Abstract
In hippocampal neurons, the scaffold protein AKAP79 recruits the phosphatase calcineurin to L-type Ca2+ channels, and couples Ca2+ influx to activation of calcineurin and of its substrate, the transcription factor NFAT. Here we show that an IAIIIT anchoring site in human AKAP79 binds the same surface of calcineurin as the PxIxIT recognition peptide of NFAT, albeit more strongly. A modest decrease in calcineurin-AKAP affinity due to an altered anchoring sequence is compatible with NFAT activation, whereas a further decrease impairs activation. Counterintuitively, increasing calcineurin-AKAP affinity increases recruitment of calcineurin to the scaffold but impairs NFAT activation, probably due both to slower release of active calcineurin from the scaffold and to sequestration of active calcineurin by “decoy” AKAP sites. We propose that calcineurin-AKAP79 scaffolding promotes NFAT signaling by balancing strong recruitment of calcineurin with its efficient release to communicate with NFAT.
Enzyme-scaffold interactions play a critical role in directing intracellular signaling. Scaffolded complexes have been described in multiple signaling pathways and serve as platforms for signal integration and propagation by localizing enzymes in the vicinity of their substrates and regulatory proteins1–3. A notable example is the A-kinase anchoring protein AKAP79, which juxtaposes the kinase PKA with the phosphatase calcineurin and other proteins, and targets them to a variety of locations in the plasma membrane4,5.
The protein serine/threonine phosphatase calcineurin (CN; also known as PP2B) is activated when cytoplasmic Ca2+ concentrations are elevated, and regulates a variety of downstream processes in cells. CN is important for cardiac development and pathophysiology, for nervous system development, and for some of the plastic changes in neurons considered to underlie learning and memory6–10. Among the best-characterized substrates of CN are the NFAT transcription factors, NFAT1–NFAT4. CN-NFAT signaling has been intensely studied for its role in immune cell activation and as the target of the immunosuppressive drugs cyclosporin A and FK506 (refs. 11,12). These studies led to the identification of a recognition sequence for CN that is shared by many CN substrates, and that was first described as having the consensus sequence PxIxIT13. We identified a high-affinity version of the PxIxIT sequence, PVIVIT, by selection from a randomized peptide library14, and used it to define the structural basis of substrate recognition by CN15,16; also see 17. CN recognition sites display considerable natural variation in their sequences and their affinities for CN10,16. The recognition sites of NFAT proteins (PRIEIT, PSIRIT and PSIQIT) display an intermediate Kd of ~25 µM for CN, whereas the sequences PQIIIS in human TRESK and PVIAVN in yeast Hph1 bind CN with Kds of 5 µM and 250 µM, respectively. The strength of individual CN-substrate interactions is an important parameter for intracellular signaling and, in particular, increasing the strength of a CN-substrate interaction above that of the wildtype proteins can lead to disordered signaling10,14,18–20.
Like many other signaling enzymes, CN is targeted to specific intracellular locations by interactions with scaffold proteins. The scaffold protein AKAP79 is of particular interest, since it balances the opposing effects of CN and PKA on neuronal voltage-gated L-type Ca2+ channels21 and coordinates the activities of CN, PKA, and PKC on a number of other channels and receptors22–28. In rat hippocampal neurons, NFAT signaling is initiated by the local elevation of Ca2+ concentration near L-type Ca2+ channels21,29. RNAi-mediated depletion of endogenous AKAP150, the rat orthologue of human AKAP79, eliminates NFAT activation in neurons, which however can be rescued by expression of human AKAP79 (ref. 21). A conserved CN-binding site has been mapped in AKAP79 (Fig. 1a), and deletion of this anchoring sequence eliminates the ability of AKAP79 to rescue NFAT activation, despite abundant CN elsewhere in the cytoplasm, emphasizing the local nature of CN-NFAT signaling and the importance of CN scaffolding by AKAP79 in these cells21,30.
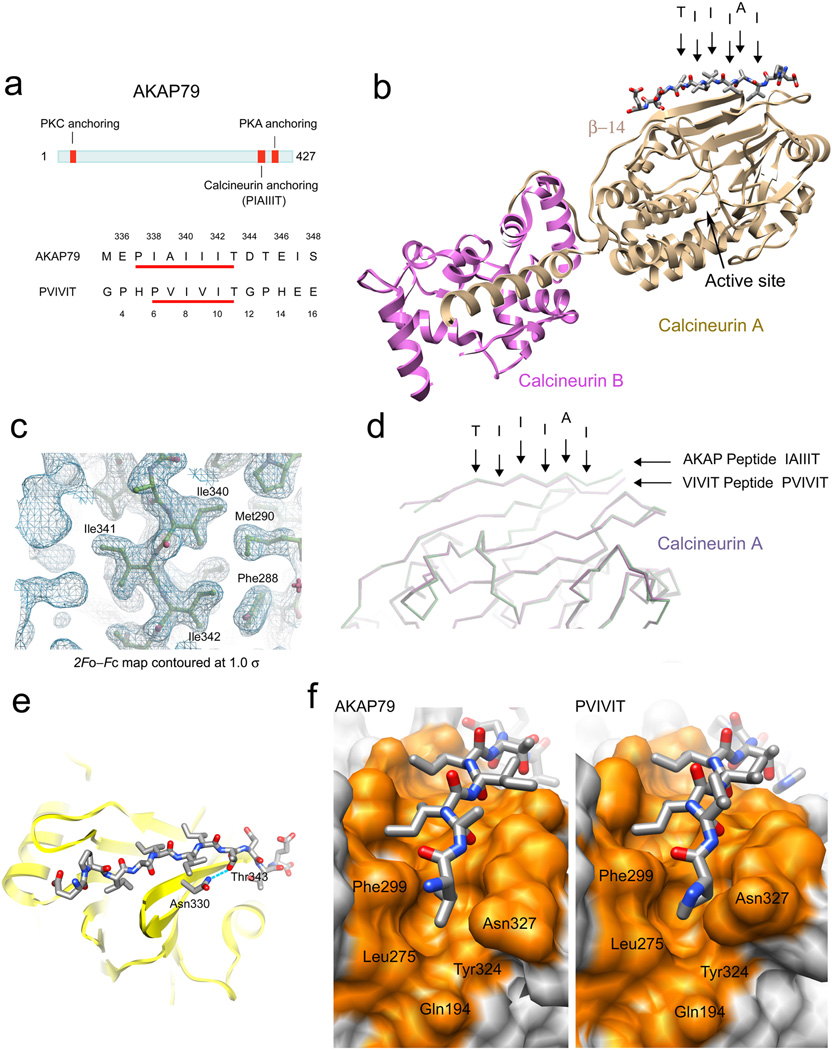
Figure 1.
Crystal structure of calcineurin in complex with AKAP79 peptide. (a) Location of the PxIxIT-like CN anchoring site within human AKAP79 protein and sequence comparison to the PVIVIT peptide. The numbering of the residues in AKAP79 and in PVIVIT peptide is shown above and below the sequences, respectively. (b) Overall structure of CN in complex with AKAP79 peptide. The key residues comprising the core CN-binding motif are labeled with arrowheads. (c) A 2Fo–Fc map contoured at 1σ showing the electron density surrounding the isoleucine stretch, Ile340–Ile342, of the IAIIIT CN-anchoring motif of AKAP79. (d) Superposition of the CN–AKAP79 peptide structure onto the previously solved CN–PVIVIT structure. The core CN-binding residues of AKAP79 are labeled as in b. (e) The hydrogen bond between Thr343 of AKAP79 and Asn330 of CN A recapitulates a critical contact13,15 in the CN–PVIVIT structure. (f) Side-by-side comparison of the CN–AKAP79 structure (left) and CN–PVIVIT structure (right) viewed from the “proline pocket” defined in the CN–PVIVIT structure. CN is shown in surface representation with residues in direct contact with the peptides colored orange, and the peptides are displayed in stick representation. The figures were prepared using Chimera38 and Coot39.
In order to gain further insight into the role of this CN-scaffold interaction in intracellular signaling, we have investigated its structural basis, the affinity and kinetics of CN-AKAP79 binding, and the effects of relatively small perturbations of the interaction on NFAT transcriptional signaling in cells. The crystal structure of a CN–AKAP79 complex shows that CN engages an atypical PxIxIT site in AKAP79. The functional studies lead to three principal conclusions: First, despite the absence of a consensus proline residue, the AKAP79 site binds CN more tightly than any of the recognition peptides identified so far in substrates; second, the site allows rapid dissociation of CN for physiological signaling; and, third, only a rather narrow window of CN-AKAP79 affinities is compatible with normal NFAT signaling. We propose that the atypical PxIxIT site of AKAP79 meets two independent requirements of intracellular signaling, in that it recruits CN efficiently to the vicinity of L-type Ca2+ channels for activation by Ca2+ entering the cell, and yet allows the disengagement of CN to couple channel activation with downstream signaling events.
RESULTS
X-ray crystallographic structure of the CN–AKAP79 complex
To define the structural basis for CN binding to AKAP79, we crystallized the complex of CN with an AKAP79 peptide and solved its structure at 2.0 Å resolution (Table 1 and Fig 1b; PDB entry 3LL8). Two CN A–CN B heterodimers and one AKAP peptide are contained in the asymmetric unit. As in the CN–PVIVIT structure16 that describes consensus CN-substrate recognition, the AKAP79 PIAIIIT peptide is in closer contact with one of the two CN heterodimers in the asymmetric unit.
