Abstract
Smac mimetics block inhibitor of apoptosis (IAP) proteins to trigger TNFα-dependent apoptosis in cancer cells. However, only a small subset of cancer cells appear to be sensitive to Smac mimetics and even sensitive cells can develop resistance. Herein, we elucidated mechanisms underlying the intrinsic and acquired resistance of cancer cells to Smac mimetics. In vitro and in vivo investigations revealed that the expression of the cell surface protein LRIG1, a negative regulator of receptor tyrosine kinases (RTKs), is downregulated in resistant derivatives of breast cancer cells sensitive to Smac mimetics. RNAi-mediated down-regulation of LRIG1 markedly attenuated the growth inhibitory activity of the Smac mimetic SM-164 in drug-sensitive breast and ovarian cancer cells. Further, LRIG1 downregulation attenuated TNFα gene expression induced by Smac mimetics and increased the activity of multiple RTKs, including c-Met and Ron. The multitargeted tyrosine kinase inhibitors Crizotinib and GSK1363089 greatly enhanced the anticancer activity of SM-164 in all resistant cell derivatives, with the combination of SM-164 and GSK1363089 also completely inhibiting the outgrowth of resistant tumors in vivo. Together, our findings show that both upregulation of RTK signaling and attenuated TNFα expression caused by LRIG1 downregulation confers resistance to Smac mimetics, with implications for a rational combination strategy.
Keywords: IAPs, small-molecule inhibitors, resistance
Introduction
Dysfunction of apoptosis is a hallmark of human cancer and confers resistance to chemo- and radio-therapy. Targeting key apoptosis regulators such as IAPs with the goal of overcoming apoptosis resistance of cancer cells is an attractive therapeutic strategy. The mammalian family of IAPs includes cellular IAP1 (cIAP1, BIRC2), cIAP2 (BIRC3), X-linked IAP (XIAP, BIRC4), survivin (BIRC5) and melanoma IAP (ML-IAP, BIRC7), which are frequently overexpressed in cancer cells and associated with chemoresistance, disease progression and poor prognosis (1–3). IAPs such as XIAP and cIAP1/2 are endogenously antagonized by Smac/DIABLO (second mitochondria-derived activator of caspases/direct IAP binding with low pI) through interaction of the N-terminal AVPI tetrapeptide of Smac with the baculovirus IAP repeat domains of IAPs (4, 5). Small molecules that have been designed to mimic the IAP-binding AVPI motif of Smac (known as Smac mimetics) function as antagonists of multiple IAPs. Smac mimetics can potently induce apoptosis in some cancer cell lines and inhibit tumor growth in xenograft tumor models (6–11). Five such compounds are currently in early clinical development as a novel class of anticancer drugs (1, 2).
Smac mimetics target cIAP1/2 and XIAP to induce TNFα-dependent apoptosis (8–10). However, only a small subset of cancer cell lines is prone to Smac mimetic-induced apoptosis (8, 12). Exogenous TNFα, TRAIL (6, 8, 12) and IL-1β (13) all significantly enhance the ability of Smac mimetics to induce apoptosis. Downregulation of c-FLIP also enhances Smac mimetic-induced apoptosis in a variety of human cancer cell lines (12). Preclinical studies showed that xenograft tumors initially responding to Smac mimetic treatment regrow after treatment ends, indicating emergence of Smac mimetic-tolerant cells in the tumors (8). In lung cancer cells, cIAP2 protein levels rebound following Smac mimetic treatment, and inhibition of cIAP2 expression sensitizes resistant cells to Smac mimetic treatment in vitro, suggesting one mechanism for tumor cells to evade Smac mimetic-induced apoptosis (14).
Hence, the clinical benefit of Smac mimetic-based cancer therapy will be limited by intrinsic and/or acquired drug resistance. An in-depth understanding of the resistant mechanisms to Smac mimetics will help to identify the patients most likely to respond to this new class of drugs and to develop mechanistic-based approaches to overcome drug resistance. In this study, we investigated the mechanisms of resistance to Smac mimetics in breast and ovarian cancer cells using a highly potent Smac mimetic SM-164, which contains two non-peptidic mimetics of the AVPI tetrapeptide binding motifs and mimics the natural dimeric Smac protein (7, 15).
Materials and Methods
Reagents and Antibodies
Bivalent SM-164 and monovalent SM-406/AT-406 were described previously (7, 16). PF2341066 (Crizotinib), GSK1363089 (XL-880), AT-7867, U0126 and SKI-606 were purchased from Selleck Chemicals. A detailed list of reagents and antibodies used are in SI Materials and Methods.
Cell Culture, Cell Viability and Apoptosis Assays
MDA-MB-231, SKOV-3, MDA-MB-453, HCC38, HCC70 and OVCAR-4 cell lines were purchased from American Type Culture Collection (ATCC) and cultured as recommended. All these cell lines were authenticated by ATCC and were passaged fewer than 6 months after purchase for all the experiments. Cell viability was evaluated by a WST-8 assay (Dojindo (7). Apoptosis was analyzed using an Annexin V/propidium iodide (PI) detection kit (Roche). In combination studies, drugs were added simultaneously to the cells and combination index (CI) was calculated using the Chou-Talalay method (17).
Microarray Analysis and Quantitative RT-PCR
Total RNA was isolated with an RNA miniprep kit (Sigma). Affymetrix human U133 Plus 2.0 chips were used for the hybridization. For real-time PCR, cDNA was synthesized using cDNA reverse transcription kits (Applied Biosystems). Taqman gene expression assays were performed with TNFα (Hs00174128_m1), LRIG1 (Hs00394267_m1) and GAPDH (Hs99999905_ml) gene specific primers/probe sets (Applied Biosystems) using an ABI 7900HT PCR machine. GAPDH was used for normalization. Relative quantitation of mRNA was calculated by the comparative cycle threshold (Ct) method. All microarray data are available to the reviewers and have been deposited into the GEO repository (GEO accession number: GSE33827).
RNA Interference, Immunoblotting and TNFα ELISA
ON-TARGETplus SMARTpools for LRIG1, TNFR1, Ron, c-Met and non-targeting negative control siRNAs were from Dhamarcon. Transfection was performed using Lipofectamine RNAiMAX (Invitrogen). Immunoblotting was performed as described (7). Concentrations of secreted Tα in culture medium were determined using the Quantikine HS Human Tα ELISA kit (R&D Systems) (7).
In vivo Xenograft Studies
In vivo studies were performed under a protocol approved by the University Committee on Use and Care of Animals of the University of Michigan. SCID mice bearing xenograft tumors were treated with SM-164 as described before (7). Tumor sizes were measured 2–3 times per week.
Statistical Analyses
For the cell viability assay, data were plotted as mean ± SD, and sigmoid fitted (variable slope). Differences in mean values of cell viability among different groups were analyzed by two-way analysis of variance. For in vivo studies, significance (P) was calculated by Student’s t test, with a P value of <0.05 being considered as significant. All statistical tests were two-sided, and all statistical analyses were performed using GraphPad Prism 5.
