Abstract
Yeast cells entering into stationary phase decrease rRNA synthesis rate by decreasing both the number of active genes and the transcription rate of individual active genes. Using chromatin immunoprecipitation assays, we found that the association of RNA polymerase I with the promoter and the coding region of rDNA is decreased in stationary phase, but association of transcription factor UAF with the promoter is unchanged. Similar changes were also observed when growing cells were treated with rapamycin, which is known to inhibit the Tor signaling system. Rapamycin treatment also caused a decrease in the amount of Rrn3p-polymerase I complex, similar to stationary phase. Because recruitment of Pol I to the rDNA promoter is Rrn3p-dependent as shown in this work, these data suggest that the decrease in the transcription rate of individual active genes in stationary phase is achieved by the Tor signaling system acting at the Rrn3p-dependent polymerase recruitment step. Miller chromatin spreads of cells treated with rapamycin and cells in post-log phase confirm this conclusion and demonstrate that the Tor system does not participate in alteration of the number of active genes observed for cells entering into stationary phase.
INTRODUCTION
In most organisms, the synthesis of rRNA is highly regulated by cellular states of growth. For example, in a culture of the yeast Saccharomyces cerevisiae (called “yeast” in this article), the rate of transcription of rRNA by RNA polymerase (Pol) I begins to decrease in late log phase and remains at a low rate in the subsequent stationary phase (Ju and Warner, 1994). Normal yeast cells carry ∼150 tandemly repeated rDNA copies per haploid genome; each 9.1-kb rDNA repeat contains the gene for the 35S precursor rRNA transcribed by Pol I and the gene for the 5S rRNA transcribed by Pol III. These two coding regions are separated by two nontranscribed regions (NTS1 and NTS2) (see Figure 1A). In exponentially growing yeast cells, approximately one-half of these ∼150 copies of the 35S rRNA genes are actively transcribed (i.e., are in an active or “open” state) and the other one-half are not transcribed (i.e., are in an inactive or “closed” state) (Dammann et al., 1993; Sandmeier et al., 2002; French et al., 2003). Previous studies using yeast cells have shown that the decrease in Pol I transcription that occurs in the post-log to stationary phase involves both a decrease in the number of active rRNA genes and a decrease in the activity of each active gene (Dammann et al., 1993; Sandmeier et al., 2002). Regarding the former, it was recently discovered that Rpd3p histone deacetylase is required for the decrease in the number of active rRNA genes upon entering into the post-log to stationary phase (Sandmeier et al., 2002), but the functional role of Rpd3p in this process remains unclear. Regarding the latter, i.e., the decrease in the activity of each active gene, studies using an in vitro transcription system have demonstrated that the amount of the initiation-competent form of Pol I, the Pol I-Rrn3p complex, is small or absent in the stationary phase cells (Milkereit and Tschochner, 1998), thus explaining the decrease in Pol I transcription of rRNA in stationary phase.
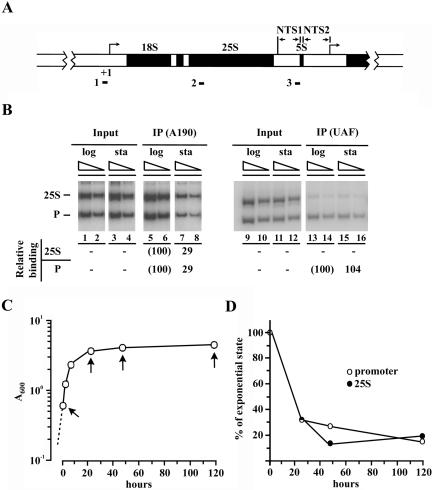
Figure 1.
Association of Pol I and UAF with rDNA as analyzed by ChIP. (A) Structure of yeast rDNA, which consists of repeats of 9.1-kb-long DNA, showing the initiation site of Pol I (defined as +1) and the positions of PCR products in the ChIP analysis (shown as 1, 2, and 3 for the promoter, the 25S coding region and the NTS region, respectively). (B) Two strains, the wild-type (NOY388, left) and the one carrying HA-tagged RRN5 (NOY798, right) were used for ChIP analyses. Exponentially growing cells (log) and cells in the 5 d (NOY388) and 7 d (NOY798) stationary phase cultures (sta) were treated with formaldehyde. The association of the A190 subunit of Pol I (left) and of the Rrn5p subunit of UAF (right) with the promoter (P) and the 25S rRNA coding region (25S) was then analyzed by ChIP by using anti-A190 and anti-HA antibodies, respectively. PCR reactions were carried out using two different concentrations of DNA and products analyzed by agarose gel electrophoresis are shown. Their amounts were quantified and values for the promoter or the 25S region DNAs bound to A190 or UAF (data indicated as IP or immunoprecipitation) were divided by corresponding values for the DNAs before IP (Input). The values for stationary phase cells obtained in this way were then normalized to the corresponding values (taken as 100%) obtained for log-phase cells and averages of the normalized values for two DNA concentrations are shown. (C) Growth curve of yeast strain carrying HA-tagged RPA135 (NOY654) in YEPD. Cell density (A600) was measured at various times after the first sampling (time 0; A600 0.6). (D) Samples were taken from the culture shown in C at times indicated by arrows and ChIP analyses were done as per the experiment shown in B by using anti-HA antibodies. The degrees of association of Pol I with the promoter and the 25S coding region were calculated as in B, and the normalized values for the degree of association are shown.
RRN3 was originally identified together with other RRN genes as an essential gene specifically required for transcription of rRNA by Pol I (Nogi et al., 1991a; Yamamoto et al., 1996). The encoded protein Rrn3p was shown to bind Pol I in the absence of DNA template, forming an initiation-competent Rrn3p-Pol I complex (Yamamoto et al., 1996; Milkereit and Tschochner, 1998; Keener et al., 1998; Fath et al., 2001). The initiation of rRNA transcription by yeast Pol I involves at least four different transcription factors, upstream activation factor (UAF), core factor (CF), TATA binding protein (TBP), and Rrn3p (reviewed by Nomura, 1998; Reeder, 1999; Nomura, 2001). UAF is a multiprotein complex composed of Rrn5p, Rrn9p, Rrn10p, Uaf30p, and histones H3 and H4 (Keys et al., 1996; Keener et al., 1998; Siddiqi et al., 2001). CF is also a multiprotein complex consisting of three proteins: Rrn6p, Rrn7p, and Rrn11p (Keys et al., 1994; Lalo et al., 1996; Lin et al., 1996). In vitro studies have demonstrated that UAF binds tightly to the upstream element of the yeast Pol I promoter and commits the template for transcription. The UAF-template complex, with the help of TBP, then recruits CF to the promoter followed by joining of the Pol I-Rrn3p complex (Keys et al., 1996; Steffan et al., 1996; Keener et al., 1998; Aprikian et al., 2001). Although Pol I transcription in mammalian systems was thought to be very different from the yeast system in the past and the transcription factor corresponding to UAF has not been found, recent studies have started to reveal considerable similarity to the yeast system. TIF-IA, which was studied by Grummt and coworkers as the factor associated with Pol I, was known to be inactivated (or degraded) in quiescent states (Buttgereit et al., 1985; Schnapp et al., 1990). It is presumably the same as the factor C* studied by others (Brun et al., 1994) and was subsequently concluded to be a homolog of Rrn3p (Bodem et al., 2000). In the yeast system, Rrn3p was shown to interact with both the A43 subunit of Pol I and the Rrn6p subunit of CF in vitro, suggesting that these interactions may play an important role for the function of Rrn3p in the initiation of rRNA transcription (Peyroche et al., 2000; Fath et al., 2001). Similarly, TIF-IA/Rrn3p in mammalian systems was shown to interact with both Pol I and SL1 (also called TIF-IB), a transcription factor that is functionally homologous to CF with bound TBP (Miller et al., 2001; Cavanaugh et al., 2002; Yuan et al., 2002). Thus, the mechanism of the final step in forming a transcription-competent preinitiation complex containing Pol I is similar among most eukaryotic organisms from yeast to human, and this step, which requires Rrn3p/TIF-IA, may be the target for growth-dependent regulation of Pol I transcription in these organisms.
