Abstract
The nuclear pore complex has long been viewed as a point-like entry and exit channel between the nucleus and the cytoplasm. New data supports a different view whereby the complex displays distinct spatial dynamics of variable duration ranging from milliseconds to events spanning the entire cell cycle. Discrete interaction sites outside the central channel become apparent, and transport regulation at these sites seems to be of greater importance than currently thought. Nuclear pore components are highly active outside the nuclear pore complex or impact the fate of cargo transport away from the nuclear pore. The nuclear pore complex is a highly dynamic, crowded environment—constantly loaded with cargo while providing selectivity based on unfolded proteins. Taken together, this comprises a new paradigm in how we view import/export dynamics and emphasizes the multiscale nature of nuclear pore complex-mediated cellular transport.
Introduction
Compartmentalization is a uniform principle of cellular function in higher evolved organisms. The nuclear pore complex (NPC) mediates the exchange of matter, energy, and information between the two major compartments in eukaryotic cells, the nucleus and the cytoplasm [1]. Traffic between these two compartments is heavy; transport rates of individual NPCs have been found to be as high as 1000 transport complexes with cargo per second [2, 3]. That such numbers are physiologically relevant can be ascertained by estimating the number of histones and ribosomal proteins that need to enter the nucleus every cell cycle. A cell with an average number of NPCs (~3000) requires a transport rate of approximately 150 import events per NPC per second for histones and ribosomal proteins and two export events per NPC per second for ribosomal subunits [4]. This calculation, however, does not account for general mRNA export or proteins needed to maintain transcription, replication, or other nuclear processes. Interestingly, the transport capacity of NPCs is not rate limiting in vivo [5], although formation of transport complexes might very well be [6]. While small molecules below ~60kDa can passively diffuse through the central channel without transport receptor adaptation [7], however, a smaller cutoff size of 40kDa was also reported [8]. Many small nuclear proteins possess a NLS and use NPC mediated transport to shuttle into the nucleus, although they are below the size limit for passive diffusion [9]. At the same time NPCs can export cargo of substantial size such as mRNAs and ribosomal subunits, which range from ~15 nm (ribosomal subunits) to ~50 nm (Balbiani Ring mRNP) [4, 10–12]. Using gold particles bound to transport receptors, channel diameters of 24 nm were found to be readily accessible [13], and even ~30 nm Qdot transport complexes were located within the central channel [14]. Importantly, Balbiani Ring particles unfold for transport across the NPC, and the transport of flexible mRNAs is much faster than that of rigid artificial cargo of similar size [10–12, 14]. Unlike other channels, however, the NPC does not mandatorily require unfolding of its cargo, like it is the case, e.g., for transport about the membrane of the endoplasmic reticulum.
Spatial Symmetry within the NPC
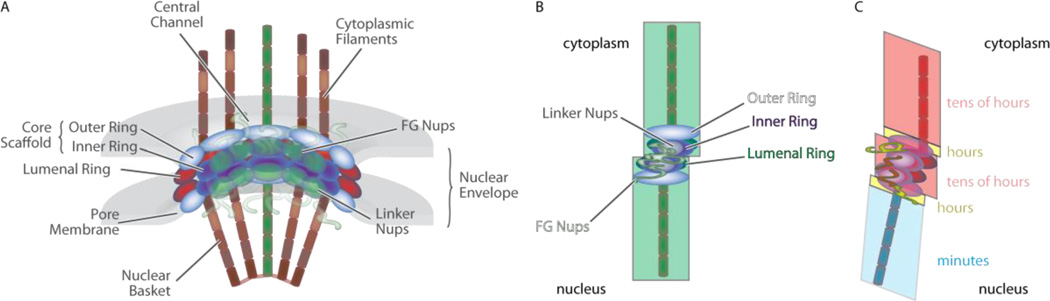
NPCs are super-protein complexes with an overall diameter of ~120 nm and a channel diameter of ~50 nm and up to 90 nm in length [15–17]. Electron microscopic images of the nuclear pore complex reveal a strict symmetry composed of eight spokes extending to the nuclear and cytoplasmic surfaces [16]. These spokes have three distinct regions, cytoplasmic filaments, a core structure, and the nuclear basket (Figure 1a). This, together with its shear mass (up to 120 MDa in frog oocytes and ~60 MDa in yeast) make NPCs arguably the largest nano-machine in the eukaryotic cell. Interestingly however, the NPC is built from a relative small number of parts. The NPC is built from approximately 30 different proteins comprising six structural motives, among them the natively unfolded phenylalanine–glycine (FG) repeat domains (concentrating towards the central channel) that form the permeability barrier [18]. While the radial symmetry of the NPC spokes is clearly maintained on the surfaces, the scaffold forming the actual pore in the membrane is a four-layered ring with the base number of eight and multiples thereof being maintained (Figure 1b) [19]. A trans-membrane domain anchors the NPC in the nuclear membrane, and three layers of rings, classified as coat, adaptor, and channel nucleoporins form the scaffold that hosts the FG-repeat in its center. While the exact orientation of subcomplexes in these rings is a matter of debate (two alternative models are termed fence pole and lattice [21, 22]), it is clear that the radial spoke symmetry of NPCs is at least supplemented by a rotational symmetric ring structure of the different layers in a central channel [20–22]. A large scale modeling approach combining available structural data on NPC subcomplexes as well as biochemical interaction data suggests that the radial symmetry might be partly broken by the ring like symmetry of nucleoporins in the central scaffold (Figure 1b) [19].
Figure 1.