Results
Lack of TNFα production contributes to Smac mimetic resistance in the sublines from MDA-MB-231 xenograft tumors
As a potent Smac mimetic, SM-164 displays a strong in vitro anticancer activity against a subset of human cancer cell lines (7). It also effectively inhibits tumor growth in the xenograft model of the MDA-MB-231 human breast cancer cell line (7). Although SM-164 achieved partial tumor regression, all the regressed tumors started to regrow after treatment ended (Fig. 1A), similar to a previous observation that HCC461 non-small cell lung xenograft tumors treated with another potent Smac mimetic (8). These data indicate that resistant cancer cells emerge from initially sensitive tumors following Smac mimetic treatment.
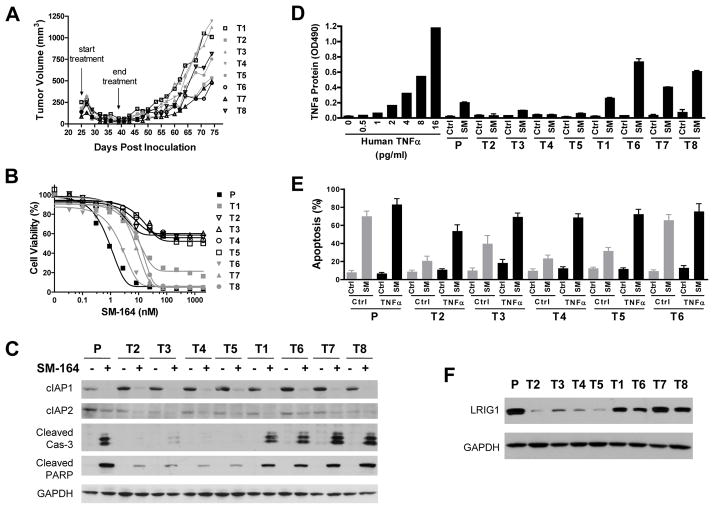
Fig. 1.
Characterization of tumor cells isolated from MDA-MB-231 xenograft tumors treated with SM-164. (A) MDA-MB-231 xenograft tumors were treated with 5 mg/kg of SM-164 intravenously 5 days/week for 2 weeks. (B) Tumor cells (T1–8) isolated from the MDA-MB-231 xenografts were treated with SM-164 for 2 days and cell viability was evaluated by WST assay. Data are representative from three independent experiments. (C) Cells were treated with 5 nM of SM-164 for 16 h for immunoblotting. (D) Cells were treated with 5 nM of SM-164 for 5 h for ELISA. Data (mean± SD) are from two independent experiments. (E) Cells were treated with 5 nM of SM-164 for 16 h and stained with Annexin V/PI for flow cytometry. Data (mean± SD) are from triplicate, including both early (Annexin V-positive/PI-negative) and late (Annexin V-positive/PI-positive) apoptotic cells. (F) Immunblotting for LRIG1 and GAPDH. P=MDA-MB-231 parental cells.
To elucidate the resistant mechanisms, we isolated MDA-MB-231 tumor cells from the regrown tumors and established 8 sublines. These 8 sublines exhibited polarized sensitivity to SM-164 treatment, with T2, T3, T4 and T5 sublines resistant (IC50>200 nM) and T1, T6, T7 and T8 sublines sensitive (IC50<20 nM) to SM-164 (Fig. 1B). Extended treatment of resistant sublines only modestly increased their susceptibility to SM-164 (Supplementary Fig. S1A), suggesting the resistance is not due to a delayed response to SM-164. The four SM-164 resistant sublines were also cross-resistant (IC50>5 μM) to a monovalent Smac mimetic AT-406 (16) (Supplementary Fig. S1B). Consistently, SM-164 effectively induced cleavage of caspase-3 and PARP, two biochemical markers for apoptosis, in parental and the sensitive sublines but not in the resistant sublines (Fig. 1C).
SM-164, like other Smac mimetics, targets cIAP1/2 and induces TNFα-dependent apoptosis following cIAP1/2 degradation and NF-κB activation (7). However, there was no significant difference in SM-164-induced degradation of cIAP1/2 and key molecular events in canonical and non-canonical NF-κB activation (upregulation of NF-κB-inducing kinase (NIK), phosphorylation of p65(Ser536) and processing of p100) among the parental cells, sensitive and resistant sublines (Fig. 1C, Supplementary S2A). Interestingly, while SM-164 induced robust production of TNFα in the parental cells and sensitive sublines in culture medium, it only weakly increased the levels of TNFα protein in all the resistant sublines (Fig. 1D, Supplementary S2B). Real time RT-PCR revealed that although TNFα mRNA levels were reduced in all the MDA-MB-231 tumor sublines compared with the parental cells, there was no significant difference between the sensitive and resistant sublines (Supplementary Fig. S2C). However, SM-164 significantly enhanced TNFα mRNA expression in the parental cell line and all the sensitive sublines but not in the resistant sublines (Supplementary Fig. S2D). Of note, all the resistant sublines still respond to TNFα-induced autocrine upregulation (Supplementary Fig. S2D), suggesting that SM-164 and TNFα induce TNFα mRNA expression via distinct mechanisms.
To determine whether lack of SM-164-induced TNFα production is responsible for the resistance, we treated cells with SM-164 together with recombinant human TNFα. While exogenous TNFα only modestly enhanced the activity of SM-164 in the parental cells and T6 sensitive subline, it dramatically potentiated SM-164-induced apoptosis in all the resistant sublines (Fig. 1E), indicating that the TNFα-mediated apoptotic pathway was intact in these resistant cells. Indeed, the parental cells as well as the T2 resistant and T6 sensitive sublines expressed comparable levels of TNFR1, RIPK1, FADD, TRADD and TRAF2, key components for TNFα-mediated signaling (Supplementary Fig. S2A). Although the expression of cIAP2 was enhanced by exogenous TNFα, SM-164 still efficiently reduced cIAP2 levels in these cells (Supplementary Fig. S2A). Recombinant TNFα or TRAIL both also restored the sensitivity of all the resistant sublines to SM-164 (Supplementary Fig. S2E). However, conditioned medium from MDA-MB-231 cells treated with SM-164 only modestly enhanced the activity of SM-164 in these resistant sublines (Supplementary Fig. S2E), suggesting that these resistant cells have a higher threshold for TNFα-mediated apoptosis upon Smac mimetic treatment and there must be additional factor(s) contributing to the resistance in these cells.
