Abstract
The most profound primary immunodeficiency disease, severe combined immunodeficiency (SCID), is fatal in infancy unless affected infants are provided with an adaptive immune system through allogeneic hematopoietic cell transplantation, enzyme replacement or gene therapy. However, most infants with SCID lack a family history or any clinical clues prior to the onset of infections, making this serious, but treatable disease a candidate for population based newborn screening. Of several approaches considered for SCID screening, testing for the DNA biomarker of normal T cell development, T cell receptor excision circles (TRECs), has proven successful. TREC numbers can be measured in DNA isolated from the dried blood spots already routinely collected for newborn screening. Infants with low or absent TRECs can thus be identified and referred for confirmatory testing and prompt intervention. TREC testing of newborns is now being performed in several states, indicating that this addition to the newborn screening panel can be successfully integrated into state public health programs.
Keywords: primary immunodeficiency, T cell receptor excision circle (TREC), severe combined immunodeficiency (SCID), lymphocytopenia, dried blood spot, newborn screening
Introduction: Why screen for SCID
Every medical textbook and every publication from the 1950’s to the last 3 years discussed severe combined immunodeficiency (SCID) in terms of how the absence of cellular and humoral immunity in this rare disorder causes affected infants to come to medical attention with infections—thrush, pneumonia, diarrhea—and failure to thrive.1 The diagnosis of SCID, and its very definition, have revolved around the devastating bacterial, viral and fungal infections of affected infants, infections often caused by weakly pathogenic, opportunistic organisms. Prior to 1968, when the first successful bone marrow transplant was performed,2 SCID was always fatal, but now it can be treated by allogeneic hematopoietic cell transplantation (HCT),2–11 enzyme replacement12 or even gene therapy,13–14 provided infections can be controlled.
Recognition of how SCID is inherited15–33 has permitted some families, often following tragic loss of an affected infant due to infection, to have the diagnosis made in subsequent affected children at birth, or even before birth.34–35 In these fortunate circumstances, early treatment of healthy infants with SCID who have avoided infections has afforded a very high likelihood of survival free of complications.35–37 Development of population based newborn screening for SCID has been motivated by the recognition that pre-symptomatic identification and treatment greatly improves survival and by the desire to give all infants born with SCID the chance for an optimal outcome. This review will describe the methodology now available for early SCID screening and diagnosis.
Differences between screening tests and diagnostic tests for individual cases
Population based newborn screening, which is mandated and performed by the public health departments of individual states, differs from testing undertaken by immunologists confronted with a known or suspected case of immunodeficiency in their practice. For an individual patient who might have SCID or a related disorder, a complete blood count, differential white blood count and lymphocyte subset determination are obtained immediately, with any T cell abnormalities followed up by further testing, such as serum antibody concentrations, in vitro lymphocyte proliferation, and molecular diagnosis.38 This approach requires liquid blood samples that are perishable and cannot be stored or re-run if there are problems or as a quality control at a later time. Furthermore, these diagnostic tests must be performed soon after the blood is obtained, they are not always consistently performed or interpreted from one laboratory to another, and they are prohibitively expensive to be used in every infant.
In contrast, screening whole populations is not targeted to a few individuals recognized to be at potentially increased risk, either because of family history or clinical considerations such as infections.39 Screening tests must be amenable to performance on a large scale in centralized state public health laboratories. These laboratories already perform multiple tests using blood, generally from a heelstick, that is spotted onto filter paper and air dried, as first developed by Robert Guthrie for population based testing of newborns for phenylketonuria.40,41 Dried blood spots (DBS) have been thoroughly standardized. They can be stored (for many years if desiccated and at −20°C), sent through the mail, and tracked by automated bar coding for generation of reports. The typical newborn screening test is done on a 3.2 mm disc, corresponding to about 3 uL of liquid blood, that is punched out of the DBS, which is roughly a 1 cm diameter circle. If at all possible, newly developed screening tests should take advantage of the DBS sample to avoid the costs of getting a separate sample and to facilitate integration into existing screening programs.39,42
Unlike an individual clinical test, a screening test represents a single, isolated opportunity to pick up a rare, but serious condition. Therefore false negative results, or the failure to identify true cases, must be kept to an absolute minimum. On the other hand, having a limited number of false positive results is viewed as a necessary price to pay to avoid missing affected infants. Thus the biomarkers that are successful in newborn screening need to be very sensitive, but not completely specific. Public health laboratories have excellent outreach systems to contact infants for additional testing when screening results are not normal, and the follow-up individual testing is used to sort out false positives so that families can be reassured if their infant is unaffected. But parental anxiety is an important factor, as is provider frustration when each test added to the screening panel brings more chances for infants to have to undergo more tests.43
Analytes and platforms considered for SCID newborn screening other than T cell receptor excision circles (TRECs)
The first suggestion that all newborns be screened for SCID was by the teams of Buckley and Puck in 1997, who made the suggestion that the great majority of cases could be identified by a complete blood count and differential to determine the absolute lymphocyte count (ALC) per mL of blood.4 Lymphocytes and several other analytes, defined as substances that are measured, have been considered, to be used either alone or in combination (Table 2). As alluded to above, the ALC is an informative test in the clinical realm, since T cells make up approximately 70% of lymphocytes in healthy infants.7,39 However, because the ALC includes all lymphocyte subsets, patients with T-B+ SCID who have high numbers of B and possibly NK cells could have normal, or false negative, results on this test. In addition, maternal cells may be found in substantial numbers in the circulation of infants with SCID, who are unable to recognize and eliminate allogeneic cells; these maternal cells also increase the ALC value and could potentially lead to a false negative result. Rare infants with Omenn syndrome, characterized by impaired T cell development with immune dysregulation and expansion of oligoclonal T cells may also have a normal to high ALC, but in addition have clinical features of erythroderma, eosinophilia and organ involvement that bring them to medical attention. Although the mean ALC of newborns is around 5,000/uL, the range is broad, so that setting the cut-off value for normal ALC at a level to capture SCID patients with high numbers of B cells or maternal cells would result in unacceptably frequent false positive tests. Thus ALC, while a valuable and often overlooked clinical tool for individual patients and high-risk settings, exhibits too much overlap between SCID and normal infants to be suitable for population-based SCID screening.