A plausible model to explain the above-mentioned observations is that Rrn3p is essential for recruitment of Pol I to the promoter, as initially suggested (Yamamoto et al., 1996; Peyroche et al., 2000). This model is supported by in vitro experiments with immobilized DNA template for the human Pol I system (Miller et al., 2001). Nevertheless, there are conflicting reports regarding the functional role of Rrn3p/TIF-IA in the recruitment of Pol I to the promoter. Earlier studies on mouse TIF-IA showed that Pol I can bind to the promoter in the absence of TIF-IA, but to become transcriptionally competent, the subsequent addition of the two factors TIF-IC and TIF-IA is required (Schnapp and Grummt, 1991; Schnapp et al., 1993). The experiments suggested that the interaction of Pol I with TIF-IA/Rrn3p takes place on the DNA template, converting the inactive state of Pol I to an active state. More recent experiments using yeast extracts and immobilized DNA template also showed that Pol I can be recruited to the promoter in the absence of Rrn3p, but the complex formed in this way cannot be activated by subsequent addition of Rrn3p, at least in vitro (Aprikian et al., 2001). The question of whether Pol I binds to the promoter in the absence of Rrn3p is addressed in this study by using chromatin immunoprecipitation (ChIP) analysis, and the results demonstrate that Pol I recruitment to the promoter is Rrn3p-dependent in vivo.
The entry into stationary phase is known to be regulated by at least two signal transduction pathways, the RAS/protein kinase A (reviewed by Broach, 1991; Thevelein and deWinde, 1999) and the Tor (reviewed by Schmelzle and Hall, 2000; Rohde et al., 2001; Jacinto and Hall, 2003) pathways. These two signaling pathways are apparently both involved in the regulation of expression of various genes required for the entry into stationary phase, but how exactly these two signaling pathways are coordinated is presently unclear (Herman, 2002). Regarding the down-regulation of Pol I in the stationary phase in yeast, it was previously shown that treatment of yeast cells with rapamycin, which inhibits Tor functions and leads to a state resembling stationary phase (Heitman et al., 1991; Barbet et al., 1996), also inhibits Pol I transcription of rRNA genes (Zaragoza et al., 1998; Powers and Walter, 1999). Therefore, one likely target of the Tor signaling pathway seems to be Rrn3p and/or Pol I, regulating formation of the Rrn3p-Pol I complex. Similarly, the inhibition of rRNA synthesis by rapamycin might suggest that the decrease in the number of active rRNA genes observed in stationary phase is also regulated by the Tor function. The experiments described in this article were designed to address these questions. We found that the Tor signaling pathway indeed regulates rRNA transcription at the step of Rrn3p-dependent Pol I recruitment to the promoter but that conversion of the open state of rRNA genes to the closed inactive state as observed in stationary phase is apparently not directly regulated by the Tor signaling pathway.
MATERIALS AND METHODS
Media, Strains, and Plasmids
YEPD medium contains 1% yeast extract, 2% peptone, and 2% glucose. Synthetic glucose (SD) medium (2% glucose, 0.67% yeast nitrogen base) was supplemented with amino acids and required bases as described by Sherman et al. (1986) and is called SD complete medium. For [methyl-3H]methionine incorporation experiments, methionine was omitted. For [3H]uridine incorporation experiments, prototrophic URA3 strains were used and the SD complete medium did not contain uracil.
The yeast strains and plasmids used in this study are described in Table 1. NOY1075 carrying rrn3 mutation S213P was constructed in the following way. First, a CEN plasmid (pNOY452) carrying LEU2 and RRN3 was cleaved by restriction enzymes, making a gap at specific segments within the RRN3 coding region. Second, DNA fragments covering each of these gaps were prepared by error-prone polymerase chain reaction (PCR). A linearized CEN plasmid with a gapped rrn3 and a corresponding PCR fragment covering the gap were mixed and introduced into NOY604 (Yamamoto et al., 1996), which has a deletion in the chromosomal RRN3 gene and grows on galactose, but not on glucose, by using helper plasmid pNOY103. Leu+ transformants obtained by gap repair at 25°C were screened for temperature-sensitive (ts) growth on glucose. One of the ts mutants thus obtained was shown to have a mutational alteration (S213P) in RRN3 recovered on the CEN plasmid. The mutant gene, rrn3 (S213P), was introduced into a URA3 integration plasmid, cleaved with PmeI within the rrn3 gene, and integrated into NOY388 at the RRN3 locus, followed by selection of recombinants on 5-fluoroorotic acid-containing plates and screening for temperature-sensitive (ts) recombinants. NOY1075 was obtained as one of these ts recombinants carrying the rrn3 (S213P) replacing the chromosomal RRN3.
Table 1.
Yeast strains and plasmids used in this study
| Strain or plasmid | Description |
|---|---|
| Strains | |
| W303-1a (NOY388) | MATaade2-1 ura3-1 trp1-1 leu2-3,112 his3-11,15 can1-100 |
| NOY654 | Mat ade2-1 ura3-1 trp1-1 leu2-3,112 his3-11,15 can1-100 rpa135Δ::LEU2, pNOY302 |
| NOY798 | MATα ade2 ura3 trp1 leu2 his3 can1 rrn5::TRP1, pNOY402 |
| NOY886 | MATα rpa135Δ::LEU2 ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 fob1Δ::HIS3, pNOY117 [CEN, RPA135, TRP1]; rDNA copy number ∼40 (French et al., 2003) |
| NOY1075 | MATaade2-1 ura3-1 trp1-1 leu2-3,112 his3-11,15 can1-100 rrn3 (S213P) |
| NOY1170 | MATα ade2-1 ura3-1 trp1-1 leu2-3,112 his3-11,15 can1-100 rrn3Δ::HIS3, pNOY452 |
| Plasmids | |
| pNOY302 | A derivative of pRS314 (CEN6 ARSH4 TRP1) carrying triple HA-tagged RPA135; the DNA sequence inserted after the initiation codon AUG encodes the following amino acids: GGRIFYPYDVPDYAGYPYDVPDYAGSYPYDVPDYAAQCGR (HA peptide underlined). |
| pNOY402 | A derivative of pRS315 (CEN6 ARSH4 LEU2) carrying triple HA- and (His)6-tagged RRN5 [RRN5-(HA)3-His)6]; see Keener et al. (1997). |
| pNOY452 | A derivative of pRS315 (CEN6 ARSH4 LEU2) carrying RRN3 tagged with (HA)7 at the N terminus. |
ChIP Analysis
ChIP analysis was carried out as described by Kuras and Struhl (1999) with some modifications. Briefly, cells treated with 1% formaldehyde were collected by centrifugation, washed, and disrupted with glass beads. Glass beads were removed from the lysate and cross-linked chromatin was recovered by centrifugation. The chromatin was washed and then sonicated to yield an average DNA fragment size of ∼500 base pairs. After removing cell debris by centrifugation, the protein concentration of chromatin samples was adjusted to 0.5 mg/ml. Antibodies were added and the mixture was incubated overnight at 4°C. Protein A-Sepharose beads were then added and incubation was continued for 2 h at 4°C. Beads were recovered, washed, and protein with cross-linked DNA was eluted from beads by incubating the beads in a buffer containing 1% SDS at 65°C for 20 min. The eluates containing protein and DNA were further incubated overnight at 65°C to reverse cross-linking. After proteinase K treatment (400 μg/ml; 2 h at 56°C) and RNaseA treatment (200 μg/ml; 1 h at 37°C), DNA was isolated by phenol-chloroform extraction followed by ethanol precipitation. “Input DNA” was prepared in the same way, but without antibody immunoprecipitation steps. The amounts of DNA at the promoter, a 25S rRNA coding region, and an NTS region were determined by PCR with three pairs of primers (each primer was ∼30 nucleotides in length), covering the regions from 8896 to -24 (229 base pairs), from 3586 to 3876 (291 base pairs), and from 7331 to 7674 (344 base pairs), respectively (see Figure 1A; numbering is with respect to the Pol I start site as +1 and the 9137-base pair SmaI fragment starts at -206 and ends at 8931). The amounts of samples for input DNA used for PCR were usually equivalent to ∼1% of those obtained after immunoprecipitation. Several concentrations of DNA samples were used to obtain concentration-dependent signals. [32P]dCTP or [32P]dATP was used to label PCR products, and the products were quantified by a PhosphorImager analysis. Normalization of the results is explained in the figure legends. To assess contribution of background, “beads only” control DNA was usually prepared in parallel to immunoprecipitated samples but by omitting antibodies. In the experiments using hemagglutinin (HA)-tagged proteins and anti-HA antibodies, a strain without the HA tagging was used as a control to assess background PCR signals. The results of these control ChIP experiments are not shown, but background values were insignificant. We note that several independent experiments were carried out for each ChIP analysis discussed in this article and that even after discarding the data sets that did not show concentration dependency, quantitative data still showed some variation among multiple experiments. However, the variation was not significant enough to alter conclusions described in this article. An example of this variation can be seen in the association of Pol I with rDNA in stationary phase at 5 d. In the experiment shown in Figure 1B that used anti-A190 antibodies, the values were 29% of the log-phase value for both promoter and 25S regions, whereas the corresponding values shown in Figure 1D, which were obtained using cells carrying the HA-tagged RPA135 gene, were ∼20%. Another example of variation is regarding the data shown in Figure 3B, namely, the association of a) Rrn3p with the Pol I promoter, b) A190 with the Pol I promoter, and c) A190 with the 25S coding region. In this case, the values (percentage of controls without rapamycin) varied in three independent experiments. The actual values were a) 37, 19, and 33; b) 30, 42, and 32; and c) 35, 28, and 38. The averages of these values, 30 ± 8, 35 ± 5, and 34 ± 4, are indicated (without standard deviations) below the autoradiogram in Figure 3B. We believe that the ChIP technique used does not allow very accurate quantification but is sufficient for making conclusions described in this article.