LRIG1 is down-regulated in the in vivo-selected MDA-MB-231 resistant sublines
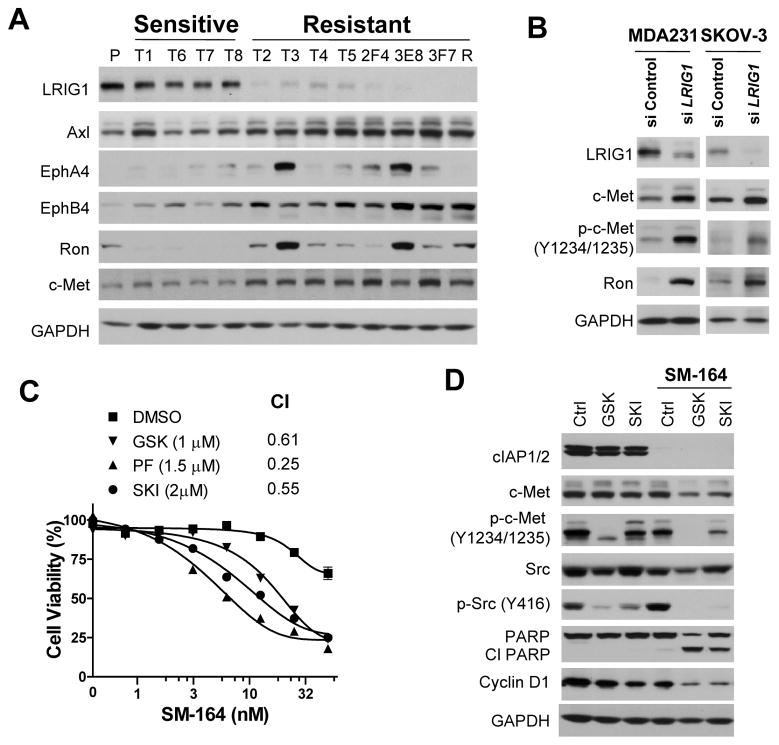
To further dissect the resistant mechanisms, we employed genome-wide gene expression profiling to identify genes that may be associated with the resistance using T2 and T3 resistant sublines, T6 and T8 sensitive sublines and the parental cells. Surprisingly, only 10 genes were consistently differentially expressed by more than 1.5-fold in T2 and T3 resistant sublines compared with T6 and T8 sensitive sublines and the parental cells in Affymetrix microarray analysis (Supplementary Table S1, all raw data will be deposited into the GEO repository). Real time RT-PCR and immunoblotting analyses of 9 genes with known functions in all four resistant sublines, four sensitive sublines and the parental cells showed that only LRIG1 (leucine-rich repeats and immunoglobulin-like domains 1) and WT1 (Wilms Tumor 1) are consistently differentially expressed between the resistant and sensitive groups of cells (Fig. 1F, Supplementary S3A, S3B).
Functional validation showed that knockdown of LRIG1, but not WT1 and four other genes, by small interfering RNAs (siRNAs) significantly attenuated the growth-inhibitory activity of SM-164 in MDA-MB-231 cells (Supplementary Fig. S3C and 3D). LRIG1 is a putative tumor suppressor whose ectopic expression inhibits the growth of a variety of cancer cell lines in vitro, and negatively regulates multiple receptor tyrosine kinases (RTKs) by enhancing their ubiquitination and degradation (18–27). LRIG1 also regulates epidermal stem cell quiescence and its expression defines a distinct multi-potent stem cell population in mammalian epidermis (28, 29).
Downregulation of LRIG1 confers resistance to SM-164
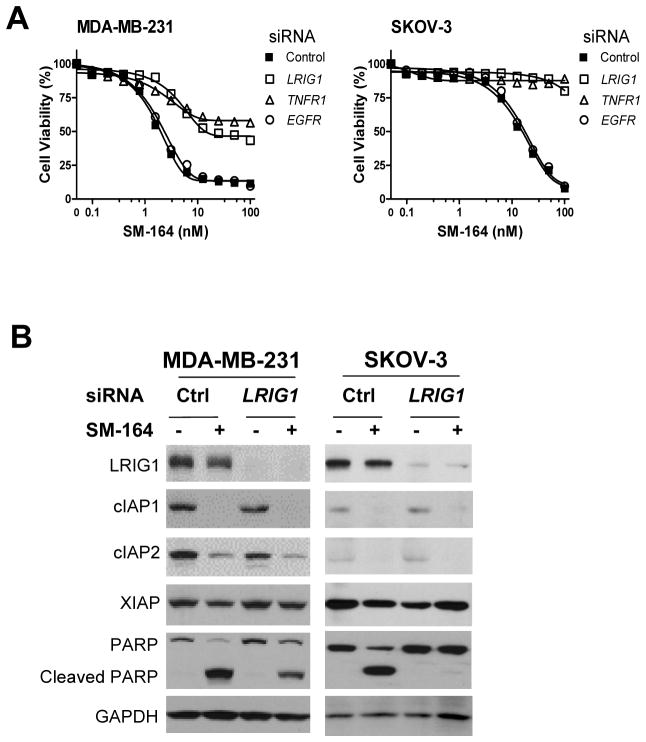
We further characterized the role of LRIG1 in the regulation of Smac mimetic activity. Knockdown of LRIG1 as well as TNFR1 markedly attenuated the growth-inhibitory activity of SM-164 in MDA-MB-231 and SKOV-3 ovarian cancer cells (Fig. 2A). However, knockdown of EGFR, a known target of LRIG1 (18, 23, 27), had no significant effect on the activity of SM-164 (Fig. 2A). Silencing LRIG1 greatly reduced apoptosis induction by SM-164 in both cell lines (Supplementary Fig. S4A) and also significantly attenuated SM-164 induced cleavage of PARP, but had no significant effect on SM-164-induced degradation of cIAP1 and cIAP2 (Fig. 2B). Neither the protein levels nor surface expression of TNFR1 were significantly altered by the depletion of LRIG1 (Fig. 2B, Supplementary S4B). The possible off-target effects of the siRNA against LRIG1 were excluded by employing multiple siRNAs targeting different regions of LRIG1 mRNA (Supplementary Fig. S4C, S4D). The growth-inhibitory activity of SM-164 was also reduced upon silencing LRIG1 in four additional SM-164-sensitive breast and ovarian cancer cell lines (Supplementary Fig. S5), suggesting that LRIG1 may be a common regulator of the activity of Smac mimetics.
Fig. 2.
Downregulation of LRIG1 confers resistance on SM-164-mediated cell growth inhibition in MDA-MB-231 and SKOV-3 cells. (A) Cells were transfected with siRNAs for 2 days, and then treated with SM-164 for 24 h. Cell viability was determined by WST assay. Data are representative of three independent experiments. (B) Cells were transfected with negative control or LRIG1-specific siRNAs for 2 days, then treated with 5 nM of SM-164 for 6 h (MDA-MB-231) or 16 h (SKOV-3) before being collected for immunoblotting.
LRIG1 expression is down-regulated in SM-164 resistant cells derived from MDA-MB-231 and SKOV-3 cells treated with SM-164 in vitro
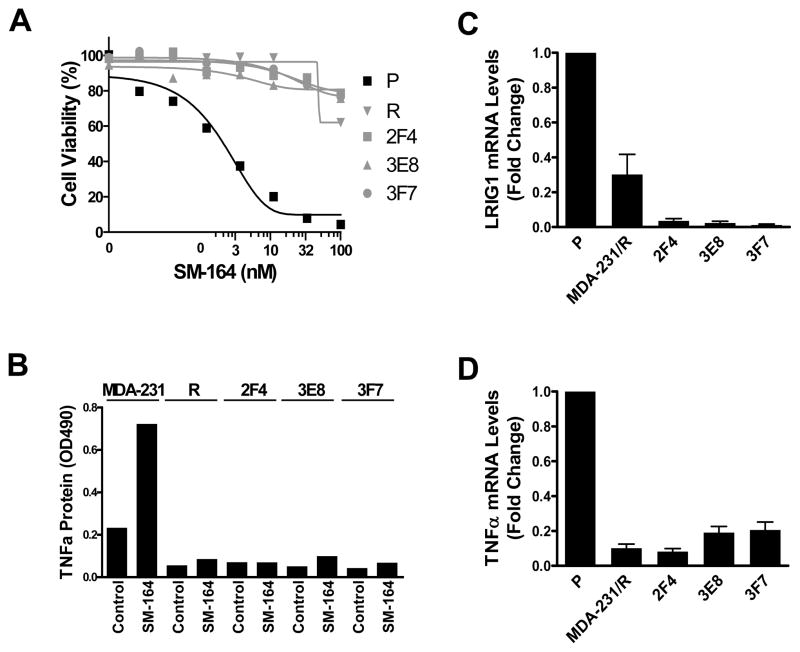
To determine whether the resistance observed in the SM-164-treated MDA-MB-231 tumors in mice can be recaptured by treating the parental cells in vitro, we obtained SM-164 tolerant cells (MDA-MB-231/R) by treating MAD-MB-231 cells with SM-164 at 50 nM, 10-times of its IC50 value, for >3 weeks. The MDA-MB-231/R cells became highly resistant to SM-164 (Fig. 3A) and also failed to produce TNFα upon SM-164 treatment (Fig. 3B). Real time RT-PCR showed that both LRIG1 and TNFα expression were reduced in MDA-MB-231/R cells compared with the parental cells (Fig. 3C, 3D).