Table 1.
Data collection and refinement statistics (molecular replacement)
| CNA–CNB–AKAP79 complex |
|
|---|---|
| Data collection | |
| Space group | P212121 |
| Cell dimensions | |
| a, b, c (Å) | 86.28, 89.70, 158.91 |
| α, β, γ (°) | 90.00, 90.00, 90.00 |
| Resolution (Å) | 50.0–2.00 (2.07–2.00) * |
| Rsym or Rmerge | 7.1 (50.3) |
| I / σI | 16.2 (1.9) |
| Completeness (%) | 95.7 (91.5) |
| Redundancy | 3.1 (2.9) |
| Refinement | |
| Resolution (Å) | 50.0–2.00 |
| No. reflections | 75,502 |
| Rwork / Rfree | 17.8 / 22.8 |
| No. atoms | |
| Protein | 8363 |
| Ligand/ion | 22 |
| Water | 583 |
| B-factors | |
| Protein | 38.6 |
| Ligand/ion | 33.4 |
| Water | 49.7 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.017 |
| Bond angles (°) | 1.6 |
Values in parentheses are for highest-resolution shell.
The electron density in the CN–AKAP complex is well defined throughout the core of the AKAP recognition peptide and the apposed surface of CN (Fig. 1c). The AKAP peptide assumes an extended configuration as a β strand aligned along β-strand 14 of CN A on a surface distant from the catalytic site (Fig. 1b–d). Seven backbone H-bonds serve to position the peptide strand between AKAP residues Ala339 and Thr345 as part of an extended β sheet.
In the core region of CN-AKAP peptide contact, Ile340 and Ile342 of AKAP make contacts on a nonpolar surface of CN in the trough between β-strand 11 and β-strand 14 (Fig. 1c), and Thr343 forms an H-bond with Asn330 of CN A (Fig. 1e). These contacts are identical to those of the corresponding Ile and Thr side chains in the CN–PVIVIT structure. Strikingly, however, Ile338 of AKAP takes the position of Pro6 in the CN–PVIVIT complex (Fig. 1f). The Ile338 sidechain is buried in the pocket to approximately the same depth as Pro6 in the CN–PVIVIT complex, resulting in a slight displacement of the backbone at Ile338 away from the CN surface compared to PVIVIT (Fig. 1d). In the N terminal flanking region of AKAP peptide, the electron density of Glu336 and Pro337 is poorly defined, and does not contact CN A. The C-terminal flanking region of AKAP peptide makes van der Waals contacts on the surface of CN, and the H-bond network anchoring the flanking region of the CN–PVIVIT complex is supplanted by Thr343-Asn333 and Asp344-Lys318 contacts. There is no sequence conservation among PxIxIT sites from different CN-binding proteins in the segments flanking the core recognition peptide, and the idiosyncratic contacts of AKAP are consistent with the view16 that PxIxIT flanking regions arrange themselves in a low-energy configuration, but may not contribute appreciably to ΔGbinding.
Solution studies of the CN–AKAP79 complex
We sought to complement the picture from X-ray crystallography by experimental characterization of CN-AKAP79 binding in solution. Size exclusion chromatography coupled with multi-angle light scattering (SEC-MALS) showed a shift in migration of the CN heterodimer peak upon formation of a complex with AKAP79(333–408) and an increase in mass of ~8 kDa, corresponding to a 1:1 complex of CN with AKAP79(333–408) (Fig. 2a).
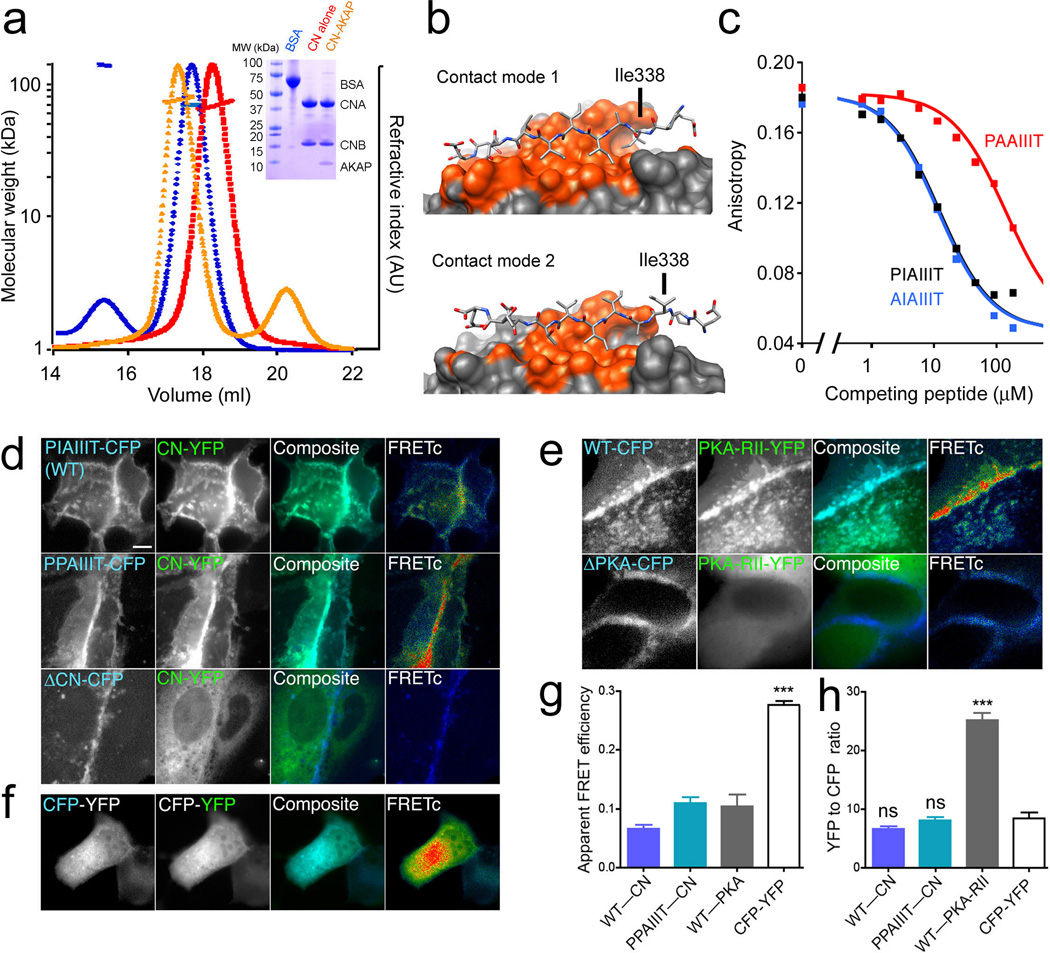
Figure 2.
The CN–AKAP79 complex in solution. (a) SEC-MALS analysis of CN alone (red), CN–AKAP complex (orange), and BSA standard (blue). Inset, corresponding SDS-polyacrylamide gel. (b) Alternative 1:1 complexes based on the two CN molecules in the crystal asymmetric unit. The figures were prepared using Chimera38. (c) Competition of the indicated GST-AKAP79(333–348) fusion proteins with fluorescent PVIVIT peptide for binding to CN. The estimated Kis are AIAIIIT, 3.7 µM; PIAIIIT, 3.9 µM; PAAIIIT, 47 µM. Total competitor concentration (not free concentration) is plotted. Data shown are representative of three experiments. (d) and (e) Images of living MDCK cells showing membrane colocalization (turquoise in composite panels) and corrected CFP donor to YFP acceptor FRET gated to the CFP channel (FRETc; pseudocolor, blue=no FRET to red=high FRET) for the indicated AKAP79-CFP proteins (blue) and (d) CNA-YFP (green) or (e) PKA-RII-YFP (green). Scale bar=5 µm. (f) CFP, YFP, and FRETc images as above for a linked CFP-YFP construct. (g) Measurements of apparent FRET efficiency, used to correct CFP fluorescence intensity for quenching. (h) Corrected YFP/CFP fluorescence intensity ratios for experiments in panels d–f. Statistical comparisons were by one-way ANOVA with a Bonferroni post-hoc test, ***p<0.001 and nsp>0.05 compared to the linked CFP-YFP standard (n=10–41 cells).
The crystal structure offers two possible modes of contact in a 1:1 complex, and in both the AKAP79 peptide occupies roughly the same surface of CN (Fig. 2b). The peptide is aligned in each case along β strand 14 of CN, and is stabilized by backbone H-bonds to β strand 14. Contact mode 1 buries 1332 Å2 of surface and makes seven backbone H-bonds, whereas contact mode 2 buries 820 Å2 and makes four backbone H-bonds. The two contact modes differ most prominently in whether the Ile338 side chain is in contact with CN (Fig. 2b). In the CN–AKAP peptide structure already described, Ile338 is buried in the “proline pocket” of the PxIxIT-binding surface, whereas in the alternative complex it extends away from CN. An I338A substitution substantially reduced the affinity of the CN-AKAP peptide interaction in a competitive binding assay (Fig. 2c), indicating that Ile338 contributes to CN-AKAP binding, consistent with contact mode 1. Finally, although neither CN-peptide contact in the crystal structure involves Pro337, the substantial energetic contribution of the Pro6 contact in the CN–PVIVIT complex16 led us to introduce a P337A substitution to rule out alternative modes of binding that bury Pro337. The P337A mutation had no impact on CN-AKAP affinity in the competitive binding assay (Fig. 2c). The combined SEC-MALS and competitive binding data establish that CN binding at the IAIIIT site, in solution, gives a 1:1 complex, and support the structural model depicted in Figures 1b and 1f.
