Abstract
Successful mitosis depends on the stable, yet regulated attachment of chromosomes to spindle microtubules. The kinetochore, a large macromolecular structure assembled at sites of centromeric heterochromatin, is responsible for generating and regulating these essential attachments. Over the last several years, concerted experimental efforts have brought the structural view of the kinetochore-microtubule interface more clearly into focus. Here, we review important recent advancements and discuss several unresolved questions regarding how kinetochores dynamically bridge mitotic chromosomes to spindle microtubules.
Keywords: kinetochore, cell cycle, cell division, chromosome segregation, KMN network, centromere, Ndc80, Hec1, Mis12, microtubule
General concepts
Mitotic cells face the challenge of equally distributing their duplicated chromosomes into two daughter cells. Anything other than equal distribution is unacceptable, since the inheritance of too many or too few chromosomes is catastrophic for the progeny [1]. The fidelity of this process relies on the specialized attachment between chromosomes and spindle microtubules. Such attachment is mediated by a protein structure called the kinetochore, built specifically atop sites of centromeric heterochromatin at the onset of each mitotic cycle (Fig. 1). In humans, 12-30 microtubules eventually bind a single kinetochore [2,3], and their coordinated plus-end dynamics are used to generate the forces required for both chromosome movements and to silence the spindle assembly checkpoint, allowing for mitotic exit.
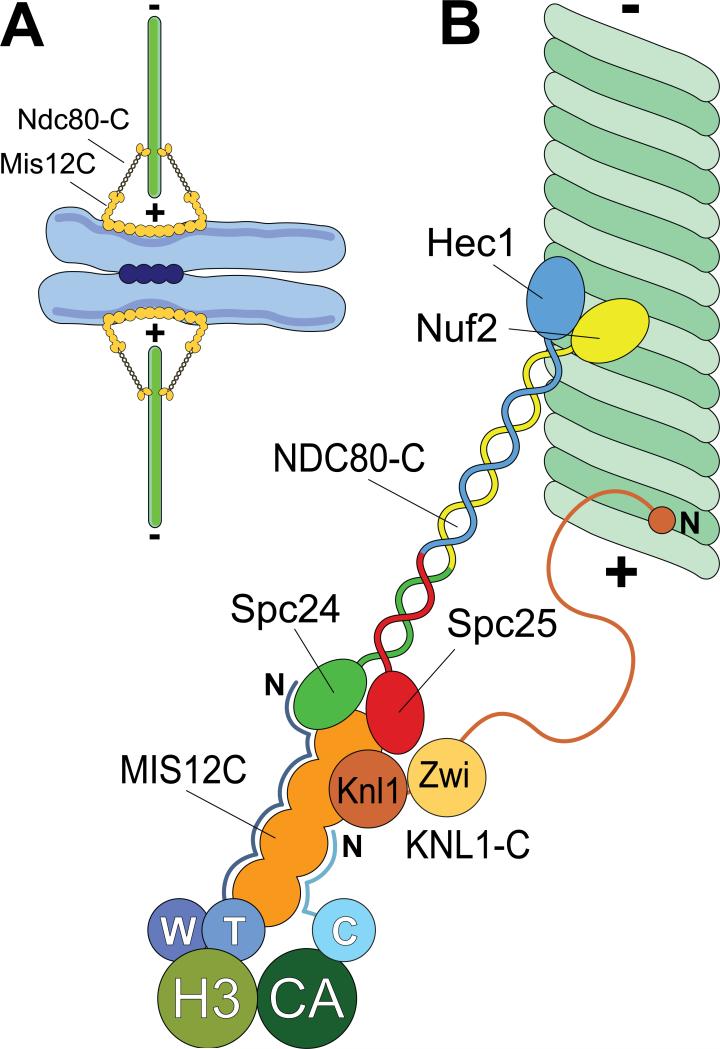
Figure 1. Schematic view of kinetochores.
A) During mitosis, sister chromatids are held together at centromeres by a cohesion complex (dark blue circles). Kinetochores (orange) assemble on centrometic chromatin and create a contact with microtubules (green). The plus (+) and minus (-) ends of microtubules are indicated. B) A close-up of kinetochores showing some of its components required for end-on microtubule binding. With the exception of the CENP-T/W complex (abbreviated as W and T) and CENP-C (abbreviated as C), all subunits of the constitutive centromere associated network (CCAN) have been omitted. CENP-T/W associates with histone H3-containing nucleosomes (H3), whereas CENP-C associates with nucleosomes containing the H3 variant CENP-A. The N-terminal region of CENP-T is an extended, largely disordered polypeptide chain that makes contacts with the 4-subunit Mis12 complex (MIS12-C) and with NDC80-C [11]. The N-terminal region of CENP-C is probably also disordered and makes contacts with MIS12-C [9,10]. The Knl1 complex (KNL1-C), which comprises Knl1 and Zwint-1 (Zwi), might contain a microtubule-binding site in the N-terminal region of Knl1 [28,56]. The C-terminal region of Knl1 interacts directly with the MIS12-C [70]. The NDC80-C is a tetramer. The Spc24 and Spc25 subunits interact with the MIS12-C, whereas the Hec1/Ndc80 and Nuf2 subunits face the microtubule.
A likely near-comprehensive kinetochore parts list, which includes over 100 individual proteins, has emerged [4,5]. Together, these components build the linkages between mitotic chromosomes and spindle microtubules. The elucidation of the foundations of kinetochore assembly on centromeric chromatin is rapidly progressing [6-11]. Here, we concentrate on the opposite end of the kinetochore, where microtubule binding takes place (Fig. 1). Combined biochemical, cell biological, and structural data support the idea that the kinetochore-microtubule binding interface is primarily composed of a highly conserved group of 10 kinetochore proteins referred to as the KMN “network”. The KMN network is comprised of the KNL1 complex (KNL1-C, comprised of Knl1/Spc105/Blinkin/CASC5/AF15q14 and Zwint-1), the MIS12 complex (MIS12-C, comprised of Mis12/Mtw1, Dsn1, Nsl1, and Nnf1), and the NDC80 complex (NDC80-C, comprised of Ndc80/Hec1, Nuf2, Spc24, and Spc25) [4,5] (Fig. 1).
Many aspects of kinetochore-microtubule attachment in early mitosis, in the phases that precede bi-orientation at the metaphase plate, remain obscure [12,13]. The KMN network might be dispensable for chromosome movement to the newly characterized “equatorial belt”, which precedes bi-orientation [12-14]. Such movements are probably achieved through association of kinetochores with the lateral surface of spindle microtubules. On the other hand, the KMN network is crucially required for end-on kinetochore-microtubule interactions. In vivo, depletion of any of the KMN protein components reduces the ability of cells to form functional kinetochore-microtubule attachments, with the most severe defects observed in cells depleted of NDC80-C components, suggesting that the NDC80-C is largely responsible for directly generating the attachments [15,16].