Fig. 3.
Characterization of resistant sublines derived from MDA-MB-231. (A) Cells were treated with SM-164 for 24 h, and the sensitivity of parental (P) and SM-164 resistant derivative (MDA-MB-231/R, 2F4, 3E8 and 3F7) were evaluated by WST assay. Data are representative of three independent experiments. (B) Cells were treated with 5 nM of SM-164 for 16 h and TNFα production was detected by ELISA. Data are representative of two independent experiments. (C & D) The mRNA levels of LRIG1 and TNFα were measured by quantitative RT-PCR. Data shown are mean ± SEM (n=3). R=MDA-MB-231/R.
Similarly, SM-164-resistant SKOV-3 cells (SKOV-3/R) were obtained by treatment of SKOV-3 cells with SM-164 at concentrations 10-times of its IC50 for >3 weeks (Supplementary Fig S6A). Compared with the parental cells, the SKOV-3/R cells had reduced levels of LRIG1 protein, and SM-164 induced TNFα production and apoptosis were much reduced in the resistant cells (Supplementary Fig. S6B, S6C).
To investigate whether the resistance to SM-164 obtained from in vitro and in vivo is acquired during treatment or results from a selection of pre-existing resistant subpopulations, we isolated and evaluated single clones of MDA-MB-231 cells. Of 106 clones evaluated, 3 of which (2F4, 3E8 and 3F7) were highly resistant to SM-164 (Fig. 3A). The remaining 103 clones were equally sensitive to SM-164 as the parental cell line. The three resistant clones had greatly reduced mRNA expression for both LRIG1 and TNFα (Fig. 3C, 3D) and failed to produce TNFα upon treatment with SM-164 (Fig. 3B).
Collectively, these data suggest that Smac mimetic resistant cells pre-exist in the bulk population of parental cells, and SM-164 treatment selectively enriches the resistant subpopulations, which share a common attribute of reduced expression of LRIG1 and are unable to produce TNFα upon treatment with SM-164.
LRIG1 regulates SM-164 induced TNFα expression
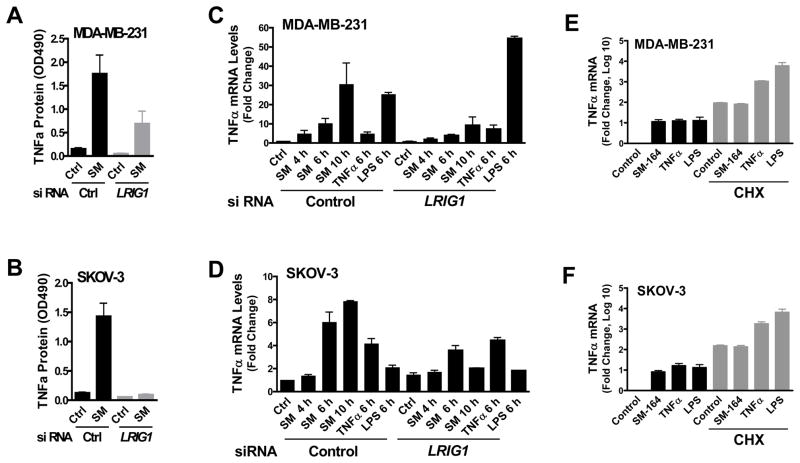
We next investigated if there is a potential direct link between LRIG1 and SM-164 induced TNFα production. Upon knockdown of LRIG1, SM-164-induced TNFα production was significantly attenuated in both MDA-MB-231 and SKOV-3 cell lines (Fig. 4A, 4B) and SM-164 induced TNFα mRNA expression was also attenuated (Fig. 4C, 4D). In contrast, TNFα- or lipopolysaccharide (LPS)-induced TNFα mRNA expression was either marginally affected or even enhanced upon knockdown of LRIG1 in these cell lines (Fig. 4C, 4D). These observations are consistent with our findings that SM-164-, but not TNFα-induced TNFα mRNA, expression was diminished in the LRIG1-low-expressing resistant cells (Supplementary Fig. S2D). Furthermore, depletion of LRIG1 significantly attenuated SM-164-induced TNFα promoter activity in MDA-MB-231 cells (Supplementary Fig. S7A). Protein synthesis inhibitor cycloheximide (CHX) efficiently blocked SM-164- but not TNFα- or LPS-induced TNFα mRNA expression (Fig. 4E, 4F), suggesting that SM-164-induced TNFα mRNA expression requires de novo protein synthesis. Thus, knockdown of LRIG1 in Smac-mimetic sensitive cancer cells recapitulates the phenotypic features of in vivo- and in vitro-selected resistant cells, and LRIG1 modulates the activity of SM-164, at least in part, by directly regulating SM-164-induced TNFα expression.
Fig. 4.
LRIG1 knockdown attenuates SM-164-induced TNFα expression. (A & B) Cells were transfected with siRNAs for 2 days, and then treated with 5 nM of SM-164 for 16 h and TNFα production was measured by ELISA. Data are mean± SEM (n=3). (C & D) Cells were transfected with siRNAs for 2 days, then treated with 5 nM of SM-164, 2 ng/ml of TNFα or 5 μg/ml of LPS for the indicated times. TNFα mRNA expression was measured by real time RT-PCR. Data are mean± SEM (n=3). Ctrl=control; SM=SM-164. (E & F) Cells were treated with 5 nM of SM-164, 2 ng/ml of TNFα and 5 μg/ml of LPS in the absence or presence of 20 μg/ml of CHX for 6 h. TNFα mRNA expression was measured by quantitative RT-PCR. Data are shown as mean ± SEM on a log10 scale (n=3).
Since Smac mimetics induce NF-kB-stimulated production of TNFα (9), we assessed the effects of LRIG1 knockdown on the NF-kB components. Depletion of LRIG1 had no significant effect on SM-164-induced phosphorylation of IkBα(S32/36) and p65(S536) (indicators of the activation of the canonical NF-kB pathway), and upregulation of NIK and processing of p100 protein (indicators of the activation of the noncanonical NF-kB pathway) in MDA-MB-231 cells (Supplementary Fig. S7B). Depletion of p65 (Rel A), a subunit of NF-kB complex, indeed diminished the constitutive expression of TNFα in MDA-MB-231 cells (Supplementary Fig. S7C-E). Collectively, these data suggest that, while constitutive expression of TNFα requires NF-kB, LRIG1 specifically regulates Smac mimetic-induced TNFα expression.