Stoichiometry of the CN–AKAP79 complex in living cells
In order to determine whether CN and full-length AKAP79 also form a 1:1 complex in living cells, we compared YFP/CFP fluorescence intensity ratios measured in MDCK cells expressing CN-YFP and AKAP79-CFP to the ratios measured in cells expressing a reference CFP-YFP protein covalently linking CFP to YFP or in cells expressing RII-YFP and AKAP79-CFP (Fig. 2d–h). As shown in our previous work21,31,32, coexpression of CN-YFP (Fig. 2d) or PKA-RII-YFP (Fig. 2e) with wildtype AKAP79-CFP resulted in colocalization of the YFP-labeled proteins with AKAP79 at the plasma membrane, and in FRET between CFP and YFP. Both the plasma membrane colocalization of CFP and YFP and the FRET signals were eliminated by deletion of the respective anchoring sites in AKAP79 (ΔCN, Fig. 2d; ΔPKA, Fig. 2e)21. In the cases of CN and RII, we applied a mask to the images in order to determine the YFP/CFP ratio specifically at membrane sites of colocalization and FRET. After correcting CFP fluorescence intensity for the quenching due to FRET, the YFP/CFP ratio of the CN–wildtype AKAP79 pair was 6.6 ± 0.5, not significantly different from the YFP/CFP ratio of 8.5 ± 1.0 observed for 1:1 linked CFP-YFP (Fig. 2h). Replacing wildtype AKAP79 with the I338P variant discussed below, to ensure high binding site occupancy, increased the YFP/CFP ratio only to 8.1 ± 0.6, still not significantly different from that of linked CFP-YFP. The YFP/CFP ratio of the RII-wildtype AKAP pair (25.2 ± 1.2) was, as expected, significantly greater than that of the 1:1 construct, in fact exceeding the reference value by more than a factor of two. One explanation for the excess YFP signal is that a minor fraction of RII-YFP is free or in complexes not involving AKAP79-CFP. There is no specific evidence whether or not there is such an excess YFP signal in the CN experiments, but any CN-YFP not complexed with AKAP79-CFP would lead to an overestimate, rather than an underestimate, of CN in the CN–AKAP79 complex. The YFP/CFP ratio measurements are most consistent with the formation of a 1:1 CN–AKAP79 complex and a 2:1 RII–AKAP79 complex in cells, and are in agreement with our SEC-MALS data for the CN–AKAP79 complex (Fig. 2a) and the known structure of the RII–AKAP79 complex33.
CN binding to and dissociation from the IAIIIT site
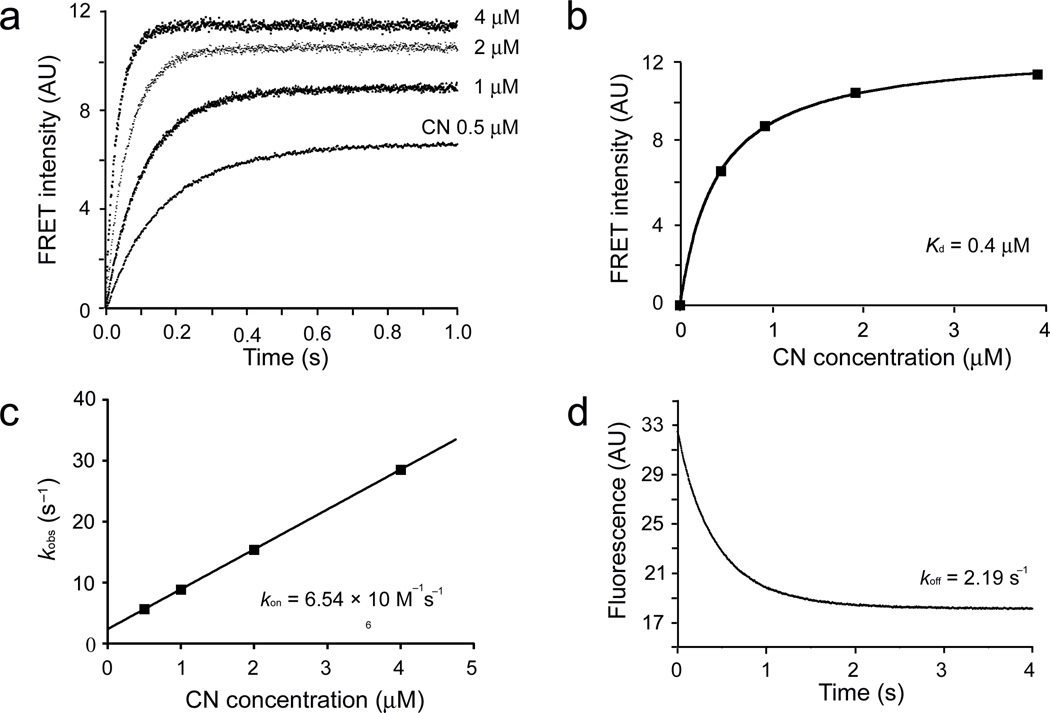
The IAIIIT anchoring site is essential for NFAT signaling in hippocampal neurons because it underlies recruitment of CN to AKAP79 complexes adjacent to the L-type Ca2+ channel21. Recruitment of CN to AKAP-scaffolded complexes in neurons will depend on the concentration of free CN and on the Kd of the binding interaction. To obtain a direct estimate of CN-AKAP79 Kd, we made quantitative measurements of FRET between an ATTO425-AKAP79 (333–408) donor and rhodamine-CN acceptor (Supplementary Fig. 1a, 1b). Titration with increasing concentrations of CN yielded a binding curve at room temperature that could be fitted with Kd 0.4 µM (Fig. 3a, 3b; Supplementary Fig. 1c, 1d), supporting the Kd estimated independently from kinetic measurements, ~0.3 µM (Fig. 3c, 3d). We conclude that interaction at the IAIIIT site can effectively recruit CN in neurons if the free CN concentration is in the high hundreds of nanomolar to low micromolar range.
Figure 3.
Equilibrium and kinetic measurements of CN binding to AKAP79. (a) CN binding to AKAP79(333–408) was monitored by stopped-flow FRET measurements. The observed time course of binding at 22°C is shown for four CN concentrations. (b) A plot of plateau FRET intensities against CN concentrations. (c) The association rate constant kon estimated from the data in panel a based on a model of reversible binding to a single class of sites. (d) Dissociation of the CN–AKAP79(333–408) complex at 22°C. In this AKAP79(333–408) protein, the RIIα-anchoring site was replaced by the AKAP-IS sequence33,40. Data shown are representative of two experiments.
The rapid dissociation of CN–AKAP79 peptide complex is particularly relevant to the transmission of intracellular signals. Even though AKAP79 and NFAT utilize the same surface to dock on CN, recruitment of CN to AKAP79 need not be in conflict with productive signaling because of the rapid equilibration between free CN and AKAP-bound CN. Experiments reported below establish that dissociation at physiological temperature is even more rapid than in the experiment of Figure 3.
Scaffold interaction and CN-NFAT signaling
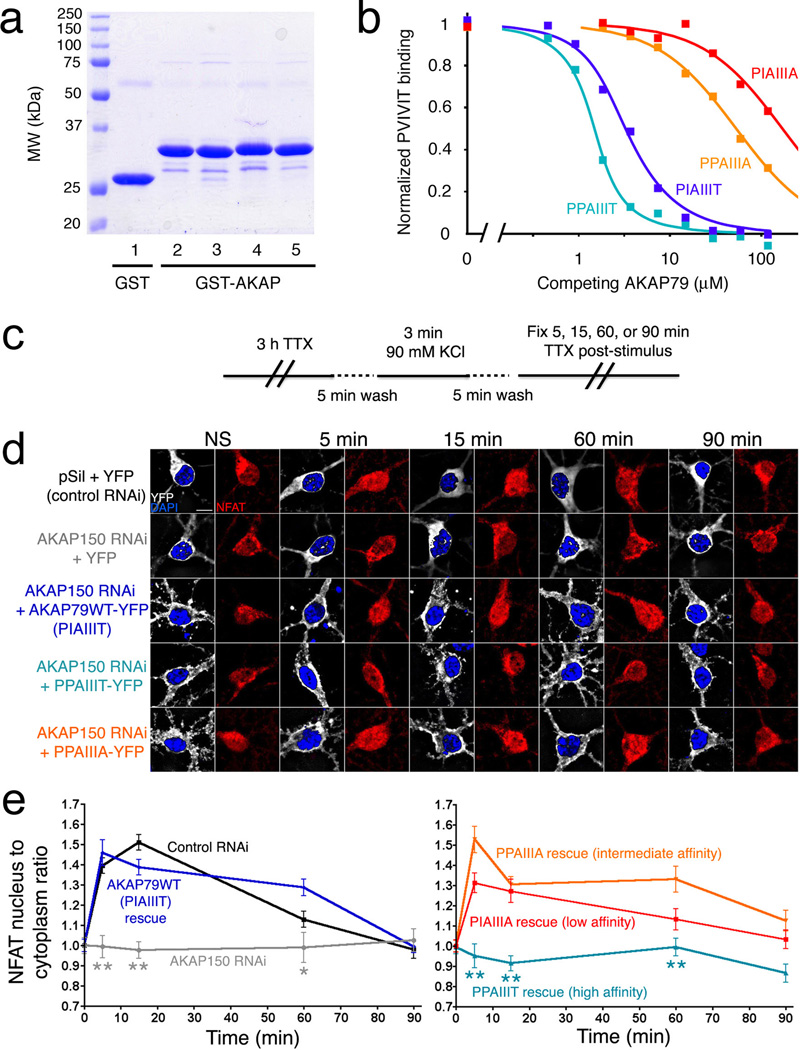
Knowledge of the CN–AKAP79 structure opens an opportunity to investigate the effect of systematically varying the affinity of this enzyme-scaffold interaction. Building on our earlier studies of the CN–PVIVIT structure and the rules that connect changes in the sequence of CN recognition sites in substrates to variations in affinity16, we designed a small panel of AKAP79 anchoring peptides whose affinities for CN span a wide range. The peptides contained the replacements I338P, T343A, or both. Wildtype and variant AKAP proteins are referred to by 7mer sequences to facilitate comparison with variant sequences of the variations introduced— for instance, a peptide containing the wildtype anchoring sequence is designated PIAIIIT, even though Pro337 is not part of the contact interface defined in the crystal structure. Competitive binding experiments in which these unlabeled AKAP variants, as GST fusion proteins, were used to displace labeled PVIVIT demonstrated that the relative affinities were PPAIIIT > PIAIIIT > PPAIIIA > PIAIIIA, covering more than a 100-fold range in Kd (Fig. 4a, 4b). Notably, the PPAIIIT variant, in which Ile338 was substituted with Pro, bound ~4-fold more strongly to CN than wildtype AKAP79.
