Abstract
Aleutian Mink Disease Virus (AMDV) is the only virus in the genus Amdovirus of family Parvoviridae. In adult mink, AMDV causes a persistent infection associated with severe dysfunction of the immune system. Cleavage of AMDV capsid proteins has been previously shown to play a role in regulating progeny virus production (Fang Cheng et al, J. Virol. 84:2687–2696, 2010). The present study shows that AMDV has evolved a second strategy to limit expression of capsid proteins by preventing processing of the full-length capsid protein-encoding mRNA transcripts. Characterization of the cis-elements of the proximal polyadenylation site [(pA)p] in the infectious clone of AMDV revealed that polyadenylation at the (pA)p site is controlled by an upstream element (USE) of 200 nts in length, the AAUAAA signal, and a downstream element (DSE) of 40 nts. A decrease in polyadenylation at the (pA)p site, either by mutating the AAUAAA signal or the DSE, which does not affect the encoding of amino acids in the infectious clone, increased the expression of capsid protein VP1/VP2 and thereby increased progeny virus production approximately 2-3-fold. This increase was accompanied by enhanced replication of the AMDV genome. Thus, this study reveals correlations among internal polyadenylation, capsid production, viral DNA replication and progeny virus production of AMDV, indicating that internal polyadenylation is a limiting step for parvovirus replication and progeny virus production.
Introduction
Aleutian Mink Disease Virus (AMDV) is the only virus in the genus Amdovirus, in the family Parvoviridae (Fauquet et al., 2005). Disease manifestations caused by AMDV vary in animals of different ages. In neonatal mink kits, AMDV infection causes acute, rapidly progressing interstitial pneumonia with a high mortality rate (Alexandersen, 1990). In adult mink, however, AMDV infection is characterized by glomerulonephritis, plasmacytosis, hypergammaglobinemia, arteritis, decreased fertility, and spontaneous abortion (Alexandersen et al., 1994; Gorham et al., 1976). Aleutian mink disease is the most significant infectious disease affecting mink farming worldwide and it has becoming a limiting factor in the production of mink in some areas (Newman and Reed, 2006). Notably, a recent study reported that AMDV may be a zoonotic disease capable of infecting humans (Jepsen et al., 2009).
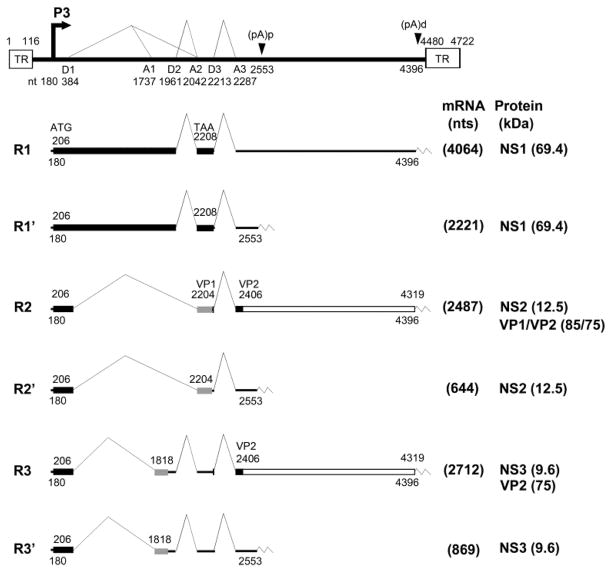
AMDV shares many features with other parvoviruses, including members of the genera Erythrovirus [human parvovirus B19 (B19V)] and Bocavirus [minute virus of canines (MVC), and human bocavirus (HBoV)]. AMDV has a single-stranded DNA (ssDNA) genome of 4.7 kb. A single pre-mRNA is transcribed from the P3 promoter and this generates six mRNA transcripts through alternative splicing and alternative polyadenylation (Qiu et al., 2006a) (Fig. 1). Only the mRNA transcripts that read through the proximal polyadenylation site [(pA)p] are capable of encoding the capsid proteins VP1 and VP2. The R2 mRNA, which is spliced from the D1 donor to the A2 acceptor and from the D3 donor to the A3 acceptor, encodes both the VP1 and VP2 capsid protein (Qiu et al., 2007b). Alternative polyadenylation is utilized commonly during pre-mRNA processing of parvoviruses (Qiu et al., 2006b). It is especially important in members of the genera Amdovirus, Erythrovirus and Bocavirus in that only one pre-mRNA is transcribed, which has to read through the internal polyadenylation site in order to generate full-length capsid protein-encoding mRNA transcripts (Liu et al., 2004; Ozawa et al., 1987; Qiu et al., 2007a; Sun et al., 2009). Block of the full-length mRNA transcript production has been suggested to be a limiting step in B19V infection of permissive cells (Liu et al., 1992). We have shown that replication of the B19V genome in permissive cells or “artificial” replication of the B19V genome supported by a heterogeneous replication origin can overcome this block, resulting in a significantly increased level of B19V capsid protein-encoding mRNA transcripts (Guan et al., 2008). However, the exact function of internal polyadenylation in parvovirus replication has not been investigated in the context of an infectious clone.
Fig. 1. Genetic map of AMDV.
The genome of AMDV-G is depicted to scale with indicated transcription units, including the hairpin terminal repeats (TR), P3 promoter, splice donor site (D1, D2, and D3), splice acceptor sites (A1, A2, and A3), internal proximal polyadenylation site [(pA)p], and internal distal polyadenylation site [(pA)d]. Six major mRNA transcripts are shown with R1-3 polyadenylated at the (pA)d site and R1’-3’ polyadenylated at the (pA)p site. The AMDV-G proteins NS1, NS2, NS3 (only putative) and VP1/VP2 encoded from each RNA are indicated.
Aleutian mink disease is an immune complex-mediated disease associated with persistent infection in adult mink (Best and Bloom, 2005b; Bloom et al., 1975; Eklund et al., 1968). Control of capsid protein production has been shown to govern the persistence of AMDV in infected host animals (Alexandersen et al., 1988; Christensen et al., 1993; Storgaard et al., 1993; Storgaard et al., 1997), which likely requires controlled low levels of capsid proteins. We have previously demonstrated that capsid proteins of AMDV are cleaved at a specific site of 417DLLD/G421 by caspases during infection (Cheng et al., 2009). Mutations of this site increase expression levels of VP1 and VP2, as well as progeny virus production. We hypothesize that the cleavage (or reduction) of capsid proteins limits packaging of the viral ssDNA genome, and that this cleavage is likely to be important for the maintenance of persistent infection and restriction of AMDV replication in infected mink, in which a low level of viral DNA replication is balanced during in vivo AMDV infection of mink. This mechanism contributes to persistent infection of AMDV in adult mink in addition to the fact that AMDV is poorly neutralized by antibodies generated during in vivo infection (Best and Bloom, 2005a).
The current study explores the relationships among internal polyadenylation, virus DNA replication, capsid protein expression and progeny virus production of AMDV in the context of an AMDV infectious clone. The cis-elements located in the internal polyadenylation site were characterized and it was found that the prevention of internal polyadenylation increased capsid protein expression as well as the replication of the AMDV genome, increasing the production of progeny virus. This is the first study to show such a relationship in the context of a parvovirus infectious clone.