NPC architecture – structural and dynamic features. A) General structure of the NPC. An NPC sliced through its center is shown revealing the octameric symmetry of its major features. The ring stack in the center extends outwards into the nucleus and cytoplasm by means of filaments resulting in eight spokes transversing the nuclear membrane. There is possibly a slight shift between the upper and lower rings as shown in B. The FG nups, anchored in the center, form the permeability barrier. C) Nucleoporins display a wide range of turnover rates within the NPC ranging from seconds to tens of hours. Core components are generally more stably associated compared to the asymmetric nucleoporins. Nup98 (yellow), the nucleoporin with the most FG repeats, has a medium association time (hours), while gb210 (see text), although part of the luminal ring shows fast turnover at the minutes time scale. (Adapted from Grünwald et al. 2011, Nature)
With respect to the transport of large cargo, the structure of the different ring layers in the central channel is of outstanding interest [23]. This is because “breathing” of the core structure could contribute to the translocation of large cargo. This concept has fueled the discussion about the existence of specialized pores; however, experimental evidence is limited [24–27].
The current transport models need to account for activity outside the central channel
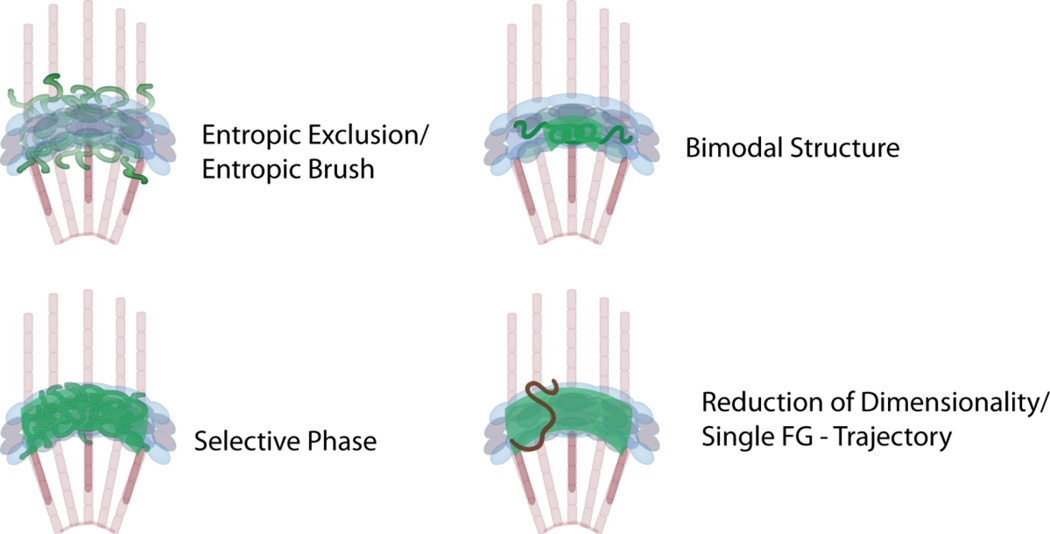
Cargo localizes to and from the nucleus across the NPC via a “ticket sequence”, i.e., the nuclear localization sequence (NLS) or nuclear export sequence (NES) [28]. This is mediated by a specific class of proteins and pathways (transport factors termed importins/exportins or karyopherins). Since the discovery of NLS and NES, different transport models have been discussed (Figure 2). While the translocation step does not require energy per se, gradients of specific metabolic energy are maintained across the nuclear membrane [29, 30]. For example, the RanGTPase gradient, by means of GTP to GDP dephosphorylation disassembles the transport complex, thereby rendering translocation irreversible for the cargo [31]. Transport receptors are recycled by RanGDP (and other factors) thereby maintaining the Ran gradient [32–34]. Based on the selectivity of transport in model systems like frog oocytes and digitonin permeabilized cells [35, 36], combined with evidence of multiple transport pathways [37–39], models for the function of the NPC have evolved over time [1, 40]. All current transport models postulate that a specific class of nucleoporins, rich in intrinsically unfolded FG repeat domains, provide an energy landscape that reduces the energy costs of translocation, while not interfering with cargo release from the NPC [41, 42]. The NPC acts as a molecular pump, which in its steady state supports a balanced flux of cargo resulting in a continuous back and forth of cargo across the NPC [43]. Net directionality of transport is the result of cargo release from the NPC resulting in an effective enrichment of concentration on one side of the NPC [29]. Unfolded nucleoporins, localized within the center of the central scaffold of the NPC form a permeability barrier sufficient for the sorting capability of the NPC [44, 45]. Interestingly, most models explicitly assume a transient interaction between transport complex and FG repeats, based on modeling of stochastic transport through a narrow channel. This would be sufficient to explain exclusion and enrichment of molecules [46]. This core structure of FG repeats has been a center of attention, based on groundbreaking work on the physical properties of the filaments that form the permeability barrier located in the central channel [19, 47–50]. Accordingly, the focus for differences in the current models is on the physical behavior of nucleoporins in the permeability barrier. These models (Figure 2) include: 1) “Entropic exclusion” based on volume occupied by FG repeat domains [41], 2) “Entopic Brush” like collapse of the FG repeats upon interaction with transport molecules [50], 3) formation of a “selective gel phase” by polymerization of FG repeats [48, 49], 4) a “Bimodal Structure” of FG repeats resulting in distinct transport regions within the central channel [51], 5) a “reduction of dimensionality” of transport by sliding of transport receptors and transport complexes along the channel wall coated with collapsed FG-repeat domains [52], and 6) a “single FG-repeat trajectory” of transport molecules along the NPC [53]. A key argument of the “reduction of dimensionality” model is that within the living cell it is likely that the NPC is always loaded with transport receptors, which may or may not be loaded with cargo [52].