Besides being required for bi-orientation, end-on attachment is generally believed to be required for poleward kinetochore movement at anaphase upon microtubule depolymerization [17]. In vitro force measurements with purified NDC80-C are consistent with the hypothesis that this complex couples chromosome movement to depolymerizing microtubules [18,19]. To be able to maintain a firm grip that is compatible with fluid tracking on the microtubule lattice, kinetochores must regulate the strength of binding to microtubules. Strong evidence has emerged that this is achieved via regulated phosphorylation of kinetochore proteins [20,21]. Additionally, mechanical tension at the kinetochore-microtubule interface might by itself directly contribute to the stabilization of kinetochore microtubules [23], whose half-life is consequently longer than that of spindle microtubules [22]. The ability of kinetochores to regulate the strength of their grip on microtubules is probably at the basis of error correction, the phenomenon whereby improperly connected microtubules are released so that the kinetochore can “reset” and try again to form correct attachments [24,25]. Aurora B kinase, which has been unequivocally implicated in error correction [26,27], phosphorylates several targets at the kinetochoremicrotubule interface, most notably the NDC80-C [21,28].
The NDC80 complex, the primary link to microtubules
The NDC80-C is a dumb-bell shaped molecule ~60 nm long [29-31]. The C-terminal globular domains of Spc24 and Spc25, at one end of the central shaft, are connected with the centromere (Fig. 1). The N-terminal globular domains of Hec1 and Nuf2, at the opposite end of the NDC80-C structure, form a tight arrangement that directly binds the microtubule lattice. The two globular ends are connected by a long coiled-coil domain, with contributions of alpha-helices from all 4 NDC80-C subunits. Within the rod-like region linking Nuf2 and Hec1 there is a short interruption in the coiled-coil where a stretch of amino acids within the Hec1 protein that are not associated with Nuf2 helices “loop out” to recruit additional microtubule-binding proteins [32,33].
X-ray crystallographic studies of the Hec1 N-terminus alone [34] and of NDC80bonsai, an engineered version of NDC80-C in which much of the internal coiled-coil domain was removed [30], revealed that the N-terminal regions of Hec1 and Nuf2 fold into calponin homology (CH) domains. CH domains mediate direct microtubule binding in other microtubule associated proteins, such as EB1. In EB1, a homodimeric protein, the CH domains are connected by a partly flexible dimerization domain, which allows the formation of an asymmetric dimeric assembly that involves extensive interactions between the CH domains [35]. The two CH domains in the NDC80-C head also form a tight dimeric assembly that is stabilized by a large hydrophobic interface, suggesting that they form an inseparable dimeric pair in which the CH domains adopt a reciprocally well-defined orientation (Fig 2).
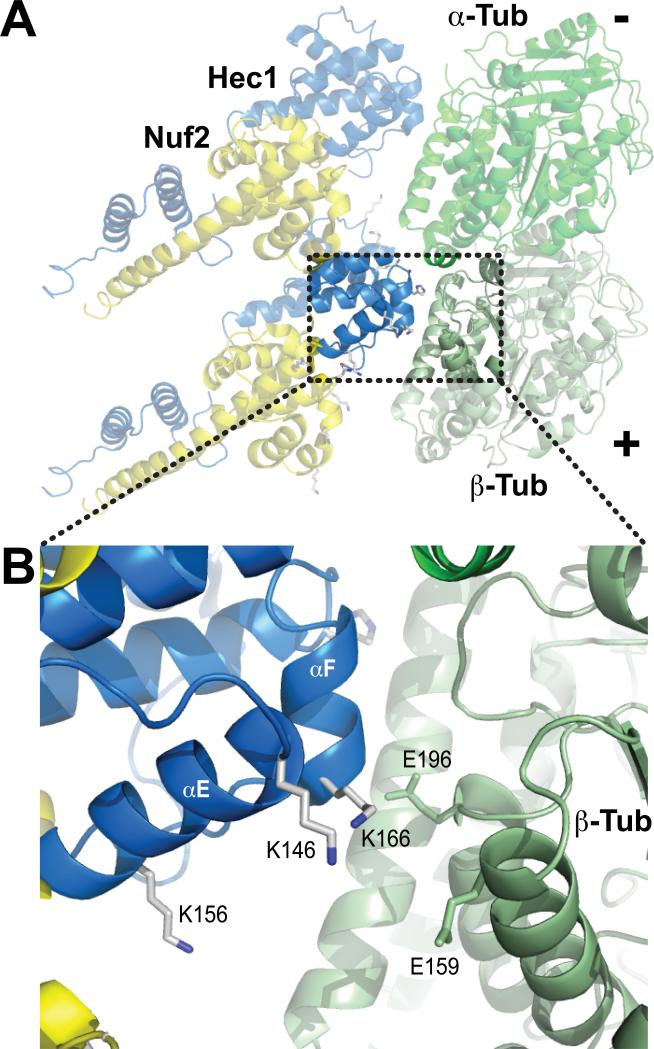
Figure 2. The “toe” and the “toe-print”, part I.
A) Cartoon showing the CH domains from a pair of NDC80-C bound to the α-tubulin/β-tubulin dimer [37]. The model was created by fitting the high-resolution structures of the NDC80Bonsai complex [30] and of the α-tubulin/β-tubulin dimer [43] in a cryo-EM 3D reconstruction of NDC80Bonsai-decorated microtubules [37]. The lower NDC80-C contacts microtubules at the intra-dimer interface. The upper NDC80-C docks at the inter-dimer interface. Changes in the relative orientation at these interfaces might modify the binding affinity for NDC80-C [37]. B) Close-up of the area boxed in A and showing residues in the “toe” and “toe-print” discussed in the text. K146 and K166 from Hec1 form a tight pair that faces a negative patch on β-tubulin. The cartoon models were created with PyMol (www.pymol.org) and assembled in Adobe Illustrator.
NDC80-microtubule interaction: the main interface
Cryo-EM reconstructions of microtubules decorated with NDC80-C recently provided a view of the NDC80-C-microtubule interface [36,37]. A crucial conclusion brought about by these analyses is that the NDC80-C binds tubulin monomers at both the inter- and intra-tubulin dimer interfaces. By fitting the high-resolution structures of the NDC80-Cbonsai construct and of the α- and β-tubulin dimer in an 8.6-Å reconstruction, a microtubule interaction domain within the Hec1 CH domain, named the “toe”, was identified (Figs. 2 and 3) [37]. The toe contains positively charged Hec1 residues previously shown to be important for microtubule binding in vitro [30], including K123, K166, K146, and H176 (in helix αC, in helix αF, in the αC-αE loop, and in the αF-αG loop, respectively) (Fig. 2B). Near the toe, K89 (αA helix) and K115 (αB helix), which have also been implicated in microtubule binding in vitro [30], appear to be positioned for a hypothetical interaction with the E-hooks, the C-terminal tails of tubulin. The latter, however, are invisible in the 3D reconstructions [36,37] (Fig. 3B).