Results
The transcription profile of the CMV promoter-driven non-replicative AMDV-G genome is comparable to that of infectious virus in CrFK cells
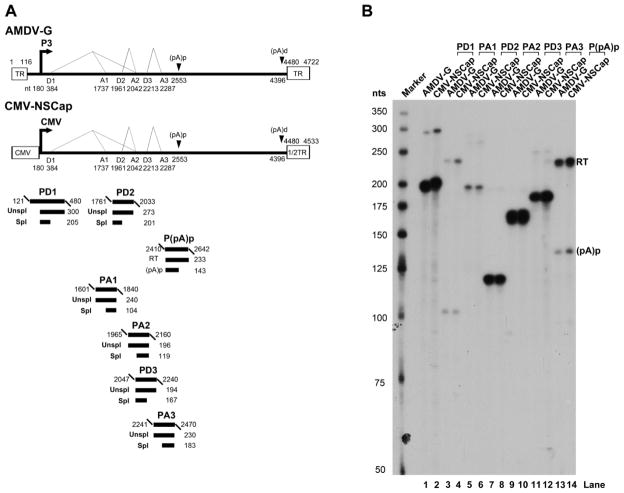
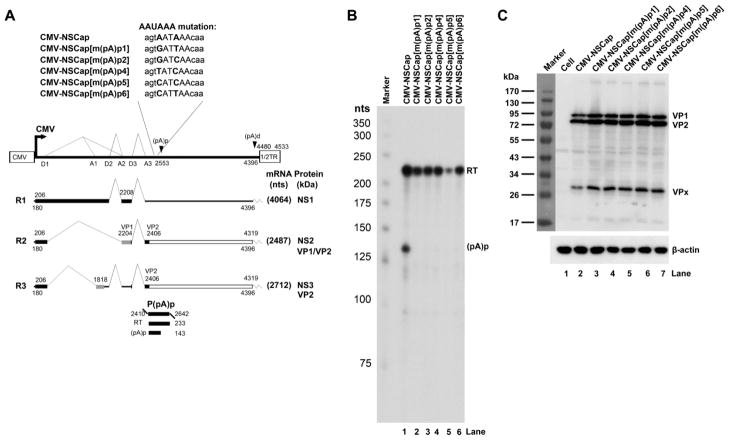
To characterize the internal polyadenylation site of AMDV, the P3 promoter was replaced with a CMV promoter. This initiated transcription of nt 180-4533 of the AMDV genome, which includes NS1- and capsid protein-encoding regions and the distal polyadenylation site [(pA)d)] (Fig. 2A). RNAs were generated from the transfection of the construct CMV-NSCap and compared with those generated from AMDV-G infection in CrFK cells. Comparisons were carried out through RNase protection assays using seven anti-sense RNA probes (Fig. 2A) (Qiu et al., 2006a). The levels of protection of AMDV mRNA transcripts in the CMV-NSCap– transfected and AMDV-G-infected CrFK were similar to those determined previously (Fig. 2B) (Qiu et al., 2006a). For example, the probe P(pA)p probe protected read-through (RT) RNA vs. RNA polyadenylated at the (pA)p site [(pA)p] in transfected cells at a ratio of ~3:1, which is approximately the same as the ratio seen during virus infection (Fig. 2B, compare lanes 13 to 14). Thus, it was decided to use transfection with the CMV-NSCap construct for identifying cis-elements lying in the (pA)p site.
Fig. 2. RNAs produced from the transfection of a non-replicative AMDV genome are processed similarly to those produced during AMDV-G infection.
(A) Schematic diagram of the AMDV genome and the probes used for the RNase protection assay (RPA). The landmarks of transcription are shown. The RPA probes PD1, PA1, PD2, PA2, PD3, PA3 and p(pA)p are shown along with their nucleotide numbers (numbering is according to Genbank accession no. JN040434). Bands from each probe that were expected to be protected are shown with sizes under their corresponding region in the genome. Spl, spliced RNAs; Unspl, unspliced RNAs; RT, RNAs resulting from read-through of the (pA)p site; (pA)p, RNAs polyadenylated RNAs at (pA)p. (B) RPA analysis. Ten μg of total RNA isolated from CMV-NSCap-transfected and AMDV-infected CrFK cells was used for the RPA analysis, along with the above described probes, in individual reactions as indicated. Lane 1, 32P-labeled RNA markers (Qiu et al., 2002), with sizes indicated to the left.
Characterization of cis-elements in the internal polyadenylation site of AMDV
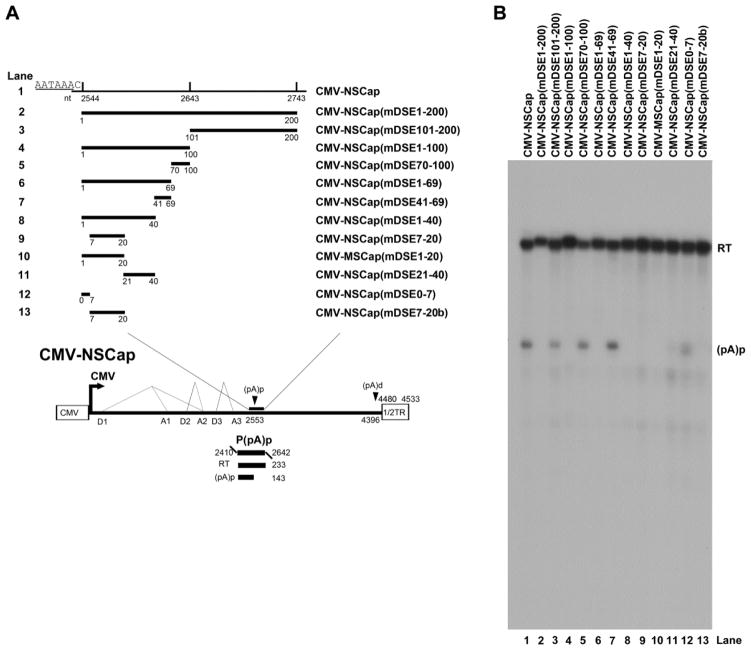
A core polyadenylation signal is comprised of an upstream element (USE), the core AAUAAA site, and a downstream element (DSE) (Zhao et al., 1999). To identify the DSE at the AMDV (pA)p site, a series of mutations were introduced in the region downstream of the (pA)p site, spanning nt 2544-2743. In CMV-NSCap, deletion of the whole region (DSE1-200) abolished polyadenylation at the (pA)p site, as did deletion of the first 100 nts (DSE 1-100), but deletion of the second 100-nts (DSE101-200) did not affect polyadenylation. This narrowed the location of the DSE down to the first 100 nts (Fig. 3B, lanes 3&4). The sequence of nt 2583-2612 was further mutated (41 and 69 nts after nt 2544, respectively) and nt 2613-2643 was mutated (70 and 100 nts after nt 2544, respectively). Neither of these mutations affected polyadenylation at the (pA)p site, which remained at a level comparable to that of the wild type in both CMV-NSCap(mDSE41-69) and CMV-NSCap(mDSE70-100) (Fig. 3B, lanes 5&7 vs. 1). However, further mutations in the region 40 nts downstream of the (pA)p site either abolished or significantly reduced polyadenylation at (pA)p (Fig. 3A, lanes 9-13). We conclude that the cis- sequence of nt 2544-2583 (40-nts), immediately downstream of the (pA)p site, is critical for polyadenylation of AMDV mRNA transcripts to be polyadenylated at the (pA)p site, which functions as a DSE (Zarudnaya et al., 2003).
Fig. 3. Identification of the DSE of the AMDV internal polyadenylation site.
(A) Schematic diagram of mutations downstream of the (pA)p site. The AMDV genome is shown, along with its transcription units. Mutations in the region of nt 2544-2473 are diagrammed, with detailed sequences described in the Materials and Methods. The P(pA)p probe is shown. (B) RPA analysis. Ten μg of total RNA isolated after two days from CMV-NSCap- or its mutant-transfected CrFK cells was used for protection by the probe P(pA)p or its homology probes. The origins of the two protected bands in the lanes are indicated to the right.
