N-CoR 51,52 and non-labeled wild type or mutant LBD. Steady state fluorescence assays showed that, when increasing concentrations of LBD were titrated into solutions of rhodamine-labeled N-CoR peptide, the anisotropy increased from 0.12 to 0.27, indicating the formation of a stable LBD-peptide complex (Figure 4). Control experiments employing a conjugated peptide with a scrambled sequence did not exhibit an anisotropy change, showing that interaction is sequence specific (Figure 4 inset).
Figure 4. Fluorescence anisotropy assay used to measure peptide binding to RARα ligand binding domain.
The binding measurements were performed at 20 °C and pH 8.1, as described in “Materials and Methods”. All measurements were repeated with at least two independent protein preparations. The results from a representative experiment are shown. The main plot depicts an experiment with wild type RARα LBD and a peptide based on the sequence of the N-CoR nuclear co-repressor. The inset depicts a fluorescence anisotropy experiment employing a scrambled sequence of the N-CoR peptide as ligand.
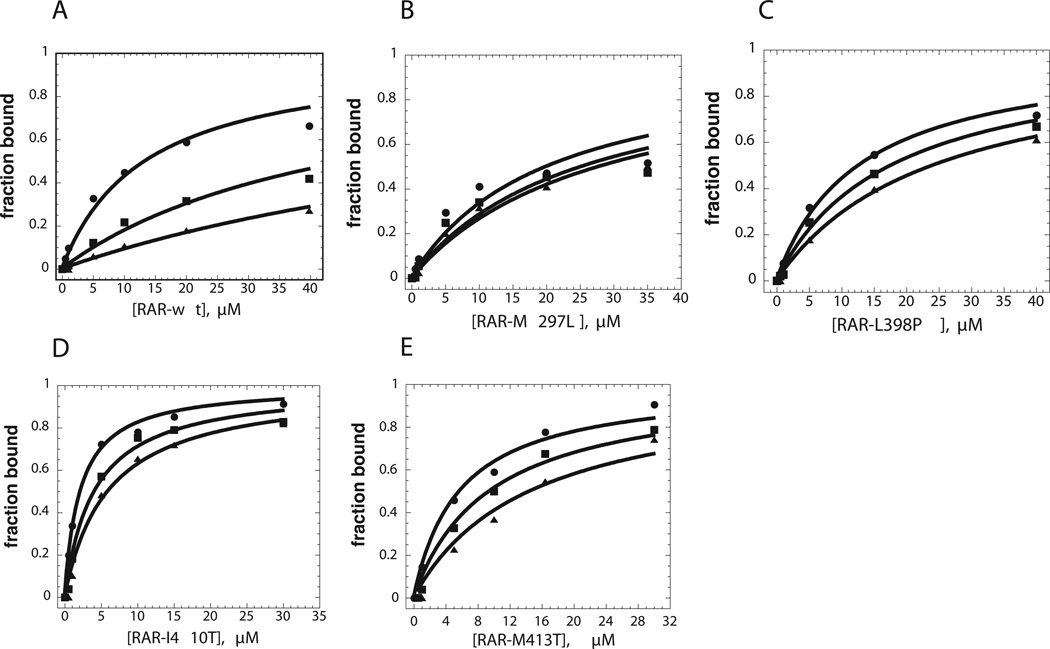
Conversion of these anisotropy changes into fraction bound (as described in “Materials and Methods”) and then plotting against increasing concentration of receptor allowed an apparent dissociation constant of 18.9 ±5 µM to be determined (Figure 5A). When the LBD was pre-incubated in the presence of 10 and 20 nM ATRA, N-CoR binding was decreased (Figure 5A). The change in dissociation constant for N-CoR in the presence of increasing concentrations of ATRA is depicted in bar form in Figure 6A. As ATRA was titrated from 0 to 209 nM, the KD for the N-CoR peptide increased from 18.9 ± 5 µM to 74.2 ± 3.6 µM. The assay could not be performed at higher concentrations of ATRA owing to ATRA-dependent decreases in fluorescence intensity. Measurements were also performed using a rhodamine-conjugated peptide based on the SMRT ID1 sequence (H2N-CASTNMGLEAIIRKALMG-COOH). Anisotropy measurements performed using this peptide showed that it did not bind significantly tighter than the N-CoR peptide (data not shown).
Figure 5. Binding of co-repressor peptides to wild type and mutant RARα as measured by fluorescence anisotropy.
The measurements were performed at 20 °C and pH 8.1, and the fraction of bound was determined as described in “Materials and Methods”. All measurements were repeated using at least two independent protein preparations, with representative plots shown. Binding experiments were carried out for the wild-type LBD and each of the four mutants, in the presence of different ATRA concentrations as described in “Materials and Methods”. A, wild type; B, M297L C, L398P; D, I410T; E., M413T. ➂, Binding isotherms for N-CoR peptide in the absence of ATRA, plotted as a function of LBD concentration. ➄, Binding isotherms for N-CoR peptide binding in the presence of 10 nM ATRA, plotted as a function of LBD concentration. ➉, Binding isotherms for N-CoR peptide binding in the presence of 20 nM ATRA, plotted as a function of LBD concentration.
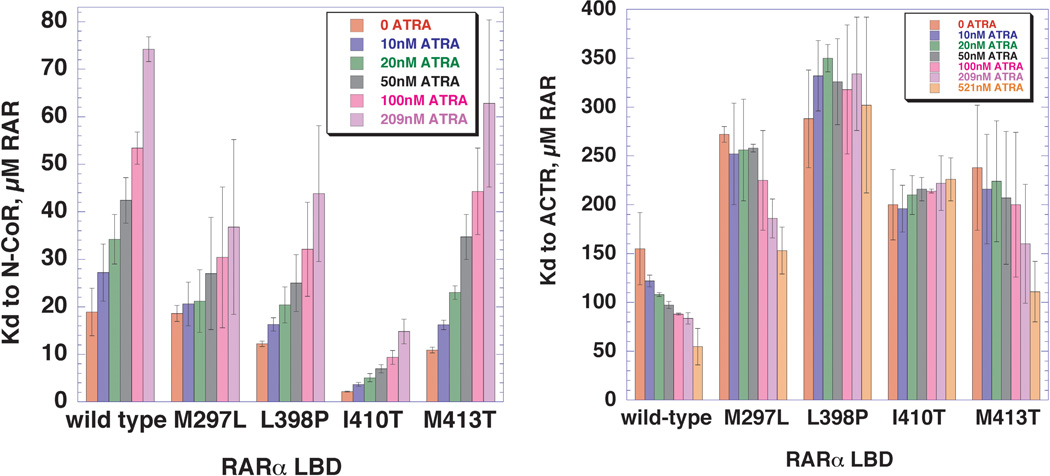
Figure 6. Variation of the dissociations constant for binding of N-CoR and ACTR peptides to wild type and mutant RARα LBDs, as a function of increasing ATRA.