Multiple RTKs are upregulated in MDA-MB-231 resistant cells
LRIG1 negatively regulates several RTKs by enhancing their degradation (18–27). Indeed, the levels of multiple phospho-RTKs were increased in the resistant cells compared with the parental cells based upon phospho-RTKs profiling of the MDA-MB-231 parental and its T2 resistant cells (Supplementary Fig. S8A). The crosstalk between different members of the RTK superfamily is prevalent in cancer cells, and the RTK coactivation is an important mechanism by which cancer cells achieve drug resistance (30). Phospho-tyrosine profiling revealed that the levels of total tyrosine phosphorylation, particularly those proteins between 80 and 200 kDa, were much higher in all the resistant sublines cells than in parental and sensitive sublines (Supplementary Fig. S8B). Immunoblotting demonstrated that, while there was no major difference in the protein levels of IAPs and TNF receptors, the levels of multiple RTKs were much higher in the resistant sublines than in the sensitive sublines and parental cells (Fig. 5A, Supplementary S8C1).
Fig. 5.
Multiple RTKs are up-regulated in MDA-MB-231 resistant sublines, and TKIs enhance the growth-inhibitory activity of SM-164. (A) Protein expression profiling of RTKs in MDA-MB-231 parental and sublines by immunoblotting. (B) Cells were transfected with negative control or LRIG1-specific siRNAs for 2 days, and cell lysates were immunoblotted to detect the indicated proteins. (C) T2 cells were treated with SM-164 and/or the TKIs for 2 days and cell viability was assessed by WST assay. Data are representative of three independent experiments. (D) T2 cells were treated with 20 nM of SM-164 and/or 1.5 μM of GSK or SKI for 40 h for immunoblotting. CI=combination index.
RTK signaling activates multiple downstream effector pathways, including Ras-Raf-MEK-ERK and PI3K-Akt-mTOR-p70S6K (31, 32). RTKs also activate Src family kinases to propagate signaling cascade (33). Phosphorylation at certain residues of the signaling nodes indicates the activation status of these pathways. The levels of p-Src(Y416), p-p70S6K(T389) and p-Akt(S473) (Fig. 5A, Supplementary Fig. S8C2) were increased in the resistant sublines compared with the sensitive sublines, indicating an enhanced activation of Src and PI3K-Akt pathways in the resistant sublines. No significant difference was observed in the levels of p-ERK1/2(S202/Y204) among the parental, sensitive and resistant sublines (Supplementary Fig. S8C2). Furthermore, silencing LRIG1 also increased the levels of multiple RTKs, such as c-Met and Ron, in MDA-MB-231 and SKOV-3 (Fig. 5B), indicating a direct role of LRIG1 in regulating these RTKs.
Synergy between tyrosine kinase inhibitors (TKIs) and SM-164 in resistant sublines
Ron and c-Met were frequently, though not uniformly, upregulated in the resistant MDA-MB-231sublines (Fig. 5A). Knockdown of c-Met and Ron individually had no profound effect on TNFα expression and modestly enhanced the sensitivity of the T2 resistant cells to SM-164 (Supplementary Fig S9), suggesting that multiple RTKs contribute to Smac mimetic resistance and simultaneous blockade of multiple RTK signaling is necessary to sensitize the resistant cells to SM-164. We therefore tested three multi-targeted TKIs on the activity of SM-164 in the resistant cells.
GSK1363089 (GSK) and PF2341066 (PF) are potent inhibitors of c-Met, Ron and several other RTKs (34–36). SKI-606 (SKI) is a dual-inhibitor of Src and Abl kinases with activity against other tyrosine kinases (37, 38). While these TKIs alone had a modest growth inhibitory effect (IC50 > 5 μM) against the T2 resistant cells (Supplementary Fig. S10A), they greatly enhanced the activity of SM-164, with a combination index (CI) (17) ranging from 0.25 to 0.61 (Fig. 5C). U0126, a selective inhibitor of MEK1/2, or AT-7867 (AT), a dual inhibitor of Akt and p70S6 kinase, minimally or modestly enhanced the activity of SM-164, respectively (Fig. 5C). Combination treatment of SM-164 with GSK or SKI led to a significant increase in PARP cleavage and a decrease in cyclin D1 protein compared to single agents (Fig. 5D), suggesting that both induction of apoptosis and inhibition of cell proliferation contribute to the synergy between SM-164 and these TKIs. Interestingly, either GSK or SKI had no major effect on TNFα expression or production (Supplementary Fig. S10B, S10C). Synergy was also observed between SM-164 and GSK, PF or SKI in other MDA-MB-231 resistant sublines (Supplementary Fig. S10D). No obvious synergy was observed when TNFα or TRAIL was combined with any of these kinase inhibitors against MDA-MB-231 parental or the T2 resistant cells (Supplementary Fig. S11).
Combination of SM-164 with GSK1363089 effectively inhibits the growth of MDA-MB-231 resistant tumors
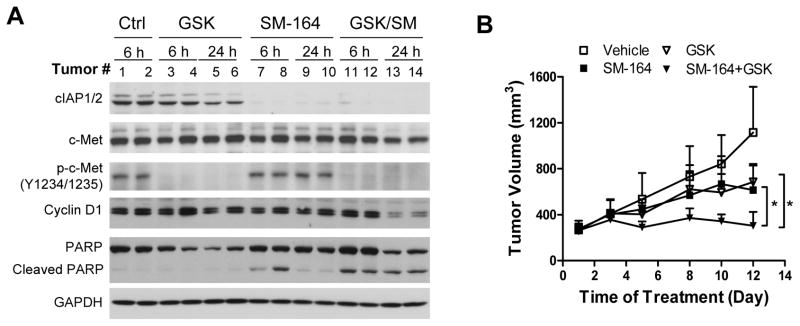
We next investigated the activity of SM-164 in combination with GSK against resistant tumors in vivo. SCID mice bearing T2 tumors were treated with 5 mg/kg of SM-164 intravenously and/or 10 mg/ml of GSK orally. The levels of p-c-Met (Y1234/1235) were diminished 6 hours after GSK was administered and remained undetectable at 24 hour post treatment (Fig. 6A). The reduction of p-c-Met (Y1234/1235) was accompanied with a decrease in the levels of p-Src (Y416), p-Akt (S473) and p-ERK (T202/Y204), mediators of RTK downstream effector pathways (Supplementary Fig. S12A). The combination also led to significant decrease of cyclin D1 and increase of PARP cleavage in the tumors (Fig. 6A). We next determined the tumor growth inhibitory activity of the combination. While SM-164 or GSK alone had a modest anti-tumor activity, their combination effectively repressed tumor growth (Fig. 6B). Of note, the T2 resistant tumors were very aggressive and one mouse each from the vehicle control and SM-164 groups died of complications of tumor invasion on days 7 and 9, respectively. The doses tested were well tolerated, and body weight was not significantly affected with the treatments (Supplementary Fig. S12B).
Fig. 6.
Antitumor activity of SM-164 and GSK1363089 alone and in combination against T2 resistant tumors. (A) SCID mice bearing T2 resistant tumors were treated with 5 mg/kg of SM-164 intravenously and/or 10 mg/kg of GSK orally for the indicated time points. Tumor lysates were immunoblotted to detect the indicated proteins. (B) SCID mice bearing T2 resistant tumors were treated with 5 mg/kg of SM-164 intravenously and/or 10 mg/kg of GSK orally for 5 days/week for 2 weeks. Data shown are mean±SEM for 6–8 mice. Asterisks indicate significant difference (P < 0.05).