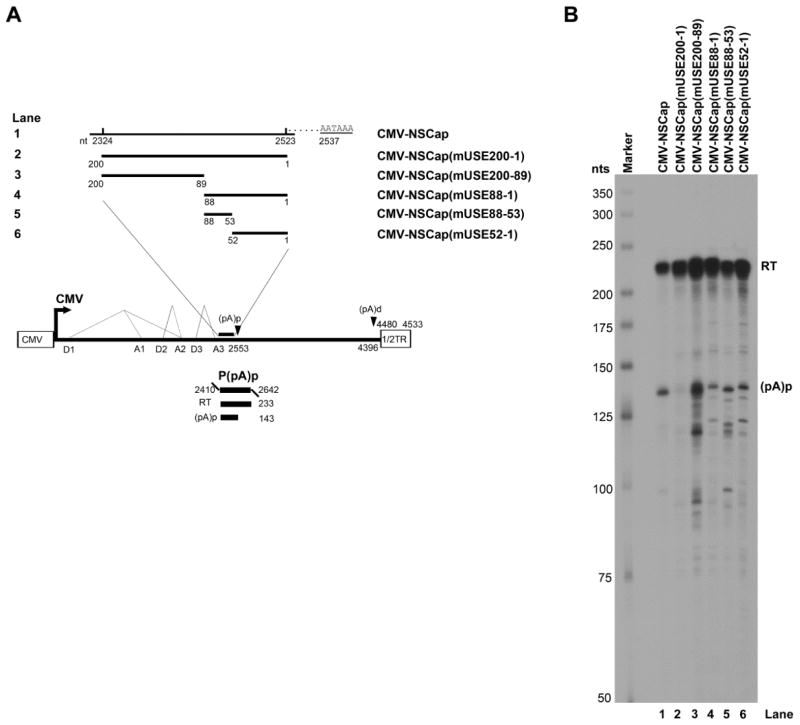
To identify the USE, we made a series of mutations in the region spanning nt 2324-2523 upstream of the (pA)p site. Only the complete mutation of all 200-nts abolished the polyadenylation of AMDV pre-mRNA at the (pA)p site (Fig. 4B, lane 2). Further mutations in this 200 nts sequence, in the downstream half of the region, reduced polyadenylation at the (pA)p site only slightly, from ~40% to ~10–20% (Fig. 4B, lane 2 vs. lanes 4–6). Mutations in the upstream half of the region did not reduce polyadenylation at the (pA)p site significantly (Fig. 4B, lane 3). We conclude that the cis-sequence at nt 2324-2523 (200-nts), upstream of the (pA)p site, plays a role as the USE for efficient polyadenylation of AMDV mRNA transcripts at the (pA)p site.
Fig. 4. Identification of the USE of the AMDV internal polyadenylation site.
(A) Schematic diagram of mutations upstream of the (pA)p site. The AMDV genome is shown, along with its transcription units. Mutations in the region of nt 2324–2523 are diagrammed, with detailed sequences described in the Materials and Methods. The P(pA)p probe is also shown. (B) RPA analysis. Ten μg of total RNA isolated from CMV-NSCap- or its mutant- transfected CrFK cells was used for the RPA, along with the P(pA)p probe. The origins of the two protected bands in the lanes are indicated to the right.
Knockdown of the core polyadenylation signal AAUAAA in an AMDV-G infectious clone increases replication of the AMDV genome and progeny virus production
The core polyadenylation signal AAUAAA, which binds to cleavage and polyadenylation specific factor 1 (CPSF160), is the key element in mammalian mRNA polyadenylation (Zhao et al., 1999). A series of mutations in this hexanucleotide were made and five mutant constructs were created on CMV-NSCap. None of the five mutants produced detectable mRNA transcripts polyadenylated at the (pA)p site (Fig. 5B, lane 1. vs. 2–6), supporting the key role of this hexanucleotide in the internal polyadenylation of AMDV mRNA transcripts. It was also observed that the levels of capsid proteins VP1 and VP2 expressed from these mutants were increased ~2-fold compared with those of the wild type (Fig. 5C, lane 2. vs. 3–7). These same mutations of the AAUAAA were then introduced into an AMDV infectious clone (pIAMDV) (Cheng et al., 2009). Transfection of the pIAMDV-based AAUAAA mutants also failed to produce AMDV mRNA transcripts polyadenylated at the (pA)p site (data not shown). This suggested that only three AMDV mRNA transcripts (R1-3) that are polyadenylated at the (pA)d site are produced (Fig. 5A).
Fig. 5. Mutagenesis analysis of the AAUAAA site.
(A) Schematic diagram of mutations of the AAUAAA site. The AMDV genome is shown, along with its transcription units. Mutations in the AAUAAA site are shown, as is the P(pA)p probe. (B) RPA analysis. Ten μg of total RNA isolated two days after transfection of CrFK cells with CMV-NSCap or mutant was used for the RPA analysis, along with the probe p(PA)p. Lane 1, 32P-labeled RNA markers. The origins of the two protected bands in the lanes are indicated to the right. (C) Western blot analysis. CrFK cells were transfected with CMV-NSCap or its mutants. At two days post-transfection, whole cell lysates were prepared and analyzed. The blot was probed using a polyclonal antibody against VP2 (#2788). β-actin was probed as a loading control.
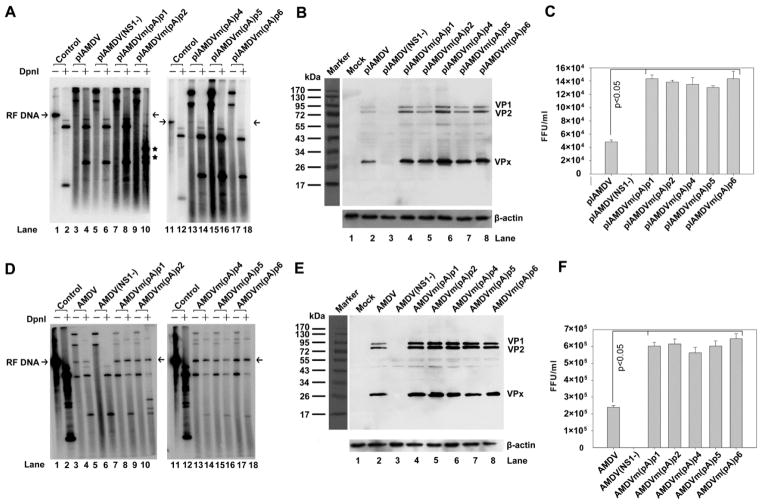
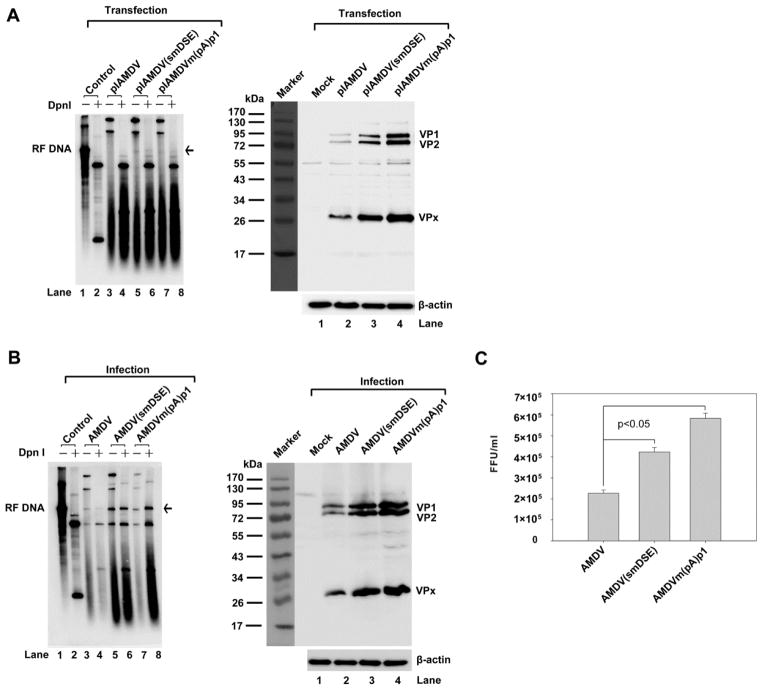
Next, viral DNA replication, capsid protein expression, and progeny virus production from transfected pIAMDV-based AAUAAA mutants were examined. AMDV-G replicates optimally in CrFK cells maintained at a low temperature of 31.8ºC, and a fully productive infection takes places over the course of six-seven days (Bloom et al., 1994; Qiu et al., 2006a). Therefore, we transfected the mutants and maintained the cells for seven days at 31.8ºC. Hirt DNA was treated with DpnI to reveal newly synthesized viral DNA. Transfection of pIAMDV showed barely any replicative form (RF) DNA from the AMDV genome (Fig. 6A, lane 4). RF DNA was detectable in samples from AAUAAA-mutated pIAMDV clones (Fig. 6A, lanes 7–10 and lanes 13–18), and was not detected in the NS1 knockout mutant (Fig. 6A, lane 6). This result suggests that the activity of (pA)p has no significant effect on the initial rounds of viral DNA replication following transfection. However, for all the (pA)p knockout mutants, there was an increase in capsid protein expression of, on average, ~2–3 fold (Fig. 6B, lane 3 vs. lanes 5–9), as well as an increase in progeny virus production of ~3-fold (Fig. 6C). Further passaging of the viruses produced from transfections revealed a clearer picture than that seen during transfection. Approximately 3-fold of increases in RF DNA, VP1/VP2, and progeny infectious virus were consistently observed in infections of the AAUAAA-mutated AMDV-G viruses (Fig. 6D-F).