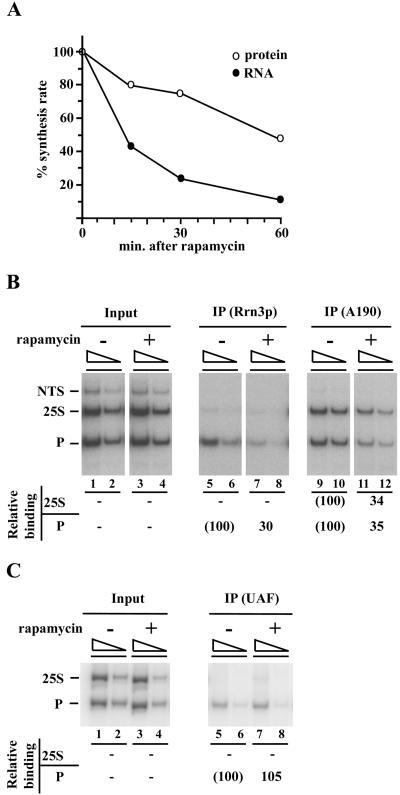
Figure 3.
Rapamycin treatment decreases association of Rrn3p and Pol I but not UAF, with rDNA. (A) Strain NOY1170 carrying HA-tagged RRN3 was grown in SD complete medium at 30°C and treated with rapamycin (0.2 μg/ml). Aliquots were taken at indicated times, mixed with [methyl-3H]methionine, and incubated for 5 min. Incorporation of the 3H label into the TCA-insoluble fraction (“protein”; corrected for small amounts of 3H label in RNA) and the RNA fraction (obtained after phenol extraction) was measured. The values normalized for those at the time of rapamycin addition are shown. (B) ChIP analysis of NOY1170 cells was carried out for samples with and without 1-h rapamycin treatment by using anti-HA antibody to measure association of Rrn3p with rDNA and by using anti-A190 to measure association of Pol I with rDNA. Relevant portions of an autoradiogram from a representative experiment are shown. Quantification of PCR products by a PhosphorImager and calculations were done as described in Figure 1. The values obtained for the cells treated with rapamycin were compared with those for the control without rapamycin and values obtained from three independent experiments were averaged (see MATERIALS AND METHODS for variation) and are shown as percentage of control values. (C) Strain NOY798 carrying HA-tagged RRN5 was used and the effects of 1-h rapamycin treatment on the association of UAF with the promoter were examined as described in B.
Coimmunoprecipitation of Rrn3p and Pol I
Strain NOY1170 was grown in 1 liter of SD complete medium to a cell density of A600 ∼0.35. The culture was divided and grown for another hour in the presence and absence of 0.2 μg/ml rapamycin. After harvesting, the cell pellets were washed with cold lysis buffer [20 mM Tris pH 7.9, 0.1 M potassium acetate, 10% (vol/vol) glycerol, and 0.1% (vol/vol) Tween 20] and stored at -70°C. The cell pellets were thawed on ice in 5 ml of cold lysis buffer supplemented with 1 mM phenyl methyl sulfonyl fluoride. The cells were broken open with a French Press (20,000 psi, two times). The resulting lysate was clarified by centrifugation (Ti80 rotor, 20,000 rpm, 20 min; Beckman Coulter, Fullerton, CA). For immunoprecipitation, 1.8 ml of the supernatant was incubated with rabbit polyclonal antiserum against the Pol I A190 subunit for 1.5 h at 4°C in the presence of 200 μl of protein A-immobilized magnetic beads (Dynal Biotech, Lake Success, NY). The beads were recovered and washed four times with lysis buffer. The immunoprecipitates were eluted by heating the washed beads at 65°C in the presence of 100 μl of SDS sample buffer. Immunoprecipitates were analyzed by standard Western blotting (8% SDS-PAGE, transfer to Immobile-P membrane; Millipore, Billerica, MA) by using monoclonal antibodies against the HA epitope (12CA5; Convance Research Products, Berkeley, CA) and against the Pol I subunit A135. Quantification of the Western blot was performed by scanning the membrane and analyzing the image using Image Quant (Amersham Biosciences, Piscataway, NJ). Various amounts of samples were analyzed, and values in the range showing a linear concentration dependence were used for calculation of the amount of Rrn3p relative to the Pol I in the precipitates.
Electron Microscopy
Yeast cells were grown in YEPD plus 1 M sorbitol at 30°C to mid-(A600 ≅ 0.4) or post (A600 ≅ 2.8)-log phase. Miller chromatin spreads were prepared as described previously (French et al., 2003) from culture volumes of 1 or 0.2 ml for mid- and post-log phase, respectively. Briefly, yeast cell walls were partially digested for 3 min in zymolyase (5 and 25 mg/ml for mid- and post-log stages, respectively) at 30°C with shaking, followed by pelleting, and then resuspension in 4 ml of 0.025% Triton X-100, pH 9, which results in hypotonic lysis. The 4-ml lysate was swirled to facilitate dispersal, and cellular contents were fixed by the addition of 0.4 ml of a solution containing 0.1 M sucrose, 3.7% formaldehyde, followed by centrifugation of the lysate onto carbon-coated electron microscopy (EM) grids. Grids were stained with phosphotungstic acid and uranyl acetate, rotary-shadow cast with platinum as described previously (Osheim and Beyer, 1989), and viewed in a JEOL 100CX electron microscope. In experiments to examine the effects of rapamycin, the drug was added to log-phase cells at a concentration of 0.2 μg/ml for either 10 or 30 min. In this case, the 3-min zymolyase treatment was started 3 min before the end of a total of 10- or 30-min exposure to rapamycin, i.e., at 7 and 27 min, respectively.
Analysis of EM Data
Chromatin spreads from multiple cultures of yeast cells were visualized, and two or more Miller spread preparations were quantitatively analyzed for each condition. For quantitative analysis, entire EM grids were scanned and all yeast chromatin regions displaying rRNA genes were photographed. For the post-log and rapamycin-treated cells, the polymerase number per gene was determined by counting the number of RNA polymerases (or nascent rRNA transcripts) on all rRNA genes that could be unambiguously followed from the 5′ to 3′ end for the entire grid database, yielding sample sizes from 76 to 132. For log-phase NOY886 cells, the polymerase number per gene was counted on 174 genes randomly selected from several different preparations. This same control population was included in our previous study (French et al., 2003).
Other Methods
Analysis of RNA labeled with [3H]uridine (46 mCi/μmol; 50 μCi/ml final) by electrophoresis on a 2% polyacrylamide/0.5% agarose composite gel was performed as described previously (Nogi et al., 1991b). Incorporation of [methyl-3H]methionine (85 mCi/μmol; 40 μCi/ml final) into protein and RNA was used to measure synthesis rates of protein and RNA by using SD complete medium without methionine (for labeling of RNA by using [methyl-3H]methionine, see Warner, 1991). Pulse labeling was done for 5 min. Aliquots were taken, and total amounts of trichloroacetic acid (TCA)-insoluble, 3H-labeled material consisting mostly of protein and a small amount of RNA were determined. RNA was prepared from the remaining portions by phenol extraction followed by ethanol precipitation. The amounts of 3H-labeled methyl group incorporated into total RNA were then determined. The amount of 3H-labeled protein was then calculated from total TCA-precipitable 3H values after small correction for 3H-labeled RNA.
Analysis of the 5′ end of precursor rRNA by primer extension was carried out as described previously (Keener et al., 1998; Vu et al., 1999), by using the [32P]5′-end-labeled primer 5′-ACACGCTGTATAGACTAGGC-3′, which hybridizes to 35S precursor rRNA 130 nucleotides downstream of the Pol I start site.