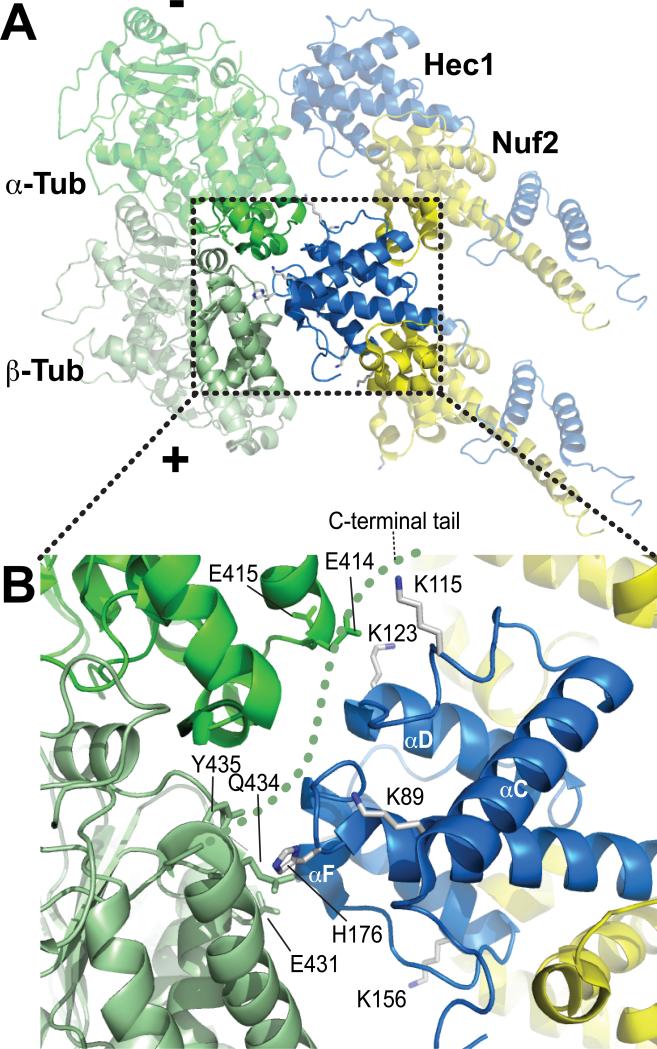
Figure 3. The “toe” and the “toe-print”, part II.
A) The view was rotated ~180° relative to the view in Fig. 2A. B) Close-up of the area boxed in A. The C-terminal tail of β-tubulin was invisible in the cryo-EM reconstructions. A hypothetical path for the C-terminal tail of tubulin (so-called E-hook) is shown as a dotted green line. K89 and K115 of Hec1 are potentially positioned for an interaction with the E-hook.
Cells expressing the Hec1 CH domain charge reversal mutants K166D or K166E fail to form stable kinetochore-microtubule attachments [38,39]. Additional mutants, such as K89E and K115E, fail to sustain full tension but cause less prominent chromosome alignment phenotypes [39]. S165, which neighbors the crucial residue for microtubule binding K166, is a site of Hec1 phosphorylation by the Nek2A kinase [40-42]. At the interface with the microtubule, this residue has the potential to play a critical role in the regulation of kinetochore-microtubule attachment and in coordinating it with the spindle checkpoint.
The ‘toe’ recognizes a site between α- and β-tubulin monomers, a ‘toe-print’ present at both intra- and inter-dimer interfaces. The toe-prints are highly negatively charged. At the intra-dimer interface, K146 and K166 on the Hec1 monomer face E159 and E196 of β-tubulin, located at the ends of the H4 and H5 helices, respectively [43] (Fig. 2B and 3B). H176 of Hec1 faces hydrophilic residues at the end of the H12 helix of β-tubulin, including E431, Q434, and Y435. And finally, K123 of Hec1 faces E414 and E415 in the H11-H12 loop of α-tubulin. Similar interactions are formed at the inter-dimer interface, where E155, E196, D431, and E434 of α-tubulin and D414 and E415 of β-tubulin are the likely functional homologs of the above-mentioned residues at the intra-dimer interface. Thus, sequence conservation between α- and β-tubulin at exposed residues of the toe-print likely explains why Hec1 binding can occur equivalently at intra- and inter-dimer regions [37].
Binding of NDC80-C to microtubules was visualized upon decoration of the straight filaments present in growing or stabilized microtubules. The interface between tubulin monomers, however, is as a hinge point along a microtubule's protofilaments. For instance, protofilaments undergo significant bending in the outward direction during disassembly of microtubules. It was therefore predicted, and experimentally demonstrated, that such conformational changes alter the accessibility of the toe-print to the NDC80-C [37]. Thus, the toe-print might act as a conformational sensor that limits the binding of NDC80-C to straight filaments. The preference of NDC80-C for straight filaments might favor sliding in the poleward direction on the lattice of depolymerizing microtubules. Furthermore, the NDC80-C does not bind tubulin dimers in solution [30].
NDC80-microtubule interaction: secondary interfaces and binding cooperativity
The interaction of residues in the Hec1 toe with the tubulin toe-print is essential for the interaction of the NDC80-C with microtubules [30,38,39]. However, two additional portions of the NDC80-C have also been implicated in an interaction with microtubules, the Nuf2 subunit and the disordered N-terminal region of Hec1 [21,30,44,45]. The precise function of both these structural elements remains elusive despite the growing structural information.
The Hec1 N-terminal “tail” is a highly positively charged 80-residue segment preceding the Hec1 CH domain. It was not included in either of the X-ray structures of Hec1 [30,34]. The N-terminal tail is required for high affinity binding of NDC80-C to microtubules in vitro [30,34] and for stable kinetochore-microtubule attachment in vivo [44-46]. However, its precise role in microtubule binding remains controversial. In one model, this positively-charged domain directly interfaces the negatively-charged microtubule lattice to contribute to high affinity binding [21,44,45] (Fig. 4A’). In agreement with a direct role in microtubule binding, the isolated Hec1 tail domain was reported to bind microtubules in vitro with affinities similar to those of the entire N-terminal region of Hec1 or of Hec1/Nuf2 dimers [44,47]. In an alternative model, further discussed below, the tail domain serves to oligomerize adjacent NDC80-C to promote cooperative association along the microtubule (Fig. 4A”). Such cooperative interactions are believed to be important for high affinity binding of NDC80 complexes to microtubules in vitro [19,30,37,39,47].