A, Bar graph representation of the equilibrium dissociation constants (Kds) for N-CoR co-repressor binding to wild type and mutant RARα LBDs, determined over a range of ATRA concentrations. Each equilibrium dissociation constant was determined from a binding isotherm of the type shown in Figure 2, performed as described in Experimental Procedures. The error bars represent standard error of the mean. B, Bar graph representation of the equilibrium dissociation constants (KDs) for ACTR co-activator binding to wild type and mutant RARα LBDs, determined over a range of ATRA concentrations.
Anisotropy experiments with the N-CoR peptide were also performed using the mutant LBDs. As indicated from the plots in Figure 5 and the representation of the data in bar graph form in Figure 6, the mutations affect the KD for the N-CoR peptide to different extents. Notably, the affinity of the N-CoR peptide for the M297L mutant LBD was essentially equal to wild type (18.6 ± 1.7µM), while the affinities of the N-CoR peptide for the L398P, M413T, and I410T mutants were significantly increased (12.2 ± 1.7µM, 10.9 ± 1.7µM, and 2.2 ± 1.7µM, respectively). Hence, one component of ATRA resistance appears to be a higher intrinsic affinity for nuclear co-repressor.
Experiments in which increasing concentrations of ATRA were titrated into the mutant LBDs provided information about how the presence of the agonist altered the LBD: N-CoR interaction. The four mutants could be divided into two different classes on the basis of how ATRA affect the KD for N-CoR. Relative to wild type, M297L and L398P both exhibited smaller fold increases in KD. The conversion of these changes into ΔΔG gave values of 0.4 and 0.74 kcal/mol, respectively. In contrast, I410T and M413T exhibited larger fold increases in KD that corresponded to 1.12 and 1.02 kcal/mol, respectively. These larger changes in KD relative to the 0.8 kcal/mol for wild type LBD suggest that both I410T and M413T retain interactions with ATRA that can influence the strength of interactions with the N-CoR peptide. The observation that ATRA was able to bring about a change in the strength of binding of N-CoR to each of the four LBDs strongly suggests that none has lost the ability to interact with ATRA.
Measurement of dissociation constants of co-activator peptide models by fluorescence anisotropy
The results in the previous section suggest that one component of the mechanism of ATRA resistance may be the decreased ability of ATRA to bring about dissociation of co-repressor. Alternatively or additionally, ATRA resistance might involve a reduced ability of ATRA to promote interaction with co-activator. This hypothesis was investigated by performing the fluorescence anisotropy experiment using a rhodamine-conjugated peptide whose sequence corresponds to the receptor interacting domain 3 of the ACTR co-activator. Relative to the change in anisotropy seen with the binding of the N-CoR peptide, the anisotropy change with ACTR was more modest, starting from an initial value of 0.12 and reaching a plateau of 0.16. As shown in Figure 6B, the affinity of ACTR for the RARα LBD in the absence of ATRA is weak (155 µM), but increases progressively as ATRA is titrated in. At the highest ATRA concentrations tested (521 nm), the KD decreased approximately three-fold, to 55 µM. Relative to wild type, all of the ATRA-resistant mutants exhibited an approximately two-fold elevated KD. In comparison to the wild type RARα LBD, the mutants required a higher concentration of ATRA to elicit a change in the KD for the ACTR peptide. As in the case of interactions with the N-CoR peptide, all four mutants did not behave identically. While M297L and M413T both exhibited modest decreases in the KD for ACTR at the highest ATRA concentration, the interactions of L398P and I410T with ACTR were essentially unresponsive to ATRA. Based on these results, a second mechanism of ATRA resistance appears to be an inability of ATRA to bring about an increase in affinity for co-activator.
DISCUSSION
Here, we employed a minimal protein domain approach to study the complex phenomenon of ATRA resistance in PML-RARα. The rationale for the selection of the isolated LBD as a model for ATRA-resistant APL is threefold: (1) many of the ATRA-resistance mutations are located in this region; (2) the LBD constitutes the site of ligand binding; and (3) the LBD contains the AF-2 activation region where nuclear co-regulators bind. Moreover, the use of small proteins and carefully defined biochemical and biophysical assays allows the consequences of the mutations to be examined in an in-depth quantitative context. Our studies suggest three major conclusions with respect to the influence of the ATRA resistance mutations on PML-RARα. First, the mutations appear not to be associated with a gross overall change in α-helical structure (Figure 2). Secondly, our results suggest that the mutations alter a major functional characteristic of the complete receptor, namely the ability to undergo co-regulator switching function in response to ATRA (Figure 6). Thirdly, our results suggest that, in at least two cases, the mutations alter the proteolytic sensitivity of the LBD (Figure 3).
The minimal biochemical system of the LBD and peptide models described here is admittedly unable to recapitulate all of the important features of PML-RARα function, and its role in promoting APL. None of the assays described here involve a cellular context, meaning that the influence of other potential cellular factors on PML-RARα function is missing. PML is known to associate in nuclear subdomains, the formation of which is disrupted by the fusion oncoprotein 53. Our system has no transcriptional readout, which is also an important metric for understanding global properties of the system. The measurements reported here do not reflect either the complex thermodynamics associated with the interaction of the full-length PML-RARα protein and its DNA target, or the influence of other factors that might regulate the interaction strength. These are clearly important, because the affinity of PML-RARα for SMRT/NCOR in the presence of a specific DNA is in the nanomolar concentration range 54, while the affinity of LBD for co-regulator peptides is only in the micromolar range (this work). Previously, the mutant substitutions studied here were characterized in the context full-length PML-RARα proteins in nuclear extracts that included other cellular factors, and it was reported that interactions with nuclear co-regulators are altered 35,49. The availability of such data influenced the selection of the mutants studied here, and provides an opportunity to validate our observations in context of a more complete system.