RESULTS
Association of UAF and Pol I with the rDNA Promoter in Stationary Phase Cells
Yeast cells were grown in YEPD medium, and samples were taken from growing cultures (A600 0.5-0.6) and from cultures kept for an additional 5-7 d, i.e., long after cessation of cell density increase (cf. Figure 1C). The association of Pol I and UAF with the promoter and the 25S rRNA coding region was analyzed by ChIP by using the primer pairs described in MATERIALS AND METHODS (Figure 1A). A yeast strain without an HA-tag (NOY388) or a strain carrying an HA-tag on RPA135 was used for ChIP analysis of Pol I, and a strain carrying an HA-tag on RRN5 was used for ChIP analysis of UAF. For the growing cells, Pol I was found to be associated with both the promoter and the coding regions (“log” in Figure 1B, left), and UAF was associated with the promoter but not with the coding region, as expected. For the stationary phase cells (Figure 1B, “sta”), it was found that the association of Pol I with both the promoter and the 25S coding region was reduced to ∼10-30% relative to the values for the growing cells (Figure 1, see legend). Large and parallel reductions (∼65-80% reduction) were also evident for these associations at earlier time points in the post-log phase or the stationary phase (Figure 1, C and D). In contrast to Pol I, however, association of UAF with the promoter was the same in the 7-d stationary cells as it was in growing cells (Figure 1B, right). Association of UAF with the promoter was also analyzed using cells at 4 and 10 d of incubation, and again, no decrease in association of UAF with the promoter was observed relative to growing cells (our unpublished data).
Association of Pol I with the rDNA Promoter in an rrn3 Temperature-sensitive Mutant
As described in INTRODUCTION, there are published articles reporting that Pol I can be recruited to the rDNA promoter in the absence of Rrn3p in in vitro experiments. To study the mechanism responsible for the decrease of Pol I association with the promoter in the stationary phase, we first designed experiments to test whether Pol I can be recruited to the promoter in the absence of active Rrn3p in vivo.
We have previously isolated several rrn3 ts mutants (Claypool and Nomura, unpublished experiments). One of them showed a tight ts phenotype resulting from an amino acid alteration of serine to proline at position 213 (S213P). As shown in Figure 2A, this mutant grows almost like the wild type in SD complete medium at 25°C, but after a temperature shift to 37°C, growth (measured by cell density) gradually decreases and eventually stops. By analyzing RNA labeled with [3H]uridine between 2.5 and 3.5 h after the temperature shift, we found that incorporation into RNAs (25S, 18S, and 5.8S) transcribed by Pol I is preferentially inhibited compared with incorporation into 5S RNA or tRNA transcribed by Pol III (Figure 2B, compare lane 2 with 1). The inhibition of Pol I activity (per cell density) measured in this way was ∼98% (Figure 2B, see legend). In another experiment using cells growing in YEPD medium, the amounts of the unstable 5′ end of the 35S precursor rRNA were measured by primer extension at 3 h after temperature shift to 37°C. The amount in the mutant was ∼5% of that observed for the wild-type control cells, suggesting that the inhibition of Pol I activity is ∼95% (Figure 2C). These experiments indicate that Rrn3p is required for Pol I function and that Rrn3p is largely inactivated by 3 h after temperature shift to 37°C in this mutant.
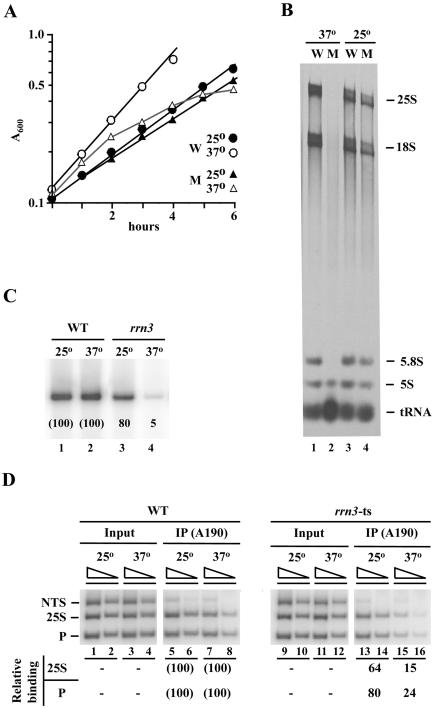
Figure 2.
A temperature-sensitive rrn3 mutation (S213P) decreases association of Pol I with the promoter at nonpermissive temperature. (A) Representative growth curves of the rrn3 (S213P) mutant (NOY1075; M) and the control RRN3 (NOY388; W) strains. Cells exponentially growing in SD complete medium at 25°C were divided into two, one shifted to 37°C (time 0) and the other kept at 25°C, and increases in cell density were followed. (B) CEN plasmid pRS316 was introduced into both NOY1075 (M) and NOY388 (W) to make them URA3. The resultant strains were grown as in A, and [3H]uridine was added at 2.5 h after the temperature shift and incubated for 1 h. Control cultures kept at 25°C were also treated in the same way. Aliquots were taken to determine the amounts of [3H]uridine incorporated into the total TCA-insoluble RNA fraction and the remaining cells were used to isolate RNA. RNA samples containing 1 × 105 cpm 3H counts were analyzed by electrophoresis on a polyacrylamide/agarose composite gel. An autoradiogram is shown. The amounts of radioactive RNA corresponding to 25S, 18S, 5.8S, 5S, and tRNA were determined and the ratios of the sum of Pol I transcripts (25S + 18S + 5.8S) to the total (25S + 18S + 5.8S + 5S +tRNA) were calculated. These ratios and the amounts of [3H]uridine incorporated into total RNA were used to calculate the levels of Pol I transcription, yielding the values of 96 and 2.2% for the mutant relative to the wild-type (100%) at 25°C and 37°C, respectively. (C) Both the WT (NOY388) and rrn3 (S213P) mutant (NOY1075) strains were grown in YEPD at 25°C to a cell density of A600 0.2, and the cultures were divided into two, one shifted to 37°C and the other kept at 25°C. Three hours later, portions were used for ChIP analysis shown in D, and the remaining portions were used to isolate RNA. Primer extension analysis was carried out using equal amounts of RNA and the amounts of the 5′ end of 35S pre-rRNA were visualized and quantified by a PhosphorImager. The values normalized to the wild type (WT) at respective temperatures are shown below the radioactive bands. (D) Samples described above were subjected to ChIP analysis by using anti-A190 antibodies to determine the degree of association of Pol I with the promoter, the 25S coding region and NTS. PCR products were quantified by a PhosphorImager, and the values were normalized to those for input were first calculated as % Bound as described in Figure 1. Those values obtained for the rrn3-ts strain were then compared with the corresponding values obtained for WT and are shown as percentage of WT.
If Rrn3p-independent recruitment of Pol I to the promoter takes place in yeast cells in vivo, as suggested by some published in vitro experiments, one should be able to detect Pol I association with the promoter by using ChIP analysis. Contrary to this expectation, the results of ChIP analysis carried out at 3 h after the temperature shift showed a great decrease (∼76% reduction) in Pol I association with the promoter in the mutant relative to the control wild-type cells (Figure 2D). A corresponding large reduction (∼85%) was also observed for the association of Pol I with the 25S RNA coding region (Figure 2D). At 25°C, differences in these associations were much smaller between the wild-type and the mutant strains (Figure 2D). Thus, we conclude that Pol I recruitment to the promoter is largely Rrn3p-dependent in yeast cells in vivo.
Association of Pol I, Rrn3p, and UAF with the Promoter after Rapamycin Treatment
Previous experiments showed that rapamycin treatment of yeast cells leads to inhibition of rRNA transcription by Pol I (Zaragoza et al., 1998; Powers and Walter, 1999). We confirmed this conclusion by measuring the total RNA synthesis rate using [methyl-3H]methionine pulse labeling at various times after rapamycin treatment (Figure 3A). In addition, [3H]uridine labeling of cells between 30 and 60 min after rapamycin was carried out, and a large inhibition (∼90% inhibition) was observed in the incorporation of [3H]uridine into large rRNAs (25S rRNA, 18S rRNA, and their precursors) as well as 5S rRNA and tRNAs, as judged by the analysis of residual radioactive RNAs by using gel electrophoresis (our unpublished data). Inhibition of synthesis of 5S RNA and tRNA by rapamycin was reported previously (Zaragoza et al., 1998). [It should be noted that rapamycin might cause inhibition of uptake of methionine or uridine, and this defect in uptake could affect measurements of rRNA synthesis rates based on isotope labeling. However, inhibition of incorporation of [methyl-3H]methionine into protein was much weaker than that into total RNA (Figure 3A). Therefore, any potential inhibition of methionine uptake would be at most those seen for incorporation into protein, and cannot explain the large inhibition of incorporation of [methyl-3H]methionine shown in Figure 3A. In fact, Powers and Walter (1999) reported that rapamycin did not inhibit uptake of methionine to any significant extent.]