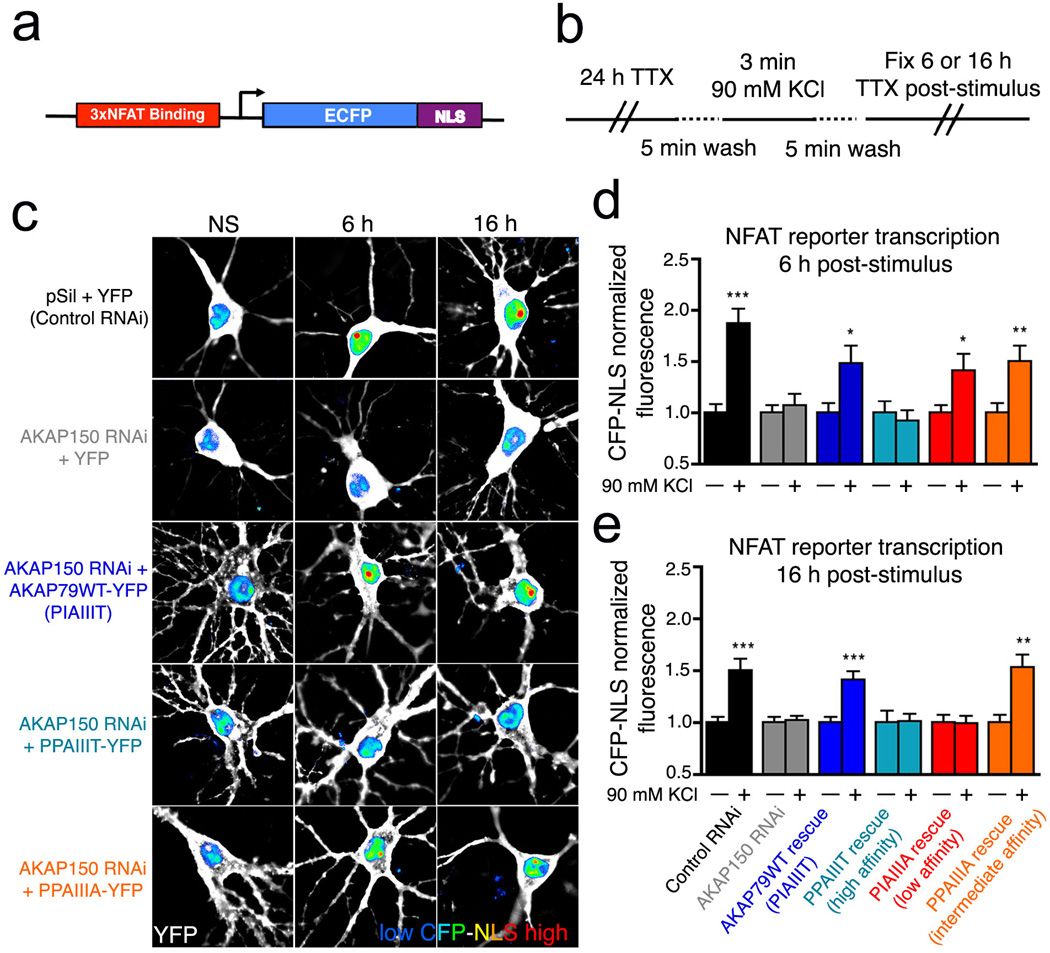
Figure 4.
A high-affinity AKAP79 variant, PPAIIIT, does not support NFAT nuclear translocation in hippocampal neurons. (a) Recombinant GST, GST-tagged wildtype AKAP79(333–408) (PIAIIIT, lane 2), and the variants PIAIIIA, PPAIIIA, and PPAIIIT (lanes 3–5) were analyzed by SDS-polyacrylamide gel electrophoresis and staining with Coomassie Brilliant Blue. (b) Kis estimated in a competitive binding assay are PPAIIIT, 0.08 µM; wildtype, 0.36 µM; PPAIIIA, 12 µM; and PIAIIIA, 39 µM. Total competitor concentration (not free concentration) is plotted. Data shown are representative of three experiments. (c) KCl stimulus protocol previously shown to activate L-type Ca2+ channel signaling through CN in hippocampal neurons21,29. (d) Summed intensity projection images of neuronal cell bodies and proximal dendrites in nonstimulated (NS) cultures and in cultures fixed at the indicated times after KCl stimulation. Transfection with control RNAi plasmid (pSil), AKAP150 RNAi plasmid, and RNAi-resistant expression plasmids is indicated. The paired images show YFP or AKAP-YFP (white), DAPI-stained nuclei (blue), and endogenous NFAT (red). (e) Time course of NFAT nuclear import after KCl stimulation, from experiments as in panel d, quantified as nucleus-to-cytoplasm mean fluorescence intensity ratios21. Each point represents n=12–25 neurons, and in each case the data have been normalized to the value for nonstimulated cultures (t = 0 min). Statistical comparisons were by one-way ANOVA with a Bonferroni post-hoc test, *p<0.05 and **p<0.01 compared to AKAP79WT rescue.
To explore the biological effect of varying the affinity of CN-scaffold interactions, we expressed full-length AKAP79 proteins with either the wildtype PIAIIIT anchoring sequence or one of the sequences PPAIIIT, PPAIIIA, and PIAIIIA in rat hippocampal neurons. The neurons were depolarized by elevating KCl concentration, using a stimulus protocol that activates L-type Ca2+ channel signaling through CN and triggers NFAT nuclear translocation and NFAT-dependent gene expression21,29. Endogenous NFAT3 (also known as NFATc4) was localized immunocytochemically. As shown previously, RNAi-resistant wildtype human AKAP79 rescued NFAT nuclear translocation in neurons after depletion of endogenous AKAP150 by RNAi (Fig. 4c–e). AKAP79 bearing the lower-affinity sequences also rescued NFAT nuclear translocation, AKAP79 with PPAIIIA (intermediate affinity) as efficiently as wildtype AKAP79, and AKAP79 with PIAIIIA (lowest affinity) somewhat less efficiently (Fig. 4d, 4e). Previous work has shown that AKAP79 lacking the CN-anchoring site does not rescue CN-NFAT signaling21. Surprisingly, AKAP79 bearing the higher-affinity sequence PPAIIIT was also completely unable to rescue signaling (Fig. 4d, 4e).
The differences in NFAT nuclear localization have practical consequences for NFAT-dependent transcription (Fig. 5). Endogenous AKAP150 in rat hippocampal neurons was depleted by RNAi as for the nuclear import experiments, the cells were reconstituted with human AKAP79 and its variants, and NFAT-dependent transcription was monitored as expression of a 3×NFAT-ECFP reporter. Wildtype AKAP79 and AKAP79 with PPAIIIA (intermediate affinity) were equivalent in rescuing transcription (Fig. 5c–e). Consistent with its effect on NFAT nuclear translocation, the low affinity PIAIIIA AKAP was partially functional, as evidenced by rescue of NFAT transcription only at an early time (Fig. 5d) but not a later time after stimulation (Fig. 5e). PPAIIIT was again completely unable to rescue NFAT signaling (Fig. 5c–e).
Figure 5.