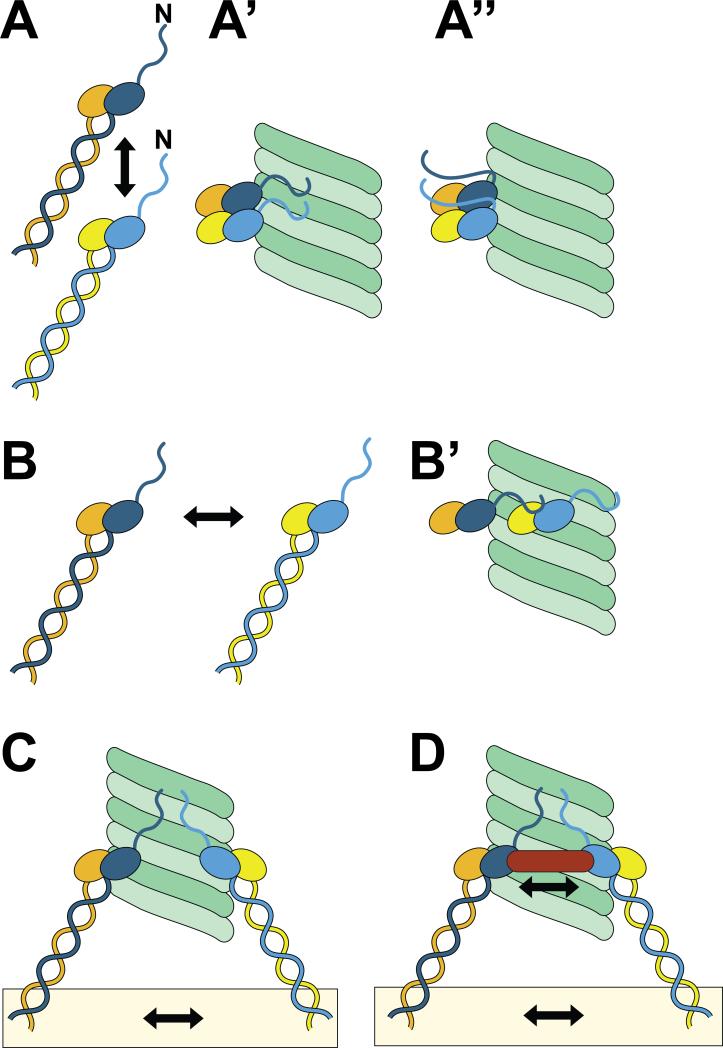
Figure 4. Hypothetical binding mechanisms.
A) The arrow indicates longitudinal interactions between two NDC80 complexes. Two possible implementations of this configuration that incorporate binding cooperativity are shown in panels A’ and A”. The model in A” recapitulates structural findings that the N-terminal tail of Hec1 packs between two NDC80 complexes in the longitudinal direction [37]. In A’, the interaction between NDC80 complexes does not involve the N-terminal tails, which are rather engaged directly in microtubule binding. B) The arrow indicates lateral interactions between two NDC80 complexes. B’) A possible implementation in which the C-terminal tails form contacts with NDC80 complexes on laterally neighboring protofilaments. There is no experimental evidence for this model. C) Embedding of individual NDC80 complexes within kinetochores (details not shown, see Fig. 1) allows multiple NDC80 complexes to form interactions with microtubules without any additional contacts between NDC80 complexes at the microtubule-binding interface. D) Molecular cross-linking of NDC80 complexes, as it might be implemented by factors such as the Dam1 complex or the SKA complex.
Interestingly, the Nuf2 CH domain does not directly interface the microtubule lattice [36,37]. Cells expressing Nuf2 CH domain mutants exhibit very mild phenotypes, contrarily to mutations in Hec1 [38]. The mutants, however, have considerable effects on microtubule binding in microtubule co-sedimentation experiments in vitro [30]. In such experiments, the ratio of NDC80-C to microtubules is much more elevated than at kinetochores, where only a handful of NDC80 complexes (7-9 complexes) per microtubule are available and sufficient to build force-bearing attachments [48-50]. The implications of this important distinction are currently incompletely understood, and it is possible that the observed role of Nuf2 in vitro reflects the formation of low-affinity interactions that have little relevance at kinetochores but significantly influence binding in vitro.
The advocated role of cooperativity in the binding of NDC80-C to microtubules needs to be understood in this framework, as it was so far measured at relatively high ratios of NDC80-C to microtubules [19,30,37,39,47]. It is less clear whether cooperativity plays a role at the low NDC80-C to microtubule ratios present at kinetochores. What information do the available structural data convey with regard to possible molecular mechanisms of cooperativity at the NDC80-C-microtubule interface? By calculating the differences between experimental density maps of NDC80Bonsai-decorated microtubules, which contained the Hec1 tails, and maps calculated from the docked crystal structures, which lacked the Hec1 tails, significant densities that ran longitudinally between adjacent NDC80 complexes were identified and attributed to the Hec1 tail [37]. Thus, the tail may not directly interface the MT lattice, but oligomerize adjacent NDC80 complexes together (Fig. 4A”).
The NDC80 complexes form clusters along the microtubule, and it is predicted that in the clustered arrangement, NDC80-C might stably bind microtubules and dynamically track growing and shortening ends [37]. In vitro, clusters of wild-type NDC80 along the microtubule lattice were found to contain a wide distribution of individual complexes, with cluster sizes of 4 or >10 being most probable [37]. In support of the model in Fig. 4A”, in which the tail promotes clustering of NDC80 complexes, cluster size is somewhat diminished when tail-less NDC80Bonsai complexes are tested [30,37].
Cluster size is also diminished when NDC80Bonsai complexes containing phospho-mimetic Hec1 (NDC80bonsai-7D) are tested [30,37]. The Hec1 N-terminal domain is phosphorylated both in vitro and in vivo by Aurora B kinase [21,30,47]. This kinase is known to regulate the stability of kinetochore-microtubule attachments in mitosis by increasing kinetochore-microtubule turnover [26,27,51,52]. Aurora B kinase-mediated Hec1 tail phosphorylation is maximal in early mitosis [20]. Thus, Aurora B may control the ability of the tail domain to oligomerize NDC80 complexes, so to progressively increase the affinity of complexes for the microtubule lattice [37].
More complex binding models are also possible. Because the tail may extend up to 12 nm in length if fully extended, it is conceivable that it could both directly contact the microtubule lattice and facilitate oligomerization (Fig. 4A’ and 4A”), depending on precisely how neighboring complexes are aligned along the microtubule lattice. It is therefore crucial to determine if certain domains within the tail mediate direct binding to the microtubule lattice and if others mediate complex oligomerization. Furthermore, since Aurora B kinase phosphorylation sites are peppered throughout the length of the Hec1 tail, it will also be important to determine which sites govern phospho-regulation of microtubule binding affinity. Finally, it will also be important to determine the role of other kinases, most notably Mps1 and Nek2A, in phosphoregulation of the function of the NDC80-C [41,42,53].
The arrangement of NDC80 complexes must facilitate dynamic kinetochore movements
The possibility that individual NDC80 complexes might become aligned in a linear array along a single protofilament, i.e. longitudinally (Figs. 2-3), was an unanticipated revelation of the high-resolution structural analysis of NDC80-C/microtubule complex [37]. In the longitudinal stacking of NDC80 complexes, the N-terminal tail is predicted to establish low-affinity contacts with Nuf2 [37], which may justify the role of this subunit in microtubule binding, at least in vitro [30]. Besides its merits, the model also presents shortcomings. For instance, super-resolution mapping of kinetochore components indicates that at metaphase, the projection length of NDC80 complexes along the inter-kinetochore axis is constant, and the measured lengths do not deviate from the average value by more than +/- 5 nm [54]. It is difficult to reconcile this with a scenario in which the NDC80 complexes are differentially positioned along a single protofilament to allow for such clusters to form, as this would encompass a distance of 16 nm for an oligomer of 4 complexes.