Fig. 6. Analysis of AAUAAA mutants in the AMDV-G infectious clone.
(A) Southern blot analysis. Hirt DNA was isolated from CrFK cells infected with AMDV or transfected with pIAMDV or its mutant. Samples were digested with or left undigested and subjected to Southern blotting. The blots were probed with a full-length AMDV probe. Arrow indicates replicative form of the AMDV genome (RF DNA). Asterisk indicates digestion of an introduced DpnI site (GATC) in the (pA)p2 mutation (Fig. 5A). (B) Western blotting analysis. Whole-cell lysates were prepared from the same CrFK cells infected with AMDV, or transfected with pIAMDV or its mutant, followed by Western blotting. The blots were probed with an anti-VP2 polyclonal antibody (#2788). β-actin was used as a loading control. (C) Titration of progeny virus. CrFK cells were infected with AMDV, or transfected with pIAMDV and its mutants, and were harvested at seven days post-treatment. The cells were frozen and thawed three times, and briefly centrifuged. The supernatant was collected, serially diluted, and used to infect CrFK cells in order to titrate FFU. FFU per ml is shown for infection from each virus preparation. (D-F) CrFK cells were infected with virus preparation of AMDV-G and its mutants. At seven days post-infection, infected cells were collected. Hirt DNA, total protein, and virus production were analyzed by Southern blotting (D), Western blotting (E), and virus titration (F), respectively, as described in panels A-C. Results shown in panels C&F represent the averages and S.D. from at least three independent experiments. P < 0.05 as assessed based on the Student’s t test.
Collectively, these results suggest that the knockout of the AAUAAA signal in the (pA)p site significantly increases the production of infectious virus after a round of reinfection, probably due to increased levels of VP1/VP2 resulting from increased production of VP1/VP2-encoding mRNAs as well as from increased DNA replication.
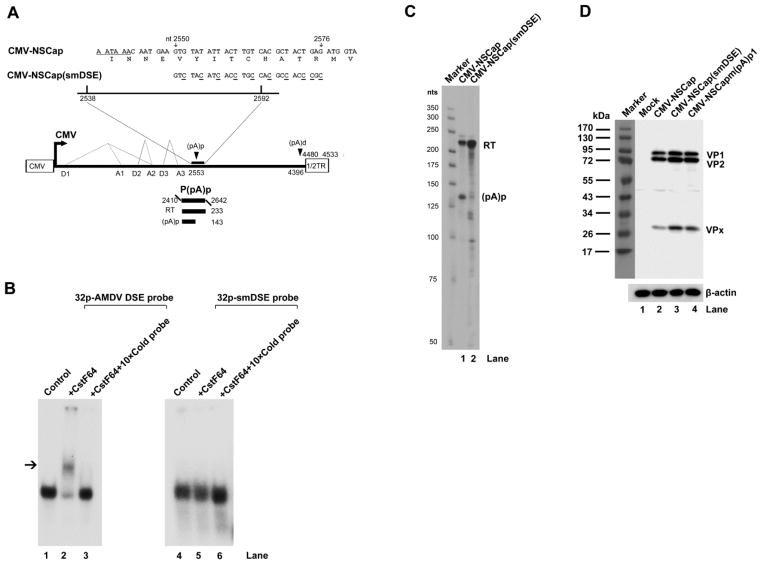
Silent mutation of the DSE, which abrogates binding to CstF64, in the AMDV infectious clone increases progeny virus production
An alternative approach to decreasing the polyadenylation of AMDV pre-mRNA at the (pA)p site is to knock out its DSE. We introduced ten silent mutations (one that would not affect the encoded amino acids of VP1/2) in the identified 40 nt DSE (Fig. 7A). The cellular protein CstF64 binds the DSE at the polyadenylation site and often plays a role in regulating alternative polyadenylation (Zhao et al., 1999). Thus, we examined the binding capability of the CstF64 to the AMDV DSE and the silent mutant (smDSE) in vitro. The binding of CstF64 to the AMDV DSE RNA shifted the location of the 32p-labelled DSE RNA to a position upstream from that of the free DSE RNA (Fig. 7B, lane 2), but this shift (binding) could be competitively inhibited by incubation with incubation of a 10 times excess of unlabeled AMDV DSE RNA (Fig. 7B, lane 3). This result suggests that the AMDV DSE but not the mutant smDSE (Fig. 7B, lanes 4–6) binds CstF64 specifically.
Fig. 7. Analysis of the DSE silent mutant.
(A) Sequence of the DSE silent mutation. A schematic diagram of the DSE silent mutation is shown above the AMDV genome and its transcription units. The P(pA)p probe is shown. (B) In vitro gel shift analysis of CstF64 binding. AMDV DSE and smDSE RNAs were in vitro transcribed and labeled as described in the Materials and Methods. The labeled RNA was incubated with purified human CstF64 or further incubated with 10 times more unlabelled RNA. The final incubation mixtures were analyzed on a 5% native polyacrylamide gel. The gel was autoradiographed using X-ray film. Arrow indicates the shifted band. (C) RPA analysis. Ten μg of total RNA isolated from CrFK cells transfected with CMV-NSCap and CMV-NSCap(smDSE) was protected using the P(pA)p probe. (D) Western blot analysis. CrFK cells were transfected with either CMV-NSCap or the smDSE mutant. At two days post-transfection, whole-cell lysates were prepared and analyzed. The blot was probed using the anti-VP2 polyclonal antibody #2788. β-actin was probed as a loading control.
We next examined the polyadenylation efficiency of the smDSE mutant in CrFK cells. Transfection of CMV-NSCap(smDSE) nearly abolished polyadenylation of AMDV pre-mRNA at the (pA)p site (Fig. 7C, lane 3 vs. lane 2), and produced ~2 times more VP1/VP2 than its non-mutated counterpart (Fig. 7D, lane 2). Furthermore, this DSE silent mutation was introduced into pIAMDV and compared with pIAMDV and pIAMDV[m(pA)p1] in terms of its effects on DNA replication, VP1/VP2 expression and virus production. After transfection, we observed that pIAMDV(smDSE) replicated as well as pIAMDV[m(pA)p1] (Fig. 8A, left panel, lanes 6&8). Furthermore, pIAMDV(smDSE) produced ~4 times more VP1/VP2 than pIAMDV (Fig. 8A, right panel, lane 3 vs. lane 2). We passaged the produced viruses in CrFK cells once. An average of ~3–4 times higher levels of RF DNA and VP1/VP were detected in AMDV(smDSE)- and AMDV[m(pA)p1]-infected cells than in AMDV-infected cells (Fig. 8B). In turn, the number of progeny virus produced in AMDV(smDSE)- and AMDV[m(pA)p1]-infected cells was ~2–3-fold that produced in AMDV-infected cells (Fig. 8C). Again, these results strongly support the above conclusion that preventing the polyadenylation of AMDV pre-mRNA at the (pA)p site increases progeny virus production.