Figure 2.
Nuclear pore complex transport models. In the entropic exclusion/entropic brush model the FG-Nups organize as a repulsing entropic barrier against non-specific cargo. Transport receptors mediate interaction of cargo complexes with this barrier thereby facilitating transport. In the selective phase model the FG-Nups form a physical gel-like barrier by dense FG–FG interaction. Transport receptors locally “melt” the gel allowing entrance and transition. In the bimodal structure model FG-Nups fold into different zones within the central channel resulting in a more gel-like center with less dense peripheries. This model predicts different spatial routes for different cargo translocation. The reduction of dimensionality model predicts that FG-Nups coat the wall of the central channel allowing for transport receptors to “slide” on a 2-dimensional surface across the NPC. The single trajectory hypothesis can be easily pictured as a discreet path across such a landscape but applies to all models presented.
While the NPC has long been viewed as a static, stable installation within the nuclear membrane, it is now clear that there is stratification among the NPC composed of different binding constants and binding times [54]. Interestingly, this dynamic nature is found not only for cargo in transit but also for nucleoporins themselves (Figure 1c). The building blocks of the NPC show turnover times covering five orders of magnitude ranging from seconds to days. In general, scaffold nucleoporins were found to be associated relatively stably (~10 h to 3 days) with the NPC (with the exception of gb210, an anchoring protein with a residence time of ~4 minutes) while peripheral nucleoporins exhibited shorter interaction times in the range of seconds to ~10 minutes [54]. The interaction times of cargo with the NPC is generally short; dwell times at the NPC of import complexes were between 1 and 100 milliseconds [5, 55, 56], while the dwell time of β-actin mRNA was between 180 milliseconds and more than 2 seconds [10]. The dwell time of mRNA from an engineered dystrophin gene was 5 to ~40 minutes [57], while quantum dots used as cargo exhibited dwell times from 2 seconds to 15 minutes [14]. The dynamic presence of mobile nucleoporins, transport complexes, and transport factors adds substantial mass to an already crowded environment [52]. Crowding has effects on the physical properties of the FG repeat domains [58], such as entropic changes caused for instance by “depletion attraction” a change in free volume due to molecular interactions of transport complexes, receptors, and FG repeat domains [59, 60]. While current transport models are elegant and account for the available data regarding transport selectivity, newly emerging spatial information on cargo interaction sites with the NPC in the living cell makes it necessary to re-think how transport is mediated outside the core structure of the central channel.
Differences between the dynamic profile of proteins and mRNA indicate cargo-specialization within the NPC topography
The extensions of the NPC into the nucleus and cytoplasm have been described as places of cargo modification [61, 62]. When artificial, fluorescently tagged protein cargos, Qdots and transport receptors were tracked from the cytoplasm to the nucleus, a single-peak distribution was found corresponding to the central channel (30–70 nm depending on the substrate) [5, 14, 56, 63]. Import times ranged from a few milliseconds to tens of milliseconds leading to the interpretation that the central channel is the determinant of the translocation process [3, 5, 14, 55, 56, 63, 64]. These data led to the widely held notion that cargo modification is largely uncoupled from translocation with the permeability barrier located in the central channel.
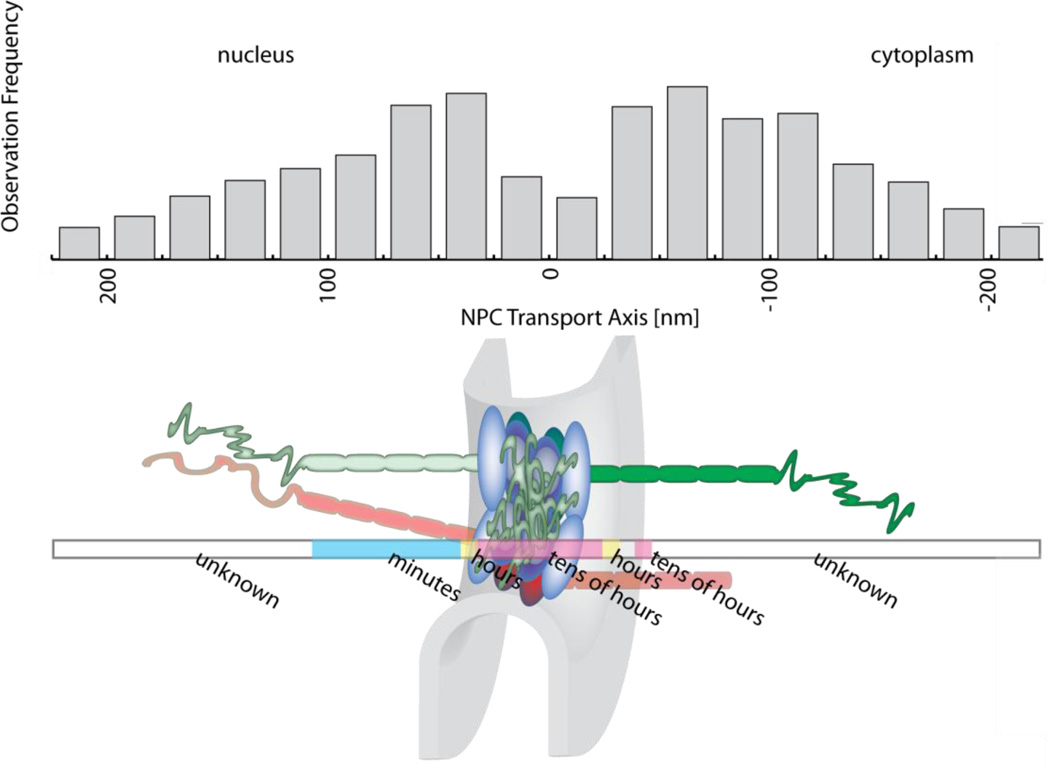
A very different picture, both spatially and dynamically, emerged when export of an endogenously tagged mRNA was followed from the nucleus to the cytoplasm [10]. A bimodal distribution with peaks located at the nuclear surface of the NPC and in the cytoplasm towards the outer edge of the nuclear filaments was observed (Figure 3). These sites, docking within the nucleus and release at the cytoplasm, showed kinetics of ~100 milliseconds for the majority of exporting mRNAs. A sub-population of mRNAs was found at these sites for at least a few seconds. Interestingly, in both cases the transit through the central channel was very fast (~5–20 msec) and comparable to the times for protein import. Using a modeling approach, a bimodal distribution of cargo was also predicted for import, but the separation between the peaks was found to be narrower than the experimental data on mRNA