Abstract
Neurons have highly dynamic cellular processes for their proper functions such as cell growth, synaptic formation, or synaptic plasticity by regulating protein synthesis and degradation. Therefore, the quality control of proteins in neurons is essential for their physiology and pathology. Autophagy is a cellular degradation pathway by which cytosolic components are sequestered in autophagosomes and degraded upon their fusion with lysosomal components. Thus, the autophagic pathway may play important roles in neuronal cell survival and neuronal function under physiological condition and pathological conditions. Recent several findings suggest that the loss of basal autophagy or imbalance of autophagic flux leads to neurodegeneration. Autophagosomes accumulate abnormally in affected neurons of several neurodegenerative diseases such as Alzheimer's disease (AD), Huntington's disease (HD), Parkinson's disease (PD), or Frontotemporal dementia (FTD). Thus, the understanding how autophagy is associated with several neurological diseases would be the first step for new therapeutic intervention in neurological disorders. In this review, I will discuss the molecular mechanism of autophagy in neurons and autophagy-associated neurodegenerative diseases.
Keywords: neuron, autophagy, neurodegenerative disease, homeostasis
INTRODUCTION
Autophagy is an intracellular degradation pathway characterized by autophagosome containing cytosolic components delivered into lysosome for degradation [1, 2].
Autophagy is highly conserved from yeast to mammals. It generally has three major types; macroautophagy, chaperon-mediated autophagy (CMA), and microautophagy. Although all three forms of autophagy occur degradation of cellular substrates within lysosomes, each pathway has unique features. Among three types of autophagy, macroautophagy has been mostly well characterized. Macroautophagy sequesters cytosolic components including proteins, lipids, sugar, RNA, and organelles such as mitochondria into autophagosomes which can be fused with lysosome for degradation [3-5]. Intriguingly, based on the type of organelles degraded in lysosomes, there are some examples of organelle specific autophagy such as mitophagy, ribophagy, reticulophagy, and pexophagy [6]. Although autophagy is generally known as a nonselective degradative process, chaperone-mediated autophagy is a kind of selective degradative pathways. The proteins containing KFERQ-like peptide motif are targeted into lysosomes for their degradation [7]. The third form of autophagy, known as microautophagy, involves in pinocytosis of small quantities of cytoplasmic regions by lysosomes [8-10]. Although autopahgy is conserved from yeast to human, some of the types, such as CMA, have been characterized in higher eukaryotes but not in yeast [11].
Autophagy has been detected and known as a type II programmed cell death, autophagic cell death, which was characterized as accumulation of autophagic vacuoles when cells were dying [12]. Excessive activity of autophagy may lead to self destruction and cell death or insufficient autophagic activity (or imbalanced autophagic flux) may contribute to cell death.
Autophagy has been extensively studied in stress condition, such as starvation or accumulation of toxic components including proteins or damaged organelles. The primary role of autophagy under starvation is to provide the energy to the starved cells by degrading cellular substrates for cell survival [12].
Recent studies have demonstrated that autophagy pathway is involved in several physiological processes such as normal development, cellular homeostasis, life span expansion, tumor suppression, and immunity as well as stress associated conditions [13, 14]. Therefore, malfunction of autophagy causes several human diseases including infectious diseases, cancer, cardiovasicular diseases, and neurodegenerative diseases. Moreover, it has been also demonstrated that in several diseases autophagy is induced and activated to protect cells against neuronal cell death by removing toxic components in damaged cells [14].
Although all cell types have autophagy pathway, growing body of evidence suggest the importance of autophagic regulation in a cell type specific manner. Neurons have highly specialized structures, axon and dendrites in which a lot of molecules including proteins, RNA, and lipids are synthesized, delivered, and degraded for their dynamic functions in synaptic growth and synaptic activity. Another feature of neurons is that they are non dividing cells which are more sensitive to accumulation of toxic components than dividing ones. Therefore, the tight control for degradation of cytoplasmic components under physiological and pathological conditions must be important for their survival and maintenance of specialized functions. Moreover, the brain requires a high energy depending on an external supply of nutrients may have a highly regulated mechanism to provide nutrients under even extreme starvation or diabetes [15]. Therefore, quality control of neuronal components including proteins and oraganelles by autophagy would be essential for their cell survival in specialized post-mitotic neurons. Indeed, inhibition of autophagy is known to cause neurodegneration in mature neurons indicating that autophagy may regulate neuronal homeostasis [16, 17]. In this review, I will briefly summarize the molecular mechanism of autophagy in mammals and focus on autophagy pathway in neuronal physiology and pathology. I will also discuss the implication of autophagy in neurodegenerative diseases.