Table 2. Molecular Causes of Severe Combined Immunodeficiency.
| Disease Gene | Defective Protein, Function, Features | % of Cases of SCIDa | Lymphocyte Profile | ||
|---|---|---|---|---|---|
| Tb | B | NK | |||
| IL2RG (X-linked) | Interleukin-2 receptor, γ-chain or common γ-chain (γc), part of the lymphocyte receptors for IL-2, -4, -7, -9, -15, and -21 | 48% (only males) | − | + | − |
| ADA | Adenosine deaminase, purine salvage pathway enzyme | 16% | − | + | − |
| IL7R | α chain of IL-7 receptor | 10% | − | + | + |
| JAK3 | Janus kinase 3, intracellular signaling kinase coupled to γc | 6% | − | + | − |
| RAG1, RAG2 | Recombinase activating genes required for T and B cell antigen receptor gene rearrangement | 6% | − | − | + |
| DCLRE1C (Artemis) | Part of T and B cell antigen receptor gene rearrangement complex, also required for DNA repair; deficiency causes radiation sensitivity | 5% | − | − | + |
| TCRD, TCRE, TCRZ | CD3 δ, ε, and ζ chains of the T cell receptor complex, required for T cell positive selection in the thymus and activation by antigen | rare | −/low | + | + |
| CD45 | Protein tyrosine phosphatase receptor (PTPRC), required for T and B cell activation by antigen | rare | −/low | + | +/low |
| LCK | Lymphocyte tyrosine kinase p56lck, required for T cell development and activation | rare | −/low | + | + |
| PNP | Purine nucleoside phosphorylase enzyme, purine pathway enzyme, deficiency also causes neurological impairment | rare | low | low | +/low |
| LIG4 | DNA ligase IV required for antigen receptor gene rejoining and DNA repair; deficiency causes radiation sensitivity | rare | − | + | + |
| DNAPKCS | DNA protein kinase catalytic subunit, required for T and B cell antigen receptor rearrangement and DNA repair | rare | − | − | + |
| NHEJ1 (Cernunnos) | Nonhomologous end joining of DNA during T cell receptor rearrangement and DNA repair; deficiency causes microcepahaly and radiation sensitivity | rare | − | − | + |
| AK2 | Adenylate kinase 2; deficiency causes reticular dysgenesis with granulocytopenia, lymphocytopenia and deafness | rare | − | − | − |
| FOXN1 | Forkhead box N1, required for thymus and hair follicle development; ortholog of nude mouse, also causes alopecia | rare | −/low | + | + |
| STAT5a | Signal transducer and activator of transcription 5, phosphorylated after cytokine receptor engagement; deficiency also causes growth hormone resistant growth failure | rare | −/low | + | − |
| CORO1A | Coronin-1A, protein mediating lymphocyte migration, including T-cell emigration from the thymus | rare | −/low | + | + |
| Currently unknown | Unknown defects, including SCID and congenital anomalies, SCID with multiple bowel atresias and others | ~10% | −/low | +/− | +/− |
Immunoassays for IL-7 and for T-cell specific proteins have been evaluated by McGhee et al.44 and by Janik et al.45 High serum IL-7 levels are associated with T-lymphocytopenic states including SCID in a limited number of cases that have been tested, but this association has not been developed into a robust newborn screening tool for SCID. Lability of IL-7 and technical problems with its detection in eluates from DBS would need to be overcome even if the biological basis for IL-7 elevation is generalizable to newborns across all SCID genotypes and presentations. Detection of proteins of the T cell receptor CD3 complex in combination with CD45 has been achieved using a high-throughput bead capture technology.45 However, the quantity of protein has been so low as to require polyclonal rabbit antisera, rather than the more standardized monoclonal antibodies, making scale-up of this assay for widespread screening problematic at this time.