Altered switching of co-regulator binding
Conceptually, the inability of ATRA resistant PML-RARα oncoproteins to respond to ligands could result from the inability to bind ligands, failure to dissociate co-repressor, or failure to recruit co-activator. Indeed, cellular extracts expressing the L398P mutant fail to exhibit co-regulator switching in the presence of ATRA 34. This led Cote et al. to conclude that failure of the mutant bind ligand largely accounts for the mutant’s inability to activate transcription 35. A priori, one would expect that the protease resistance of a mutant that completely lacked the ability to bind ligand would be unchanged by the presence of ligand, and that the dissociation constants for co-repressor or co-activator peptide would be similarly ligand independent. Significantly, none of the mutants tested in our in vitro assays conformed precisely to this behavior. The L398P mutant was phenotypically the most similar to a ligand binding null mutant, in that it appeared to be insensitive to the presence of ATRA in both protease resistance (Figure 3) and co-activator recruitment experiments (Figure 6). However, the L398P LBD still exhibited an approximately three-fold increase in the KD for co-repressor peptide at 209 nM ATRA (Figure 6), arguing that interaction with agonist may not be completely abolished. (Higher concentrations could not be examined, owing to interference of ATRA with the fluorescence signal.) Essentially all of the mutants showed some response to ATRA with regard to KD for the co-repressor peptide (Figure 6A and Figure 7A,) suggesting that in none of the examples studied was ATRA binding completed ablated.
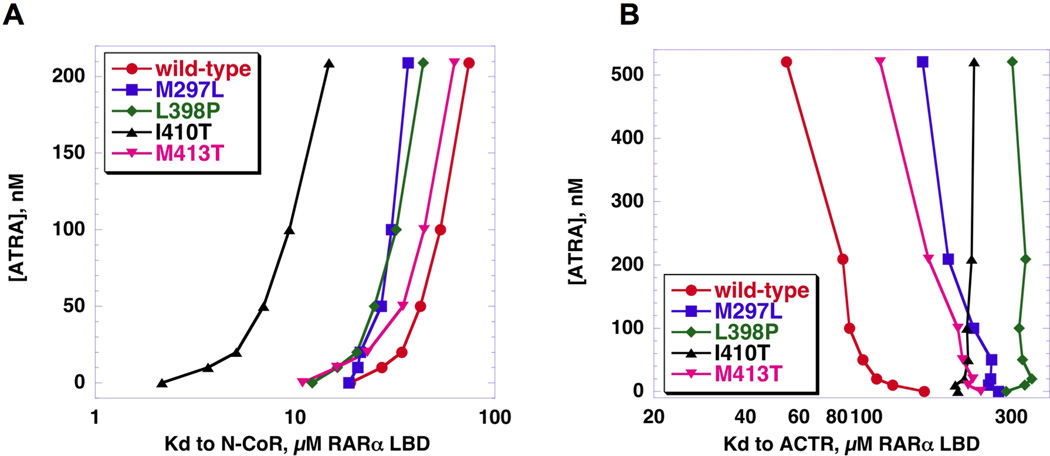
Figure 7. Thermodynamic effect of co-factor binding on mutant ATRA affinity.
ATRA concentration is plotted versus the log of the KD for each RARα LBD to N-CoR or ACTR. A, ATRA binding affinity is largely unaffected by N-CoR binding. With the exception of M297L, affinity to ATRA does not differ significantly amongst the N-CoR-bound receptors, visualized by the similarly shaped ATRA-response curves. B, The presence of ACTR, however, has a large effect on ATRA binding affinity. The mutations, particularly L398P and I410T, weaken the ATRA affinity of ACTR-bound receptors, visualized by the diverse shapes of the ATRA-response curves.
Studies on co-activator binding presented a different picture. Compared to wild type, all four mutants exhibited higher dissociation constants, signifying weaker affinity (Figure 6B). In two of the mutants, (L398P and I410T) the presence of ATRA produced no improvement in the affinity for the co-activator peptide (Figure 6B and 7B). For the other two mutants (M297L and M413T), ATRA increased the affinity for co-activator peptide, but not to a level equivalent to wild type (Figure 6B and 7B). The mutant substitutions therefore had a differential effect on co-factor switching characterized by a relatively slight diminution in the ability of ATRA to mediate co-repressor release, but a relatively large decrease in the ability of ATRA to mediate co-activator recruitment. It is possible that, in the context of the full length PML-RARα and SMRT/NCOR co-repressor, the effects we observed with the LBDs and peptide constructs are magnified. This might explain why even at micromolar concentrations of ATRA, full length L398P PML-RARα is unable to dissociate co-repressor and attract co-activator 34.
Superposition of the closely related crystal structures of the antagonist bound form of RARα with the agonist 55 and co-activator bound form of RARβ56 supports the hypothesis that all four mutants influence the switching process at the molecular level. The substituted residues M297 and L398 both directly abut the ligand-binding site in the RARα and RARβ structures, while I410 and M413 are localized in Helix12 (Figure 1). In the antagonist bound complex 55, M297 cooperates with neighboring residues Met283, Phe302, Phe 228, and Phe286 to create the hydrophobic walls of the ligand binding pocket, particularly for the C and D rings of the BMS614 antagonist 55. As part of the switch from antagonist to agonist, Phe302 undergoes a 90° rotation to remodel the pocket. The M297L substitution is appropriately positioned to influence the structural consequences of the ring rotation. L398, which is located at the end of helix 112, also contacts the C ring of the antagonist ligand, but participates in a slightly different conformational change. In the switch from bound agonist to antagonist, the last two helical turns of H11 undergo a helix to turn transition. This change extends up the length of the random coil between H11 and H12, allowing H12 to switch into its antagonist configuration. As proposed in the “mouse trap model” 57, introduction of a proline at position 398 introduces a discontinuity in H12, facilitating the transition to the antagonist configuration and preventing AF-2 from executing its proper functions. Leu398 also packs against Trp225, another important contributor to ligand binding. The last two mutants, I410T and M413T, alter residues on the surface of helix 12 that mimic the leucine residues of the LXXLL motif of co-activator. These mutations may serve to enforce positioning of helix 12 in its co-repressor orientation, such that ATRA binding cannot bring about helix 12 reorientation, and co-activator recruitment 35.