export [53]. The existence of translocation-limiting binding sites outside of the central channel for mRNA exports extends the transport process well beyond the permeability barrier in the central channel. A time-consuming release step is well in agreement with speculation that mRNA needs to be loaded with multiple transport factors that need to be removed to prevent re-import after export from the nucleus [65]. Structure data showing the interaction between DBP5 (DEAD box helicase active in RNA export), Nup214 (cytoplasmic nucleoporin) and mRNA are consistent with the low number of import events found for β-actin mRNA [10, 66, 67]. A common picture is emerging that the release of DBP5 from the mRNA functions in an ATP-dependent manner in conjunction with Gle1 (mRNA export factor) and Nup214 as a de facto ratchet for mRNA export [68–70]. The bimodal binding site distribution contrasts with the interaction of transport receptors with the NPC, which has been found to center closely on the central channel of the NPC. A symmetric binding site distribution is presumed based on 2D imaging of translocation of transport receptors through NPC in the equatorial plane of the nucleus and 3D interpolation of tracing data [5, 71]. Interestingly, an EM study based on ultrafast freezing of cells showed a distinct spatial distribution of cargo within the central channel [72]. Deletion of nucleoporin sections and labeling of different transport markers (either primary transport receptors or cofactors like DBP5 or Gle1) were used to determine if cargo traveled closer to the rim or the center of the central channel. These data strongly supported spatially separate transport pathways within the NPC; mRNA is likely to travel more centrally while smaller cargo might travel more towards the periphery of the central channel [72]. A similar prediction was made based on the biophysical interpretation of hydrodynamic diameters of nucleoporins contributing to the permeability barrier [51].
Figure 3.
NPC-mediated mRNA export. mRNA docks (~80 msec), translocates (>20 msec) and releases (~80 msec) in a three-step kinetic process across the NPC. The grey bars indicate the binding site distribution of exporting mRNA along the NPC axis and are combined with the turnover rates shown in Figure 1. The roles of individual nucleoporins on nuclear and cytoplasmic processes away from the NPC add to in a multiscale space and time topography of NPC function. Addition of more data on import cargoes, genetic mutants and disease related transport defects will open a new view on many subcellular processes in the future.
Understanding of transport regulation will rely on how the extended binding site distribution along the transport axis of the NPC and the spatial organization of transport zones orthogonal to the transport axis interact for different cargo types. This is especially interesting as the formation of the receptor-cargo complex and access of the complex to the central channel are two rate-limiting steps for the transport process.
The life of nucleoporins away from the NPC
Multiple nucleoporins have been found to have additional functions outside the NPC. The nucleoporins Sec13, Nup88 (which is overexpressed in tumors [73]) and Nup98 were shown to localize to chromatin in the absence of NPCs in Drosophila; Nup98 and Sec13 were also identified as transcription factors [74]. Among ciliates (a unicellular organism) Nup98 distinguishes the transcriptionally active macronucleus from the inactive micronucleus, and is responsible for differences in import specificity of the NPC in the two nuclei [75]. Also, in Drosophila, Nup153 was found to be involved in transcription [76]. Nup98 together with Nup50 and Nup62 were shown to impact gene expression in S2 cells by acting as transcription factors [77]. In yeast the TREX2 complex links transcription of at least a subset of genes to the NPC [78]. Various other aspects of the relationship of nucleoporins to the nuclear structure of the genome have recently been reviewed (cell differentiation, [79]; regulatory functions, [79]; cancer and nuclear structure, [80]). Tpr, the major constituent of the nuclear basket, has been reported to function not only as a docking site in import and export, but also to be required for the formation of heterochromatin-free areas close to NPCs, which are thought to be needed to regulate accessibility of NPCs to cargo [81, 82]. The interaction of nuclear Nup60 with localized mRNA is, however, necessary to maintain the proper delivery of the mRNA (shown for ASH1 and IST2 mRNA) to the bud of the yeast cell, placing nucleoporins not only in a context of nuclear interactions but extending their reign into the cytoplasm [83].