The high-affinity AKAP79 variant PPAIIIT does not couple Ca2+ influx to NFAT-dependent transcription in hippocampal neurons. (a) Diagram of the 3×NFAT-AP1-CFP-NLS transcriptional reporter construct used for single-cell imaging of NFAT-dependent transcription. (b) Modified KCl stimulation protocol used to assay L-type Ca2+ channel activation of NFAT-dependent reporter gene transcription in hippocampal neurons. (c) Summed intensity projection images of neuronal cell bodies and proximal dendrites in nonstimulated (NS) cultures and in cultures fixed at the indicated times after KCl stimulation for neurons transfected with the 3×NFAT-AP1-CFP-NLS reporter along with the indicated RNAi, YFP, and AKAP79-YFP constructs as in Figure 4b. YFP fluorescence is in white and nuclear-localized CFP fluorescence is in pseudocolor with a relative scale from blue (low intensity) to red (high intensity). (d) and (e) Quantification of CFP reporter gene expression (d) 6 hr or (e) 16 hr after KCl stimulation (+) from experiments as in panel c, normalized to the nonstimulated condition (−). *p<0.05, **p<0.01, and ***p<0.001 by Student’s t-test compared to the respective nonstimulated condition (n=11–50 neurons).
Altered CN interactions with the PPAIIIT site
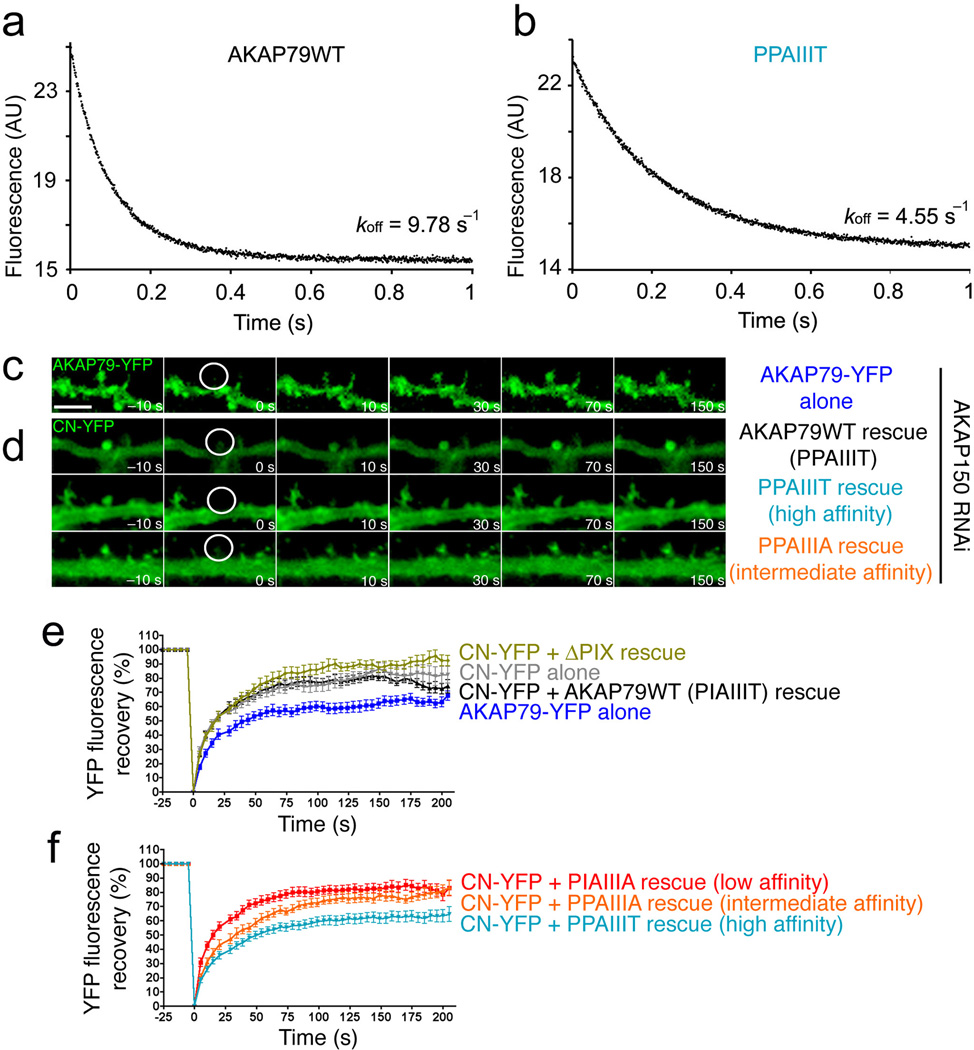
Among the possible reasons for the failure of PPAIIIT to support signaling, we first considered availability of CN to bind NFAT after CN activation at the scaffold complex associated with the L-type Ca2+ channel. We compared dissociation of CN from its complexes with wildtype AKAP79(333–408) (PIAIIIT) and with the PPAIIIT variant in stopped-flow FRET experiments at room temperature and at 36 °C (Fig. 6a, 6b, and Supplementary Fig. 2). Preformed CN–AKAP79 complex was mixed with an excess of unlabelled PVIVIT peptide to prevent rebinding of CN to AKAP79, and complex dissociation was monitored as the decrease in the FRET signal. The dissociation of CN from wildtype AKAP79 was rapid at both temperatures, with the mean dissociation rate at 36 °C, 9.96 s−1 (range 8.84–11.84 s−1, n = 4), corresponding to a half-time of ~70 ms (Fig. 6a). The implication is that CN can rapidly expose its NFAT-binding surface even if that surface is initially engaged in a physiological complex with wildtype AKAP79. Dissociation of the higher-affinity CN–PPAIIIT AKAP79 complex was appreciably slower, with mean dissociation rate 4.24 s−1 (range 3.90–4.55 s−1, n = 4) and corresponding half-time ~163 ms (Fig. 6b), consistent with a modest reduction in availability of the docking surface to interact with NFAT after CN activation in cells.
Figure 6.
The high-affinity AKAP79 PPAIIIT variant decreases the rate of CN dissociation in vitro and reduces the mobility of CN in dendritic spines. (a) and (b) Dissociation of CN from (a) wildtype AKAP79(333–408) and (b) its I338P variant at 36 °C. (c) Time-lapse images of wildtype AKAP79-YFP fluorescence recovery in single dendritic spines of rat hippocampal neurons in culture at the indicated times after photobleaching. A prebleach image (t = −10 s) is shown for comparison. (d) YFP fluorescence recovery in neurons cotransfected with CN A-YFP (CN-YFP) and CFP-tagged wildtype AKAP79 (AKAP79WT) or CFP-tagged AKAP79 variants (PPAIIIT, PPAIIIA) as indicated. CFP fluorescence is not shown. (e) and (f) Percent YFP fluorescence recovery plotted against time for the FRAP experiments illustrated in panels c and d and for similar experiments with the other AKAP79 variants PIAIIIA and ΔPIX (deletion of PIAIIIT). In the experiment with CN-YFP alone, transfection with the RNAi plasmid was omitted. Maximal percent recovery (mobile fraction) values stated in the text were calculated from the curves in panels e and f by fitting a single exponential function. Statistical p values stated in the text were determined by one-way ANOVA with a Bonferonni post-hoc test (n=10–28 neurons).
We undertook a second line of experiments to gain insight into the anchoring of CN by AKAP79 in living cells. We examined the dynamics, and more specifically the mobile fraction, of CN-YFP in rat hippocampal neuron dendritic spines using fluorescence recovery after photobleaching (FRAP). Endogenous AKAP150 was depleted by RNAi and replaced with wildtype or mutant AKAP79. Coexpression of CN-YFP with the ΔPIX variant (deletion of PIAIIIT), which cannot bind CN, led to a significant increase in the mobile fraction of CN (89.5 ± 0.7%, p<0.001), compared to rescue with wildtype AKAP79 (77.5 ± 0.6%) or the intermediate affinity PPAIIIA variant (77.1 ± 0.8%) (Fig. 6c–f). Coexpression with the low-affinity PIAIIIA variant also gave a modest, but still significant, increase in mobile fraction (81.3 ± 0.6%, p<0.01) consistent with reduced anchoring of CN to this variant (Fig. 6f). In contrast, AKAP with the high-affinity site PPAIIIT decreased the mobile fraction of CN (61.6 ± 0.7%, p<0.001 compared to wildtype) (Fig. 6d, 6f) to a level that was very similar to that of the less mobile AKAP79 itself (61.8 ± 0.6%) (Fig. 6c, 6e). The variant AKAP79 proteins were all expressed at the same levels across experiments, as was CN-YFP, and there was no change in the mobile fraction of AKAP79 with any variant tested (Supplementary Fig. 3). These results indicate that CN-AKAP binding contributes perceptibly to the immobilization of CN in dendritic spines, and that an increase in CN-AKAP affinity that blocks NFAT signaling is reflected in an increase in CN anchored by AKAP in the spine. Note that the CN immobilization detected in cells extends to a much longer time scale than CN-AKAP79 dissociation in vitro and thus probes a different aspect of CN interaction with PPAIIIT, as discussed below.
DISCUSSION
Our studies provide strong evidence that the IAIIIT recognition peptide makes a principal contribution to CN-AKAP79 anchoring in vitro and in cells. In fact, the sequence IAIIIT exhibits the highest affinity so far observed for a natural CN recognition site, with a measured Kd less than 1 µM. Competitive binding assays with the PAAIIIT, PIAIIIA and AIAIIIT variant peptides and larger AKAP fragments document the importance of the Ile338 and Thr343 contacts for CN-AKAP79 binding in solution, and support the structural model presented here as the basis for CN-AKAP79 binding. Comparison of CN binding to the 14mer peptide and to AKAP79(333–408) (Figures 2c and 4b) suggests a small additional energetic contribution from AKAP79 residues outside the IAIIIT site, but the essential interaction is with the core anchoring site. The evidence that CN forms a 1:1 complex with the primary anchoring site does not rule out a 2:1 complex of CN with AKAP79 (ref. 34) if the complex is stabilized by other interactions.