The relationship between ATRA resistance and proteolysis resistance, and its influence on the effectiveness of APL therapeutics
Initially, PML-RARα was thought to promote APL based on its ability to act as a dominant repressor of RARα’s gene activation function in hematopoietic cells, thereby causing a block in differentiation 5. This model was rationalized by the significant role that the RARα repressor plays in hematopoietic development, and its role as a regulator of many genes. An important feature of PML-RARα in this scenario is its failure to exhibit ATRA response at physiological concentrations 34. Instead, at physiological RA concentrations, PML-RARα is suggested to occupy the response elements normally employed by RARα in a predominantly repressive mode. Possibly as a consequence of tighter interactions with the nuclear co-repressors SMRT and NCoR (as well as histone deacetylases) PML-RARα requires higher concentrations of ATRA to undergo the switch from repressor to activator 58. These, in turn, are linked to increased interactions with the DNA methyl transferase machinery, which brings about gene silencing, particularly at the RARβ promoter. Indeed, APL shows a characteristic pattern of gene silencing as measured by genome wide methylation59. Separately, PML-RARα may also interfere with PML’s normal functions, which include enhancing resistance to apoptosis and regulating the extent of stem cell character 60. Under this model, raising ATRA to pharmacological concentrations in the course of therapy would lead to a release of the differentiation block, by virtue of the de-repression of the genes under PML-RARα control.
Despite the initial attractiveness of this “differentiation block” model, many key observations are inconsistent with its predictions, leading to a re-assessment of PML-RARα mechanism of action. First, no direct linage between transcriptional repression and the differentiation block has ever been established. Second, PML-RARα has been shown to regulate many more genes than just those regulated by RARα, an increase in DNA binding promiscuity that is likely due to the formation of heterotetramers composed of PML-RARα and RXR, which can then bind to many non-canonical RARα sites 61,62. Lastly, combination treatments involving the administration of both ATRA and arsenic trioxide (ATO) produce a high frequency of durable remissions by a mechanism distinct from induced differentiation10. Rather, the mechanism by which these agents promote remission appears to be based on their ability to enhance the degradation of the PML-RARα oncoprotein. This latter property is associated with the re-localization of PML to nuclear bodies, and a cellular subpopulation of leukemic cells (LICs), whose survival may be essential to the persistence of the disease.
RA and ATO both promote PML-RARα degradation, but by different mechanisms 48. ATRA itself elicits at least two distinct degradative mechanisms. The first, which appears to be linked hematopoietic cell differentiation, is mediated by caspase 3, occurs at the C-terminal of the PML region, and is independent of the proteosome 63. By contrast, the second mechanism requires the participation of both ubiquitin and the proteosome, features the DNA-bound RARα/RXR heterodimer as its substrate, and is dependent on the AF-2 region of RARα 64. Notably, the latter segment has been shown to interact with SUG-1, a component of the proteosome 65. ATO mediated PML-RARα degradation occurs by a third mechanism 66 that involves competition by arsenate for the zinc sites within the ring finger motif of PML 60. Arsenic substitution appears to elicit a number of uncharacterized conformational changes in PML-RARα that lead to sumoylation, ubiquitinylation and, ultimately, recruitment to the 11S proteosome. The significance of these observations for treatment of APL has been underscored by the recent demonstration that destruction of PML-RARα by ATRA and ATO strongly inhibits the survival of leukemia initiating cells, and causes regression of APL 67. Thus, the real efficacy of anti-APL treatments may depend on their ability to prevent the growth of these “cancer stem cells”, as opposed to the ability of therapeutics to re-initiate the myeloid cell differentiation program.
In the context of this new appreciation for the clinical significance of PML-RARα degradation, the differences in proteolytic sensitivity we observed among the various ATRA-resistant LBDs may be noteworthy. The wild type, I410T, and M297L RARα LBDs all exhibited sensitivity to trypsin in the absence of ATRA, but less so in the presence of agonist. By contrast, the L398P and M413T LBDs both showed altered proteolytic patterns, with a significant amount of LBD remaining even at the end of the incubation period. Of course, these observations must be tempered by the caveats that the true biological molecule of interest is the complete PML-RARα fusion protein, not the LBD, and that trypsin is not implicated in any of cleavage/degradation mechanisms described above. At best, trypsin represents a probe for detecting structural changes in the LBD (e.g. of the AF-2 region) that might impinge on a degradation process involving other agents 64. With that stated qualification, there are some notable correlations between the results in our simple system, and more complex cellular and animal models. In the original isolation of the L398P mutant as an ATRA resistant subclone of NB4 cells (NB4-R4), it was noted that this mutant does not undergo ATRA-mediated degradation, whereas the otherwise unmutated PML-RARα does so in the parent line 44. Thus, resistance to degradation and a mutation in the AF-2 region are linked in the context of the full-length oncoprotein. A more striking correlation is seen with a mouse model containing a PML-RARα L902P (equivalent to L398P) transgene. Here, the combination of ATRA and cAMP was observed to promote differentiation (possibly by the isomerization of RA to 9-cis-RA, a ligand for RXR). Yet, no decrease in the population of LIC was observed, and the transgene-derived PL-RAR L902P oncoprotein did not undergo degradation. Comparable results are not available for the M413T LBD, but the results of both the experiments performed here and reported previously suggest that its agonist binding and nuclear co-regulator properties are minimally affected 46,28. Hence, its phenotype as a degradation resistant mutant is plausible.