To account for the invariant distribution of NDC80 positioning in the super-resolution study [54] one would need to hypothesize that the NDC80 complexes may be clustered laterally along adjacent protofilaments (Fig. 4B-B’). However, no contacts between NDC80 complexes in neighboring protofilaments are evident [37]. It is also possible that the kinetochore itself organizes the KMN network components in a manner that does not require explicit oligomerization of the microtubule-binding head of the NDC80-C (Fig. 4C). In this model, the mere co-existence of multiple NDC80 complexes on a substrate such as the kinetochore would cause the individual low-affinity individual contributions of each complex to add up to create considerable binding affinity, as predicted by Hill's implementation of the biased-diffusion model [55].
Kinetochore-microtubule attachment does not rely on KMN alone
In summary, binding cooperativity has been advocated for its potential contributions to NDC80-C binding to microtubules, but whether it plays a role at the kinetochore, and if so, precisely trough which mechanism, is currently unclear. The picture is further complicated by the consideration that the kinetochore is disseminated of additional microtubule-binding activities. Within the KMN network itself, the N-terminal region of the Knl1 subunit has been proposed to host a second microtubule-binding domain [28,56] (Fig. 1). Acting in concert with the microtubule-binding domain in the NDC80-C, this region of Knl1 might enhance the overall binding affinity of the KMN network for microtubules.
Furthermore, although the KMN network generates the primary contacts between kinetochores and microtubules, it does not accomplish the task alone. In budding yeast, the NDC80-C works in concert with the DAM1 complex to facilitate the formation of functional kinetochoremicrotubule attachments in cells [57]. In vitro, DAM1 complexes can form rings and oligomeric assemblages around or along the length of the microtubule lattice, both of which can support the coupling of microtubule dynamics to force generation for cargo movement [58]. NDC80 complexes directly associate with DAM1 complexes in vitro, possibly through an involvement of the Ndc80 loop region [33]. Cooperation between the two complexes appears to be essential in budding yeast for the formation of stable, regulatable kinetochore-microtubule attachments (as shown schematically in Fig. 4D).
No clear homolog of the budding yeast DAM1 complex has been identified in vertebrates, thus the DAM1/NDC80-C coupling mechanism is likely not conserved throughout evolution. Vertebrate kinetochore components have been identified, however, that are required for microtubule attachment in addition to the NDC80-C. Most notably, cultured cells depleted of the trimeric SKA complex fail to form stable kinetochore-microtubule attachments, although NDC80-C kinetochore localization is not perturbed [59-63]. Interestingly, the SKA complex can track depolymerizing microtubules in vitro and can support the coupling of this movement to force generation for cargo movement, raising the possibility that it is the functional homolog of the DAM1 complex [60]. It is therefore important to address if the SKA complex interacts directly with subunits of the KMN complex.
Conclusions and Perspectives
Upon microtubule binding, significant structural rearrangements occur within the kinetochore. Electron microscopy reveals dramatic reorganization of fibers within the outer domain that likely represent changes in the positions of the microtubule binding elements within the kinetochore [64]. Super-resolution protein mapping experiments have also demonstrated dramatic changes in the positioning of many proteins within the kinetochore as it transitions from an unbound to microtubule-bound state [54,65-68]. Recent structural studies using reconstituted protein components have moved us significantly closer to understanding how these large-scale changes in the kinetochore are related to the physical changes in individual protein complexes that allow for direct binding to the microtubule lattice and for the coupling of microtubule dynamics to the generation of forces required for chromosome movements [31,34,37,69-73].
Direct measures of changes in protein-protein interactions within the kinetochore will no doubt further our understanding of the kinetochore-microtubule interface and of how it dynamically changes throughout mitosis. The main limitation towards testing different binding models in living cells is that we lack a comprehensive view of all the relevant players in kinetochoremicrotubule attachment, and accurate maps of their interactions. Under these conditions, developing strategies for selective interference of desired interactions is unfeasible. For instance, cooperative interactions among NDC80 complexes might be important for microtubule binding, as explained above. But in the absence of a complete census of the interactions of the NDC80-C and of its parts, our incomplete understanding might significantly bias our interpretation of the effects of structural perturbations. We envision that research in the kinetochore area will proceed through an iterative combination of structural and biochemical analyses, in vivo reconstitution and analysis of appropriately modified mutants, and modeling.
Acknowledgments
Work in JGD's laboratory is funded by the National Institutes of Health (NIH-GM088371 and NIH-K01CA125051) and the Pew Scholars Program in the Biomedical Sciences. Work in AM's laboratory is funded by the European Union's 7th Framework Program ERC agreement KINCON and the Integrated Project MitoSys, the Italian Association for Cancer Research (AIRC), the Fondo di Investimento per la Ricerca di Base (FIRB), the Cariplo Foundation, the Human Frontier Science Program, and Programmi Integrati di Oncologia (PIO) Strategici 7/07.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Holland AJ, Cleveland DW. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol. 2009;10:478–487. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wendell KL, Wilson L, Jordan MA. Mitotic block in HeLa cells by vinblastine: ultrastructural changes in kinetochore-microtubule attachment and in centrosomes. J Cell Sci. 1993;104(Pt 2):261–274. doi: 10.1242/jcs.104.2.261. [DOI] [PubMed] [Google Scholar]