We asked whether inhibition of Tor signaling with rapamycin leads to a decrease in Pol I association with the promoter, as predicted by the model that the observed decrease of Pol I association with the promoter in the stationary phase cells is caused mainly by a decrease in Tor signaling. In the experiment shown in Figure 3B, a yeast strain carrying an HA-tagged RRN3 was used and the associations of Rrn3p and Pol I with the promoter and with the 25S rRNA coding region were measured simultaneously by ChIP after 1-h treatment of exponentially growing cells with rapamycin. First, in contrast to Pol I, which shows association with both the promoter and the 25S rRNA coding region (but not the NTS region), Rrn3p showed association mostly with the promoter and only slightly, if any, with the 25S coding region in the control growing cells (lanes 5 and 6 compared with 9 and 10). This observation is consistent with the results of in vitro experiments showing that Rrn3p is released from the DNA template soon after the initiation of transcription (Milkereit and Tschochner, 1998; Aprikian et al., 2001). Second, rapamycin treatment caused decreases in the association of both Rrn3p and Pol I with the promoter to lower levels, 30 and 35% relative to the controls, respectively, in this experiment. The association of Pol I with the 25S rRNA coding region also decreased to a lower level, 34% of the control. We conclude that Rrn3p-dependent recruitment of Pol I to the rDNA promoter is regulated by the Tor signaling system.
We also studied the effects of rapamycin on the association of UAF with the promoter using a strain carrying an HA-tagged RRN5. As shown in Figure 3C, no significant reduction in the association was observed in the presence of rapamycin. Thus, the situation resembles that observed in the stationary phase. We conclude that the association of UAF with the promoter is independent of the Tor signal transduction system that regulates Pol I recruitment to the promoter.
Association of Pol I with Rrn3p in Cells Treated with Rapamycin
We next examined whether the inhibition of Pol I recruitment to the promoter observed in rapamycin-treated cells is due to the decrease in the amount of the initiation competent Pol I-Rrn3p complex. Strain NOY1170 carrying HA-tagged RRN3 was used. Extracts were prepared from cells treated with rapamycin for 1 h and from control cells without rapamycin treatment. Pol I was immunoprecipitated with anti-A190 antibodies and the amount of Rrn3p that was coprecipitated with Pol I was quantified by Western blot analyses and normalized to the amount of the Pol I A135 subunit in the precipitate. An example of such experiments is shown in Figure 4. The calculation showed that the amount of Rrn3p complexed with Pol I in the extracts from rapamycin treated cells decreased to ∼38% relative to the control. In another experiment, this value was ∼50%. Thus, it is likely that the inhibition of Pol I (and Rrn3p) recruitment to the promoter by rapamycin is caused largely, if not entirely, by a decrease in the amount of the initiation-competent Pol I-Rrn3p complex.
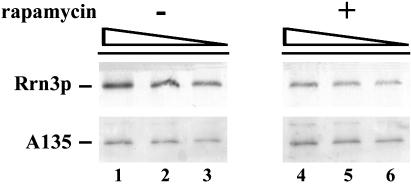
Figure 4.
Decrease in the association of Rrn3p with Pol I after rapamycin treatment as analyzed by coimmunoprecipitation. Cellular extracts were prepared from NOY1170 cells carrying HA-tagged RRN3 treated with rapamycin for 1 h and from untreated cells. Pol I was immunoprecipitated with anti-A190 antibodies. Increasing amounts of the immunoprecipitated proteins (4, 6, and 8 μl) were subjected to Western blot analysis by using antibodies against the Pol I A135 subunit and the HA tag. Quantitation indicated that the amount of Rrn3p relative to Pol I in the immunoprecipitates for the rapamycin treated cells was 38% of that for the untreated cells.
Transcription of rRNA Genes after Rapamycin Treatment Analyzed by the EM Miller Spreading Method
The presence of two kinds of rRNA genes, the active (or open) form and the inactive (or closed) form, were most convincingly demonstrated by EM visualization of rRNA genes by the Miller spreading method (McKnight and Miller, 1976; Osheim et al., 1996; French et al., 2003). We used this method to study whether the Tor pathway is used to regulate rRNA synthesis rate by altering the number of active genes in response to nutritional states. Specifically, we asked whether treatment of yeast cells with rapamycin causes a decrease in the number of active genes as observed for cells shifting from an exponentially growing state to a stationary phase state. For this purpose, we used a yeast strain (NOY886) with only 42 rRNA genes. There are two reasons for using this strain. First, all or nearly all of the rRNA genes are transcribed in NOY886. In ordinary yeast strains, only about one-half (∼75) of their ∼150 rRNA genes are transcribed, whereas the other one-half remains in an inactive state, even in exponentially growing cells (French et al., 2003). In Miller spreads, active rRNA genes occur in the familiar Christmas tree configuration, whereas inactive rRNA genes can be identified only as transcript-free chromatin strands ∼9.1 kb in length (or multiples thereof) adjacent to active genes. Thus, in typical strains, detection of an increase in the number of inactive genes relative to active genes is technically difficult because one-half of the genes are already inactive even without rapamycin. A second reason for using NOY886 is that its genes are very highly transcribed. It synthesizes rRNA at the same rate as its control strain with ∼140 gene copies and hence its density of Pol I (∼100 polymerases per gene) is twice that seen for active genes in the ∼140 copy strain (50 polymerases per gene) (French et al., 2003). The appearance of inactive rRNA genes should, therefore, be easily detected, if rapamycin causes a conversion of the active form of rRNA genes to an inactive form. In addition, a change in polymerase density should also be easily detected if rapamycin causes a change in transcription initiation rate.
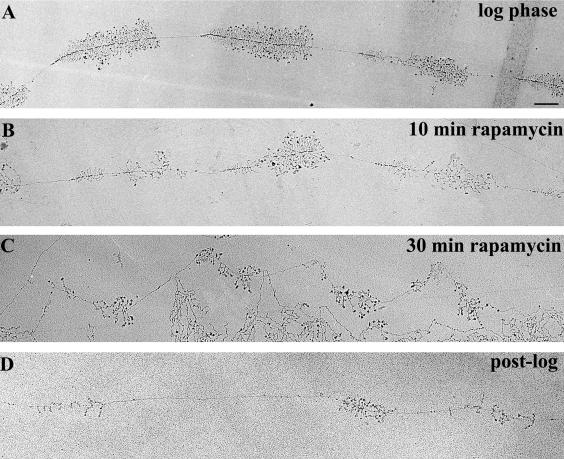
The ∼40 copy strain was treated with rapamycin and samples were taken before (“log phase”) and at 10 and 30 min after rapamycin treatment. Samples were processed and rRNA genes were visualized by the EM Miller spread method. Examples of micrographs are shown in Figure 5, A-C. Using these micrographs, we determined 1) the average number of polymerases per gene, and 2) whether genes became inactive due to rapamycin treatment. The results are summarized in Figure 6 and Table 2.
Figure 5.
Representative electron micrographs of rRNA genes from various growth conditions. (A) Log-phase cells, A600 ∼0.4. (B) Log phase cells exposed to 0.2 μg/ml rapamycin for 10 min. (C) Log-phase cells exposed to 0.2 μg/ml rapamycin for 30 min. (D) Post-log phase cells, A600 ∼2.8. Bar, 0.5 μm (all at same magnification). Note that the polymerase density decreases with exposure to rapamycin (A-C) but that individual genes remain active. In post-log phase cells, inactive genes are sometimes seen, as shown by the transcript-free interval in D. Although the genes shown in D exhibit relatively short transcripts, this is not a general observation for post-log phase cells. Rather, many transcripts seem to be normal length with characteristic terminal balls (our unpublished data).
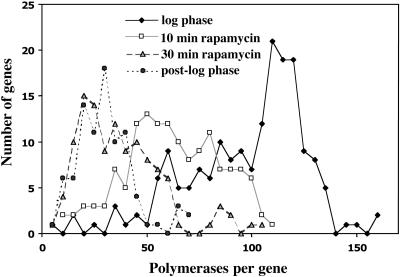
Figure 6.
Decrease in polymerase density per gene in rapamycin-treated and post-log phase cells. The number of polymerases per gene is shown for four conditions: log phase, 10-min rapamycin, 30-min rapamycin, and post-log phase. Note the shift to lower average polymerase density with the increasing time of rapamycin treatment and also as cells leave logarithmic growth phase (post-log phase). Quantitative data for the four samples are shown in Table 2.
Table 2.
Transcription of rRNA genes in the ≈40 copy yeast strain (NOY886) after rapamycin treatment and in post-log phase as analyzed by the EM Miller spreading technique
| Polymerase density
|
Inactive genes (inactive/active)
|
|||
|---|---|---|---|---|
| Samples | Ave. | Mode | N | |
| Log phase (A600 0.42) | 100 ± 29 | 114 | 174 | <1%* |
| 10-min rapamycin | 60 ± 23 | 57 | 132 | 2/233 |
| 30-min rapamycin | 36 ± 20 | 25 | 114 | 2/202 |
| Post-log phase (A600 2.8) | 30 ± 13 | 27 | 76 | 22/229 |
Although rare occurrences of inactive genes were observed, > 99% of rRNA genes were active as described previously (French et al., 2003).