By contrast, I410T and M297L both exhibited proteolytic resistance patterns that were reminiscent of the wild type LBD, which does undergo ATRA-mediated degradation. The I410T substitution was originally isolated from an NB-4 (PML-RARα expressing) cell line that had been treated with successively higher concentrations of ATRA 49, while the M297L mutant was derived from a relapsed patient 35. In addition to its increased proteolytic sensitivity, the I410T mutant differed from L398P in several notable respects. While the full length I410T protein did show reduced ATRA binding in cellular extracts, binding activity was recovered in the presence of the SMRTβ isoform, indicating that the ligand binding domain is not inherently compromised for binding activity 50. In experiments studying the effect of HDAC inhibitors, the combination of ATRA plus trichostatin A (TSA) was able to rescue the transcription of I410T, but not of L398P 50. Our data indicated that while I410T RARα LBD bound co-repressor peptide with higher affinity than wild type, the interaction was sensitive to ATRA. This suggests a fundamental distinction between I410T and L398P; namely that the ATRA resistance of the former originates principally from the defect in cofactor switching, which is at least partially alleviated by the combination of ATRA and TSA.
SUMMARY
Our work suggests that ATRA resistance mutations that map to the LBD of PML-RARα can, in principle, influence at least two distinct functional properties. The first is the ability to exchange nuclear co-repressor for co-activator, which was exemplified by the I410T mutant. Despite the increase in affinity for co-repressor and poor affinity for co-activator, mutants with these properties appear to be easier to “rescue” at the transcriptional level, particularly in the presence of HDAC inhibitors. The second property is the ability to undergo ligand-stimulated degradation that, as exemplified by ATRA-resistant L398P LBD, may be critical for the effectiveness of ATRA/ATO combination therapeutics. Further analysis of the specific contributions of individual domains of PML-RARα to therapeutic-stimulated breakdown may help to refine clinical strategies in ATRA-resistant relapsed patients.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Wilson Miller for providing the original PML-RARα clones, Tim Hunter and the University of Vermont DNA Analysis Facility for DNA sequencing, and David Jameson at the University of Hawaii for advice and suggestions related to the anisotropy analysis.
Footnotes
This work was supported by the Children’s Leukemia Research Association. Additional support was provided by the Vermont Genetics Network through Grant Number P20 RR16462 from the INBRE Program of the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH).
Its contents are solely the responsibility of the authors and do not necessarily represent the official views of NCRR or NIH.
Abbreviations: ACTR, nuclear co-activator; APL, acute promyelocytic leukemia; ATRA, all trans retinoic acid; CD, circular dichroism; HAC, histone acetylase complex; HDAC, histone deacetylase complex; LIC, leukemia initiating cells; LBD, ligand binding domain; MALDI-TOF, matrix associated laser desorption time of flight [spectroscopy]; N-CoR, nuclear co-repressor; PML, promyelocytic leukemia gene; RARα, retinoic acid receptor alpha; SMRT, silencing mediator for retinoid and thyroid receptors; TFL, tri-fluoroacetic acid; TSA, trichostain A.
REFERENCES CITED
- 1.Larson RA, Kondo K, Vardiman JW, Butler AE, Golomb HM, Rowley JD. Evidence for a 15;17 translocation in every patient with acute promyelocytic leukemia. Am J Med. 1984;76(5):827–841. doi: 10.1016/0002-9343(84)90994-x. [DOI] [PubMed] [Google Scholar]
- 2.Warrell RP, Jr, de The H, Wang ZY, Degos L. Acute promyelocytic leukemia [see comments] N Engl J Med. 1993;329(3):177–189. doi: 10.1056/NEJM199307153290307. [DOI] [PubMed] [Google Scholar]
- 3.Slack JL, Gallagher RE. The molecular biology of acute promyelocytic leukemia. Cancer Treat Res. 1999;99:75–124. doi: 10.1007/978-0-585-38571-6_4. [DOI] [PubMed] [Google Scholar]
- 4.Borrow J, Goddard AD, Sheer D, Solomon E. Molecular analysis of acute promyelocytic leukemia breakpoint cluster region on chromosome 17. Science. 1990;249:1576–1580. doi: 10.1126/science.2218500. [DOI] [PubMed] [Google Scholar]
- 5.de The H, Lavau C, Marchio A, Chomienne C, Degos L, Dejean A. The PML-RAR alpha fusion mRNA generated by the t(15;17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell. 1991;66(4):675–684. doi: 10.1016/0092-8674(91)90113-d. [DOI] [PubMed] [Google Scholar]
- 6.Kakizuka A, Miller WH, Jr, Umesono K, Warrell RP, Jr, Frankel SR, Murty VV, Dmitrovsky E, Evans RM. Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RAR alpha with a novel putative transcription factor, PML. Cell. 1991;66(4):663–674. doi: 10.1016/0092-8674(91)90112-c. [DOI] [PubMed] [Google Scholar]
- 7.Grisolano JL, Wesselschmidt RL, Pelicci PG, Ley TJ. Altered myeloid development and acute leukemia in transgenic mice expressing PML-RAR alpha under control of cathepsin G regulatory sequences. Blood. 1997;89(2):376–387. [PubMed] [Google Scholar]