THE CORE MACHINERY OF AUTOPHAGY
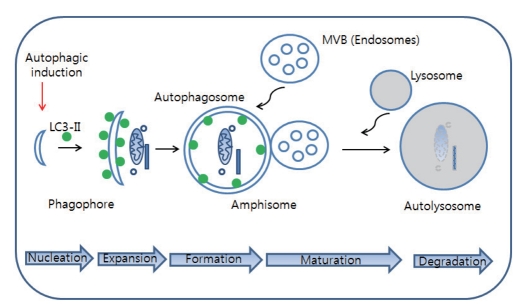
The detailed autophagic machinery has been already extensively reviewed [1, 5]. Thus, only major autophagic components for understanding of basic concept of autophagy will be described in this review. Autophagy has several key steps for final degradation of cytosolic component in lysosomes: initiation and nucleation of phagophore (isolation membrane), expansion of autophagosomes, maturation of autopahgosomes into amphisome/autolysosomes, and execution of autophagy (degradation) (Fig. 1). Several autophagy-related "atg" proteins and non atg proteins are known to regulate these processes [5].
Fig. 1.
The cellular processes during autophagy. Autophagic process follows distinct stages: vesicle nucleation (formation of phagophore), vesicle expansion (autophagosome formation), maturation (fusion of autophagosome with MVB (multivesicular body)/lysosome, degradation (acidification). Once autophagy is induced by autophagic stimuli such as inhibition of mTOR, phagophore (isolation membrane) begin to be formed and then cytosolic components are sequestered by autophagosomes characterized by LC3-II-positive double membrane structure. Endosome such as MVB or lysosome can be fused with autophagosome to form amphisome or autolysosome, respectively. In final step, cytosolic components are degraded in autolysosome.
Initiation and nucleation of phagophore
Autophagy can be constitutively activated at a basal level to maintain cellular homeostasis. Autophagy can be also induced and activated by specialized stimuli for the cell to be adapted in cellular environment.
Once autophagy is activated, it begins with formation of phagophore (a precursor of autophagosomes) whose source was still under a considerable debate whether it comes from endoplasmic reticulum (ER), Golgi complex, mitochondria, or plasma membrane via clathrin mediated endocytosis [18-21]. This process depends on the stepwise recruitment of specific proteins including atg proteins into newly forming autophagosomal membranes. There are two major essential components to regulate this process [22, 23]. The first one is the class III phosphatidylinositol 3-kinase (PI3K) Vps 34 which can generate phosphatidylinositol-3-phosphate (PI-3-P) necessary for recruitment of autophagy specific proteins (ATG17, ATG13) in the region of phagophore formation [24]. Vps 34 could associate with other molecules involved in autophagosome formation such as Beclin1 (the mammalian orthologue of yeast Atg6), p150, UVRAG, Atg14, or Ambra1 [25-27]. The second component involved in the biogenesis of atuophagosomes is ULK1 which associates with FIP200 and ATG13. Their association is known to lead to proper localization of ULK1 and stimulate its kinase activity [28].
The expansion of autophagosomal membranes
The elongation of phagophores requires two ubiquitin-like conjugating systems. 1) ATG12-ATG5-ATG16L system: ATG12 is conjugated into ATG5 via ATG7 (E1-like enzyme) and ATG10 (E2-like enzyme) and then the conjugated ATG12-ATG5 complex associates with ATG16L [29-32]. The ATG12-ATG5-ATG16L complex is localized to outer membrane of elongating autophagosomes but it dissociates before the completion of autophagosome formation. However, recently it has been reported that there is ATG5/ATG7-independent alternative macroautophagy under a certain stress condition suggesting that there is an alternative pathway to form autophagosomes [33]. 2) Phosphatidylethanolamine (PE)-LC3 system: As the other system, microtubule-associated protein 1 light chain 3 (MAP1-LC3, simply LC3), mammalian orthologue of ATG8, is conjugated to PE. Cytosolic form of LC3, LC3-I is generated by cleavage of pro-LC by ATG4B and further processed by ATG7 and ATG3 to be conjugated to PE (LC3-II) [34]. LC3-II specifically associates with autophagosome membranes. Therefore, the number of autophagosomes correlates with the level of LC3-II. LC3 has been known to promote membrane tethering [35, 36].