Fig. 8. Analysis of the DSE silent mutant in the context of an AMDV infectious clone.
(A) Southern blot and Western blot analysis of transfected cells. CrFK cells were transfected with pIAMDV, pIAMDV(smDSE) and pIAMDV[m(pA)p1]. At seven days post-transfection, transfected cells were collected and used to prepare Hirt DNA and total protein. Hirt DNA was digested with DpnI or left undigested for Southern blotting. The blots were probed with a full-length AMDV probe. Whole-cell lysates were used for Western blotting. The blots were probed with the anti-VP2 polyclonal antibody #2788. β-actin was used as a loading control. (B) Southern blot and Western blot analysis of infected cells. CrFK cells were infected with AMDV-G, AMDV-G(smDSE) and AMDV-G[m(pA)p1]. At seven days post-infection, infected cells were collected and used to prepare Hirt DNA and whole-cell lysate. Hirt DNA samples were digested with DpnI or left undigested for Southern blotting. The blots were probed with a full-length AMDV probe. Whole-cell lysates were used for Western blotting. The blots were probed with an anti-VP2 polyclonal antibody. β-actin was used as a loading control. (C) Titration of progeny virus. CrFK cells were infected with AMDV, AMDV(smDSE), or AMDV[m(pA)p1], and were harvested at seven days post-treatment. The cells were frozen and thawed three times, and briefly centrifuged. The supernatant was collected, serially diluted, and used to infect CrFK cells in order to titrate FFU. FFU per ml is shown for each infection. Results shown represent the averages and S.D. from at least three independent experiments. P < 0.05 as assessed based on the Student’s t test.
Discussion
In this study, we have identified the USE and DSE that control internal polyadenylation of the AMDV pre-mRNA at the (pA)p site. Silent mutations of either the DSE or the AAUAAA hexanucleotide in the context of an AMDV infectious clone abolished polyadenylation of AMDV pre-mRNA at this site and facilitated progeny virus production. Thus, our study reveals for the first time the biological function of internal polyadenylation during parvovirus infection. This has implications for other parvoviruses, e.g., B19V and MVC, which also use internal polyadenylation in the processing of their pre-mRNA.
The established B19V infectious clone is not efficient, and production of progeny virus is extremely low in the current culture system (Zhi et al., 2006; Zhi et al., 2004). This makes it practically impossible to examine the function of internal polyadenylation during B19V infection. In MVC, a member of the genus Bocavirus, silent mutations at the AAUAAA or the downstream region of the (pA)p site failed to knock out polyadenylation at this site (Sun Y & Qiu J, unpublished observation), which also made it impossible to examine the function of MVC internal polyadenylation during infection. The successful knockout of the (pA)p site in the AMDV infectious clone and identification of its function in progeny virus production may imply a common mechanism underlying infection by parvoviruses with functional (pA)p sites. Regulation of polyadenylation can be achieved in cis through binding of CPSF160 to the AAUAAA hexanucleotide, binding of CstF64 to the DSE, and binding of several factors to the USE (Zhao et al., 1999). The AMDV (pA)p site contains a consensus AAUAAA site, in contrast to the nonconsensus AUUAAA in B19V pre-mRNA, indicating that it might be difficult to regulate internal polyadenylation through binding of the CPSF160 to this site. AMDV may have evolved in such way so that it can regulate the internal polyadenylation through factors binding to the DSE or USE site in different tissues or animals at various ages during virus infection. We had tried to improve the DSE to a consensus CstF64-binding sequence, for example, 5’GUUGUGGUGU3’ (Zarudnaya et al., 2003), at 15-nts downstream of the AAUAAA site in the context of the non-replicative CMV-NSCap construct. This mutation resulted in the polyadenylation of almost all of the AMDV pre-mRNA at the (pA)p site (data not shown). However, we were not able to introduce such a silent mutation into the genome of the infectious clone. Notably, we found that the DSE is conserved among various AMDV isolates (data not shown). Collectively, these observations suggest that AMDV could have evolved to sustain its replication at a moderate level in order to maintain a status of persistent infection through a conserved and functional DSE sequence that binds to CstF64. The USE spans a large sequence of 200 nts, and a small region was not defined (Fig. 4). We speculate there are two or more redundant positive control elements, or mixed positive and negative control elements located in the USE, which warrants further characterization.
A hallmark of AMDV infection in adult mink is persistent infection. Viral production during such infection is restricted (Best and Bloom, 2005a), which may be beneficial in terms of reduced neutralization by neutralization antibodies and formation of immune complexes. Permissive replication of AMDV in CrFK cells has been shown to require the activation of caspases (Best et al., 2002), which can be activated by the expression of AMDV capsid proteins (Cheng et al., 2009). Caspases have been shown to cleave the large nonstructural protein NS1 at two sites, D227 and D285, respectively. These cleavages events were required for nuclear localization of the NS1 (Best et al., 2003). In addition, active caspase cleaves VP1/VP2 at site of the 417DLLD/G421 site, reducing production of full length of VP1 and VP2 suitable for capsid assembly (Cheng et al., 2009). Therefore, there must be a balance of capsid protein production, active caspase, and functional NS1 for DNA replication. AMDV may finely tune the expression level of capsid proteins to activate caspases and assemble progeny virions through the evolution of the (pA)p signal to an optimal level for this expression.
The AMDV infectious clone replicates DNA and produces progeny virus inefficiently after a single transfection (Bloom et al., 1990; Cheng et al., 2009). Production of progeny virus requires at least one passage of the virus produced from transfection but will eventually reach a level akin to that from virus infection (Bloom et al., 1990; Cheng et al., 2009). The exact mechanism underlying this phenomenon is unknown. RF DNA is very hard to detect from transfection of the infectious clone. The AUG translation initiation site for VP1/VP2 in the infectious clone was knocked out, and we hoped to observe a single burst of DNA replication from the AMDV genome (without the potential for reinfection). Such a single burst of DNA replication has been clearly shown with the MVC infectious clone (Sun et al., 2009). However, RF DNA was not detectable after transfection with the AMDV VP1/VP2-knockout mutant (data not shown). Thus, we believe that mutations of the AAUAAA and DSE sites do not directly affect replication of AMDV RF DNA. An ~3–4-fold increase in VP1/VP2 expression was observed in response to preventing polyadenylation at the (pA)p site, which in turn increased the level of DNA replication. This strongly suggests that increased levels of VP1/VP2 result in increased assembly of empty virions to encapsidate the ssDNA genome generated from the replication of RF DNA, and this in turn produces more progeny virus. The rolling-hairpin replication model of parvoviral DNA (Cotmore and Tattersall, 2005) and several recent studies (Cheng et al., 2009; Plevka et al., 2011) support the idea that the assembly of empty capsids is a prerequisite for parvoviral ssDNA production. It is highly possible that there is a feedback mechanism between assembled empty capsids and ssDNA production.
In conclusion, we have provided direct evidence for the first time that internal polyadenylation is a limiting step for parvovirus AMDV production. This control mechanism might also be utilized by other parvoviruses that use internal polyadenylation for pre-mRNA processing. These parvoviruses have evolved to maintain a persistent (or regulated) infection. The genome of HBoV contains a (pA)p site that is used during infection (Dijkman et al., 2009) and this site may prevent excessive production of virus, according to results from specimens from lower respiratory tract infections (Allander, 2008; Lin et al., 2008). B19V may also use this post-transcriptional regulatory approach to control progeny virus production (Guan et al., 2008). Further investigations are warranted to examine these possibilities.