- 8.He LZ, Tribioli C, Rivi R, Peruzzi D, Pelicci PG, Soares V, Cattoretti G, Pandolfi PP. Acute leukemia with promyelocytic features in PML/RARalpha transgenic mice. Proc Natl Acad Sci U S A. 1997;94(10):5302–5307. doi: 10.1073/pnas.94.10.5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Westervelt P, Lane AA, Pollock JL, Oldfather K, Holt MS, Zimonjic DB, Popescu NC, DiPersio JF, Ley TJ. A high penetrance mouse model of acute promyelocytic leukemia with very low levels of PML-RAR{alpha} expression. Blood. 2003 doi: 10.1182/blood-2002-12-3779. [DOI] [PubMed] [Google Scholar]
- 10.Sanz MA, Lo-Coco F. Modern approaches to treating acute promyelocytic leukemia. J Clin Oncol. 2011;29(5):495–503. doi: 10.1200/JCO.2010.32.1067. [DOI] [PubMed] [Google Scholar]
- 11.Bernardi R, Pandolfi PP. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol. 2007;8(12):1006–1016. doi: 10.1038/nrm2277. [DOI] [PubMed] [Google Scholar]
- 12.Salomoni P, Bellodi C. New insights into the cytoplasmic function of PML. Histol Histopathol. 2007;22(8):937–946. doi: 10.14670/HH-22.937. [DOI] [PubMed] [Google Scholar]
- 13.Lallemand-Breitenbach V, de The H. PML nuclear bodies. Cold Spring Harb Perspect Biol. 2010;2(5):a000661. doi: 10.1101/cshperspect.a000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernardi R, Papa A, Pandolfi PP. Regulation of apoptosis by PML and the PML-NBs. Oncogene. 2008;27(48):6299–6312. doi: 10.1038/onc.2008.305. [DOI] [PubMed] [Google Scholar]
- 15.Ito K, Bernardi R, Morotti A, Matsuoka S, Saglio G, Ikeda Y, Rosenblatt J, Avigan DE, Teruya-Feldstein J, Pandolfi PP. PML targeting eradicates quiescent leukaemia-initiating cells. Nature. 2008;453(7198):1072–1078. doi: 10.1038/nature07016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Regad T, Bellodi C, Nicotera P, Salomoni P. The tumor suppressor Pml regulates cell fate in the developing neocortex. Nat Neurosci. 2009;12(2):132–140. doi: 10.1038/nn.2251. [DOI] [PubMed] [Google Scholar]
- 17.Chambon P. A decade of molecular biology of retinoic acid receptors. Faseb J. 1996;10(9):940–954. [PubMed] [Google Scholar]
- 18.Bourguet W, Germain P, Gronemeyer H. Nuclear receptor ligand-binding domains: three-dimensional structures, molecular interactions and pharmacological implications. Trends Pharmacol Sci. 2000;21(10):381–388. doi: 10.1016/s0165-6147(00)01548-0. [DOI] [PubMed] [Google Scholar]
- 19.McKenna NJ, O'Malley BW. Nuclear receptors, coregulators, ligands, and selective receptor modulators: making sense of the patchwork quilt. Ann N Y Acad Sci. 2001;949:3–5. doi: 10.1111/j.1749-6632.2001.tb03997.x. [DOI] [PubMed] [Google Scholar]
- 20.Kastner P, Mark M, Chambon P. Nonsteroid nuclear receptors: what are genetic studies telling us about their role in real life? Cell. 1995;83(6):859–869. doi: 10.1016/0092-8674(95)90202-3. [DOI] [PubMed] [Google Scholar]
- 21.He LZ, Guidez F, Tribioli C, Peruzzi D, Ruthardt M, Zelent A, Pandolfi PP. Distinct interactions of PML-RARalpha and PLZF-RARalpha with co-repressors determine differential responses to RA in APL. Nat Genet. 1998;18(2):126–135. doi: 10.1038/ng0298-126. [DOI] [PubMed] [Google Scholar]
- 22.Ordentlich P, Downes M, Evans RM. Corepressors and nuclear hormone receptor function. Curr Top Microbiol Immunol. 2001;254:101–116. doi: 10.1007/978-3-662-10595-5_5. [DOI] [PubMed] [Google Scholar]
- 23.McKenna NJ, O'Malley BW. Minireview: nuclear receptor coactivators--an update. Endocrinology. 2002;143(7):2461–2465. doi: 10.1210/endo.143.7.8892. [DOI] [PubMed] [Google Scholar]
- 24.Nagy L, Kao HY, Chakravarti D, Lin RJ, Hassig CA, Ayer DE, Schreiber SL, Evans RM. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89(3):373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 25.Collingwood TN, Urnov FD, Wolffe AP. Nuclear receptors: coactivators, corepressors and chromatin remodeling in the control of transcription. J Mol Endocrinol. 1999;23(3):255–275. doi: 10.1677/jme.0.0230255. [DOI] [PubMed] [Google Scholar]
- 26.Warrell RP, Jr, Frankel SR, Miller WH, Jr, Scheinberg DA, Itri LM, Hittelman WN, Vyas R, Andreeff M, Tafuri A, Jakubowski A, et al. Differentiation therapy of acute promyelocytic leukemia with tretinoin (all-trans-retinoic acid) N Engl J Med. 1991;324(20):1385–1393. doi: 10.1056/NEJM199105163242002. [DOI] [PubMed] [Google Scholar]
- 27.Degos L, Dombret H, Chomienne C, Daniel MT, Miclea JM, Chastang C, Castaigne S, Fenaux P. All-trans-retinoic acid as a differentiating agent in the treatment of acute promyelocytic leukemia.[see comment] Blood. 1995;85(10):2643–2653. [PubMed] [Google Scholar]
- 28.Gallagher RE. Retinoic acid resistance in acute promyelocytic leukemia. Leukemia. 2002;16(10):1940–1958. doi: 10.1038/sj.leu.2402719. [DOI] [PubMed] [Google Scholar]
- 29.Tsimberidou AM, Tirado-Gomez M, Andreeff M, O'Brien S, Kantarjian H, Keating M, Lopez-Berestein G, Estey E. Single-agent liposomal all-trans retinoic acid can cure some patients with untreated acute promyelocytic leukemia: an update of The University of Texas M. D. Anderson Cancer Center Series. Leuk Lymphoma. 2006;47(6):1062–1068. doi: 10.1080/10428190500463932. [DOI] [PubMed] [Google Scholar]