The maturation of autophagosomes
After completion of autophagosome formation, autophagosomes can be fused with endosomes or lysosomes resulting in the formation of amphisome or autolysosome, respectively [37, 38]. Fusion firstly requires movement of autophagosome into endosomes or lysosomes along microtubules using dynein-dynactin complex [39, 40]. Therefore, cytoskeletal complexes are important facilitators of autophagic process. In addition, for fusion of autophagosomes with endosome/lysosomes, non atg components such as endosomal sorting complex required for transport (ESCRT), soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs), Rab proteins, ATPase, are required [41-44]. However, how exactly each component can contribute to each step of maturation of autophagosome remains elusive.
The execution of autophagy: degradation
In final step, cytoplasmic components are actually degraded in autolysosomes. Therefore, the activity of lysosomes is necessary for degradation. Acidification by proton pump or V-ATPase is essential for fusion with lysosomes and degradation in autolysosomes [45]. The defects of lysosomal enzymes such as cathepsin induce blockage of degradation in autophagy pathway [46]. Intriguingly, recent study showed that post-translational modification of substrates such as acetylation or phosphorylation could regulate the efficiency of autophagic degradation [47, 48].
In addition, specific defects in selective autophagy or in cargo selection process could induce neurodegeneration. Indeed, this hypothesis was supported by the studies using p62, NBR1, or ALFY involved in cargo recognition and degradation [49, 50]. The defects of any step during autophagy process can lead to abnormal accumulation of cytosolic components leading to disease states. Therefore, each step between autophagic processes should be tightly regulated for efficient autophagic degradation.
AUTOPHAGY AS A HOUSEKEEPER IN NEURONAL HOMEOSTASIS
Previous studies in vivo and in vitro using GFP-LC3, an autophagosome marker, showed that autophagosmes are hardly detectable in healthy neurons under nutrient rich condition [51, 52]. There are two possibilities to explain the scarcity the autophagosomes in healthy neurons. One possibility is that autophagic activity is maintained at a low level in normal brain. The other possibility is that autophagic degradation is so efficient that autophagosomes could not be accumulated in healthy neurons at a detectable level. This interesting idea was supported by recent study which showed that inhibition of lysosomal degradation caused rapid accumulation of autophagosomes in primary cortical neurons suggesting the active possible role of constitutive autophagy even under nutrient rich condition [53, 54]. Therefore, basal autophagy in healthy neurons seems to be relatively active. High efficiency of autophagic degradation was also supported by the observation that the intermediate forms of autophagosomes (immature autophagosomes) are relatively low in healthy brain [41, 55].
If autophagic activity is highly maintained in normal healthy neurons, what is the primary role of basal autophagy in neurons? The study using neuron specific atg5 or atg7 deficient mice showed the abnormal protein accumulation and eventual neurodegeneration in central nervous system indicating that autophagy is constitutively active and essential for neuronal cell survival [16, 17]. Basically, the dysfunction of any molecule involved in autophagic process may cause neurodegeneration. Interestingly, increasing evidence suggests that neuronal constitutive autophagy may be an important regulatory pathway for axonal homeostasis. The loss of basal autophagy by either deletion of atg gene or inhibition of autophagic clearance in neurons caused disruption of axonal transport of vesicles containing substrates degraded in lysosomes and axonal swelling, leading to axonal dystrophy [16, 17, 54, 56]. Another evidence as an implication of axonal autophagy comes from the study of ATG1/Unc-51 (C. elegans uncoordinated-51). Unc-51 mutants in C. elegans showed disruptions in axonal membrane structures [57]. Furthermore, Unc51.1, the murine homologue, is required for neurite extension during axonal growth indicating its possible role in homeostasis of axonal membrane network [58-60]. In addition, neural specific deletion of FIP200 involved in autophagosome biogenesis caused neuronal cell death and axon degeneration leading to cerebellar degeneration [61]. Thus, defects of basal autophagy seem to be vulnerable to affect on axonal structure and function through retrograde axonal transport. However, autophagy pathway may also have important roles in dendrites in which active degradation and synthesis of various molecules occur. Indeed, autophagosomes are found in proximal and distal region of dendrites in neurons indicating their regulatory roles in dendritic regions under physiological or pathological condition [42, our unpublished data]. Notably, mTOR has been known to regulate post-synaptic long-term potentiation (LTP) or long-term depression (LTD) suggesting that autophagic regulation may be essential for synaptic plasticity. Therefore, to dissect out the specific roles in axon or dendrites will give us a better understanding of autophagy pathway in various contexts of neuronal cell survival or neuronal signaling.