Summary
Trafficking of cargo between nucleus and cytoplasm mediated by the NPC spans time scales ranging from milliseconds to the entire duration of the cell cycle and beyond, length scales from 50 nm for the permeability barrier to multiple 100 nm for the docking and release sites, and localizations from an individual pore at the transcription site and the cytoplasm covering micrometer distances (Figure 3). These features make the NPC a multi-scale player on the cellular level. The ability to follow individual proteins and RNA complexes in real time in the living cell, and super-imposing spectrally-resolved single molecule signals with high resolution, has opened a window on the functional details of cargo translocation. Extending this technology towards single mRNA imaging in yeast promises to reveal further insights into the mechanisms underlying nucleocytoplasmic transport.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Walde S, Kehlenbach RH. The Part and the Whole: functions of nucleoporins in nucleocytoplasmic transport. Trends Cell Biol. 2010;20:461–469. doi: 10.1016/j.tcb.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 2.Ribbeck K, Gorlich D. Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 2001;20:1320–1330. doi: 10.1093/emboj/20.6.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang W, Gelles J, Musser SM. Imaging of single-molecule translocation through nuclear pore complexes. Proc Natl Acad Sci U S A. 2004;101:12887–12892. doi: 10.1073/pnas.0403675101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cook PR. Principles of nuclear structure and function. New York: J. Wiley and Sons; 2001. [Google Scholar]
- 5.Dange T, Grunwald D, Grunwald A, Peters R, Kubitscheck U. Autonomy and robustness of translocation through the nuclear pore complex: a single-molecule study. J Cell Biol. 2008;183:77–86. doi: 10.1083/jcb.200806173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Timney BL, Tetenbaum-Novatt J, Agate DS, Williams R, Zhang WZ, Chait BT, Rout MP. Simple kinetic relationships and nonspecific competition govern nuclear import rates in vivo. J. Cell Biol. 2006;175:579–593. doi: 10.1083/jcb.200608141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gorlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Bi. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 8.Kiseleva E, Goldberg MW, Allen TD, Akey CW. Active nuclear pore complexes in Chironomus: visualization of transporter configurations related to mRNP export. Journal of Cell Science. 1998;111:223–236. doi: 10.1242/jcs.111.2.223. [DOI] [PubMed] [Google Scholar]
- 9.Breeuwer M, Goldfarb DS. Facilitated nuclear transport of histone H1 and other small nucleophilic proteins. Cell. 1990;60:999–1008. doi: 10.1016/0092-8674(90)90348-i. [DOI] [PubMed] [Google Scholar]
- 10. Grunwald D, Singer R. In Vivo Imaging of Labelled Endogenous β-actin mRNA During Nucleocytoplasmic Transport. Nature. 2010;467:604–607. doi: 10.1038/nature09438.. ** This is the first study to follow a single mRNA in detail through the, NPC, showing that overall transport times are fast, ~hundreds of milliseconds, and that docking and release are visible kinetic steps.
- 11.Stevens BJ, Swift H. RNA transport from nucleus to cytoplasm in Chironomus salivary glands. The Journal of cell biology. 1966;31:55–77. doi: 10.1083/jcb.31.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mehlin H, Daneholt B, Skoglund U. Translocation of a Specific Premessenger Ribonucleoprotein Particle through the Nuclear-Pore Studied with Electron-Microscope Tomography. Cell. 1992;69:605–613. doi: 10.1016/0092-8674(92)90224-z. [DOI] [PubMed] [Google Scholar]
- 13.Kiseleva E, Allen TD, Rutherford S, Bucci M, Wente SR, Goldberg MW. Yeast nuclear pore complexes have a cytoplasmic ring and internal filaments. J Struct Biol. 2004;145:272–288. doi: 10.1016/j.jsb.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 14. Lowe AR, Siegel JJ, Kalab P, Siu M, Weis K, Liphardt JT. Selectivity mechanism of the nuclear pore complex characterized by single cargo tracking. Nature. 2010;467:600–603. doi: 10.1038/nature09285.. ** This paper presents the constraints on large cargo transport for artificial, not deformable, cargo, showing the lower time limit for NPC translocation and the upper limit for cargo diameter.