- 30.Lo-Coco F, Ammatuna E, Montesinos P, Sanz MA. Acute promyelocytic leukemia: recent advances in diagnosis and management. Semin Oncol. 2008;35(4):401–409. doi: 10.1053/j.seminoncol.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 31.Estey E, Garcia-Manero G, Ferrajoli A, Faderl S, Verstovsek S, Jones D, Kantarjian H. Use of all-trans retinoic acid plus arsenic trioxide as an alternative to chemotherapy in untreated acute promyelocytic leukemia. Blood. 2006;107(9):3469–3473. doi: 10.1182/blood-2005-10-4006. [DOI] [PubMed] [Google Scholar]
- 32.Ablain J, de The H. Revisiting the differentiation paradigm in acute promyelocytic leukemia. Blood. 2011;117(22):5795–5802. doi: 10.1182/blood-2011-02-329367. [DOI] [PubMed] [Google Scholar]
- 33.Melnick A, Licht JD. Deconstructing a disease: RARalpha, its fusion partners, and their roles in the pathogenesis of acute promyelocytic leukemia. Blood. 1999;93(10):3167–3215. [PubMed] [Google Scholar]
- 34.Lin RJ, Nagy L, Inoue S, Shao W, Miller WH, Jr, Evans RM. Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature. 1998;391(6669):811–814. doi: 10.1038/35895. [DOI] [PubMed] [Google Scholar]
- 35.Cote S, Zhou D, Bianchini A, Nervi C, Gallagher RE, Miller WH., Jr Altered ligand binding and transcriptional regulation by mutations in the PML/RARalpha ligand-binding domain arising in retinoic acid-resistant patients with acute promyelocytic leukemia. Blood. 2000;96(9):3200–3208. [PubMed] [Google Scholar]
- 36.Weber G. Polarization of the fluorescence of macromolecules. I. Theory and experimental method. Biochemistry Journal. 1952;51(2):145–155. doi: 10.1042/bj0510145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lakowicz JR. Principles of fluorescence spectroscopy. New York: Kluwer Academic/Plenum Publishers; 1999. [Google Scholar]
- 38.Wilson GM. In: Reviews in Fluorescence. Geddes CD, Lakowicz JR, editors. Volume 2. New York: Springer Science Business Media; 2005. pp. 223–243. [Google Scholar]
- 39.Moras D, Gronemeyer H. The nuclear receptor ligand-binding domain: structure and function. Curr Opin Cell Biol. 1998;10(3):384–391. doi: 10.1016/s0955-0674(98)80015-x. [DOI] [PubMed] [Google Scholar]
- 40.Egea PF, Mitschler A, Moras D. Molecular recognition of agonist ligands by RXRs. Mol Endocrinol. 2002;16(5):987–997. doi: 10.1210/mend.16.5.0823. [DOI] [PubMed] [Google Scholar]
- 41.Chen H, Lin RJ, Schiltz RL, Chakravarti D, Nash A, Nagy L, Privalsky ML, Nakatani Y, Evans RM. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90(3):569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 42.Westin S, Kurokawa R, Nolte RT, Wisely GB, McInerney EM, Rose DW, Milburn MV, Rosenfeld MG, Glass CK. Interactions controlling the assembly of nuclear-receptor heterodimers and co-activators. Nature. 1998;395(6698):199–202. doi: 10.1038/26040. [DOI] [PubMed] [Google Scholar]
- 43.Westin S, Rosenfeld MG, Glass CK. Nuclear receptor coactivators. Adv Pharmacol. 2000;47:89–112. doi: 10.1016/s1054-3589(08)60110-6. [DOI] [PubMed] [Google Scholar]
- 44.Shao W, Benedetti L, Lamph WW, Nervi C, Miller WH., Jr A retinoid-resistant acute promyelocytic leukemia subclone expresses a dominant negative PML-RAR alpha mutation. Blood. 1997;89(12):4282–4289. [PubMed] [Google Scholar]
- 45.Imaizumi M, Suzuki H, Yoshinari M, Sato A, Saito T, Sugawara A, Tsuchiya S, Hatae Y, Fujimoto T, Kakizuka A, Konno T, Iinuma K. Mutations in the E-domain of RAR portion of the PML/RAR chimeric gene may confer clinical resistance to all-trans retinoic acid in acute promyelocytic leukemia. Blood. 1998;92(2):374–382. [PubMed] [Google Scholar]
- 46.Ding W, Li YP, Nobile LM, Grills G, Carrera I, Paietta E, Tallman MS, Wiernik PH, Gallagher RE. Leukemic cellular retinoic acid resistance and missense mutations in the PML-RARalpha fusion gene after relapse of acute promyelocytic leukemia from treatment with all-trans retinoic acid and intensive chemotherapy. Blood. 1998;92(4):1172–1183. [PubMed] [Google Scholar]
- 47.Lupisella JA, Driscoll JE, Metzler WJ, Reczek PR. The ligand binding domain of the human retinoic acid receptor gamma is predominantly alpha-helical with a Trp residue in the ligand binding site. J Biol Chem. 1995;270(42):24884–24890. doi: 10.1074/jbc.270.42.24884. [DOI] [PubMed] [Google Scholar]
- 48.Zhu J, Lallemand-Breitenbach V, de The H. Pathways of retinoic acid- or arsenic trioxide-induced PML/RARalpha catabolism, role of oncogene degradation in disease remission. Oncogene. 2001;20(49):7257–7265. doi: 10.1038/sj.onc.1204852. [DOI] [PubMed] [Google Scholar]
- 49.Cote S, Rosenauer A, Bianchini A, Seiter K, Vandewiele J, Nervi C, Miller WH., Jr Response to histone deacetylase inhibition of novel PML/RARalpha mutants detected in retinoic acid-resistant APL cells. Blood. 2002;100(7):2586–2596. doi: 10.1182/blood-2002-02-0614. [DOI] [PubMed] [Google Scholar]
- 50.Cote S, McNamara S, Brambilla D, Bianchini A, Rizzo G, del Rincon SV, Grignani F, Nervi C, Miller WH., Jr Expression of SMRTbeta promotes ligand-induced activation of mutated and wild-type retinoid receptors. Blood. 2004;104(13):4226–4235. doi: 10.1182/blood-2003-10-3583. [DOI] [PubMed] [Google Scholar]