AUTOPHAGY IN NEURODEGENERATIVE DISEASES
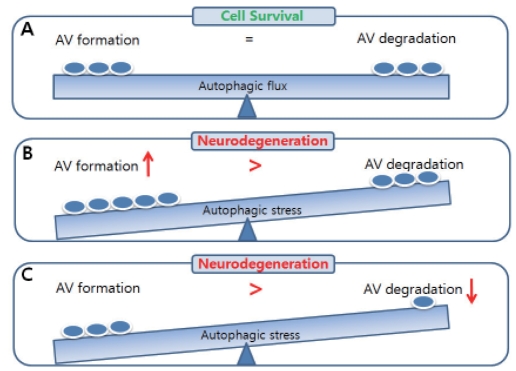
Abnormal accumulation of autophagic vacuoles including autophagosomes or autolysosmes has been observed in affected neurons of brain in several neurodegenerative diseases. It is unclear, however, whether accumulated autophagic vacuoles in degenerating neurons reflect increase in autophagic flux (as measured by actual degradation of cytosolic contents). Autophagic contents could be increased by either increased autophagic flux or impaired flux. The sustained impairment of balance between autophagosome formation and degradation is known to cause "autophagic stress" [62]. Either excessive autophagic demand cannot be supported by cellular reserves or defects of fusion or lysosomal degradation of autophagic vacuoles could cause autophagic stress which is associated with neurodegeneration (Fig. 2). To find out how autophagic stress occurs in each neurodegenerative diseases would be a first step to understand the molecular pathogenesis of autophagy-associated neurodegenerative diseases. I will briefly summarize the pathogenic mechanism associated with autophagy pathway.
Fig. 2.
The balance of autophagic flux in neuronal cell survival. Autophagic flux indicates the balance between autophagosome formation and autophagic degradation. (A) If autophagosomes can be efficiently degraded in autolysosomes either at a basal level or at an activated level, neuronal cells maintain their homeostasis leading to neuronal cell survival. (B) When the rate of autophagosome formation highly exceeds the rate of autophagic degradation or (C) when late stage of autophagic process (maturation or degradation) has defects, autophagic degradation is impaired and causes accumulation of autophagic vacuoles leading to neurodegeneration [62].
AD
AD is the most common dementia and symptoms include confusion, irritability, trouble with language, and loss of long-term memory. Autophagic vacuoles have been found to accumulate in dystrophic neuritis and in cell body of brain of AD [55]. The accumulation of extracellular plaques including aggregated amyloid-β (Aβ) peptide and intracellular tangles is associated with the pathogenesis of AD. Since Aβ is generated in endo-lysosomal pathway, the produced Aβ is normally found in autophagosomes and in lysosomes. However, in disease state, impeded turnover of increased autophagic vacuoles due to reduced fusion or altered endocytic pathway could cause autophagic stress as shown in accumulation of autophagic components [63]. Although alteration mechanism in autophagy is complex in AD, induction and activation of autophagy using autophagy-related protein such as beclin could promote degradation of Aβ and reduce AD pathology raising the possibility of inducers of autophagy as a therapeutic target of AD [64]. However, the other therapeutic modulation targeting late step in the autophagy pathway should be also considered since the impaired clearance of autophagic vacuoles has been observed in AD animal model and AD patients [53, 54].
HD
HD is one of the most common polyglutamine diseases which are a group of inherited neurodegenerative diseases caused by CAG trinucleotide expansion mutation. This disorder shows neuronal loss in striatum and cortex leading to progressive impairment of voluntary movement coordination. Protein aggregates and inclusion caused by mutant huntingtin protein have been shown to induce autophagy. Their inclusions can be degraded by autophagy pathway regardless of a debate about their role in cellular toxicity [65-67]. A growing body of evidence suggests that further stimulation and activation of autophagy are indeed beneficial for HD. Several studies suggest that pharmacological modifier of mTOR dependent or independent pathway could lead to a neuroprotection in cellular and animal HD model by reducing polyglutamine aggregation [23, 66]. Interestingly, CMA enhancement by the selective targeting of mutant huntingtin using fusion molecule consisting of polyglutamine binding peptide 1 (QBP1) and HSC70-binding motifs in vitro and in vivo facilitated degradation of mutant protein and ameliorated disease phenotype in HD animal model [68]. Therefore, induction of macroautophagy or CMA could be a new strategy for HD therapy.