- 15.Beck M, Forster F, Ecke M, Plitzko JM, Melchior F, Gerisch G, Baumeister W, Medalia O. Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science. 2004;306:1387–1390. doi: 10.1126/science.1104808. [DOI] [PubMed] [Google Scholar]
- 16.Stoffler D, Feja B, Fahrenkrog B, Walz J, Typke D, Aebi U. Cryo-electron tomography provides novel insights into nuclear pore architecture: Implications for nucleocytoplasmic transport. J. Mol. Biol. 2003;328:119–130. doi: 10.1016/s0022-2836(03)00266-3. [DOI] [PubMed] [Google Scholar]
- 17.Huve J, Wesselmann R, Kahms M, Peters R. 4Pi microscopy of the nuclear pore complex. Biophys J. 2008;95:877–885. doi: 10.1529/biophysj.107.127449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Devos D, Dokudovskaya S, Williams R, Alber F, Eswar N, Chait BT, Rout MP, Sali A. Simple fold composition and modular architecture of the nuclear pore complex. Proc Natl Acad Sci U S A. 2006;103:2172–2177. doi: 10.1073/pnas.0506345103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- 20.Kampmann M, Blobel G. Three-dimensional structure and flexibility of a membrane-coating module of the nuclear pore complex. Nat Struct Mol Biol. 2009;16:782–788. doi: 10.1038/nsmb.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boehmer T, Jeudy S, Berke IC, Schwartz TU. Structural and functional studies of Nup107/Nup133 interaction and its implications for the architecture of the nuclear pore complex. Mol Cell. 2008;30:721–731. doi: 10.1016/j.molcel.2008.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Debler EW, Ma Y, Seo HS, Hsia KC, Noriega TR, Blobel G, Hoelz A. A fence-like coat for the nuclear pore membrane. Mol Cell. 2008;32:815–826. doi: 10.1016/j.molcel.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 23.Hoelz A, Debler EW, Blobel G. The structure of the nuclear pore complex. Annual review of biochemistry. 2011;80:613–643. doi: 10.1146/annurev-biochem-060109-151030. [DOI] [PubMed] [Google Scholar]
- 24.Wolf C, Mofrad MR. On the octagonal structure of the nuclear pore complex: insights from coarse-grained models. Biophys J. 2008;95:2073–2085. doi: 10.1529/biophysj.108.130336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lezon TR, Sali A, Bahar I. Global motions of the nuclear pore complex: insights from elastic network models. PLoS computational biology. 2009;5:e1000496. doi: 10.1371/journal.pcbi.1000496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iborra FJ, Jackson DA, Cook PR. The path of RNA through nuclear pores: apparent entry from the sides into specialized pores. J Cell Sci. 2000;113(Pt 2):291–302. doi: 10.1242/jcs.113.2.291. [DOI] [PubMed] [Google Scholar]
- 27.Perez-Terzic C, Behfar A, Mery A, van Deursen JM, Terzic A, Puceat M. Structural adaptation of the nuclear pore complex in stem cell-derived cardiomyocytes. Circ Res. 2003;92:444–452. doi: 10.1161/01.RES.0000059415.25070.54. [DOI] [PubMed] [Google Scholar]
- 28.Macara IG. Transport into and out of the nucleus. Microbiol Mol Biol Rev. 2001;65:570–594. doi: 10.1128/MMBR.65.4.570-594.2001. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gorlich D, Pante N, Kutay U, Aebi U, Bischoff FR. Identification of different roles for RanGDP and RanGTP in nuclear protein import. Embo Journal. 1996;15:5584–5594. [PMC free article] [PubMed] [Google Scholar]
- 30.Lyman SK, Guan T, Bednenko J, Wodrich H, Gerace L. Influence of cargo size on Ran and energy requirements for nuclear protein import. J Cell Biol. 2002;159:55–67. doi: 10.1083/jcb.200204163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith AE, Slepchenko BM, Schaff JC, Loew LM, Macara IG. Systems analysis of Ran transport. Science. 2002;295:488–491. doi: 10.1126/science.1064732. [DOI] [PubMed] [Google Scholar]
- 32.Gorlich D, Seewald MJ, Ribbeck K. Characterization of Ran-driven cargo transport and the RanGTPase system by kinetic measurements and computer simulation. Embo Journal. 2003;22:1088–1100. doi: 10.1093/emboj/cdg113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kalab P, Pralle A, Isacoff EY, Heald R, Weis K. Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature. 2006;440:697–701. doi: 10.1038/nature04589. [DOI] [PubMed] [Google Scholar]
- 34.Melchior F, Guan T, Yokoyama N, Nishimoto T, Gerace L. GTP hydrolysis by Ran occurs at the nuclear pore complex in an early step of protein import. The Journal of cell biology. 1995;131:571–581. doi: 10.1083/jcb.131.3.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cassany A, Gerace L. Reconstitution of nuclear import in permeabilized cells. Methods Mol Biol. 2009;464:181–205. doi: 10.1007/978-1-60327-461-6_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keminer O, Siebrasse JP, Zerf K, Peters R. Optical recording of signal-mediated protein transport through single nuclear pore complexes. Proc Natl Acad Sci U S A. 1999;96:11842–11847. doi: 10.1073/pnas.96.21.11842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Izaurralde E. A novel family of nuclear transport receptors mediates the export of messenger RNA to the cytoplasm. Eur J Cell Biol. 2002;81:577–584. doi: 10.1078/0171-9335-00273. [DOI] [PubMed] [Google Scholar]
- 38.Camblong J, Stutz F. Analysis of mRNA export pathways by genome-wide studies of mRNA export mutants. Yeast. 2003;20:S345–S345. [Google Scholar]
- 39.Fried H, Kutay U. Nucleocytoplasmic transport: taking an inventory. Cell Mol Life Sci. 2003;60:1659–1688. doi: 10.1007/s00018-003-3070-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grunwald D, Singer RH, Rout M. Nuclear export dynamics of RNA-protein complexes. Nature. 2011;475:333–341. doi: 10.1038/nature10318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rout MP, Aitchison JD, Magnasco MO, Chait BT. Virtual gating and nuclear transport: the hole picture. Trends Cell Biol. 2003;13:622–628. doi: 10.1016/j.tcb.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 42.Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao YM, Chait BT. The yeast nuclear pore complex: Composition, architecture, and transport mechanism. J. Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kopito RB, Elbaum M. Nucleocytoplasmic transport: a thermodynamic mechanism. HFSP J. 2009;3:130–141. doi: 10.2976/1.3080807.. * This paper presents a detailed picture of transport directionality based on fluorescence fluctiation spectroscopy, showing how equilibrium states imoact nucleocytoplsmic transport.