- 51.Cohen RN, Brzostek S, Kim B, Chorev M, Wondisford FE, Hollenberg AN. The specificity of interactions between nuclear hormone receptors and corepressors is mediated by distinct amino acid sequences within the interacting domains. Molecular Endocrinology. 2001;15(7):1049–1061. doi: 10.1210/mend.15.7.0669. [DOI] [PubMed] [Google Scholar]
- 52.Cohen RN, Putney A, Wondisford FE, Hollenberg AN. The nuclear corepressors recognize distinct nuclear receptor complexes. Molecular Endocrinology. 2000;14(6):900–914. doi: 10.1210/mend.14.6.0474. [DOI] [PubMed] [Google Scholar]
- 53.Heun P. SUMOrganization of the nucleus. Curr Opin Cell Biol. 2007;19(3):350–355. doi: 10.1016/j.ceb.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 54.Zhou J, Peres L, Honore N, Nasr R, Zhu J, de The H. Dimerization-induced corepressor binding and relaxed DNA-binding specificity are critical for PML/RARA-induced immortalization. Proc Natl Acad Sci U S A. 2006;103(24):9238–9243. doi: 10.1073/pnas.0603324103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bourguet W, Vivat V, Wurtz JM, Chambon P, Gronemeyer H, Moras D. Crystal structure of a heterodimeric complex of RAR and RXR ligand-binding domains. Mol Cell. 2000;5(2):289–298. doi: 10.1016/s1097-2765(00)80424-4. [DOI] [PubMed] [Google Scholar]
- 56.Pogenberg V, Guichou JF, Vivat-Hannah V, Kammerer S, Perez E, Germain P, de Lera AR, Gronemeyer H, Royer CA, Bourguet W. Characterization of the interaction between retinoic acid receptor/retinoid X receptor (RAR/RXR) heterodimers and transcriptional coactivators through structural and fluorescence anisotropy studies. Journal of Biological Chemistry. 2005;280(2):1625–1633. doi: 10.1074/jbc.M409302200. [DOI] [PubMed] [Google Scholar]
- 57.Renaud JP, Rochel N, Ruff M, Vivat V, Chambon P, Gronemeyer H, Moras D. Crystal structure of the RAR-gamma ligand-binding domain bound to all-trans retinoic acid. Nature. 1995;378(6558):681–689. doi: 10.1038/378681a0. [DOI] [PubMed] [Google Scholar]
- 58.Grignani F, De Matteis S, Nervi C, Tomassoni L, Gelmetti V, Cioce M, Fanelli M, Ruthardt M, Ferrara FF, Zamir I, Seiser C, Lazar MA, Minucci S, Pelicci PG. Fusion proteins of the retinoic acid receptor-alpha recruit histone deacetylase in promyelocytic leukaemia. Nature. 1998;391(6669):815–818. doi: 10.1038/35901. [DOI] [PubMed] [Google Scholar]
- 59.Di Croce L, Raker VA, Corsaro M, Fazi F, Fanelli M, Faretta M, Fuks F, Lo Coco F, Kouzarides T, Nervi C, Minucci S, Pelicci PG. Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor. Science. 2002;295(5557):1079–1082. doi: 10.1126/science.1065173. [DOI] [PubMed] [Google Scholar]
- 60.Chen SJ, Zhou GB, Zhang XW, Mao JH, de The H, Chen Z. From an old remedy to a magic bullet: molecular mechanisms underlying the therapeutic effects of arsenic in fighting leukemia. Blood. 2011;117(24):6425–6437. doi: 10.1182/blood-2010-11-283598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu J, Nasr R, Peres L, Riaucoux-Lormiere F, Honore N, Berthier C, Kamashev D, Zhou J, Vitoux D, Lavau C, de The H. RXR is an essential component of the oncogenic PML/RARA complex in vivo. Cancer Cell. 2007;12(1):23–35. doi: 10.1016/j.ccr.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 62.Figueroa ME, Lugthart S, Li Y, Erpelinck-Verschueren C, Deng X, Christos PJ, Schifano E, Booth J, van Putten W, Skrabanek L, Campagne F, Mazumdar M, Greally JM, Valk PJ, Lowenberg B, Delwel R, Melnick A. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell. 17(1):13–27. doi: 10.1016/j.ccr.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nervi C, Ferrara FF, Fanelli M, Rippo MR, Tomassini B, Ferrucci PF, Ruthardt M, Gelmetti V, Gambacorti-Passerini C, Diverio D, Grignani F, Pelicci PG, Testi R. Caspases mediate retinoic acid-induced degradation of the acute promyelocytic leukemia PML/RARalpha fusion protein. Blood. 1998;92(7):2244–2251. [PubMed] [Google Scholar]
- 64.Zhu J, Gianni M, Kopf E, Honore N, Chelbi-Alix M, Koken M, Quignon F, Rochette-Egly C, de The H. Retinoic acid induces proteasome-dependent degradation of retinoic acid receptor alpha (RARalpha) and oncogenic RARalpha fusion proteins. Proc Natl Acad Sci U S A. 1999;96(26):14807–14812. doi: 10.1073/pnas.96.26.14807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.vom Baur E, Harbers M, Um SJ, Benecke A, Chambon P, Losson R. The yeast Ada complex mediates the ligand-dependent activation function AF-2 of retinoid X and estrogen receptors. Genes Dev. 1998;12(9):1278–1289. doi: 10.1101/gad.12.9.1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lallemand-Breitenbach V, Jeanne M, Benhenda S, Nasr R, Lei M, Peres L, Zhou J, Zhu J, Raught B, de The H. Arsenic degrades PML or PML-RARalpha through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat Cell Biol. 2008;10(5):547–555. doi: 10.1038/ncb1717. [DOI] [PubMed] [Google Scholar]
- 67.Nasr R, Guillemin MC, Ferhi O, Soilihi H, Peres L, Berthier C, Rousselot P, Robledo-Sarmiento M, Lallemand-Breitenbach V, Gourmel B, Vitoux D, Pandolfi PP, Rochette-Egly C, Zhu J, de The H. Eradication of acute promyelocytic leukemia-initiating cells through PML-RARA degradation. Nat Med. 2008;14(12):1333–1342. doi: 10.1038/nm.1891. [DOI] [PubMed] [Google Scholar]
- 68.DeLano WL. PyMOL, Version 0.95. San Carlos, CA: Delano Scientific LLS; 2004. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.