PD
PD is characterized by progressive degeneration of dopaminergic neurons of substantia nigra and the symptoms are tremor, rigidity and impaired balance and coordination [69]. α-synuclein whose mutations cause familiar PD is major protein component of Lewy bodies found in PD. Interestingly, recent studies showed that mutant α-synuclein protein was degraded via macroautophagy instead of CMA in which wild type protein was degraded. It has been demonstrated that inhibition of CMA by mutant protein was accompanied by compensatory activation of macroautophagy [70]. Therefore, autophagic modulation for the restoration of CMA function will be an effective therapeutic strategy in PD.
The enhancement of autophagy pathway for the efficient turnover of dysfunctional mitochondria could be also considered as a therapeutic target in PD. Recent studies implicate that the defects in mitophagy may be an important pathogenic mechanism of PD. Parkin and PINK1 whose mutations cause autosomal recessive form of PD play important roles in mitophagy. The selective targeting of parkin into damaged mitochondria during mitophagy is dependent on wild type PINK1 but not mutant PINK1 suggesting the regulatory role of parkin and PINK1 in mitophagy [71, 72].
FTD
FTD is the second most common dementia under 65 and it is characterized by progressive degeneration of frontal and temporal lobe. The symptoms include the defects in personality, alteration of social behavior, aggressiveness. The rare mutation in the ESCRT subunit CHMP2B caused accumulation of ubiquitinated protein aggregates and autophagosomes by impairment of fusion of autophagosomes with lysosomes, eventually leading to neurodegeneration [42, 73]. The reduction of autohagic stress by inhibiting formation of autophagosome delayed neuronal cell loss indicating that excessive autophagosomes can contribute to disease pathogenesis [74]. Although reduction of autophagic stress by inhibiting autophagy temporally delay the neurodegeneration, the selective way to facilitate the delivery of autophagic components into lysosomes under pathological condition should be further investigated.
CONCLUSION AND PERSPECTIVES
Neuronal autophagy functions as a housekeeper to maintain their cellular homeostasis such as protein or organelle quality control. In contrast, neuronal autophagy could acts as a fighter under disease state such as accumulation of protein aggregates or damaged organelles to protect the cell. Either defect of autophagic flux or excessive activity of autophagy can contribute to neurodegeneration and neuronal cell death. Therefore, therapeutic consideration for each autophagic failure type in each neurodegenerative disease would be essential for the development of therapeutic intervention through autophagic modulation. Thus, further studies to explore alternative approach to modulate autophagy are necessary. The target point to identify drug candidates could be the selective targeting of substrates into lysosomes, enhancement of autophagosome-lysosome fusion or facilitation of delivery of autophagic vacuoles to lysosomes. This will provide a broad spectrum of potential drug targets in various neurodegenerative diseases as well as a better understanding of signaling pathway in autophagic processes.
ACKNOWLEDGEMENTS
I apologize to colleagues whose work could not be cited due to space limitation and thank lab members for careful reading for an earlier draft. This work was supported by the Hannam Research Program (2011).
References
- 1.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 2.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Majeski AE, Dice JF. Mechanisms of chaperone-mediated autophagy. Int J Biochem Cell Biol. 2004;36:2435–2444. doi: 10.1016/j.biocel.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 8.Uttenweiler A, Mayer A. Microautophagy in the yeast Saccharomyces cerevisiae. Methods Mol Biol. 2008;445:245–259. doi: 10.1007/978-1-59745-157-4_16. [DOI] [PubMed] [Google Scholar]
- 9.Mijaljica D, Prescott M, Devenish RJ. Microautophagy in mammalian cells: revisiting a 40-year-old conundrum. Autophagy. 2011;7:673–682. doi: 10.4161/auto.7.7.14733. [DOI] [PubMed] [Google Scholar]