- 44.Patel SS, Belmont BJ, Sante JM, Rexach MF. Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell. 2007;129:83–96. doi: 10.1016/j.cell.2007.01.044. [DOI] [PubMed] [Google Scholar]
- 45.Strawn LA, Shen TX, Shulga N, Goldfarb DS, Wente SR. Minimal nuclear pore complexes define FG repeat domains essential for transport. Nature Cell Biology. 2004;6:197–206. doi: 10.1038/ncb1097. [DOI] [PubMed] [Google Scholar]
- 46. Zilman A, Di Talia S, Jovanovic-Talisman T, Chait BT, Rout MP, Magnasco MO. Enhancement of transport selectivity through nano-channels by non-specific competition. PLoS computational biology. 2010;6:e1000804. doi: 10.1371/journal.pcbi.1000804.. * This paper presents a theoretical framework for modeling nucleocytoplasmic transport and explain selectivity therein by properties of the translocation channel.
- 47.Jovanovic-Talisman T, Tetenbaum-Novatt J, McKenney AS, Zilman A, Peters R, Rout MP, Chait BT. Artificial nanopores that mimic the transport selectivity of the nuclear pore complex. Nature. 2008 doi: 10.1038/nature07600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frey S, Richter RP, Gorlich D. FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science. 2006;314:815–817. doi: 10.1126/science.1132516. [DOI] [PubMed] [Google Scholar]
- 49.Frey S, Gorlich D. A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell. 2007;130:512–523. doi: 10.1016/j.cell.2007.06.024. [DOI] [PubMed] [Google Scholar]
- 50.Lim RY, Fahrenkrog B, Koser J, Schwarz-Herion K, Deng J, Aebi U. Nanomechanical basis of selective gating by the nuclear pore complex. Science. 2007;318:640–643. doi: 10.1126/science.1145980. [DOI] [PubMed] [Google Scholar]
- 51. Yamada J, Phillips JL, Patel S, Goldfien G, Calestagne-Morelli A, Huang H, Reza R, Acheson J, Krishnan VV, Newsam S, et al. A bimodal distribution of two distinct categories of intrinsically disordered structures with separate functions in FG nucleoporins. Mol Cell Proteomics. 2010;9:2205–2224. doi: 10.1074/mcp.M000035-MCP201.. ** This paper shows that the unfolded FG-repeat domains can assume different global shapes and postulates how this can impact the physical structure of the permeability barrier.
- 52.Peters R. Translocation through the nuclear pore complex: selectivity and speed by reduction-of-dimensionality. Traffic. 2005;6:421–427. doi: 10.1111/j.1600-0854.2005.00287.x. [DOI] [PubMed] [Google Scholar]
- 53. Mincer JS, Simon SM. Simulations of nuclear pore transport yield mechanistic insights and quantitative predictions. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:E351–E358. doi: 10.1073/pnas.1104521108.. * This modeling study predicts a general bi-modal distribution of binding sites for cargo at the NPC.
- 54.Rabut G, Doye V, Ellenberg J. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nature cell biology. 2004;6:1114–1121. doi: 10.1038/ncb1184. [DOI] [PubMed] [Google Scholar]
- 55.Sun C, Yang W, Tu LC, Musser SM. Single-molecule measurements of importin alpha/cargo complex dissociation at the nuclear pore. Proc Natl Acad Sci U S A. 2008;105:8613–8618. doi: 10.1073/pnas.0710867105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kubitscheck U, Grunwald D, Hoekstra A, Rohleder D, Kues T, Siebrasse JP, Peters R. Nuclear transport of single molecules: dwell times at the nuclear pore complex. J Cell Biol. 2005;168:233–243. doi: 10.1083/jcb.200411005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mor A, Suliman S, Ben-Yishay R, Yunger S, Brody Y, Shav-Tal Y. Dynamics of single mRNP nucleocytoplasmic transport and export through the nuclear pore in living cells. Nat Cell Biol. 2010;12:543–552. doi: 10.1038/ncb2056.. * This study uses various large exogenous mRNP cargos and follows them in vivo. Their progress from the transcription site to the NPC is shown to be slow (minutes), whereas nuclear transport is observed to be slow but modeled to have a fast translocation step.
- 58.Kim YC, Best RB, Mittal J. Macromolecular crowding effects on protein-protein binding affinity and specificity. The Journal of chemical physics. 2010;133:205101. doi: 10.1063/1.3516589. [DOI] [PubMed] [Google Scholar]
- 59.Marenduzzo D, Finan K, Cook PR. The depletion attraction: an underappreciated force driving cellular organization. J Cell Biol. 2006;175:681–686. doi: 10.1083/jcb.200609066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chari A, Fischer U. Cellular strategies for the assembly of molecular machines. Trends Biochem.Sci. 2010;35:676–683. doi: 10.1016/j.tibs.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 61.Hutten S, Flotho A, Melchior F, Kehlenbach RH. The Nup358-RanGAP complex is required for efficient importin alpha/beta-dependent nuclear import. Mol Biol Cell. 2008;19:2300–2310. doi: 10.1091/mbc.E07-12-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 63. Kahms M, Lehrich P, Huve J, Sanetra N, Peters R. Binding site distribution of nuclear transport receptors and transport complexes in single nuclear pore complexes. Traffic. 2009;10:1228–1242. doi: 10.1111/j.1600-0854.2009.00947.x.. * A detailed study on the interaction sides of multiple cargos and transport receptors with the NPC.