First, it is clear that rapamycin treatment caused a significant decrease in polymerase density, reflecting inhibition of rRNA synthesis at the step of initiation; for cells treated with rapamycin for 30 min, polymerase density was, on the average, 36 ± 20, whereas polymerase density without rapamycin treatment was 100 ± 29. Yet, we found only two inactive genes among 233 active genes observed as tandem linear stretches for the 10-min rapamycin samples and two inactive genes among 202 active genes in linear stretches for the 30-min rapamycin samples (Table 2). The data on polymerase density indicate that the rate of Pol I transcription decreased to 60% at 10 min and 36% at 30 min after rapamycin treatment in this experiment, and this decrease did not involve any significant decrease in the number of active genes. Although for technical reasons we did not measure the rate of Pol I transcription under the same condition used for the EM Miller spread analysis, the degrees of decrease in Pol I transcription calculated from the polymerase density data are comparable with those measured by [methyl-3H]methionine incorporation by using a ∼150 copy strain growing in SD complete medium (Figure 3A). It should be noted that although there is a wide distribution of the polymerase density for the 30-min rapamycin sample with an average polymerase density of 36 ± 20, the distribution is skewed to lower rather than higher numbers, giving the modal value of 25 (Figure 6 and Table 2). (We note that the wide variation in polymerase density in this sample is mainly due to variation in individual cells' apparent sensitivity to rapamycin rather than to variation between individual rRNA gene repeats in a given cell; our unpublished data.) It is clear that inhibition of Tor by rapamycin led to a significant inhibition of Pol I transcription initiation without any significant conversion of active genes to an inactive state.
We next analyzed the same ∼40 copy strain in the post-log phase (“post-log” cells; Figures 5D and 6, and Table 2) at a cell density of A600 ∼2.8. This cell density was reached ∼7.5 h after the time point where log-phase cells were sampled (A600 ∼0.4 in these experiments) and before the entrance into the “true” stationary phase (usually a cell density reaching A600 ∼4.0 in rich media; cf. Figure 1C). (True stationary phase cells have been refractory to chromatin spreading.) The results of EM analysis of these post-log phase cells (Figure 5D) showed a narrower polymerase density distribution with the average density of 30 ± 13 polymerases per active gene and a modal value of 27 (Figure 6 and Table 2). Therefore, disregarding a small fraction of cells in the 30-min rapamycin sample that had an unusually high polymerase density as explained above, the decrease in polymerase density in this sample (modal value of 25) was very similar to that found in the sample taken at the post-log phase (modal value of 27). In the latter, we found the presence of significant numbers of inactive genes (22) among active genes (229; Table 2 and Figure 5D). Thus, cells at the post-log phase had switched ∼10% of rRNA genes from the active state to an inactive state; the value (10%) for inactive gene fraction must represent a minimum value because inactive genes without adjacent recognizable active genes cannot be counted as inactive genes and would have been missed. If the conversion of active genes to an inactive state observed in the postexponential phase is a result of the Tor signaling pathway, we would have observed inactive genes in the 30-min rapamycin sample to an extent comparable with that observed for the post-log phase cells. Because this was not the case, we conclude that the switch of rRNA genes from active to inactive states observed in post-log to stationary phases is not caused by regulatory pathways involving Tor.
DISCUSSION
The Tor pathway regulates the activity of rRNA genes in the active form but does not alter the number of active genes
Decreases in rRNA synthesis rates in the stationary phase have been known to involve both a decrease in the number of active genes and a decrease in the transcription rate of each active gene. A previous study showed that Rpd3p histone deacetylase is required for the decrease in the number of active genes, but it is not required for the decrease in the activity of each active gene observed upon entry into the stationary phase, and suggested that the two processes are regulated differently (Sandmeier et al., 2002). This observation, however, did not rule out the possibility that both processes are achieved through the Tor pathway, which senses nutritional states and signals to various targets. It is known that inhibition of the Tor protein kinase by rapamycin affects various targets, but the alterations of the targets are achieved through several divergent pathways. For example, the recent study by Broach and coworkers (Duvel et al., 2003) has demonstrated that prolonged activation of stress response genes seen after rapamycin addition or nutritional starvation is achieved through effects of Tor on the complexes of the type 2A-related phosphatases with Tap42p, but inhibition of ribosomal protein gene expression by rapamycin or nutritional starvation is achieved independently of the Tap42p-phosphatase complexes. In fact, this inhibition was reported to require the presence of Rpd3p histone deacetylase, leading to the conclusion that the Tor pathway links nutrient sensing with histone acetylation, apparently by altering the local chromatin structure at ribosomal protein gene promoters (Rohde and Cardenas, 2003). Thus, regulation of switching between active and inactive forms of rRNA genes was a potential regulatory target of Tor.
The results of analyses of yeast cells after rapamycin treatment and cells in the post-log phase presented in this article demonstrate that the inhibition of rRNA synthesis by rapamycin reported by previous workers (Zaragoza et al., 1998; Powers and Walter, 1999) is explained by a decrease in the activity of each active gene, but not by a decrease in the number of active genes. Thus, decline in the activity of each active gene in yeast cells upon entering the stationary phase seems to be achieved mostly, if not entirely, by sensing of nutritional status by the Tor pathway. In contrast, decrease in the number of active genes apparently does not involve the Tor pathway. How this regulation is achieved remains unknown.
Regarding the decrease in the activity of each active rRNA gene caused by inhibition of Tor, the EM Miller spread analysis clearly indicates that regulation takes place at the step of transcription initiation and not at the elongation step. If the elongation step rather than the initiation step were the target, polymerase density per gene would be expected to increase, but such an increase was not observed. In fact, the decrease in polymerase density observed after rapamycin treatment seems to parallel the decrease in rRNA synthesis rate, suggesting that regulation by Tor acts at the step of transcription initiation. This conclusion is consistent with the conclusion obtained from the ChIP analyses that rapamycin inhibits Pol I transcription at the step of Rrn3p-dependent recruitment of Pol I to the promoter, which is discussed below.
Rrn3p Is Essential for Recruitment of Pol I to the Promoter In Vivo
As described in INTRODUCTION, there were conflicting reports regarding the question of whether Pol I is recruited to the promoter in the absence of Rrn3p/TIF-IA. The experiments using the rrn3 (S213P) mutant described in the present article clearly demonstrate a requirement of Rrn3p for the recruitment of Pol I to the promoter in exponentially growing yeast cells. This conclusion is consistent with the results obtained in the human in vitro system by using an immobilized DNA template carrying SL1 bound at the promoter (Miller et al., 2001), but it is not consistent with the results obtained for the yeast in vitro system by using an immobilized DNA template and crude cell extracts from various yeast mutants (Aprikian et al., 2001). In the latter in vitro system, an efficient and apparently specific binding of Pol I to the promoter was observed in the complete absence of Rrn3p, although the resultant complex containing UAF, TBP, CF, and Pol I was converted to an initiation-competent complex only by a subsequent addition of extracts containing both Pol I and Rrn3p, and not by Rrn3p alone (Aprikian et al., 2001). Such an Rrn3p-independent binding of Pol I might possibly take place under certain conditions in vivo, but it does not seem to take place in growing yeast cells or in stationary phase cells analyzed in the present work.
The present experiments also do not exclude the possibility that, in addition to its role in Pol I recruitment, Rrn3p plays an important role in some steps subsequent to the formation of a preinitiation complex in vivo, for example, promoter escape. In mammalian Pol I transcription systems, phosphorylation of Rrn3p/TIF-1A was demonstrated to be required for its interaction with Pol I and its stimulation of Pol I transcription (Cavanaugh et al., 2002; Zhao et al., 2003). Furthermore, it was suggested that dephosphorylation of Rrn3p might be required for promoter escape (Cavanaugh et al., 2002). Although yeast Rrn3p can apparently function in stimulating in vitro transcription without being phosphorylated (Keener et al., 1998; Fath et al., 2001; see discussion below), it is also subject to phosphorylation both in vivo and in vitro (Fath et al., 2001; our unpublished experiments). Thus, the significance of Rrn3p phosphorylation deserves further studies, especially because Rrn3p-dependent Pol I transcription initiation is regulated by the Tor pathway, which is known to involve protein phosphorylation-dephosphorylation cascades.
Pol I Recruitment to the Promoter Is Likely a Key Target of Regulation by the Tor Signaling Pathway
We have shown that rapamycin, which inhibits Tor functions, causes a decrease in the association of Pol I with the promoter and the coding region. This result is consistent with previous observations that the amount of the initiation-competent Rrn3p-Pol I complex decreases in quiescent yeast cells (Milkereit and Tschochner, 1998) and that inactive extracts from both yeast and mammalian quiescent cells can be reactivated by addition of Rrn3p or an Rrn3p-Pol I complex (Milkereit and Tschochner, 1998; Bodem et al., 2000; Cavanaugh et al., 2002; Yuan et al., 2002).