- 10.Nixon RA. Autophagy in neurodegenerative disease: friend, foe or turncoat? Trends Neurosci. 2006;29:528–535. doi: 10.1016/j.tins.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 11.Chen Y, Klionsky DJ. The regulation of autophagy - unanswered questions. J Cell Sci. 2011;124:161–170. doi: 10.1242/jcs.064576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741–752. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 13.Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12:823–830. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Todde V, Veenhuis M, van der Klei IJ. Autophagy: principles and significance in health and disease. Biochim Biophys Acta. 2009;1792:3–13. doi: 10.1016/j.bbadis.2008.10.016. [DOI] [PubMed] [Google Scholar]
- 15.Boland B, Nixon RA. Neuronal macroautophagy: from development to degeneration. Mol Aspects Med. 2006;27:503–519. doi: 10.1016/j.mam.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 16.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 17.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 18.Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geng J, Nair U, Yasumura-Yorimitsu K, Klionsky DJ. Post-Golgi Sec proteins are required for autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2010;21:2257–2269. doi: 10.1091/mbc.E09-11-0969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, Lippincott-Schwartz J. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol. 2010;12:747–757. doi: 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kundu M, Thompson CB. Autophagy: basic principles and relevance to disease. Annu Rev Pathol. 2008;3:427–455. doi: 10.1146/annurev.pathmechdis.2.010506.091842. [DOI] [PubMed] [Google Scholar]
- 23.Jimenez-Sanchez M, Thompson F, Zavodsky E, Rubinsztein DC. Autophagy and polyglutamine diseases. Prog Neurobiol. 2011 doi: 10.1016/j.pneurobio.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blommaart EF, Krause U, Schellens JP, Vreeling-Sindelárová H, Meijer AJ. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur J Biochem. 1997;243:240–246. doi: 10.1111/j.1432-1033.1997.0240a.x. [DOI] [PubMed] [Google Scholar]
- 25.Kihara A, Kabeya Y, Ohsumi Y, Yoshimori T. Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep. 2001;2:330–335. doi: 10.1093/embo-reports/kve061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang C, Sir D, Lee S, Ou JH, Jung JU. Beyond autophagy: the role of UVRAG in membrane trafficking. Autophagy. 2008;4:817–820. doi: 10.4161/auto.6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, Corazzari M, Fuoco C, Ucar A, Schwartz P, Gruss P, Piacentini M, Chowdhury K, Cecconi F. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–1125. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- 28.Ganley IG, Lam du H, Wang J, Ding X, Chen S, Jiang X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284:12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y. A protein conjugation system essential for autophagy. Nature. 1998;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- 30.Mizushima N, Noda T, Ohsumi Y. Apg16p is required for the function of the Apg12p-Apg5p conjugate in the yeast autophagy pathway. EMBO J. 1999;18:3888–3896. doi: 10.1093/emboj/18.14.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mizushima N, Kuma A, Kobayashi Y, Yamamoto A, Matsubae M, Takao T, Natsume T, Ohsumi Y, Yoshimori T. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J Cell Sci. 2003;116:1679–1688. doi: 10.1242/jcs.00381. [DOI] [PubMed] [Google Scholar]
- 32.Tanida I, Tanida-Miyake E, Ueno T, Kominami E. The human homolog of Saccharomyces cerevisiae Apg7p is a Protein-activating enzyme for multiple substrates including human Apg12p, GATE-16, GABARAP, and MAP-LC3. J Biol Chem. 2001;276:1701–1706. doi: 10.1074/jbc.C000752200. [DOI] [PubMed] [Google Scholar]
- 33.Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T, Kanaseki T, Komatsu M, Otsu K, Tsujimoto Y, Shimizu S. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461:654–658. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- 34.Tanida I, Nishitani T, Nemoto T, Ueno T, Kominami E. Mammalian Apg12p, but not the Apg12p.Apg5p conjugate, facilitates LC3 processing. Biochem Biophys Res Commun. 2002;296:1164–1170. doi: 10.1016/s0006-291x(02)02057-0. [DOI] [PubMed] [Google Scholar]
- 35.Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165–178. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 36.Weidberg H, Shpilka T, Shvets E, Abada A, Shimron F, Elazar Z. LC3 and GATE-16 N termini mediate membrane fusion processes required for autophagosome biogenesis. Dev Cell. 2011;20:444–454. doi: 10.1016/j.devcel.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 37.Gordon PB, Seglen PO. Prelysosomal convergence of autophagic and endocytic pathways. Biochem Biophys Res Commun. 1988;151:40–47. doi: 10.1016/0006-291x(88)90556-6. [DOI] [PubMed] [Google Scholar]