- 64.Yang W, Musser SM. Nuclear import time and transport efficiency depend on importin beta concentration. J Cell Biol. 2006;174:951–961. doi: 10.1083/jcb.200605053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stewart M. Ratcheting mRNA out of the nucleus. Mol Cell. 2007;25:327–330. doi: 10.1016/j.molcel.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 66.von Moeller H, Basquin C, Conti E. The mRNA export protein DBP5 binds RNA and the cytoplasmic nucleoporin NUP214 in a mutually exclusive manner. Nat Struct Mol Biol. 2009;16:247–254. doi: 10.1038/nsmb.1561. [DOI] [PubMed] [Google Scholar]
- 67.Napetschnig J, Kassube SA, Debler EW, Wong RW, Blobel Gn, Hoelz A. Structural and functional analysis of the interaction between the nucleoporin Nup214 and the DEAD-box helicase Ddx19. Proceedings of the National Academy of Sciences. 2009;106:3089–3094. doi: 10.1073/pnas.0813267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Montpetit B, Thomsen ND, Helmke KJ, Seeliger MA, Berger JM, Weis K. A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP(6) in mRNA export. Nature. 2011;472:238–242. doi: 10.1038/nature09862.. ** This study presents the atomic structures of protein complexes for mRNA and factors that have been implicated in NPC-related export, and provides a model for how the release step of large cargo from the NPC is achieved.
- 69.Alcazar-Roman AR, Bolger TA, Wente SR. Control of mRNA export and translation termination by inositol hexakisphosphate requires specific interaction with Gle1. J Biol Chem. 2010;285:16683–16692. doi: 10.1074/jbc.M109.082370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hodge CA, Tran EJ, Noble KN, Alcazar-Roman AR, Ben-Yishay R, Scarcelli JJ, Folkmann AW, Shav-Tal Y, Wente SR, Cole CN. The Dbp5 cycle at the nuclear pore complex during mRNA export I: dbp5 mutants with defects in RNA binding and ATP hydrolysis define key steps for Nup159 and Gle1. Gene Dev. 2011;25:1052–1064. doi: 10.1101/gad.2041611.. ** This study presents an alternative view of the molecular steps involved in the DBP5 dependent release of cargo from the NPC after export.
- 71. Ma J, Yang W. Three-dimensional distribution of transient interactions in the nuclear pore complex obtained from single-molecule snapshots. Proc Natl Acad Sci U S A. 2010;107:7305–7310. doi: 10.1073/pnas.0908269107.. * A first 3D topography of transport traces across the NPC. Using an extensive theoretical frame work to interpolate a third spatial diemnsion in 2D tracking data this paper suggest the existence of disting transport routes accross the NPC
- 72. Fiserova J, Richards SA, Wente SR, Goldberg MW. Facilitated transport and diffusion take distinct spatial routes through the nuclear pore complex. J Cell Sci. 2010;123:2773–2780. doi: 10.1242/jcs.070730.. ** This study uses super-fast freezing of samples to capture cargo within the NPC in intact cells, demonstrating that cargo can travel along specific routes in the NPC.
- 73.Martinez N, Alonso A, Moragues MD, Ponton J, Schneider J. The nuclear pore complex protein Nup88 is overexpressed in tumor cells. Cancer research. 1999;59:5408–5411. [PubMed] [Google Scholar]
- 74.Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell. 2010;140:372–383. doi: 10.1016/j.cell.2009.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Iwamoto M, Asakawa H, Hiraoka Y, Haraguchi T. Nucleoporin Nup98: a gatekeeper in the eukaryotic kingdoms. Genes to cells : devoted to molecular & cellular mechanisms. 2010;15:661–669. doi: 10.1111/j.1365-2443.2010.01415.x. [DOI] [PubMed] [Google Scholar]
- 76.Vaquerizas JM, Suyama R, Kind J, Miura K, Luscombe NM, Akhtar A. Nuclear pore proteins nup153 and megator define transcriptionally active regions in the Drosophila genome. PLoS genetics. 2010;6:e1000846. doi: 10.1371/journal.pgen.1000846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kalverda B, Pickersgill H, Shloma VV, Fornerod M. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell. 2010;140:360–371. doi: 10.1016/j.cell.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 78.Dieppois G, Stutz F. Connecting the transcription site to the nuclear pore: a multi-tether process that regulates gene expression. Journal of Cell Science. 2010;123:1989–1999. doi: 10.1242/jcs.053694. [DOI] [PubMed] [Google Scholar]
- 79.Yasuhara N, Oka M, Yoneda Y. The role of the nuclear transport system in cell differentiation. Semin Cell Dev Biol. 2009;20:590–599. doi: 10.1016/j.semcdb.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 80.Kohler A, Hurt E. Gene regulation by nucleoporins and links to cancer. Mol. Cell. 2010;38:6–15. doi: 10.1016/j.molcel.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 81. Krull S, Dorries J, Boysen B, Reidenbach S, Magnius L, Norder H, Thyberg J, Cordes VC. Protein Tpr is required for establishing nuclear pore-associated zones of heterochromatin exclusion. The EMBO journal. 2010;29:1659–1673. doi: 10.1038/emboj.2010.54.. ** This study shows how the NPC impacts the physical structure of its spatial envioronment, creating access areas for cargo to and from the nuclear space.
- 82.Schermelleh L, Carlton PM, Haase S, Shao L, Winoto L, Kner P, Burke B, Cardoso MC, Agard DA, Gustafsson MG, et al. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science. 2008;320:1332–1336. doi: 10.1126/science.1156947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Powrie EA, Zenklusen D, Singer RH. A nucleoporin, Nup60p, affects the nuclear and cytoplasmic localization of ASH1 mRNA, in, S. cerevisiae. RNA. 2010 doi: 10.1261/rna.1210411. [DOI] [PMC free article] [PubMed] [Google Scholar]