Discussion
Five Smac mimetics are being tested in the clinic as new anticancer agents (1, 2). Preclinical studies showed that the majority of cancer cell lines is resistant to Smac mimetics, and that the lack of TNFα production upon Smac mimetic treatment is the dominant mechanism of intrinsic resistance to Smac mimetics (2, 7, 8, 12). Even when tumors initially respond to Smac mimetics, they may subsequently acquire resistance. Therefore, understanding resistant mechanisms of tumor cells to Smac mimetics can guide the rational development of Smac mimetics. In this study, using in vitro- and in vivo-selected resistant models, we identified LRIG1 as a key regulator of resistance of tumor cells to Smac mimetics in breast and ovarian cancer cell lines and demonstrated that downregulation of LRIG1 mediates Smac mimetic resistance through both attenuation of Smac mimetic-induced TNFα production and upregulation of RTKs.
LRIG1 is a negative regulator of RTKs by enhancing receptor ubiquitination and degradation (18–22). Consistent with this notion, downregulation of LRIG1 is inversely correlated with the upregulation of multiple RTKs in breast and ovarian cancer cell lines (Fig. 5A, Supplementary S8C). RTKs are promising targets for cancer treatment based on their key roles in regulating critical cellular processes; and aberrant RTK activation has been implicated in cancer resistance to molecular-targeted therapies (32). It has been reported that combining a Smac mimetic with inhibitors of PDGFR, IGF1R or EGFR increases cell death compared with monotherapy in human glioblastoma multiforme (39). Synergy between Smac mimetics and inhibitors of FLT3 and BCR-ABL was also observed against leukemia in vitro and in vivo (40, 41). In breast cancer cells, a Smac mimetic modestly increased apoptosis induction by ErbB antagonists in Her2- or EGFR-overexpressing cells (42). These data, coupled with our findings suggest that combination of Smac mimetics with TKIs can be a promising approach for the treatment of certain subtypes of cancer.
We found that inhibition of multiple RTKs by TKIs, but not the silencing of individual RTKs, significantly enhances the sensitivity of MDA-MB-231 resistant sublines to SM-164, consistent with our observation that multiple RTKs are upregulated in the SM-164 resistant cells. Importantly, SM-164 in combination with GSK1363089 effectively inhibited the tumor growth of Smac mimetic resistant cells of MDA-MB-231 (Fig. 6B), suggesting that TKIs can be used to overcome or delay the emergence of Smac mimetic resistance.
Smac mimetics induce activation of both canonical and non-canonical NF-κB pathways, regardless of whether the cells are sensitive or resistant to Smac mimetic-mediated growth inhibition (9, 10). However, only Smac mimetics-sensitive cells produce TNFα upon the activation of NF-κB, resulting in apoptosis in an autocrine manner (9, 10). In our studies, there is no significant difference in the molecular events of NF-κB activation between the sensitive and resistant sublines of MDA-MB-231 cells (Supplementary Fig. S2A). In Smac-mimetic sensitive cells, knockdown of LRIG1 attenuated TNFα mRNA expression induced by Smac mimetics, but not by TNFα or LPS, suggesting that manipulation of LRIG1 expression has minimal effect on classical NF-κB activation. Indeed, knockdown of LRIG1 expression had no significant effect on SM-164-mediated activation of canonical and non-canonical NF-κB pathways in MDA-MB-231 cells (Supplementary Fig. S7B). Instead, our data suggest that Smac mimetics induce TNFα expression through a mechanism different from that of TNFα or LPS and Smac mimetic-enhanced TNFα expression requires de novo protein synthesis (Fig. 4). Additional studies are needed to elucidate the precise mechanism of how LRIG1 regulates the TNFα expression induced by Smac mimetics.
Downregulation of LRIG1 leads to upregulation of multiple RTKs (Fig. 5A). It has been reported that activation of RTKs such as Ron and Axl suppresses TNFα expression in immune cells via downregulation of NF-κB activation and/or upregulation of the transcription factor Twist (49–51). However, we did not observe any significant differences in the expression of either Twist1 or Twist2 between parental cells, resistant and sensitive sublines of MDA-MB-231.
Recently, the rebound of cIAP2 expression upon Smac mimetic treatment of lung cancer cells was described as a mechanism of Smac mimetic resistance (14). In our studies, protein levels of cIAP2 and other key regulators of extrinsic apoptotic pathways are not significantly different between sensitive and resistant sublines (Supplementary Fig. S8C1) and cIAP2 was efficiently targeted by SM-164 (Supplementary Fig. S2A), suggesting the existence of more than one resistant mechanism to Smac mimetics. Our findings that both the in vivo- and in vitro-selected resistant cells from the MDA-MB-231 cell line have a similar attribute of low LRIG1 expression suggesting that, in the bulk population of MDA-MB-231 cells, there is a small population of cells intrinsically resistant to Smac mimetics.
In summary, our study has identified a novel mechanism of Smac mimetics resistance and provided a basis for development of rational combination strategies to overcome such resistance.
Supplementary Material
Acknowledgments
Grant support: Breast Cancer Research Foundation (S. W.), National Institutes of Health, National Cancer Institute (NCI) R01CA109025 (S. W.) and R01CA127551 (S. W.), and University of Michigan Cancer Center Core grant from the NCI P30CA046592
Footnotes
Disclosure of conflicts of interest: S. Wang is a consultant for and owns stocks and stock options in Ascenta Therapeutics, which has licensed the IAP inhibitors from the University of Michigan for clinical development. All other authors declare no conflict of interest.