Rapamycin inhibition of Pol I transcription may involve a phosphorylation-dephosphorylation cascade. In fact, formation of the Rrn3p-Pol I complex, which is clearly affected by rapamycin, is apparently regulated by phosphorylation in both yeast and mammalian systems. In the yeast system, dephosphorylation of Pol I, but not Rrn3p, inhibited formation of Rrn3p-Pol I complex in vitro (Fath et al., 2001). In contrast, studies on mammalian systems showed that a phosphorylated form of Rrn3p, and not unphosphorylated Rrn3p, is the one that is capable of association with Pol I and thus initiating transcription in vitro (Cavanaugh et al., 2002). Regardless of this discrepancy, one may speculate that inhibition of the Tor kinase by rapamycin may lead to an activation of a protein phosphatase and the eventual inhibition of Rrn3p-Pol I complex formation, perhaps by altering phosphorylation states of Pol I and/or Rrn3p, which may lead to a defect in the recruitment of Pol I (and Rrn3p) to the promoter, as observed in the present experiments. In this connection, we note that our coimmunoprecipitation experiments showed that the decrease in the association of Rrn3p with Pol I upon 1-h rapamycin treatment was 50-62%, whereas the inhibition of rRNA synthesis under the same condition was ∼80% (cf. Figures 3A and 4). Whether this difference is due to some inherent analytical problems, e.g., difficulty in estimating the in vivo amounts of “active” Rrn3p-Pol I complex by the method used, or reflects some unidentified step(s) affected by rapamycin (other than the Rrn3p-Pol I association as the cause for inhibition of rRNA transcription) remains unanswered.
UAF Is Stably Bound to the Promoter Independently of Pol I Transcription States
It is known that UAF binds specifically and tightly to the upstream element of the promoter and remains bound during multiple cycles of transcription in vitro (Keys et al., 1996; Aprikian et al., 2001). We have now found that the association of UAF with the promoter is unchanged after rapamycin treatment or after cells entered the stationary phase when rRNA transcription by Pol I is minimal. Purified UAF contains histones H3 and H4 but not histones H2A and H2B (Keener et al., 1997). The tight binding of UAF to the promoter may be related to the presence of histones H3 and H4. UAF is essential for silencing of transcription of chromosomal rRNA genes by Pol II (Oakes et al., 1999; Vu et al., 1999; Siddiqi et al., 2001) and for silencing of Pol II transcription of reporter genes inserted into rDNA (Buck et al., 2002; Cioci et al., 2003). Perhaps the presence of histones H3 and H4 might allow a stable structure resembling a mononucleosome to serve as a nucleation site to organize a chromatin structure unique to rDNA and to prevent events that are inappropriate for normal cell physiology such as Pol II transcription of chromosomal rRNA genes (Oakes et al., 1999; Vu et al., 1999) or an increase in unequal recombinational events (our unpublished experiments).
We also note that the observed constant association of UAF with the promoter irrespective of Pol I transcription states does not mean the absence of participation of UAF in Pol I regulation. For example, modification of UAF components, including histone H3 and H4, might possibly play a role(s) in regulation of Pol I transcription, for example, by changing chromatin structures responsible for the two states of rRNA genes, the active, and inactive states. Clearly, this subject deserves future investigation.
Acknowledgments
We thank Melanie Oakes and Chikako Itabashi for technical help, David Schneider for critical reading of the manuscript, and Melanie Oakes and Shelly VanAmburg for help in preparation of the manuscript. This work was supported by Public Health Service grants GM-35949 (to M.N.) and GM-63952 (to A.L.B.), and by the Human Frontier Science Program Organization grant RG-0336 (to M.N.).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-08-0594. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-08-0594.
References
- Aprikian, P., Moorefield, B., and Reeder, R.H. (2001). New model for the yeast RNA polymerase I transcription cycle. Mol. Cell Biol. 21, 4847-4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbet, N.C., Schneider, U., Helliwell, S.B., Stansfield, I., Tuite, M.F., and Hall, M.N. (1996). TOR controls translation initiation and early G1 progression in yeast. Mol. Biol. Cell 7, 25-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodem, J., Dobreva, G., Hoffmann-Rohrer, U., Iben, S., Zentgraf, H., Delius, H., Vingron, M., and Grummt, I. (2000). TIF-IA, the factor mediating growth-dependent control of ribosomal RNA synthesis, is the mammalian homolog of yeast Rrn3p. EMBO Rep. 1, 171-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broach, J.R. (1991). RAS genes in Saccharomyces cerevisiae: signal transduction in search of a pathway. Trends Genet. 7, 28-33. [DOI] [PubMed] [Google Scholar]
- Brun, R.P., Ryan, K., and Sollner-Webb, B. (1994). Factor C*, the specific initiation component of the mouse RNA polymerase I holoenzyme, is inactivated early in the transcription process. Mol. Cell. Biol. 14, 5010-5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck, S.W., Sandmeier, J.J., and Smith, J.S. (2002). RNA polymerase I propagates unidirectional spreading of rDNA silent chromatin. Cell 111, 1003-1014. [DOI] [PubMed] [Google Scholar]
- Buttgereit, D., Pflugfelder, G., and Grummt, I. (1985). Growth-dependent regulation of rRNA synthesis is mediated by a transcription initiation factor (TIF-IA). Nucleic Acids Res. 13, 8165-8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanaugh, A.H., Hirschler-Laszkiewicz, I., Hu, Q., Dundr, M., Smink, T., Misteli, T., and Rothblum, L.I. (2002). Rrn3 phosphorylation is a regulatory checkpoint for ribosome biogenesis. J. Biol. Chem. 277, 27423-27432. [DOI] [PubMed] [Google Scholar]
- Cioci, F., Vu, L., Eliason, K., Oakes, M., Siddiqi, I., and Nomura, M. (2003). Silencing in yeast rDNA chromatin: reciprocal relationship in gene expression between RNA polymerase I and II. Mol. Cell 12, 135-145. [DOI] [PubMed] [Google Scholar]
- Dammann, R., Lucchini, R., Koller, T., and Sogo, J.M. (1993). Chromatin structures and transcription of rDNA in yeast Saccharomyces cerevisiae. Nucleic Acids Res. 21, 2331-2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvel, K., Santhanam, A., Garrett, S., Schneper, L., and Broach, J.R. (2003). Multiple roles of Tap42 in mediating rapamycin-induced transcriptional changes in yeast. Mol. Cell 11, 1467-1478. [DOI] [PubMed] [Google Scholar]
- Fath, S., Milkereit, P., Peyroche, G., Riva, M., Carles, C., and Tschochner, H. (2001). Differential roles of phosphorylation in the formation of transcriptional active RNA polymerase I. Proc. Natl. Acad. Sci. USA 98, 14334-14339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French, S.L., Osheim, Y.N., Cioci, F., Nomura, M., and Beyer, A.L. (2003). In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol. Cell. Biol. 23, 1558-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman, J., Movva, N.R., and Hall, M.N. (1991). Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 253, 905-909. [DOI] [PubMed] [Google Scholar]
- Herman, P.K. (2002). Stationary phase in yeast. Curr. Opin. Microbiol. 5, 602-607. [DOI] [PubMed] [Google Scholar]
- Jacinto, E., and Hall, M.N. (2003). Tor signalling in bugs, brain and brawn. Nat. Rev. Mol. Cell. Biol. 4, 117-126. [DOI] [PubMed] [Google Scholar]
- Ju, Q., and Warner, J.R. (1994). Ribosome synthesis during the growth cycle of Saccharomyces cerevisiae. Yeast 10, 151-157. [DOI] [PubMed] [Google Scholar]
- Keener, J., Dodd, J.A., Lalo, D., and Nomura, M. (1997). Histones H3 and H4 are components of upstream activation factor required for the high-level transcription of yeast rDNA by RNA polymerase I. Proc. Natl. Acad. Sci. USA 94, 13458-13462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keener, J., Josaitis, C.A., Dodd, J.A., and Nomura, M. (1998). Reconstitution of yeast RNA polymerase I transcription in vitro from purified components. TATA-binding protein is not required for basal transcription. J. Biol. Chem. 273, 33795-33802. [DOI] [PubMed] [Google Scholar]