- 3.McEwen BF, Chan GK, Zubrowski B, Savoian MS, Sauer MT, Yen TJ. CENP-E is essential for reliable bioriented spindle attachment, but chromosome alignment can be achieved via redundant mechanisms in mammalian cells. Mol Biol Cell. 2001;12:2776–2789. doi: 10.1091/mbc.12.9.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Santaguida S, Musacchio A. The life and miracles of kinetochores. EMBO J. 2009;28:2511–2531. doi: 10.1038/emboj.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheeseman IM, Desai A. Molecular architecture of the kinetochore–microtubule interface. Nat Rev Mol Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- 6*.Guse A, Carroll CW, Moree B, Fuller CJ, Straight AF. In vitro centromere and kinetochore assembly on defined chromatin templates. Nature. 2011 doi: 10.1038/nature10379. doi:10.1038/nature10379. [Through an in vitro reconstitution approach, this paper investigates the requirements for centromeric chromatin in the assembly of kinetochore components, identifying a crucial determinant for kinetochore assembly in the C-terminal region of CENP-A.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foltz DR, Jansen LET, Black BE, Bailey AO, Yates JR, Cleveland DW. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- 8.Okada M, Cheeseman IM, Hori T, Okawa K, McLeod IX, Yates JR, Desai A, Fukagawa T. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat Cell Biol. 2006;8:446–457. doi: 10.1038/ncb1396. [DOI] [PubMed] [Google Scholar]
- 9.Screpanti E, De Antoni A, Alushin GM, Petrovic A, Melis T, Nogales E, Musacchio A. Direct binding of Cenp-C to the Mis12 complex joins the inner and outer kinetochore. Curr Biol. 2011;21:391–398. doi: 10.1016/j.cub.2010.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Przewloka MR, Venkei Z, Bolanos-Garcia VM, Debski J, Dadlez M, Glover DM. CENP-C is a structural platform for kinetochore assembly. Curr Biol. 2011;21:399–405. doi: 10.1016/j.cub.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 11*.Gascoigne KE, Takeuchi K, Suzuki A, Hori T, Fukagawa T, Cheeseman IM. Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell. 2011;145:410–422. doi: 10.1016/j.cell.2011.03.031. [The authors demonstrate that the N-terminal regions of two kinetochore proteins, CENP-C and CENP-T, promote assembly of ectopic kinetochores. Together with references 6, 9 and 10 this paper sets the foundations for further studies in the area of kinetochore assembly.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12*.Kitajima TS, Ohsugi M, Ellenberg J. Complete kinetochore tracking reveals error-prone homologous chromosome biorientation in Mammalian oocytes. Cell. 2011;146:568–581. doi: 10.1016/j.cell.2011.07.031. [An imaging tour-de-force of chromosomes undergoing meiosis. The authors identify the prometaphase belt as a crucial intermediate of chromosome alignment. The paper makes a coherent pair with reference 13.] [DOI] [PubMed] [Google Scholar]
- 13*.Magidson V, O'Connell CB, Loncarek J, Paul R, Mogilner A, Khodjakov A. The Spatial Arrangement of Chromosomes during Prometaphase Facilitates Spindle Assembly. Cell. 2011;146:555–567. doi: 10.1016/j.cell.2011.07.012. [Complete tracking of chromosome movements in mitotic cells, identifying, like in meiotic cells (see reference 12), an intermediate configuration of chromosomes, the prometaphase belt.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai S, O'Connell CB, Khodjakov A, Walczak CE. Chromosome congression in the absence of kinetochore fibres. Nat Cell Biol. 2009;11:832–838. doi: 10.1038/ncb1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alushin G, Nogales E. Visualizing kinetochore architecture. Curr Opin Struct Biol. 2011 doi: 10.1016/j.sbi.2011.07.009. doi:10.1016/j.sbi.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tooley J, Stukenberg PT. The Ndc80 complex: integrating the kinetochore's many movements. Chromosome Res. 2011;19:377–391. doi: 10.1007/s10577-010-9180-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koshland DE, Mitchison TJ, Kirschner MW. Polewards chromosome movement driven by microtubule depolymerization in vitro. Nature. 1988;331:499–504. doi: 10.1038/331499a0. [DOI] [PubMed] [Google Scholar]
- 18*.McIntosh J, Grishchuk E, Morphew M, Efremov A, Zhudenkov K, Volkov V, Cheeseman I, Desai A, Mastronarde D, Ataullakhanov F. Fibrils Connect Microtubule Tips with Kinetochores: A Mechanism to Couple Tubulin Dynamics to Chromosome Motion. Cell. 2008;135:322–333. doi: 10.1016/j.cell.2008.08.038. [Electron tomography identifies slender kinetochore fibrils that connect chromosomes to the curved ends of microtubules. The authors demonstrate that the Ndc80 complex has the potential to act as a microtubule coupler in vitro.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19*.Powers AF, Franck AD, Gestaut DR, Cooper J, Gracyzk B, Wei RR, Wordeman L, Davis TN, Asbury CL. The Ndc80 kinetochore complex forms load-bearing attachments to dynamic microtubule tips via biased diffusion. Cell. 2009;136:865–875. doi: 10.1016/j.cell.2008.12.045. [The authors demonstrate that the Ndc80 complex can sustain load-bearing attachments to dynamic microtubules, and is capable of biased diffusion on the microtubule lattice.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20*.DeLuca KF, Lens SMA, DeLuca JG. Temporal changes in Hec1 phosphorylation control kinetochore-microtubule attachment stability during mitosis. J Cell Sci. 2011;124:622–634. doi: 10.1242/jcs.072629. [After proposing a role of phosphorylation of the Hec1 N-terminal tail in the regulation of the strength of kinetochore-microtubule attachment in reference 21, the authors characterize the role of individual phosphorylation sites during mitosis.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeLuca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, Salmon ED. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–982. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- 22.Nicklas RB, Ward SC. Elements of error correction in mitosis: microtubule capture, release, and tension. J Cell Biol. 1994;126:1241–1253. doi: 10.1083/jcb.126.5.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23**.Akiyoshi B, Sarangapani KK, Powers AF, Nelson CR, Reichow SL, Arellano-Santoyo H, Gonen T, Ranish JA, Asbury CL, Biggins S. Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature. 2010;468:576–579. doi: 10.1038/nature09594. [The authors purified kinetochore particles from S. cerevisiae, and tested their properties in single-molecule assays. A striking finding is that kinetochores stabilize microtubules in vitro merely through binding, regardless of phosphorylation state.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khodjakov A, Pines J. Centromere tension: a divisive issue. Nat Cell Biol. 2010;12:919–923. doi: 10.1038/ncb1010-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nezi L, Musacchio A. Sister chromatid tension and the spindle assembly checkpoint. Curr Opin Cell Biol. 2009 doi: 10.1016/j.ceb.2009.09.007. doi:10.1016/j.ceb.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 26.Biggins S, Murray AW. The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev. 2001;15:3118–3129. doi: 10.1101/gad.934801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanaka TU, Rachidi N, Janke C, Pereira G, Gálová M, Schiebel E, Stark MJR, Nasmyth K. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 2002;108:317–329. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- 28.Welburn JPI, Vleugel M, Liu D, Yates JR, Lampson MA, Fukagawa T, Cheeseman IM. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol Cell. 2010;38:383–392. doi: 10.1016/j.molcel.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ciferri C, De Luca J, Monzani S, Ferrari KJ, Ristic D, Wyman C, Stark H, Kilmartin J, Salmon ED, Musacchio A. Architecture of the human ndc80-hec1 complex, a critical constituent of the outer kinetochore. J Biol Chem. 2005;280:29088–29095. doi: 10.1074/jbc.M504070200. [DOI] [PubMed] [Google Scholar]