Materials and Methods
Cell and virus
The CrFK cells (Crandall feline kidney cell line, ATCC CCL-94) were obtained from the American Type Culture Collection (ATCC, Manassas, VA). Cells were maintained in Dulbecco's modified Eagle's medium with 10% fetal calf serum at 37°C in 5% CO2. The AMDV-G strain of AMDV was propagated and assayed in CrFK cells as previously reported (Bloom et al., 1990; Bloom et al., 1993; Mayer et al., 1983).
Transfection and virus infection
CrFK cells were transfected using two μg of DNA per 60-mm plate with Lipofectamine and Plus reagent (Invitrogen, Carlsbad, CA) as described previously (Qiu and Pintel, 2002). CrFK cells were infected with AMDV-G at a multiplicity of infection (MOI) of 1 [1 fluorescence-focus forming unit (FFU) per cell] (Bloom et al., 1980; Porter et al., 1977).
Plasmid constructs
(i) Construction of AMDV expression plasmids
CMV-NSCap-based plasmids
CMV-NSCap has been previously described (Qiu et al., 2006a). The CMV-NSCap-based plasmids used to examine downstream- and upstream-elements (DSE and USE) were illustrated in Figs. 3&4. Detailed depictions of the mutations in each of the regions are listed in Table 1. CMV-NSCap-based mutants with mutations in the AAUAAA region are illustrated in Fig. 5. CMV-NSCap(smDSE) was constructed by silently mutating the DSE at nt 2550–2576 as shown in Fig. 7.
pIAMDV-based plasmids
pIAMDV has been previously described (Cheng et al., 2009). pIAMDV-based m(pA)p and smDSE mutants were constructed by replacing the AAUAAA and DSE sites of the pIAMDV with their respective mutations in the CMV-NSCap –based mutants. pIAMDV(NS1-) was constructed by mutating the G residues at nt 404 and 407 to T in pIAMDV, which knocked out the expression of the full-length NS1 (data not shown).
(ii) Clones used to generate probes for the RNase protection assay
RNase protection assay was used to map the transcription profile generated from transfection of CMV-NSCap. The probes used, PD1, PA1, PA2, PD3, and PA3 (see Fig. 2A), were constructed as described previously (Qiu et al., 2006a). Probe P(pA)p and its homologous probes were made by cloning nt 2410-2642 of the AMDV-G genome and the corresponding regions of AMDV-G mutants with mutations at the (pA)p site into BamHI-HindIII-digested pGEM4Z (Promega).
All of the AMDV-G nucleotide numbers (nt) used in this study refer to Genbank accession no. JN040434 (Bloom et al., 1990). All of the DNA constructs were sequenced at www.mclab.com to confirm that the mutations were as designed.
RNA isolation and RNase protection assay
Total RNA was isolated using TRIzol® Reagent (Invitrogen, Carlsbad, CA) (Guan et al., 2008). RNase protection assays were performed with 10 μg of total RNA as previously described (Naeger et al., 1992; Schoborg and Pintel, 1991). Probes were generated from linearized templates using the MAXIscript® in vitro transcription kit from Ambion (Austin, TX). RNA hybridizations were carried out in substantial probe excess, and signals were quantified with a Typhoon FLA 9000 Phosphor Imager and ImageQuant TL (version 4.2.2) imaging software (GE Heath, Piscataway, NJ). Relative molar ratios of individual species of RNAs were determined after adjustment for the number of 32P-labeled uridines in each protected fragment, as previously described (Schoborg and Pintel, 1991).
Southern blot analysis
AMDV-G replicative form (RF) DNA produced from transfection or infection was visualized by Southern blot analysis. Hirt DNA was isolated from transfected or infected cells by the Hirt extraction method, as previously described (Qiu et al., 2006a). Samples were run on a 1% agarose gel, and Southern blotting was performed as described previously (Naeger et al., 1990; Tullis et al., 1993), using the AMDV-G DNA of nt 1-4722. Blots were exposed to a GE Health Care phosphor imaging screen, and the DNA forms were quantified using a Typhoon FLA 9000 Phosphor Imager and ImageQuant TL (version 4.2.2) imaging software (GE Health Care).
Western blot analysis
Western blotting was performed on cell extracts taken at seven days post-infection or two days post-transfection. Whole-cell lysate was prepared as previously described (Qiu et al., 2006a). Samples were subjected to immunoblotting as previously described (Liu et al., 2004), using a polyclonal antibody (#2788) against AMDV VP2 amino acids 428-446 (Cheng et al., 2009). Anti-β-actin monoclonal antibody was purchased from Sigma (St. Louis, MO). Signals were developed using SuperSignal West Dura Chemiluminescent Substrate (ThermoFisher Scientific, Waltham, MA). The images were taken using the Fuji LAS-3000 imaging system and quantified by Fuji Multi Gauge v2.3 software.
Fluorescence focus-forming assay to titrate virus production
Virus titers were determined according to FFU in CrFK cells at 31.8°C as previously reported (Bloom et al., 1980; Porter et al., 1977). Briefly, 4-well chamber slides (Lab Tek II, Nunc) were seeded with CrFK cells at a density 5.0×105 cells/well, and incubated for 24 hrs at 37 °C in 5% CO2, to produce a confluent monolayer. The virus sample was titrated 10-fold and used to infect the cell monolayer in a volume of 0.1 ml. The infected monolayer was incubated at 31.8°C in 5% CO2. After 6 days of infection, an immunofluorenscence assay was carried out on the infected cells. Overlay medium was removed from the wells and replaced with cold phosphate-buffered saline (PBS). After 5 min incubation on ice, cells were fixed for 10 min in ice-cold acetone and then washed with PBS. The cells were incubated with antibody #2788 at a dilution of 1:100 in PBS containing 0.2% BSA (PBS-BSA) for 1 hr at room temperature and then washed three times with PBS-BSA. Antibody-labeled cells were detected through the incubation of the cells for 1 hr with a secondary antibody conjugated with FITC (at a dilution of 1:100 in PBS-BSA), followed by washing three times with PBS-BSA. Cells that were labeled with fluorescent antibody were visualized under a NIKON Ti S inverted fluorescence microscope with a Photometrics HQ2 dual model B&W low light CCD camera. Fluorescence foci located within each well were counted, and virus titers were calculated and expressed as FFU per ml.
Flag-tagged CstF64 protein expression and purification
Full-length cDNA encoding human cleavage stimulation factor-64 (CstF64) was provided from Dr. Clinton C. MacDonald (Dass et al., 2001). The C-terminal flag-tagged CstF64 ORF was inserted into the plasmid pLenti-MCS-IRES-GFP-WPRE, generating the plasmid pLenti-CstF64-Flag. The pLenti-MCS-IRES-GFP-WPRE was constructed by modifying sequences downstream from the CMV promoter in pLenti-GFP-Puro (#17448, Addgene, Cambridge, MA) with, in order, a multiple cloning site (MCS, BamH-EcoRI-EcoRV-SpeI-SalI), IRES-GFP (from pIRES2-EGFP, Clontech) and the WPRE (Woodchuck post-transcriptional element). FreeStyle 293-F cells were cultured in suspension in FreeStyle 293 Expression Medium and transfected with the FreeStyle MAX Reagent following the manufacturers’ instructions (Invitrogen). At two days post-transfection, the cells were collected. CstF64-Flag protein was purified using anti-Flag® M2 affinity gel (Sigma) according to the manufacturer’s instructions. Briefly, the conditioned medium containing CstF64-Flag and proteinase inhibitors was incubated overnight at 4°C with the appropriate amount of anti-Flag M2 affinity gel in the conditioned medium. After incubation, the resin was collected by filtration through a chromatography column and washed. The CstF64-Flag protein was then eluted with 3×Flag peptide in TBS (pH 7.5). BSA (1 mg/ml) was added to the protein before being storage at −80°C. The purified proteins were quantified by Western blot with Flag-BAP protein (Sigma) as a standard using anti-Flag® M2 antibody (Strategene).