The involvement of AKAP79 scaffolding in CN-NFAT signaling in hippocampal neurons poses competing requirements: CN must use its PxIxIT-interacting surface to bind AKAP79 adjacent to L-type Ca2+ channels, but the same surface must be available to recognize NFAT. The level of CN in brain is high, ~1% of soluble protein35, translating to a total CN concentration in the cell cytoplasm of ~10 µM. Allowing for CN bound to partner proteins, a plausible free CN concentration in neurons is in the high hundreds of nanomolar to low micromolar range, in which case the majority of AKAP79 sites in neuronal cells will be occupied by CN at any given time. Nevertheless the rapid dissociation of the CN–AKAP79 peptide complex in vitro indicates that the docking surface on CN is in principle available for efficient interaction with NFAT and other cellular substrates.
Only AKAP79 proteins with CN-scaffold affinities in a relatively narrow window, 0.4 to 12 µM, were as effective as wildtype AKAP79 in supporting CN-NFAT signaling. The impaired signaling seen with low-affinity AKAP79 (PIAIIIA) and the loss of signaling with AKAP lacking the CN anchoring site are readily explained by a failure to recruit sufficient CN to scaffold complexes associated with L-type Ca2+ channels (Fig. 7). Although the intermediate-affinity AKAP79 (PPAIIIA) supports NFAT signaling, it may not be biologically interchangeable with wildtype AKAP79: Nuclear import of NFAT is a relatively insensitive measure, which requires an experimental design with a fairly strong Ca2+ signal, and the same strong Ca2+ signal was used for transcriptional assays in order to drive measurable transcription with a 3-minute stimulus. The window for NFAT signaling may well be narrower at low levels of physiological stimulation.
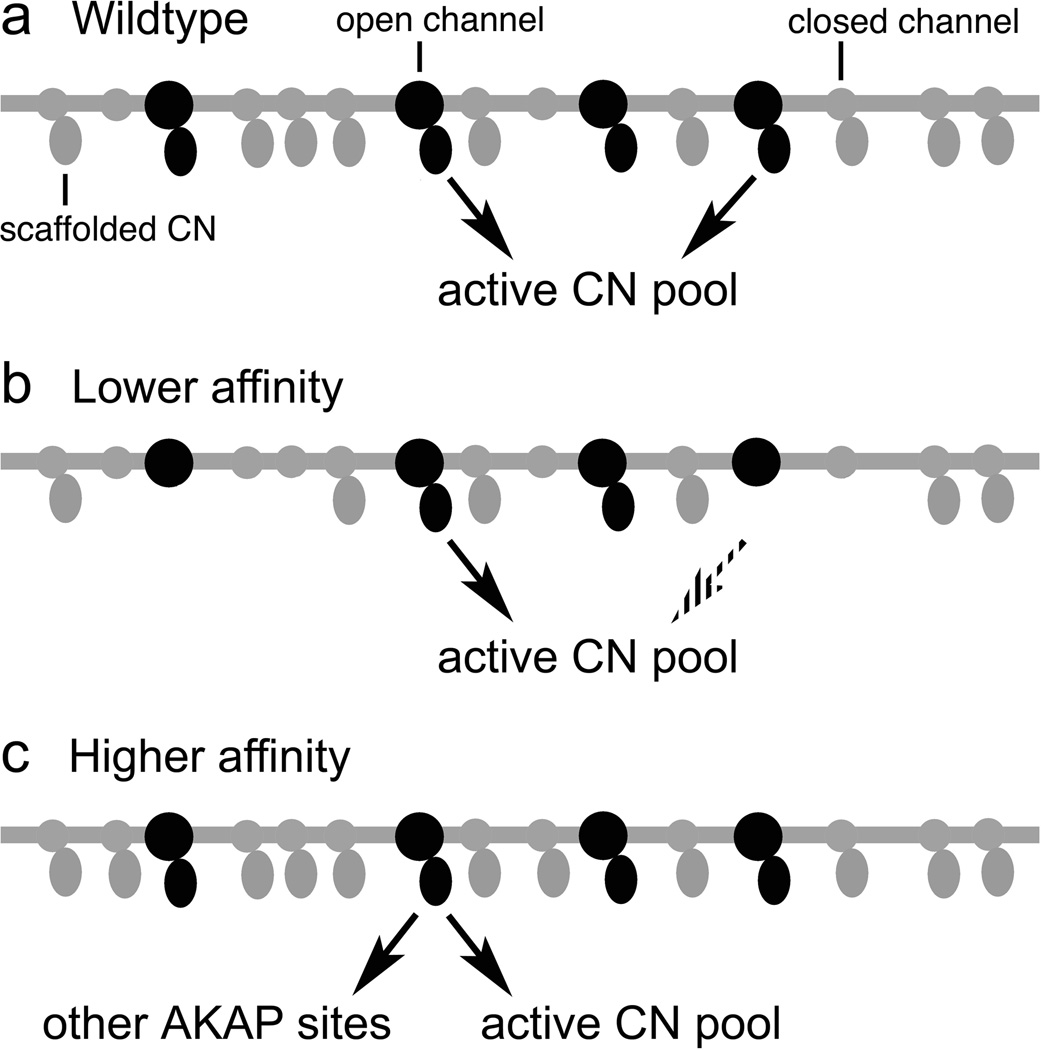
Figure 7.
Model explaining the effects of altered CN-AKAP79 anchoring interactions. (a) With wildtype AKAP79, most CN-anchoring sites at the L-type Ca2+ channel are occupied. Both inactive and active CN are continually released from the scaffold sites and replaced, so that any activated CN rapidly becomes available to interact with NFAT. Release of inactive CN is not depicted. (b) Low-affinity anchoring sites are partially occupied. CN release from the scaffold sites is efficient, but only occupied sites contribute active CN at any instant in time, and thus the rate of release of active CN is reduced compared to wildtype AKAP79. (c) High-affinity anchoring sites are fully occupied. The rate of release of CN is somewhat reduced and, at any given time, a larger fraction of the active CN is in complex with other AKAP79 scaffold sites than in the case of wildtype AKAP79.
Unexpectedly, despite its modestly increased affinity for CN, AKAP79 with the PPAIIIT sequence impaired NFAT signaling rather than further promoting it. A partial explanation is that the higher affinity decreased the release of active CN from AKAP79 into the cytoplasm (Fig. 7). Similarly, the PRIEIT>PRIEIA substitution in NFAT1 causes only a 10-fold shift in Kd but eliminates CN-NFAT signaling in T cells13. Indeed, simplified mathematical models predict a very sharp dependence of NFAT activation on CN activity36. Nevertheless, the fact that the 2- to 4-fold shifts in CN-AKAP79 affinity and dissociation rate produced by the I338P substitution are associated with a complete loss of CN-NFAT signaling suggests that a second process may be involved.
The FRAP experiments provide a strong indication of the nature of this second process. An increased fraction of CN is immobilized by binding to AKAP79 with the PPAIIIT anchoring site, and this measure may in fact understate the sequestration of CN by altered AKAP, because over half of the AKAP79 in the dendritic spines is itself mobile on the time scale examined. Thus we consider it likely that the decreased release of active CN into the cytoplasm is compounded by CN binding to “decoy” AKAP79 sites at the L-type Ca2+ channel and elsewhere, which act in competition with the NFAT substrate (Fig. 7). A parallel situation has been observed in yeast, where the engineered high-affinity substrate Crz1(PVIVIT) diverts CN from other substrates whose dephosphorylation is necessary for the response to alkaline pH18.
Our FRAP experiments indicate that a fraction of CN in hippocampal neurons is immobilized by AKAP79 on a considerably longer time scale than that required for dissociation of CN–AKAP complexes in vitro. These results imply that at least some CN–AKAP complexes are stabilized by additional interactions in cells, such as interactions through a subsidiary CN-binding site in AKAP (ref. 34), interactions with other proteins known to associate with AKAP79, and tethering of CNB to the cell membrane through its myristoyl modification. If CN is held by additional interactions, the correlate in cells of the complete dissociation of the CN–AKAP79 complex observed in vitro might be a "breathing" of the CN-IAIIIT contact that exposes the PxIxIT-binding surface of CN but does not result in full dissociation of the assembled protein complex.
In summary, the conserved IAIIIT anchoring sequence of AKAP79 provides a high-affinity binding site to recruit CN to the vicinity of the L-type Ca2+ channel, where CN can be activated by Ca2+ influx, while at the same time preserving a dynamic interaction that permits sufficient unbinding to allow active CN to recognize NFAT. We speculate that the binding affinity has been finely tuned during evolution by these competing requirements, perhaps leaving the interaction poised at the single affinity that maintains calcineurin-substrate communication across the range of physiological signal strengths. The more general conclusion is that a third element, the precise strength of docking interactions, must be included in the description of intracellular signaling networks, in addition to the connectivity of the signaling networks determined by scaffold proteins and the modulatory interactions that can occur within individual scaffold complexes1–3,37. Our experiments illustrate the precise tailoring of calcineurin-scaffold docking affinity to the requirements of signaling in hippocampal neurons, and suggest that a similar matching of affinity to signaling may occur in other pathways built upon transient protein-protein interactions.
METHODS
Additional experimental details are provided in Supplementary Methods.