References
- 1.Flygare JA, Fairbrother WJ. Small-molecule pan-IAP antagonists: a patent review. Expert Opin Ther Pat. 2010;20:251–267. doi: 10.1517/13543770903567077. [DOI] [PubMed] [Google Scholar]
- 2.Gyrd-Hansen M, Meier P. IAPs: from caspase inhibitors to modulators of NF-kappaB, inflammation and cancer. Nat Rev Cancer. 2010;10:561–574. doi: 10.1038/nrc2889. [DOI] [PubMed] [Google Scholar]
- 3.LaCasse EC, Mahoney DJ, Cheung HH, Plenchette S, Baird S, Korneluk RG. IAP-targeted therapies for cancer. Oncogene. 2008;27:6252–6275. doi: 10.1038/onc.2008.302. [DOI] [PubMed] [Google Scholar]
- 4.Du C, Fang M, Li Y, Li L, Wang X. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell. 2000;102:33–42. doi: 10.1016/s0092-8674(00)00008-8. [DOI] [PubMed] [Google Scholar]
- 5.Verhagen AM, Ekert PG, Pakusch M, Silke J, Connolly LM, Reid GE, Moritz RL, Simpson RJ, Vaux DL. Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing IAP proteins. Cell. 2000;102:43–53. doi: 10.1016/s0092-8674(00)00009-x. [DOI] [PubMed] [Google Scholar]
- 6.Li L, Thomas RM, Suzuki H, De Brabander JK, Wang X, Harran PG. A small molecule Smac mimic potentiates TRAIL-and TNFalpha-mediated cell death. Science. 2004;305:1471–1474. doi: 10.1126/science.1098231. [DOI] [PubMed] [Google Scholar]
- 7.Lu J, Bai L, Sun H, Nikolovska-Coleska Z, McEachern D, Qiu S, Miller RS, Yi H, Shangary S, Sun Y, et al. SM-164: a novel, bivalent Smac mimetic that induces apoptosis and tumor regression by concurrent removal of the blockade of cIAP-1/2 and XIAP. Cancer Res. 2008;68:9384–9393. doi: 10.1158/0008-5472.CAN-08-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petersen SL, Wang L, Yalcin-Chin A, Li L, Peyton M, Minna J, Harran P, Wang X. Autocrine TNFalpha signaling renders human cancer cells susceptible to Smac-mimetic-induced apoptosis. Cancer Cell. 2007;12:445–456. doi: 10.1016/j.ccr.2007.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vince JE, Wong WW, Khan N, Feltham R, Chau D, Ahmed AU, Benetatos CA, Chunduru SK, Condon SM, McKinlay M, et al. IAP antagonists target cIAP1 to induce TNFalpha-dependent apoptosis. Cell. 2007;131:682–693. doi: 10.1016/j.cell.2007.10.037. [DOI] [PubMed] [Google Scholar]
- 10.Varfolomeev E, Blankenship JW, Wayson SM, Fedorova AV, Kayagaki N, Garg P, Zobel K, Dynek JN, Elliott LO, Wallweber HJ, et al. IAP antagonists induce autoubiquitination of c-IAPs, NF-kappaB activation, and TNFalpha-dependent apoptosis. Cell. 2007;131:669–681. doi: 10.1016/j.cell.2007.10.030. [DOI] [PubMed] [Google Scholar]
- 11.Gaither A, Porter D, Yao Y, Borawski J, Yang G, Donovan J, Sage D, Slisz J, Tran M, Straub C, et al. A Smac mimetic rescue screen reveals roles for inhibitor of apoptosis proteins in tumor necrosis factor-alpha signaling. Cancer Res. 2007;67:11493–11498. doi: 10.1158/0008-5472.CAN-07-5173. [DOI] [PubMed] [Google Scholar]
- 12.Cheung HH, Mahoney DJ, Lacasse EC, Korneluk RG. Down-regulation of c-FLIP Enhances death of cancer cells by smac mimetic compound. Cancer Res. 2009;69:7729–7738. doi: 10.1158/0008-5472.CAN-09-1794. [DOI] [PubMed] [Google Scholar]
- 13.Cheung HH, Beug ST, St Jean M, Brewster A, Kelly NL, Wang S, Korneluk RG. Smac Mimetic Compounds Potentiate Interleukin-1{beta}-mediated Cell Death. J Biol Chem. 2010;285:40612–40623. doi: 10.1074/jbc.M110.183616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petersen SL, Peyton M, Minna JD, Wang X. Overcoming cancer cell resistance to Smac mimetic induced apoptosis by modulating cIAP-2 expression. Proc Natl Acad Sci U S A. 2010;107:11936–11941. doi: 10.1073/pnas.1005667107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun H, Nikolovska-Coleska Z, Lu J, Meagher JL, Yang CY, Qiu S, Tomita Y, Ueda Y, Jiang S, Krajewski K, et al. Design, synthesis, and characterization of a potent, nonpeptide, cell-permeable, bivalent Smac mimetic that concurrently targets both the BIR2 and BIR3 domains in XIAP. J Am Chem Soc. 2007;129:15279–15294. doi: 10.1021/ja074725f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cai Q, Sun H, Peng Y, Lu J, Nikolovska-Coleska Z, McEachern D, Liu L, Qiu S, Yang CY, Miller R, et al. A Potent and Orally Active Antagonist (SM-406/AT-406) of Multiple Inhibitor of Apoptosis Proteins (IAPs) in Clinical Development for Cancer Treatment. J Med Chem. 2011;54:2714–2726. doi: 10.1021/jm101505d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 18.Gur G, Rubin C, Katz M, Amit I, Citri A, Nilsson J, Amariglio N, Henriksson R, Rechavi G, Hedman H, et al. LRIG1 restricts growth factor signaling by enhancing receptor ubiquitylation and degradation. EMBO J. 2004;23:3270–3281. doi: 10.1038/sj.emboj.7600342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ledda F, Bieraugel O, Fard SS, Vilar M, Paratcha G. Lrig1 is an endogenous inhibitor of Ret receptor tyrosine kinase activation, downstream signaling, and biological responses to GDNF. J Neurosci. 2008;28:39–49. doi: 10.1523/JNEUROSCI.2196-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller JK, Shattuck DL, Ingalla EQ, Yen L, Borowsky AD, Young LJ, Cardiff RD, Carraway KL, 3rd, Sweeney C. Suppression of the negative regulator LRIG1 contributes to ErbB2 overexpression in breast cancer. Cancer Res. 2008;68:8286–8294. doi: 10.1158/0008-5472.CAN-07-6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shattuck DL, Miller JK, Laederich M, Funes M, Petersen H, Carraway KL, 3rd, Sweeney C. LRIG1 is a novel negative regulator of the Met receptor and opposes Met and Her2 synergy. Mol Cell Biol. 2007;27:1934–1946. doi: 10.1128/MCB.00757-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye F, Gao Q, Xu T, Zeng L, Ou Y, Mao F, Wang H, He Y, Wang B, Yang Z, et al. Upregulation of LRIG1 suppresses malignant glioma cell growth by attenuating EGFR activity. J Neurooncol. 2009;94:183–194. doi: 10.1007/s11060-009-9836-1. [DOI] [PubMed] [Google Scholar]
- 23.Segatto O, Anastasi S, Alema S. Regulation of epidermal growth factor receptor signalling by inducible feedback inhibitors. J Cell Sci. 2011;124:1785–1793. doi: 10.1242/jcs.083303. [DOI] [PubMed] [Google Scholar]
- 24.Laederich MB, Funes-Duran M, Yen L, Ingalla E, Wu X, Carraway KL, 3rd, Sweeney C. The leucine-rich repeat protein LRIG1 is a negative regulator of ErbB family receptor tyrosine kinases. J Biol Chem. 2004;279:47050–47056. doi: 10.1074/jbc.M409703200. [DOI] [PubMed] [Google Scholar]