- Keys, D.A., Lee, B.S., Dodd, J.A., Nguyen, T.T., Vu, L., Fantino, E., Burson, L.M., Nogi, Y., and Nomura, M. (1996). Multiprotein transcription factor UAF interacts with the upstream element of the yeast RNA polymerase I promoter and forms a stable preinitiation complex. Genes Dev. 10, 887-903. [DOI] [PubMed] [Google Scholar]
- Keys, D.A., Vu, L., Steffan, J.S., Dodd, J.A., Yamamoto, R.T., Nogi, Y., and Nomura, M. (1994). RRN6 and RRN7 encode subunits of a multiprotein complex essential for the initiation of rDNA transcription by RNA polymerase I in Saccharomyces cerevisiae. Genes Dev. 8, 2349-2362. [DOI] [PubMed] [Google Scholar]
- Kuras, L., and Struhl, K. (1999). Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature 399, 609-613. [DOI] [PubMed] [Google Scholar]
- Lalo, D., Steffan, J.S., Dodd, J.A., and Nomura, M. (1996). RRN11 encodes the third subunit of the complex containing Rrn6p and Rrn7p that is essential for the initiation of rDNA transcription by yeast RNA polymerase I. J. Biol. Chem. 271, 21062-21067. [DOI] [PubMed] [Google Scholar]
- Lin, C.W., Moorefield, B., Payne, J., Aprikian, P., Mitomo, K., and Reeder, R.H. (1996). A novel 66-kilodalton protein complexes with Rrn6, Rrn7, and TATA-binding protein to promote polymerase I transcription initiation in Saccharomyces cerevisiae. Mol. Cell. Biol. 16, 6436-6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight, S.L., and Miller, O.L., Jr. (1976). Ultrastructural patterns of RNA synthesis during early embryogenesis of Drosophila melanogaster. Cell 8, 305-319. [DOI] [PubMed] [Google Scholar]
- Milkereit, P., and Tschochner, H. (1998). A specialized form of RNA polymerase I, essential for initiation and growth-dependent regulation of rRNA synthesis, is disrupted during transcription. EMBO J. 17, 3692-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, G., Panov, K.I., Friedrich, J.K., Trinkle-Mulcahy, L., Lamond, A.I., and Zomerdijk, J.C. (2001). hRRN3 is essential in the SL1-mediated recruitment of RNA Polymerase I to rRNA gene promoters. EMBO J. 20, 1373-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogi, Y., Vu, L., and Nomura, M. (1991a). An approach for isolation of mutants defective in 35S ribosomal RNA synthesis in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 88, 7026-7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogi, Y., Yano, R., and Nomura, M. (1991b). Synthesis of large rRNAs by RNA polymerase II in mutants of Saccharomyces cerevisiae defective in RNA polymerase I. Proc. Natl. Acad. Sci. USA 88, 3962-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura, M. (1998). Transcription factors used by Saccharomyces cerevisiae RNA polymerase I and the mechanism of initiation. In: Transcription of Ribosomal RNA Genes by Eukaryotic RNA Polymerase I, ed. M. Paule, Austin: RG Landes & Co., 155-172.
- Nomura, M. (2001). Ribosomal RNA gene, RNA polymerases, nucleolar structures and synthesis of rRNA in the yeast Saccharomyces cerevisiae. Cold Spring Harbor. Symp. Quant. Biol. 66, 555-565. [DOI] [PubMed] [Google Scholar]
- Oakes, M., Siddiqi, I., Vu, L., Aris, J., and Nomura, M. (1999). Transcription factor UAF, expansion and contraction of ribosomal DNA (rDNA) repeats, and RNA polymerase switch in transcription of yeast rDNA. Mol. Cell. Biol. 19, 8559-8569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osheim, Y.N., and Beyer, A.L. (1989). Electron microscopy of ribonucleoprotein complexes on nascent RNA using Miller chromatin spreading method. Methods Enzymol. 180, 481-509. [DOI] [PubMed] [Google Scholar]
- Osheim, Y.N., Mougey, E.B., Windle, J., Anderson, M., O'Reilly, M., Miller, O.L., Jr., Beyer, A., and Sollner-Webb, B. (1996). Metazoan rDNA enhancer acts by making more genes transcriptionally active. J. Cell Biol. 133, 943-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyroche, G., Milkereit, P., Bischler, N., Tschochner, H., Schultz, P., Sentenac, A., Carles, C., and Riva, M. (2000). The recruitment of RNA polymerase I on rDNA is mediated by the interaction of the A43 subunit with Rrn3. EMBO J. 19, 5473-5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers, T., and Walter, P. (1999). Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol. Biol. Cell 10, 987-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder, R.H. (1999). Regulation of RNA polymerase I transcription in yeast and vertebrates. Prog. Nucleic Acid. Res. Mol. Biol. 62, 293-327. [DOI] [PubMed] [Google Scholar]
- Rohde, J., and Cardenas, M.E. (2003). The Tor pathway regulates gene expression by linking nutrient sensing to histone acetylation. Mol. Cell. Biol. 23, 629-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde, J., Heitman, J., and Cardenas, M.E. (2001). The TOR kinases link nutrient sensing to cell growth. J. Biol. Chem. 276, 9583-9586. [DOI] [PubMed] [Google Scholar]
- Sandmeier, J.J., French, S., Osheim, Y., Cheung, W.L., Gallo, C.M., Beyer, A.L., and Smith, J.S. (2002). RPD3 is required for the inactivation of yeast ribosomal DNA genes in stationary phase. EMBO J. 21, 4959-4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelzle, T., and Hall, M.N. (2000). TOR, a central controller of cell growth. Cell 103, 253-262. [DOI] [PubMed] [Google Scholar]
- Schnapp, A., and Grummt, I. (1991). Transcription complex formation at the mouse rDNA promoter involves the stepwise association of four transcription factors and RNA polymerase I. J. Biol. Chem. 266, 24588-24595. [PubMed] [Google Scholar]
- Schnapp, A., Pfleiderer, C., Rosenbauer, H., and Grummt, I. (1990). A growth-dependent transcription initiation factor (TIF-IA) interacting with RNA polymerase I regulates mouse ribosomal RNA synthesis. EMBO J. 9, 2857-2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnapp, A., Schnapp, G., Erny, B., and Grummt, I. (1993). Function of the growth-regulated transcription initiation factor TIF-IA in initiation complex formation at the murine ribosomal gene promoter. Mol. Cell. Biol. 13, 6723-6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F., Fink, G. R., and Hicks, J. B. (1986). Laboratory Course Manual for Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
- Siddiqi, I.N., Dodd, J.A., Vu, L., Eliason, K., Oakes, M.L., Keener, J., Moore, R., Young, M.K., and Nomura, M. (2001). Transcription of chromosomal rRNA genes by both RNA polymerase I and II in yeast uaf30 mutants lacking the 30 kDa subunit of transcription factor UAF. EMBO J. 20, 4512-4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan, J.S., Keys, D.A., Dodd, J.A., and Nomura, M. (1996). The role of TBP in rDNA transcription by RNA polymerase I in Saccharomyces cerevisiae: TBP is required for upstream activation factor-dependent recruitment of core factor. Genes Dev. 10, 2551-2563. [DOI] [PubMed] [Google Scholar]
- Thevelein, J.M., and de Winde, J.H. (1999). Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 33, 904-918. [DOI] [PubMed] [Google Scholar]
- Vu, L., Siddiqi, I., Lee, B.S., Josaitis, C.A., and Nomura, M. (1999). RNA polymerase switch in transcription of yeast rDNA: role of transcription factor UAF (upstream activation factor) in silencing rDNA transcription by RNA polymerase II. Proc. Natl. Acad. Sci. USA 96, 4390-4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner, J.R. (1991). Labeling of RNA and phosphoproteins in Saccharomyces cerevisiae. Methods Enzymol. 194, 423-428. [DOI] [PubMed] [Google Scholar]
- Yamamoto, R.T., Nogi, Y., Dodd, J.A., and Nomura, M. (1996). RRN3 gene of Saccharomyces cerevisiae encodes an essential RNA polymerase I transcription factor which interacts with the polymerase independently of DNA template. EMBO J. 15, 3964-3973. [PMC free article] [PubMed] [Google Scholar]
- Yuan, X., Zhao, J., Zentgraf, H., Hoffmann-Rohrer, U., and Grummt, I. (2002). Multiple interactions between RNA polymerase I, TIF-IA and TAFI subunits regulate preinitiation complex assembly at the ribosomal gene promoter. EMBO Rep. 3, 1082-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaragoza, D., Ghavidel, A., Heitman, J., and Schultz, M.C. (1998). Rapamycin induces the G0 program of transcriptional repression in yeast by interfering with the TOR signaling pathway. Mol. Cell. Biol. 18, 4463-4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J., Yuan, X., Frodin, M., and Grummt, I. (2003). ERK-dependent phosphorylation of the transcription initiation factor TIF-IA is required for RNA polymerase I transcription and cell growth. Mol. Cell 11, 405-413. [DOI] [PubMed] [Google Scholar]