- 30**.Ciferri C, Pasqualato S, Screpanti E, Varetti G, Santaguida S, Reis Dos G, Maiolica A, Polka J, De Luca JG, De Wulf P, et al. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008;133:427–439. doi: 10.1016/j.cell.2008.03.020. [A thorough structural and functional analysis of the NDC80 complex, made possible by the generation of an engineered version of the complex named Ndc80bonsai.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei RR, Sorger PK, Harrison SC. Molecular organization of the Ndc80 complex, an essential kinetochore component. Proc Natl Acad Sci USA. 2005;102:5363–5367. doi: 10.1073/pnas.0501168102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsu K-S, Toda T. Ndc80 internal loop interacts with Dis1/TOG to ensure proper kinetochore-spindle attachment in fission yeast. Curr Biol. 2011;21:214–220. doi: 10.1016/j.cub.2010.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maure J-F, Komoto S, Oku Y, Mino A, Pasqualato S, Natsume K, Clayton L, Musacchio A, Tanaka TU. The Ndc80 loop region facilitates formation of kinetochore attachment to the dynamic microtubule plus end. Curr Biol. 2011;21:207–213. doi: 10.1016/j.cub.2010.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34*.Wei RR, Al-Bassam J, Harrison SC. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat Struct Mol Biol. 2007;14:54–59. doi: 10.1038/nsmb1186. [Structural characterization of the globular head of Hec1/Ndc80, and discovery that it contains a CH domain, previously implicated in microtubule and actin binding.] [DOI] [PubMed] [Google Scholar]
- 35.Buey RM, Mohan R, Leslie K, Walzthoeni T, Missimer JH, Menzel A, Bjelic S, Bargsten K, Grigoriev I, Smal I, et al. Insights into EB1 structure and the role of its C- terminal domain for discriminating microtubule tips from the lattice. Mol Biol Cell. 2011;22:2912–2923. doi: 10.1091/mbc.E11-01-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36*.Wilson-Kubalek EM, Cheeseman IM, Yoshioka C, Desai A, Milligan RA. Orientation and structure of the Ndc80 complex on the microtubule lattice. J Cell Biol. 2008;182:1055–1061. doi: 10.1083/jcb.200804170. [A three-dimensional reconstruction by helical image analysis of the Hec1-Nuf2 heterodimer from C. elegans bound to microtubules shows an alternation of strong and weak densities with 4 nm spacing.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37**.Alushin GM, Ramey VH, Pasqualato S, Ball DA, Grigorieff N, Musacchio A, Nogales E. The Ndc80 kinetochore complex forms oligomeric arrays along microtubules. Nature. 2010;467:805–810. doi: 10.1038/nature09423. [An 8.6 Å 3D reconstruction of NDC80bonsai complex bound to microtubules. After unequivocal fitting of crystal structures of Ndc80bonsai and tubulin, a molecular view of the kinetochore-microtubule interaction is made available for the first time.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38*.Sundin LJR, Guimaraes GJ, DeLuca JG. The NDC80 complex proteins Nuf2 and Hec1 make distinct contributions to kinetochore-microtubule attachment in mitosis. Mol Biol Cell. 2011;22:759–768. doi: 10.1091/mbc.E10-08-0671. [A thorough mutational and functional characterization of the mechanism of binding of the Ndc80 complex to microtubules. The study questions a direct role of Nuf2 in microtubule binding. A coherent pair with reference 39.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39*.Tooley JG, Miller SA, Stukenberg PT. The Ndc80 complex uses a tripartite attachment point to couple microtubule depolymerization to chromosome movement. Mol Biol Cell. 2011;22:1217–1226. doi: 10.1091/mbc.E10-07-0626. [Together with reference 38, this paper probes the role of different segments of the Ndc80 complex in microtubule binding.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Y, Riley DJ, Zheng L, Chen P-L, Lee W-H. Phosphorylation of the mitotic regulator protein Hec1 by Nek2 kinase is essential for faithful chromosome segregation. J Biol Chem. 2002;277:49408–49416. doi: 10.1074/jbc.M207069200. [DOI] [PubMed] [Google Scholar]
- 41.Wei R, Ngo B, Wu G, Lee W-H. Phosphorylation of the Ndc80 complex protein, HEC1, by Nek2 kinase modulates chromosome alignment and signaling of the spindle assembly checkpoint. Mol Biol Cell. 2011 doi: 10.1091/mbc.E11-01-0012. doi:10.1091/mbc.E11-01-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Du J, Cai X, Yao J, Ding X, Wu Q, Pei S, Jiang K, Zhang Y, Wang W, Shi Y, et al. The mitotic checkpoint kinase NEK2A regulates kinetochore microtubule attachment stability. Oncogene. 2008;27:4107–4114. doi: 10.1038/onc.2008.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nogales E, Wolf SG, Downing KH. Structure of the alpha beta tubulin dimer by electron crystallography. Nature. 1998;391:199–203. doi: 10.1038/34465. [DOI] [PubMed] [Google Scholar]
- 44*.Miller SA, Johnson ML, Stukenberg PT. Kinetochore attachments require an interaction between unstructured tails on microtubules and Ndc80(Hec1). Current Biology. 2008;18:1785–1791. doi: 10.1016/j.cub.2008.11.007. [Together with reference 45, this manuscript analyses the role of the disordered N-terminal tail of Hec1 in microtubule binding.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45*.Guimaraes GJ, Dong Y, McEwen BF, DeLuca JG. Kinetochore-microtubule attachment relies on the disordered N-terminal tail domain of Hec1. Current Biology. 2008;18:1778–1784. doi: 10.1016/j.cub.2008.08.012. [As a logical pair with reference 44, this paper investigates the role of the N-terminal tail of Hec1 in microtubule binding.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mattiuzzo M, Vargiu G, Totta P, Fiore M, Ciferri C, Musacchio A, Degrassi F. Abnormal kinetochore-generated pulling forces from expressing a N-terminally modified Hec1. PLoS ONE. 2011;6:e16307. doi: 10.1371/journal.pone.0016307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 48*.Johnston K, Joglekar A, Hori T, Suzuki A, Fukagawa T, Salmon ED. Vertebrate kinetochore protein architecture: protein copy number. J Cell Biol. 2010;189:937–943. doi: 10.1083/jcb.200912022. [With references 49 and 50, the last episode of a saga to characterize copy number of kinetochores proteins in different eukaryotes.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Joglekar AP, Bouck D, Finley K, Liu X, Wan Y, Berman J, He X, Salmon E, Bloom KS. Molecular architecture of the kinetochore-microtubule attachment site is conserved between point and regional centromeres. J Cell Biol. 2008;181:587–594. doi: 10.1083/jcb.200803027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Joglekar AP, Bouck DC, Molk JN, Bloom KS, Salmon ED. Molecular architecture of a kinetochore–microtubule attachment site. Nat Cell Biol. 2006;8:581–585. doi: 10.1038/ncb1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pinsky BA, Kung C, Shokat KM, Biggins S. The Ipl1-Aurora protein kinase activates the spindle checkpoint by creating unattached kinetochores. Nat Cell Biol. 2006;8:78–83. doi: 10.1038/ncb1341. [DOI] [PubMed] [Google Scholar]