- 25.Stutz MA, Shattuck DL, Laederich MB, Carraway KL, 3rd, Sweeney C. LRIG1 negatively regulates the oncogenic EGF receptor mutant EGFRvIII. Oncogene. 2008;27:5741–5752. doi: 10.1038/onc.2008.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldoni S, Iozzo RA, Kay P, Campbell S, McQuillan A, Agnew C, Zhu JX, Keene DR, Reed CC, Iozzo RV. A soluble ectodomain of LRIG1 inhibits cancer cell growth by attenuating basal and ligand-dependent EGFR activity. Oncogene. 2007;26:368–381. doi: 10.1038/sj.onc.1209803. [DOI] [PubMed] [Google Scholar]
- 27.Rubin C, Gur G, Yarden Y. Negative regulation of receptor tyrosine kinases: unexpected links to c-Cbl and receptor ubiquitylation. Cell Res. 2005;15:66–71. doi: 10.1038/sj.cr.7290268. [DOI] [PubMed] [Google Scholar]
- 28.Jensen KB, Watt FM. Single-cell expression profiling of human epidermal stem and transit-amplifying cells: Lrig1 is a regulator of stem cell quiescence. Proc Natl Acad Sci U S A. 2006;103:11958–11963. doi: 10.1073/pnas.0601886103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jensen KB, Collins CA, Nascimento E, Tan DW, Frye M, Itami S, Watt FM. Lrig1 expression defines a distinct multipotent stem cell population in mammalian epidermis. Cell Stem Cell. 2009;4:427–439. doi: 10.1016/j.stem.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu AM, Huang PH. Receptor tyrosine kinase coactivation networks in cancer. Cancer Res. 2010;70:3857–3860. doi: 10.1158/0008-5472.CAN-10-0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moritz A, Li Y, Guo A, Villen J, Wang Y, MacNeill J, Kornhauser J, Sprott K, Zhou J, Possemato A, et al. Akt-RSK-S6 kinase signaling networks activated by oncogenic receptor tyrosine kinases. Sci Signal. 2010;3:ra64. doi: 10.1126/scisignal.2000998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aleshin A, Finn RS. SRC: a century of science brought to theclinic. Neoplasia. 2010;12:599–607. doi: 10.1593/neo.10328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Christensen JG, Schreck R, Burrows J, Kuruganti P, Chan E, Le P, Chen J, Wang X, Ruslim L, Blake R, et al. A selective small molecule inhibitor of c-Met kinase inhibits c-Met-dependent phenotypes in vitro and exhibits cytoreductive antitumor activity in vivo. Cancer Res. 2003;63:7345–7355. [PubMed] [Google Scholar]
- 35.Qian F, Engst S, Yamaguchi K, Yu P, Won KA, Mock L, Lou T, Tan J, Li C, Tam D, et al. Inhibition of tumor cell growth, invasion, and metastasis by EXEL-2880 (XL880, GSK1363089), a novel inhibitor of HGF and VEGF receptor tyrosine kinases. Cancer Res. 2009;69:8009–8016. doi: 10.1158/0008-5472.CAN-08-4889. [DOI] [PubMed] [Google Scholar]
- 36.Zou HY, Li Q, Lee JH, Arango ME, McDonnell SR, Yamazaki S, Koudriakova TB, Alton G, Cui JJ, Kung PP, et al. An orally available small-molecule inhibitor of c-Met, PF-2341066, exhibits cytoreductive antitumor efficacy through antiproliferative and antiangiogenic mechanisms. Cancer Res. 2007;67:4408–4417. doi: 10.1158/0008-5472.CAN-06-4443. [DOI] [PubMed] [Google Scholar]
- 37.Golas JM, Arndt K, Etienne C, Lucas J, Nardin D, Gibbons J, Frost P, Ye F, Boschelli DH, Boschelli F. SKI-606, a 4-anilino-3-quinolinecarbonitrile dual inhibitor of Src and Abl kinases, is a potent antiproliferative agent against chronic myelogenous leukemia cells inculture and causes regression of K562 xenografts in nude mice. Cancer Res. 2003;63:375–381. [PubMed] [Google Scholar]
- 38.Puttini M, Coluccia AM, Boschelli F, Cleris L, Marchesi E, Donella-Deana A, Ahmed S, Redaelli S, Piazza R, Magistroni V, et al. In vitro and in vivo activity of SKI-606, a novel Src-Abl inhibitor, against imatinib-resistant Bcr-Abl+ neoplastic cells. Cancer Res. 2006;66:11314–11322. doi: 10.1158/0008-5472.CAN-06-1199. [DOI] [PubMed] [Google Scholar]
- 39.Ziegler DS, Wright RD, Kesari S, Lemieux ME, Tran MA, Jain M, Zawel L, Kung AL. Resistance of human glioblastoma multiforme cells to growth factor inhibitors is overcome by blockade of inhibitor of apoptosis proteins. J Clin Invest. 2008;118:3109–3122. doi: 10.1172/JCI34120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weisberg E, Kung AL, Wright RD, Moreno D, Catley L, Ray A, Zawel L, Tran M, Cools J, Gilliland G, et al. Potentiation of antileukemic therapies by Smac mimetic, LBW242: effects on mutant FLT3-expressing cells. Mol Cancer Ther. 2007;6:1951–1961. doi: 10.1158/1535-7163.MCT-06-0810. [DOI] [PubMed] [Google Scholar]
- 41.Weisberg E, Ray A, Barrett R, Nelson E, Christie AL, Porter D, Straub C, Zawel L, Daley JF, Lazo-Kallanian S, et al. Smac mimetics: implications for enhancement of targeted therapies in leukemia. Leukemia. 2010;24:2100–2109. doi: 10.1038/leu.2010.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Foster FM, Owens TW, Tanianis-Hughes J, Clarke RB, Brennan K, Bundred NJ, Streuli CH. Targeting inhibitor of apoptosis proteins in combination with ErbB antagonists in breast cancer. Breast Cancer Res. 2009;11:R41. doi: 10.1186/bcr2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mehrotra S, Languino LR, Raskett CM, Mercurio AM, Dohi T, Altieri DC. IAP regulation of metastasis. Cancer Cell. 2010;17:53–64. doi: 10.1016/j.ccr.2009.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dogan T, Harms GS, Hekman M, Karreman C, Oberoi TK, Alnemri ES, Rapp UR, Rajalingam K. X-linked and cellular IAPs modulate the stability of C-RAF kinase and cell motility. Nat Cell Biol. 2008;10:1447–1455. doi: 10.1038/ncb1804. [DOI] [PubMed] [Google Scholar]
- 45.Brunelleschi S, Penengo L, Santoro MM, Gaudino G. Receptor tyrosine kinases as target for anti-cancer therapy. Curr Pharm Des. 2002;8:1959–1972. doi: 10.2174/1381612023393530. [DOI] [PubMed] [Google Scholar]
- 46.Haluska P, Adjei AA. Receptor tyrosine kinaseinhibitors. Curr Opin Investig Drugs. 2001;2:280–286. [PubMed] [Google Scholar]
- 47.Belasco JG, Brawerman G. Control of messenger RNA stability. xviii. San Diego: Academic Press; 1993. p. 517. [Google Scholar]
- 48.Ross J. mRNA stability in mammalian cells. Microbiol Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alciato F, Sainaghi PP, Sola D, Castello L, Avanzi GC. TNF-alpha, IL-6, and IL-1 expression is inhibited by GAS6 in monocytes/macrophages. J Leukoc Biol. 2010;87:869–875. doi: 10.1189/jlb.0909610. [DOI] [PubMed] [Google Scholar]
- 50.Nikolaidis NM, Gray JK, Gurusamy D, Fox W, Stuart WD, Huber N, Waltz SE. Ron receptor tyrosine kinase negatively regulates TNFalpha production in alveolar macrophages by inhibiting NF-kappaB activity and Adam17 production. Shock. 2010;33:197–204. doi: 10.1097/SHK.0b013e3181ae8155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sharif MN, Sosic D, Rothlin CV, Kelly E, Lemke G, Olson EN, Ivashkiv LB. Twist mediates suppression of inflammation by type I IFNs and Axl. J Exp Med. 2006;203:1891–1901. doi: 10.1084/jem.20051725. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.