Crystallization and structure determination
Purified CN–AKAP79 complex, 7 mg/mL in 20 mM TES pH 7.0, 50 mM CaCl2 1 mM DTT, was mixed 1:1 with reservoir buffer containing 50 mM TES pH 7.5, 15% PEG 20,000, 100 mM MgSO4, 2 mM DTT, and incubated as sitting drops at 20 °C. Small plate-shaped crystals appeared overnight and continued to grow to about 0.4 × 0.15 × 0.05 mm3 in a week. X-ray diffraction data were collected on NE-CAT beamline 24-ID-C at the Advanced Photon Source. Wavelength of data collection, 0.9789 Å; temperature, 105 K. Diffraction data from one single crystal were used. The structure was solved by molecular replacement using the coordinates of a CNA–CNB complex from PDB entry 2P6B as a search model in Phaser41 and refined to 2.0 Å resolution using CNS42. Ramachandran statistics: 96.8% favored, 0.2% outliers.
SEC-MALS
SEC-MALS experiments were performed on a light scattering system (Wyatt Technology Corporation, Santa Barbara, CA) comprising a DAWN EOS detector and an Optilab rEX refractive index detector coupled to an ÄKTA Purifier (GE Healthcare, Piscataway, NJ), and analyzed using Astra V software (Wyatt Technology).
Calcineurin-AKAP79 competitive binding assays
The ability of GST-AKAP79(333–408) and GST-AKAP79(333–348) fusion proteins to compete with fluorescent PVIVIT peptide for binding to CN was measured15. Competitive displacement data were analyzed with a simple competitive binding model43,44 as described in detail in Supplementary Methods.
FRET-based stopped-flow binding assays
AKAP79(333–408) was labeled with ATTO 425 donor (ATTO-TECH, Siegen, Germany), and CN with tetramethylrhodamine acceptor (Invitrogen, Carlsbad, CA). FRET-based stopped-flow experiments were performed using a HI-TECH KinetAsyst stopped-flow system (model SF-61SX2; TgK Scientific, Bradford-on-Avon, UK). Labeled AKAP79 concentration was 100 nM. In dissociation measurements, a preformed complex of labeled AKAP79 and CN was mixed with 100 µM (final concentration) unlabeled PVIVIT peptide. Kinetic data were modeled as single exponential processes, and FRET plateau data were modeled as binding at a single class of sites.
YFP/CFP fluorescence ratio mesurements in cells
Living MDCK cells expressing AKAP79-CFP, CNAα-YFP, PKA-RIIα-YFP, or linked CFP-YFP, as indicated, were imaged at room temperature. Three-filter FRET images were captured21,31,32, apparent FRET efficiency (FRETeff) values were calculated as described in Supplementary Methods, and the observed YFP/CFP fluorescence intensity ratios were corrected for CFP quenching due to FRET. Calculation of FRETeff depends on the acceptor:donor stoichiometry of the complex (Supplementary Methods). For RII, FRETeff and the corrected YFP/CFP ratio were calculated based on the known 2:1 complex. For CN, both 1:1 and 2:1 complexes were considered. Corrected YFP/CFP ratios for CN–AKAP pairs treated as 1:1 complexes (6.6±0.5 for wildtype AKAP79, 8.1±0.6 for the PPAIIIT variant) or treated as 2:1 complexes (6.1±0.4 for wildtype AKAP79, 7.1±0.5 for the PPAIIIT variant) differed only minimally, and did not exceed the ratio for the 1:1 CFP-YFP construct. Therefore, in Figures 2g and 2h, only FRETeff values and corrected YFP/CFP ratios for a 1:1 CN–AKAP79 complex are plotted.
Primary culture and transfection of rat hippocampal neurons
Neurons were cultured as described45. We have shown that the pSil-ShRNAi AKAP150 vector effectively knocks down AKAP150 expression in hippocampal neurons for 2–12 days post transfection post transfection21,25,46 and allows its replacement with human AKAP79.
NFAT translocation assays
Cultured hippocampal neurons were incubated in Tyrode's salt solution with 1µM TTX (Tocris) at 37°C, 5% CO2, for 3 hr to dampen spontaneous activity and lower basal levels of nuclear NFAT21,25,46. Neurons were stimulated by a 3-min depolarization with isotonic 90 mM KCl Tyrode's solution; then, after 5, 15, 60, or 90 min further incubation in Tyrode's solution containing 1 µM TTX at 37°C, 5% CO2, the cells were fixed, permeabilized, and stained for endogenous NFAT3 (also termed NFATc4)45. Nuclei were counterstained with DAPI.
NFAT transcriptional reporter assays
Hippocampal cultures were transfected with an NFAT-AP1 reporter plasmid that drives expression of ECFP fused to three copies of the SV40 nuclear localization sequence. TTX (1 µM) was added to the medium for 24 hr prior to stimulation to dampen spontaneous activity and decrease basal CFP expression. After a 3-min depolarization with isotonic 90 mM KCl, cells were further incubated in the presence of 1µM TTX for 6 or 16 hours before fixation, permeabilization, and counterstaining of nuclei with propidium iodide.
FRAP microscopy
A micro-point tunable laser ablation system (Photonic Instruments, St Charles, IL) focused through the epi-fluorescence optics of the Zeiss Axiovert 200M was used for photobleaching. After the capture of five pre-bleach baseline images at 5 s intervals, 515-nm laser pulses were directed at single dendritic spines for 2 repetitions at 75–85% maximum laser power, to bleach YFP selectively. Directly after bleaching, 40 time-lapse images were captured at 5 s intervals. Pre-bleached intensities were averaged and set to 100%, and the immediate post-bleach image was set to 0%47,48. The mobile fraction of CN-YFP or AKAP79-YFP was estimated by fitting averaged recovery data to a single exponential curve.
Supplementary Material
ACKNOWLEDGMENT
We are grateful to the Department of Neurobiology, Harvard Medical School, for use of their spectrofluorometer, and to Dr Sherwin Lehrer, Boston Biomedical Research Institute, for access to the stopped-flow instrument. Diffraction data were collected at the Advanced Photon Source on Northeastern Collaborative Access Team beamline 24-ID-C, which is supported by award RR15-301 from the National Center for Research Resources at the National Institutes of Health. Use of the Advanced Photon Source, an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the U.S. DOE under Contract No. DE-AC02-06CH11357. We thank Dr. Alexander Sorkin, University of Pittsburgh, for providing the tandem YFP-CFP construct. The work was supported by US National Institutes of Health grants AI40127 (to Anjana Rao and PGH), MH080291 (to MLD), and AOI090428 (to HL). MDP was supported in part by T32NS007083 and an American Heart Association Predoctoral Fellowship from the Pacific Mountain Affiliate. JGM was supported in part by T32HD041697.
Footnotes
METHODS
Methods and any associated references can be found in the online version of the paper at http://www.nature.com/nsmb/
Note: Supplementary Information is available on the Nature Structural & Molecular Biology website.
ACCESSION CODE
The crystallographic data have been deposited in the Protein Data Bank under accession code 3LL8.