RNA gel shift analysis
An RNA sequence (probe) spanning either the DSE or the silently mutated DSE (smDSE) region (nt 2487-2615) was generated from linearized templates using the MAXIscript® in vitro transcription kit from Ambion (Austin, TX). Approximately (~)0.5 μg of CstF64-Flag protein and 32p-UTP-labeled RNA probe (~15,000 cpm) were incubated in the binding buffer (20 mM Tris-HCl pH 7.6, 0.5 mM EDTA, 0.5 mM dithiothreitol, 10% glycerol, 30 mM KCl, 3mM MgCl2) at room temperature for 20 min. The samples were then separated in a 5% non- denaturing polyacrylamide gel in 0.5×TBE buffer at 120 V for 2 hrs. The gels were then dried and exposed to radiographic films.
Table.
Plasmid construction.
| Construct name | Sequence on AMDV-G (JN040434) | Sequence on Lambda DNA (J02459.1) |
|---|---|---|
| CMV-NSCap(mDSE1-200) | nt 2544-2743 | 401-600 |
| CMV-NSCap(mDSE101-200) | nt 2644-2743 | 501-600 |
| CMV-NSCap(mDSE1-100) | nt 2544-2643 | 401-500 |
| CMV-NSCap(mDSE70-100) | nt 2613-2643 | 470-500 |
| CMV-NSCap(mDSE1-69) | nt 2544-2612 | 401-469 |
| CMV-NSCap(mDSE41-69) | nt 2584-2612 | 5’ctggatctcagtgcgctgctggcgagcct3’ |
| CMV-NSCap(mDSE1-40) | nt 2544-2583 | 401-440 |
| CMV-NSCap(mDSE7-20) | nt 2550-2563 | 5’TCTCGCGCGCGCGCG3’ |
| CMV-NSCap(mDSE1-20) | nt 2544-2563 | 401-420 |
| CMV-NSCap(mDSE21-40) | nt 2564-2583 | 5’ccacccaccaaccaaacacc3’ |
| CMV-NSCap(mDSE0-7) | nt 2543-2549 | ATTACTT |
| CMV-NSCap(mDSE7-20b) | nt 2550-2563 | 5’CACAAGAGGACGGCA3’ |
| CMV-NSCap(mUSE200-1) | nt 2324-2523 | 401-600 |
| CMV-NSCap(mUSE200-89) | nt 2324-2435 | 401-512 |
| CMV-NSCap(mUSE 88-1) | nt 2436-2523 | 513-600 |
| CMV-NSCap(mUSE 88-53) | nt 2436-2471 | 513-548 |
| CMV-NSCap(mUSE 52-1) | nt 2472-2523 | 549-600 |
Acknowledgments
This work was supported by PHS grant R01 AI070723 and R21 AI085236, and was supported in part by the Intramural Research Program of the National Institutes of Health (NIH), National Institute of Allergy and Infectious Diseases (NIAID). We thank members in the Qiu lab for valuable discussions, and are indebted to Fang Cheng for excellent technical support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexandersen S. Pathogenesis of disease caused by Aleutian mink disease parvovirus. APMIS Suppl. 1990;14:1–32. [PubMed] [Google Scholar]
- Alexandersen S, Bloom ME, Wolfinbarger J. Evidence of restricted viral replication in adult mink infected with Aleutian disease of mink parvovirus. J Virol. 1988;62:1495–1507. doi: 10.1128/jvi.62.5.1495-1507.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandersen S, Larsen S, Aasted B, Uttenthal A, Bloom ME, Hansen M. Acute interstitial pneumonia in mink kits inoculated with defined isolates of Aleutian mink disease parvovirus. Vet Pathol. 1994;31:216–228. doi: 10.1177/030098589403100209. [DOI] [PubMed] [Google Scholar]
- Allander T. Human bocavirus. J Clin Virol. 2008;41:29–33. doi: 10.1016/j.jcv.2007.10.026. [DOI] [PubMed] [Google Scholar]
- Best SM, Bloom ME. Aleutian mink disease parvovirus. In: Kerr JR, Cotmore SF, Bloom ME, Linden ME, Parrish CR, editors. The parvoviruses. Hodder Arnold; London, United Kingdom: 2005a. pp. 457–471. [Google Scholar]
- Best SM, Bloom ME. Pathogenesis of aleutian mink disease parvovirus and similarities to b19 infection. J Vet Med B Infect Dis Vet Public Health. 2005b;52:331–334. doi: 10.1111/j.1439-0450.2005.00864.x. [DOI] [PubMed] [Google Scholar]
- Best SM, Shelton JF, Pompey JM, Wolfinbarger JB, Bloom ME. Caspase cleavage of the nonstructural protein NS1 mediates replication of Aleutian mink disease parvovirus. J Virol. 2003;77:5305–5312. doi: 10.1128/JVI.77.9.5305-5312.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best SM, Wolfinbarger JB, Bloom ME. Caspase activation is required for permissive replication of Aleutian mink disease parvovirus in vitro. Virology. 2002;292:224–234. doi: 10.1006/viro.2001.1238. [DOI] [PubMed] [Google Scholar]
- Bloom ME, Alexandersen S, Garon CF, Mori S, Wei W, Perryman S, Wolfinbarger JB. Nucleotide sequence of the 5'-terminal palindrome of Aleutian mink disease parvovirus and construction of an infectious molecular clone. J Virol. 1990;64:3551–3556. doi: 10.1128/jvi.64.7.3551-3556.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom ME, Berry BD, Wei W, Perryman S, Wolfinbarger JB. Characterization of chimeric full-length molecular clones of Aleutian mink disease parvovirus (ADV): identification of a determinant governing replication of ADV in cell culture. J Virol. 1993;67:5976–5988. doi: 10.1128/jvi.67.10.5976-5988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom ME, Kanno H, Mori S, Wolfinbarger JB. Aleutian mink disease: puzzles and paradigms. Infect Agents Dis. 1994;3:279–301. [PubMed] [Google Scholar]
- Bloom ME, Race RE, Hadlow WJ, Chesebro B. Aleutian disease of mink: the antibody response of sapphire and pastel mink to Aleutian disease virus. J Immunol. 1975;115:1034–1037. [PubMed] [Google Scholar]
- Bloom ME, Race RE, Wolfinbarger JB. Characterization of Aleutian disease virus as a parvovirus. J Virol. 1980;35:836–843. doi: 10.1128/jvi.35.3.836-843.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F, Chen AY, Best SM, Bloom ME, Pintel D, Qiu J. The capsid proteins of Aleutian mink disease virus (AMDV) activate caspases and are specifically cleaved during infection. J Virol. 2009;84:2687–2696. doi: 10.1128/JVI.01917-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen J, Storgaard T, Viuff B, Aasted B, Alexandersen S. Comparison of promoter activity in Aleutian mink disease parvovirus, minute virus of mice, and canine parvovirus: possible role of weak promoters in the pathogenesis of Aleutian mink disease parvovirus infection. J Virol. 1993;67:1877–1886. doi: 10.1128/jvi.67.4.1877-1886.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotmore SF, Tattersall P. A rolling-haipin strategy: basic mechanisms of DNA replication in the parvoviruses. In: Kerr J, Cotmore SF, Bloom ME, Linden RM, Parrish CR, editors. Parvoviruses. Hoddler Arond; London: 2005. pp. 171–181. [Google Scholar]
- Dass B, McMahon KW, Jenkins NA, Gilbert DJ, Copeland NG, MacDonald CC. The gene for a variant form of the polyadenylation protein CstF-64 is on chromosome 19 and is expressed in pachytene spermatocytes in mice. J Biol Chem. 2001;276:8044–8050. doi: 10.1074/jbc.M009091200. [DOI] [PubMed] [Google Scholar]
- Dijkman R, Koekkoek SM, Molenkamp R, Schildgen O, van der Hoek L. Human bocavirus can be cultured in differentiated human airway epithelial cells. J Virol. 2009;83:7739–7748. doi: 10.1128/JVI.00614-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eklund CM, Hadlow WJ, Kennedy RC, Boyle CC, Jackson TA. Aleutian disease of mink: properties of the etiologic agent and the host responses. J Infect Dis. 1968;118:510–526. doi: 10.1093/infdis/118.5.510. [DOI] [PubMed] [Google Scholar]
- Fauquet CM, Mayo MA, Desselberger U, Ball LA. Virus Taxonomy: VIIIth report of the International Committee on Taxonomy of Viruses. Academic Press; 2005. [Google Scholar]
- Gorham JR, Henson JB, Crawford TB, Padgett GA. The epizootiology of aleutian disease. Front Biol. 1976;44:135–58. 135–158. [PubMed] [Google Scholar]