- 52.Cimini D, Wan X, Hirel CB, Salmon ED. Aurora kinase promotes turnover of kinetochore microtubules to reduce chromosome segregation errors. Curr Biol. 2006;16:1711–1718. doi: 10.1016/j.cub.2006.07.022. [DOI] [PubMed] [Google Scholar]
- 53.Kemmler S, Stach M, Knapp M, Ortiz J, Pfannstiel J, Ruppert T, Lechner J. Mimicking Ndc80 phosphorylation triggers spindle assembly checkpoint signalling. EMBO J. 2009;28:1099–1110. doi: 10.1038/emboj.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54**.Wan X, O'Quinn RP, Pierce HL, Joglekar AP, Gall WE, DeLuca JG, Carroll CW, Liu ST, Yen TJ, McEwen BF, et al. Protein architecture of the human kinetochore microtubule attachment site. Cell. 2009;137:672–684. doi: 10.1016/j.cell.2009.03.035. [This thorough analysis of projection distances of kinetochore subunits along the inter- kinetochore axis provides a map of protein-protein interactions within kinetochores.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hill TL. Theoretical problems related to the attachment of microtubules to kinetochores. Proc Natl Acad Sci USA. 1985;82:4404–4408. doi: 10.1073/pnas.82.13.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pagliuca C, Draviam VM, Marco E, Sorger PK, De Wulf P. Roles for the conserved spc105p/kre28p complex in kinetochore-microtubule binding and the spindle assembly checkpoint. PLoS ONE. 2009;4:e7640. doi: 10.1371/journal.pone.0007640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lampert F, Westermann S. A blueprint for kinetochores - new insights into the molecular mechanics of cell division. Nat Rev Mol Cell Biol. 2011;12:407–412. doi: 10.1038/nrm3133. [DOI] [PubMed] [Google Scholar]
- 58.Westermann S, Drubin DG, Barnes G. Structures and functions of yeast kinetochore complexes. Annu Rev Biochem. 2007;76:563–591. doi: 10.1146/annurev.biochem.76.052705.160607. [DOI] [PubMed] [Google Scholar]
- 59.Theis M, Slabicki M, Junqueira M, Paszkowski-Rogacz M, Sontheimer J, Kittler R, Heninger A-K, Glatter T, Kruusmaa K, Poser I, et al. Comparative profiling identifies C13orf3 as a component of the Ska complex required for mammalian cell division. EMBO J. 2009;28:1453–1465. doi: 10.1038/emboj.2009.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Welburn JPI, Grishchuk EL, Backer CB, Wilson-Kubalek EM, Yates JR, Cheeseman IM. The human kinetochore Ska1 complex facilitates microtubule depolymerization-coupled motility. Dev Cell. 2009;16:374–385. doi: 10.1016/j.devcel.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hanisch A, Silljé HHW, Nigg EA. Timely anaphase onset requires a novel spindle and kinetochore complex comprising Ska1 and Ska2. EMBO J. 2006;25:5504–5515. doi: 10.1038/sj.emboj.7601426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gaitanos TN, Santamaria A, Jeyaprakash AA, Wang B, Conti E, Nigg EA. Stable kinetochore-microtubule interactions depend on the Ska complex and its new component Ska3/C13Orf3. EMBO J. 2009;28:1442–1452. doi: 10.1038/emboj.2009.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Raaijmakers JA, Tanenbaum ME, Maia AF, Medema RH. RAMA1 is a novel kinetochore protein involved in kinetochore-microtubule attachment. J Cell Sci. 2009;122:2436–2445. doi: 10.1242/jcs.051912. [DOI] [PubMed] [Google Scholar]
- 64.Dong Y, Vanden Beldt KJ, Meng X, Khodjakov A, McEwen BF. The outer plate in vertebrate kinetochores is a flexible network with multiple microtubule interactions. Nat Cell Biol. 2007;9:516–522. doi: 10.1038/ncb1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65**.Maresca TJ, Salmon ED. Intrakinetochore stretch is associated with changes in kinetochore phosphorylation and spindle assembly checkpoint activity. J Cell Biol. 2009;184:373–381. doi: 10.1083/jcb.200808130. [Together with reference 66, this paper identifies intrakinetochore stretch as a crucial factor in the control of the spindle assembly checkpoint response, associating structural changes in the kinetochore to checkpoint control.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66*.Uchida KS, Takagaki K, Kumada K, Hirayama Y, Noda T, Hirota T. Kinetochore stretching inactivates the spindle assembly checkpoint. J Cell Biol. 2009 doi: 10.1083/jcb.200811028. doi:10.1083/jcb.200811028. [Together with reference 65, this paper identifies the crucial role of intra-kinetochore stretch in the checkpoint response.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Joglekar AP, Bloom K, Salmon ED. In vivo protein architecture of the eukaryotic kinetochore with nanometer scale accuracy. Curr Biol. 2009;19:694–699. doi: 10.1016/j.cub.2009.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68*.Suzuki A, Hori T, Nishino T, Usukura J, Miyagi A, Morikawa K, Fukagawa T. Spindle microtubules generate tension-dependent changes in the distribution of inner kinetochore proteins. J Cell Biol. 2011;193:125–140. doi: 10.1083/jcb.201012050. [The authors demonstrate that the kinetochore protein CENP-T undergoes a tension- dependent conformational change that separates its N- and C-terminal ends along the kinetochore's axis.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hornung P, Maier M, Alushin GM, Lander GC, Nogales E, Westermann S. Molecular architecture and connectivity of the budding yeast Mtw1 kinetochore complex. J Mol Biol. 2011;405:548–559. doi: 10.1016/j.jmb.2010.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70*.Petrovic A, Pasqualato S, Dube P, Krenn V, Santaguida S, Cittaro D, Monzani S, Massimiliano L, Keller J, Tarricone A, et al. The MIS12 complex is a protein interaction hub for outer kinetochore assembly. J Cell Biol. 2010;190:835–852. doi: 10.1083/jcb.201002070. [A through biochemical and structural analysis of the mechanism of assembly of the human KMN network, which pairs with equivalent work on the S. cerevisiae's complex discussed in reference 71.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71*.Maskell DP, Hu X-W, Singleton MR. Molecular architecture and assembly of the yeast kinetochore MIND complex. J Cell Biol. 2010;190:823–834. doi: 10.1083/jcb.201002059. [Together with reference 70, this paper illustrates the organization of the yeast MTW1/MIND complex, equivalent to the MIS12 complex of higher eukaryotes.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang H-W, Long S, Ciferri C, Westermann S, Drubin D, Barnes G, Nogales E. Architecture and flexibility of the yeast Ndc80 kinetochore complex. J Mol Biol. 2008;383:894–903. doi: 10.1016/j.jmb.2008.08.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wei RR, Schnell JR, Larsen NA, Sorger PK, Chou JJ, Harrison SC. Structure of a central component of the yeast kinetochore: the Spc24p/Spc25p globular domain. Structure. 2006;14:1003–1009. doi: 10.1016/j.str.2006.04.007. [DOI] [PubMed] [Google Scholar]