AUTHOR CONTRIBUTIONS
HL prepared the recombinant CN and AKAP79 proteins, carried out the X-ray crystallography, developed the FRET assay for CN-AKAP interaction, and performed equilibrium binding measurements. HL and PGH analyzed data from these experiments. HL and AS carried out SEC-MALS measurements. HL and PGH performed and analyzed the stopped-flow kinetic experiments. MDP, JGM, and MLD carried out and analyzed the FRET and YFP/CFP ratio measurements in MDCK cells. MDP and MLD carried out and analyzed the NFAT localization and FRAP experiments with hippocampal neurons. JGM and MLD carried out and analyzed the transcriptional reporter experiments with hippocampal neurons. PGH and HL drafted the manuscript, with significant contributions from MLD, MDP, and JGM.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Zeke A, Lukacs M, Lim WA, Remenyi A. Scaffolds: interaction platforms for cellular signalling circuits. Trends Cell Biol. 2009;19:364–374. doi: 10.1016/j.tcb.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scott JD, Pawson T. Cell signaling in space and time: where proteins come together and when they're apart. Science. 2009;326:1220–1224. doi: 10.1126/science.1175668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Good MC, Zalatan JG, Lim WA. Scaffold proteins: hubs for controlling the flow of cellular information. Science. 2011;332:680–686. doi: 10.1126/science.1198701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coghlan VM, et al. Association of protein kinase A and protein phosphatase 2B with a common anchoring protein. Science. 1995;267:108–111. doi: 10.1126/science.7528941. [DOI] [PubMed] [Google Scholar]
- 5.Logue JS, Scott JD. Organizing signal transduction through A-kinase anchoring proteins (AKAPs) Febs J. 277:4370–4375. doi: 10.1111/j.1742-4658.2010.07866.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hogan PG, Chen L, Nardone J, Rao A. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 2003;17:2205–2232. doi: 10.1101/gad.1102703. [DOI] [PubMed] [Google Scholar]
- 7.Molkentin JD, Dorn GW., 2nd Cytoplasmic signaling pathways that regulate cardiac hypertrophy. Annu. Rev. Physiol. 2001;63:391–426. doi: 10.1146/annurev.physiol.63.1.391. [DOI] [PubMed] [Google Scholar]
- 8.Heineke J, Molkentin JD. Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat. Rev. Mol. Cell Biol. 2006;7:589–600. doi: 10.1038/nrm1983. [DOI] [PubMed] [Google Scholar]
- 9.Groth RD, Dunbar RL, Mermelstein PG. Calcineurin regulation of neuronal plasticity. Biochem. Biophys. Res. Commun. 2003;311:1159–1171. doi: 10.1016/j.bbrc.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 10.Li H, Rao A, Hogan PG. Interaction of calcineurin with substrates and targeting proteins. Trends Cell Biol. 2011;21:91–103. doi: 10.1016/j.tcb.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feske S, Okamura H, Hogan PG, Rao A. Ca2+/calcineurin signalling in cells of the immune system. Biochem. Biophys. Res. Commun. 2003;311:1117–1132. doi: 10.1016/j.bbrc.2003.09.174. [DOI] [PubMed] [Google Scholar]
- 12.Macian F. NFAT proteins: key regulators of T-cell development and function. Nat. Rev. Immunol. 2005;5:472–484. doi: 10.1038/nri1632. [DOI] [PubMed] [Google Scholar]
- 13.Aramburu J, et al. Selective inhibition of NFAT activation by a peptide spanning the calcineurin targeting site of NFAT. Mol. Cell. 1998;1:627–637. doi: 10.1016/s1097-2765(00)80063-5. [DOI] [PubMed] [Google Scholar]
- 14.Aramburu J, et al. Affinity-driven peptide selection of an NFAT inhibitor more selective than cyclosporin A. Science. 1999;285:2129–2133. doi: 10.1126/science.285.5436.2129. [DOI] [PubMed] [Google Scholar]
- 15.Li H, Rao A, Hogan PG. Structural delineation of the calcineurin–NFAT interaction and its parallels to PP1 targeting interactions. J. Mol. Biol. 2004;342:1659–1674. doi: 10.1016/j.jmb.2004.07.068. [DOI] [PubMed] [Google Scholar]
- 16.Li H, Zhang L, Rao A, Harrison SC, Hogan PG. Structure of calcineurin in complex with PVIVIT peptide: portrait of a low-affinity signalling interaction. J. Mol. Biol. 2007;369:1296–1306. doi: 10.1016/j.jmb.2007.04.032. [DOI] [PubMed] [Google Scholar]
- 17.Takeuchi K, Roehrl MH, Sun ZY, Wagner G. Structure of the calcineurin-NFAT complex: defining a T cell activation switch using solution NMR and crystal coordinates. Structure. 2007;15:587–597. doi: 10.1016/j.str.2007.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roy J, Li H, Hogan PG, Cyert MS. A conserved docking site modulates substrate affinity for calcineurin, signaling output, and in vivo function. Mol. Cell. 2007;25:889–901. doi: 10.1016/j.molcel.2007.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roy J, Cyert MS. Cracking the phosphatase code: docking interactions determine substrate specificity. Sci. Signal. 2009;2:re9. doi: 10.1126/scisignal.2100re9. [DOI] [PubMed] [Google Scholar]
- 20.Shi Y. Serine/threonine phosphatases: mechanism through structure. Cell. 2009;139:468–484. doi: 10.1016/j.cell.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 21.Oliveria SF, Dell'Acqua ML, Sather WA. AKAP79/150 anchoring of calcineurin controls neuronal L-type Ca2+ channel activity and nuclear signaling. Neuron. 2007;55:261–275. doi: 10.1016/j.neuron.2007.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dell'Acqua ML, et al. Regulation of neuronal PKA signaling through AKAP targeting dynamics. Eur. J. Cell Biol. 2006;85:627–633. doi: 10.1016/j.ejcb.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 23.Wong W, Scott JD. AKAP signalling complexes: focal points in space and time. Nat Rev Mol. Cell Biol. 2004;5:959–970. doi: 10.1038/nrm1527. [DOI] [PubMed] [Google Scholar]
- 24.Hoshi N, et al. AKAP150 signaling complex promotes suppression of the M-current by muscarinic agonists. Nat. Neurosci. 2003;6:564–571. doi: 10.1038/nn1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoshi N, Langeberg LK, Scott JD. Distinct enzyme combinations in AKAP signalling complexes permit functional diversity. Nat. Cell Biol. 2005;7:1066–1073. doi: 10.1038/ncb1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tavalin SJ, et al. Regulation of GluR1 by the A-kinase anchoring protein 79 (AKAP79) signaling complex shares properties with long-term depression. J. Neurosci. 2002;22:3044–3051. doi: 10.1523/JNEUROSCI.22-08-03044.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu G, et al. Assembly of a Ca2+-dependent BK channel signaling complex by binding to β2 adrenergic receptor. EMBO J. 2004;23:2196–2205. doi: 10.1038/sj.emboj.7600228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tunquist BJ, et al. Loss of AKAP150 perturbs distinct neuronal processes in mice. Proc. Natl. Acad. Sci. USA. 2008;105:12557–12562. doi: 10.1073/pnas.0805922105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Graef IA, et al. L-type calcium channels and GSK-3 regulate the activity of NF-ATc4 in hippocampal neurons. Nature. 1999;401:703–708. doi: 10.1038/44378. [DOI] [PubMed] [Google Scholar]
- 30.Dell'Acqua ML, Dodge KL, Tavalin SJ, Scott JD. Mapping the protein phosphatase-2B anchoring site on AKAP79. Binding and inhibition of phosphatase activity are mediated by residues 315–360. J. Biol. Chem. 2002;277:48796–48802. doi: 10.1074/jbc.M207833200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oliveria SF, Gomez LL, Dell'Acqua ML. Imaging kinase–AKAP79–phosphatase scaffold complexes at the plasma membrane in living cells using FRET microscopy. J. Cell. Biol. 2003;160:101–112. doi: 10.1083/jcb.200209127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gorski JA, Gomez LL, Scott JD, Dell'Acqua ML. Association of an A-kinase-anchoring protein signaling scaffold with cadherin adhesion molecules in neurons and epithelial cells. Mol. Biol. Cell. 2005;16:3574–3590. doi: 10.1091/mbc.E05-02-0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gold MG, et al. Molecular basis of AKAP specificity for PKA regulatory subunits. Mol. Cell. 2006;24:383–395. doi: 10.1016/j.molcel.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 34.Gold MG, et al. Architecture and dynamics of an A-kinase anchoring protein 79 (AKAP79) signaling complex. Proc. Natl. Acad. Sci. USA. 2011;108:6426–6431. doi: 10.1073/pnas.1014400108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stemmer P, Klee CB. Serine/threonine phosphatases in the nervous system. Curr. Opin. Neurobiol. 1991;1:53–64. doi: 10.1016/0959-4388(91)90010-5. [DOI] [PubMed] [Google Scholar]
- 36.Salazar C, Hofer T. Allosteric regulation of the transcription factor NFAT1 by multiple phosphorylation sites: a mathematical analysis. J. Mol. Biol. 2003;327:31–45. doi: 10.1016/s0022-2836(03)00085-8. [DOI] [PubMed] [Google Scholar]
- 37.Bashor CJ, Horwitz AA, Peisajovich SG, Lim WA. Rewiring Cells: Synthetic biology as a tool to interrogate the organizational principles of living systems. Annu. Rev. Biophys. 2010;39:515–537. doi: 10.1146/annurev.biophys.050708.133652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pettersen EF, et al. UCSF Chimera— A visualization system for exploratory research and analysis. J. Comput. Chem. 2004;13:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 39.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 40.Alto NM, et al. Bioinformatic design of A-kinase anchoring protein-in silico: A potent and selective peptide antagonist of type II protein kinase A anchoring. Proc. Natl. Acad. Sci. USA. 2003;100:4445–4450. doi: 10.1073/pnas.0330734100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCoy AJ, et al. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brunger AT, et al. Crystallography & NMR system: A new software system for macromolecular structure determination. Acta Cryst. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 43.Wang ZX. An exact mathematical expression for describing competitive binding of two different ligands to a protein molecule. FEBS Lett. 1995;360:111–114. doi: 10.1016/0014-5793(95)00062-e. [DOI] [PubMed] [Google Scholar]
- 44.Roehrl MH, et al. Selective inhibition of calcineurin-NFAT signaling by blocking protein–protein interaction with small organic molecules. Proc. Natl. Acad. Sci. USA. 2004;101:7554–7559. doi: 10.1073/pnas.0401835101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gomez LL, Alam S, Smith KE, Horne E, Dell'Acqua ML. Regulation of A-kinase anchoring protein 79/150–cAMP-dependent protein kinase postsynaptic targeting by NMDA receptor activation of calcineurin and remodeling of dendritic actin. J. Neurosci. 2002;22:7027–7044. doi: 10.1523/JNEUROSCI.22-16-07027.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robertson HR, Gibson ES, Benke TA, Dell'Acqua ML. Regulation of postsynaptic structure and function by an A-kinase anchoring protein–membrane-associated guanylate kinase scaffolding complex. J. Neurosci. 2009;29:7929–7943. doi: 10.1523/JNEUROSCI.6093-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lippincott-Schwartz J, Snapp E, Kenworthy A. Studying protein dynamics in living cells. Nat. Rev. Mol. Cell. Biol. 2001;2:444–456. doi: 10.1038/35073068. [DOI] [PubMed] [Google Scholar]
- 48.Sharma K, Fong DK, Craig AM. Postsynaptic protein mobility in dendritic spines: long-term regulation by synaptic NMDA receptor activation. Mol. Cell. Neurosci. 2006;31:702–712. doi: 10.1016/j.mcn.2006.01.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.