- Guan W, Cheng F, Yoto Y, Kleiboeker S, Wong S, Zhi N, Pintel DJ, Qiu J. Block to the production of full-length B19 virus transcripts by internal polyadenylation is overcome by replication of the viral genome. J Virol. 2008;82:9951–9963. doi: 10.1128/JVI.01162-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jepsen JR, d'Amore F, Baandrup U, Clausen MR, Gottschalck E, Aasted B. Aleutian mink disease virus and humans. Emerg Infect Dis. 2009;15:2040–2042. doi: 10.3201/eid1512.090514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F, Guan W, Cheng F, Yang N, Pintel D, Qiu J. ELISAs using human bocavirus VP2 virus-like particles for detection of antibodies against HBoV. J Virol Methods. 2008;149:110–117. doi: 10.1016/j.jviromet.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JM, Green SW, Shimada T, Young NS. A block in full–length transcript maturation in cells nonpermissive for B19 parvovirus. J Virol. 1992;66:4686–4692. doi: 10.1128/jvi.66.8.4686-4692.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Qiu J, Cheng F, Chu Y, Yoto Y, O'Sullivan MG, Brown KE, Pintel DJ. Comparison of the transcription profile of simian parvovirus with that of the human erythrovirus B19 reveals a number of unique features. J Virol. 2004;78:12929–12939. doi: 10.1128/JVI.78.23.12929-12939.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer LW, Aasted B, Garon CF, Bloom ME. Molecular cloning of the Aleutian disease virus genome: expression of Aleutian disease virus antigens by a recombinant plasmid. J Virol. 1983;48:573–579. doi: 10.1128/jvi.48.3.573-579.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naeger LK, Cater J, Pintel DJ. The small nonstructural protein (NS2) of the parvovirus minute virus of mice is required for efficient DNA replication and infectious virus production in a cell-type-specific manner. J Virol. 1990;64:6166–6175. doi: 10.1128/jvi.64.12.6166-6175.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naeger LK, Schoborg RV, Zhao Q, Tullis GE, Pintel DJ. Nonsense mutations inhibit splicing of MVM RNA in cis when they interrupt the reading frame of either exon of the final spliced product. Genes Dev. 1992;6:1107–1119. doi: 10.1101/gad.6.6.1107. [DOI] [PubMed] [Google Scholar]
- Newman SJ, Reed A. A national survey for Aleutian disease prevalence in ranch mink herds in Canada. Scientiffur. 2006;30:33–40. [Google Scholar]
- Ozawa K, Ayub J, Hao YS, Kurtzman G, Shimada T, Young N. Novel transcription map for the B19 (human) pathogenic parvovirus. J Virol. 1987;61:2395–2406. doi: 10.1128/jvi.61.8.2395-2406.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plevka P, Hafenstein S, Li L, D'Abrgamo A, Jr, Cotmore SF, Rossmann MG, Tattersall P. Structure of a Packaging-Defective Mutant of Minute Virus of Mice Indicates that the Genome Is Packaged via a Pore at a 5-Fold Axis. J Virol. 2011;85:4822–4827. doi: 10.1128/JVI.02598-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter DD, Larsen AE, Cox NA, Porter HG, Suffin SC. Isolation of Aleutian disease virus of mink in cell culture. Intervirology. 1977;8:129–144. doi: 10.1159/000148888. [DOI] [PubMed] [Google Scholar]
- Qiu J, Cheng F, Burger LR, Pintel D. The transcription profile of Aleutian Mink Disease Virus (AMDV) in CRFK cells is generated by alternative processing of pre-mRNAs produced from a single promoter. J Virol. 2006a;80:654–662. doi: 10.1128/JVI.80.2.654-662.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Cheng F, Johnson FB, Pintel D. The transcription profile of the bocavirus bovine parvovirus is unlike those of previously characterized parvoviruses. J Virol. 2007a;81:12080–12085. doi: 10.1128/JVI.00815-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Cheng F, Pintel D. The abundant R2 mRNA generated by aleutian mink disease parvovirus is tricistronic, encoding NS2, VP1, and VP2. J Virol. 2007b;81:6993–7000. doi: 10.1128/JVI.00244-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Nayak R, Tullis GE, Pintel DJ. Characterization of the transcription profile of adeno-associated virus type 5 reveals a number of unique features compared to previously characterized adeno-associated viruses. J Virol. 2002;76:12435–12447. doi: 10.1128/JVI.76.24.12435-12447.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Pintel DJ. The adeno-associated virus type 2 Rep protein regulates RNA processing via interaction with the transcription template. Mol Cell Biol. 2002;22:3639–3652. doi: 10.1128/MCB.22.11.3639-3652.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Yoto Y, Tullis GE, Pintel D. Parvovirus RNA processing strategies. In: Kerr JR, Cotmore SF, Bloom ME, Linden ME, Parish CR, editors. Parvoviruses. Hodder Arnold; London, UK: 2006b. pp. 253–274. [Google Scholar]
- Schoborg RV, Pintel DJ. Accumulation of MVM gene products is differentially regulated by transcription initiation, RNA processing and protein stability. Virology. 1991;181:22–34. doi: 10.1016/0042-6822(91)90466-o. [DOI] [PubMed] [Google Scholar]
- Storgaard T, Christensen J, Aasted B, Alexandersen S. cis-acting sequences in the Aleutian mink disease parvovirus late promoter important for transcription: comparison to the canine parvovirus and minute virus of mice. J Virol. 1993;67:1887–1895. doi: 10.1128/jvi.67.4.1887-1895.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storgaard T, Oleksiewicz M, Bloom ME, Ching B, Alexandersen S. Two parvoviruses that cause different diseases in mink have different transcription patterns: transcription analysis of mink enteritis virus and Aleutian mink disease parvovirus in the same cell line. J Virol. 1997;71:4990–4996. doi: 10.1128/jvi.71.7.4990-4996.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Chen AY, Cheng F, Guan W, Johnson FB, Qiu J. Molecular characterization of infectious clones of the minute virus of canines reveals unique features of bocaviruses. J Virol. 2009;83:3956–3967. doi: 10.1128/JVI.02569-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullis GE, Burger LR, Pintel DJ. The minor capsid protein VP1 of the autonomous parvovirus minute virus of mice is dispensable for encapsidation of progeny single-stranded DNA but is required for infectivity. J Virol. 1993;67:131–141. doi: 10.1128/jvi.67.1.131-141.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarudnaya MI, Kolomiets IM, Potyahaylo AL, Hovorun DM. Downstream elements of mammalian pre-mRNA polyadenylation signals: primary, secondary and higher-order structures. Nucleic Acids Res. 2003;31:1375–1386. doi: 10.1093/nar/gkg241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Hyman L, Moore C. Formation of mRNA 3' ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi N, Mills IP, Lu J, Wong S, Filippone C, Brown KE. Molecular and functional analyses of a human parvovirus B19 infectious clone demonstrates essential roles for NS1, VP1, and the 11-kilodalton protein in virus replication and infectivity. J Virol. 2006;80:5941–5950. doi: 10.1128/JVI.02430-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi N, Zadori Z, Brown KE, Tijssen P. Construction and sequencing of an infectious clone of the human parvovirus B19. Virology. 2004;318:142–152. doi: 10.1016/j.virol.2003.09.011. [DOI] [PubMed] [Google Scholar]