- 38.Strømhaug PE, Berg TO, Fengsrud M, Seglen PO. Purification and characterization of autophagosomes from rat hepatocytes. Biochem J. 1998;335:217–224. doi: 10.1042/bj3350217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ravikumar B, Acevedo-Arozena A, Imarisio S, Berger Z, Vacher C, O'Kane CJ, Brown SD, Rubinsztein DC. Dynein mutations impair autophagic clearance of aggregateprone proteins. Nat Genet. 2005;37:771–776. doi: 10.1038/ng1591. [DOI] [PubMed] [Google Scholar]
- 40.Jahreiss L, Menzies FM, Rubinsztein DC. The itinerary of autophagosomes: from peripheral formation to kiss-and-run fusion with lysosomes. Traffic. 2008;9:574–587. doi: 10.1111/j.1600-0854.2008.00701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Furuta N, Fujita N, Noda T, Yoshimori T, Amano A. Combinational soluble N-ethylmaleimide-sensitive factor attachment protein receptor proteins VAMP8 and Vti1b mediate fusion of antimicrobial and canonical autophagosomes with lysosomes. Mol Biol Cell. 2010;21:1001–1010. doi: 10.1091/mbc.E09-08-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee JA, Beigneux A, Ahmad ST, Young SG, Gao FB. ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Curr Biol. 2007;17:1561–1567. doi: 10.1016/j.cub.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 43.Lee JA, Gao FB. Roles of ESCRT in autophagy-associated neurodegeneration. Autophagy. 2008;4:230–232. doi: 10.4161/auto.5384. [DOI] [PubMed] [Google Scholar]
- 44.Eskelinen EL. Maturation of autophagic vacuoles in Mammalian cells. Autophagy. 2005;1:1–10. doi: 10.4161/auto.1.1.1270. [DOI] [PubMed] [Google Scholar]
- 45.Mousavi SA, Kjeken R, Berg TO, Seglen PO, Berg T, Brech A. Effects of inhibitors of the vacuolar proton pump on hepatic heterophagy and autophagy. Biochim Biophys Acta. 2001;1510:243–257. doi: 10.1016/s0005-2736(00)00354-0. [DOI] [PubMed] [Google Scholar]
- 46.Renna M, Schaffner C, Winslow AR, Menzies FM, Peden AA, Floto RA, Rubinsztein DC. Autophagic substrate clearance requires activity of the syntaxin-5 SNARE complex. J Cell Sci. 2011;124:469–482. doi: 10.1242/jcs.076489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jeong H, Then F, Melia TJ, Jr, Mazzulli JR, Cui L, Savas JN, Voisine C, Paganetti P, Tanese N, Hart AC, Yamamoto A, Krainc D. Acetylation targets mutant huntingtin to autophagosomes for degradation. Cell. 2009;137:60–72. doi: 10.1016/j.cell.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alers S, Löffler AS, Wesselborg S, Stork B. Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: cross talk, shortcuts, and feedbacks. Mol Cell Biol. 2012;32:2–11. doi: 10.1128/MCB.06159-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clausen TH, Lamark T, Isakson P, Finley K, Larsen KB, Brech A, Øvervatn A, Stenmark H, Bjørkøy G, Simonsen A, Johansen T. p62/SQSTM1 and ALFY interact to facilitate the formation of p62 bodies/ALIS and their degradation by autophagy. Autophagy. 2010;6:330–344. doi: 10.4161/auto.6.3.11226. [DOI] [PubMed] [Google Scholar]
- 50.Knaevelsrud H, Simonsen A. Fighting disease by selective autophagy of aggregate-prone proteins. FEBS Lett. 2010;584:2635–2645. doi: 10.1016/j.febslet.2010.04.041. [DOI] [PubMed] [Google Scholar]
- 51.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nixon RA, Cataldo AM, Mathews PM. The endosomal-lysosomal system of neurons in Alzheimer's disease pathogenesis: a review. Neurochem Res. 2000;25:1161–1172. doi: 10.1023/a:1007675508413. [DOI] [PubMed] [Google Scholar]
- 53.Boland B, Kumar A, Lee S, Platt FM, Wegiel J, Yu WH, Nixon RA. Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer's disease. J Neurosci. 2008;28:6926–6937. doi: 10.1523/JNEUROSCI.0800-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee S, Sato Y, Nixon RA. Primary lysosomal dysfunction causes cargo-specific deficits of axonal transport leading to Alzheimer-like neuritic dystrophy. Autophagy. 7 doi: 10.4161/auto.7.12.17956. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nixon RA. Endosome function and dysfunction in Alzheimer's disease and other neurodegenerative diseases. Neurobiol Aging. 2005;26:373–382. doi: 10.1016/j.neurobiolaging.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 56.Yue Z. Regulation of neuronal autophagy in axon: implication of autophagy in axonal function and dysfunction/degeneration. Autophagy. 2007;3:139–141. doi: 10.4161/auto.3602. [DOI] [PubMed] [Google Scholar]
- 57.Sigmond T, Fehér J, Baksa A, Pásti G, Pálfia Z, Takács-Vellai K, Kovács J, Vellai T, Kovács AL. Qualitative and quantitative characterization of autophagy in Caenorhabditis elegans by electron microscopy. Methods Enzymol. 2008;451:467–491. doi: 10.1016/S0076-6879(08)03228-X. [DOI] [PubMed] [Google Scholar]
- 58.Okazaki N, Yan J, Yuasa S, Ueno T, Kominami E, Masuho Y, Koga H, Muramatsu M. Interaction of the Unc-51-like kinase and microtubule-associated protein light chain 3 related proteins in the brain: possible role of vesicular transport in axonal elongation. Brain Res Mol Brain Res. 2000;85:1–12. doi: 10.1016/s0169-328x(00)00218-7. [DOI] [PubMed] [Google Scholar]
- 59.Komatsu M, Wang QJ, Holstein GR, Friedrich VL, Jr, Iwata J, Kominami E, Chait BT, Tanaka K, Yue Z. Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc Natl Acad Sci U S A. 2007;104:14489–14494. doi: 10.1073/pnas.0701311104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yue Z, Friedman L, Komatsu M, Tanaka K. The cellular pathways of neuronal autophagy and their implication in neurodegenerative diseases. Biochim Biophys Acta. 2009;1793:1496–1507. doi: 10.1016/j.bbamcr.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liang CC, Wang C, Peng X, Gan B, Guan JL. Neural-specific deletion of FIP200 leads to cerebellar degeneration caused by increased neuronal death and axon degeneration. J Biol Chem. 2010;285:3499–3509. doi: 10.1074/jbc.M109.072389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chu CT. Autophagic stress in neuronal injury and disease. J Neuropathol Exp Neurol. 2006;65:423–432. doi: 10.1097/01.jnen.0000229233.75253.be. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nixon RA. Autophagy, amyloidogenesis and Alzheimer disease. J Cell Sci. 2007;120:4081–4091. doi: 10.1242/jcs.019265. [DOI] [PubMed] [Google Scholar]
- 64.Pickford F, Masliah E, Britschgi M, Lucin K, Narasimhan R, Jaeger PA, Small S, Spencer B, Rockenstein E, Levine B, Wyss-Coray T. The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. J Clin Invest. 2008;118:2190–2199. doi: 10.1172/JCI33585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ravikumar B, Duden R, Rubinsztein DC. Aggregateprone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet. 2002;11:1107–1117. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- 66.Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, Scaravilli F, Easton DF, Duden R, O'Kane CJ, Rubinsztein DC. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 67.Kegel KB, Kim M, Sapp E, McIntyre C, Castaño JG, Aronin N, DiFiglia M. Huntingtin expression stimulates endosomal-lysosomal activity, endosome tubulation, and autophagy. J Neurosci. 2000;20:7268–7278. doi: 10.1523/JNEUROSCI.20-19-07268.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bauer PO, Goswami A, Wong HK, Okuno M, Kurosawa M, Yamada M, Miyazaki H, Matsumoto G, Kino Y, Nagai Y, Nukina N. Harnessing chaperone-mediated autophagy for the selective degradation of mutant huntingtin protein. Nat Biotechnol. 2010;28:256–263. doi: 10.1038/nbt.1608. [DOI] [PubMed] [Google Scholar]
- 69.Thomas B. Parkinson's disease: from molecular pathways in disease to therapeutic approaches. Antioxid Redox Signal. 2009;11:2077–2082. doi: 10.1089/ars.2009.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science. 2004;305:1292–1295. doi: 10.1126/science.1101738. [DOI] [PubMed] [Google Scholar]
- 71.Dagda RK, Cherra SJ, 3rd, Kulich SM, Tandon A, Park D, Chu CT. Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J Biol Chem. 2009;284:13843–13855. doi: 10.1074/jbc.M808515200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vives-Bauza C, Zhou C, Huang Y, Cui M, de Vries RL, Kim J, May J, Tocilescu MA, Liu W, Ko HS, Magrané J, Moore DJ, Dawson VL, Grailhe R, Dawson TM, Li C, Tieu K, Przedborski S. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci U S A. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rusten TE, Vaccari T, Lindmo K, Rodahl LM, Nezis IP, Sem-Jacobsen C, Wendler F, Vincent JP, Brech A, Bilder D, Stenmark H. ESCRTs and Fab1 regulate distinct steps of autophagy. Curr Biol. 2007;17:1817–1825. doi: 10.1016/j.cub.2007.09.032. [DOI] [PubMed] [Google Scholar]
- 74.Lee JA, Gao FB. Inhibition of autophagy induction delays neuronal cell loss caused by dysfunctional ESCRT-III in frontotemporal dementia. J Neurosci. 2009;29:8506–8511. doi: 10.1523/JNEUROSCI.0924-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]