Another approach is to detect DNA sequence variants in SCID disease genes, either previously defined or as yet unreported deleterious mutations. Many newborn screening laboratories that include cystic fibrosis in their screening test panel currently use CFTR (cystic fibrosis transmembrane conductance regulator) DNA mutation arrays as a second tier test for known gene mutations.46 Unfortunately, the situation with SCID is much more complicated. There are now 14 known SCID genes (Table 2) as well as patients with SCID whose molecular etiology remains unknown even after extensive gene sequencing.15–33 Moreover, in addition to hundreds of disease-associated mutations for the known genes, new mutations continue to be discovered, so it is not possible to rely on a catalog of mutations on an array when diagnosing a new patient.47 A re-sequencing array capable of detecting known and unknown SCID variants developed by Puck and Warrington was able to identify over 90%, but not 100%, of known mutations as well as previously unknown ones in prospective SCID cases.47 Some of the failures were in DNA samples with single base insertions and deletions. While yielding a specific gene diagnosis if positive, this approach has an unacceptable false negative rate as well as a cost that is too high to use as a primary screen. In the future, however, deep genomic sequencing technologies will improve, and whole genome sequencing may revolutionize newborn screening (see Future Prospects and Challenges, below).
The TREC test
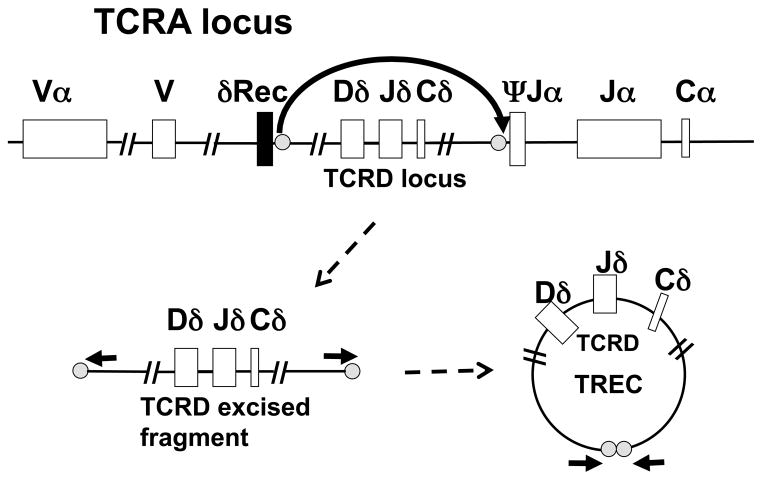
In the germ line and all non-lymphoid cells, the gene loci encoding antibody molecules and T cell antigen receptor chains are composed of large numbers of alternate segments, called variable (V), diversity (D) and joining (J) regions that lie upstream of constant regions. Recombination of the T cell receptor (TCR) genes in the thymus is the process whereby a diverse repertoire of T cells is generated from randomly chosen alternate sections to synthesize a unique rearrangement in each cell. In the TCRα locus a Vα segment becomes linked to a Jα and a Cα segment through cutting and rejoining of the DNA by a series of enzymes that produce double strand DNA breaks at specific sites and then carry out processing and repair. Only T cell progenitors in which re-ligation produces an in-frame, rearranged locus are selected to survive and mature. The excised DNA fragments that are not destined to be incorporated into the mature TCR locus can be joined at their ends to form a great variety of circular DNA byproducts, called T cell receptor excision circles, or TRECs. Late in the maturation program, 70% of the thymocytes that will ultimately express αβ T cell receptors form one specific circular DNA TREC, the δRec-ψJa signal joint TREC, from the excised TCRδ gene (TCRD) that lies within the TCRα gene (TCRA) locus (Figure 1). The DNA circles are stable and are maintained following cell divisions, but because they do not replicate they become diluted as T cells proliferate by mitotic division. A quantitative PCR reaction across the joint of the circular DNA, using primers indicated by the short arrows in Figure 1, provides the TREC copy number, an indicator of new thymic emigrant T cells being produced.
Figure 1.
Generation of the δRec-ΨJα TREC. The germ line configuration of the TCRA locus, with TCRD embedded, is shown at the top of the Figure, which also shows the points (gray dots) at which the DNA is cut to excise the TCRD locus in T lymphocyte progenitors destined to express the α and β TCRs. After excision, lower left, and ligation, lower right, of the δRec-ΨJα fragment to form a T cell receptor excision circle (TREC), PCR primers (horizontal arrows) can amplify a DNA junction fragment containing the joint.
The groups of van Dongen et al.48 and Douek et al.49 found that TRECs are a biomarker for newly produced, naïve T cells that emigrate into peripheral blood from the thymus. Douek noted that infant blood samples have the highest numbers of TRECs, around one TREC per 10 T cells, reflecting the high rate of new T cell generation early in life. Older children and adults have 10-fold and 100-fold lower TRECs, respectively, reflecting increased peripheral T cell expansion as compared to new production of naïve T cells. TREC copy number varies widely between individuals, so a single measurement is not a useful clinical test in most settings, although serial measurements in a patient over time can document T cell reconstitution after HCT or increased TRECs following institution of anti-retroviral therapy in patients with HIV-AIDS.
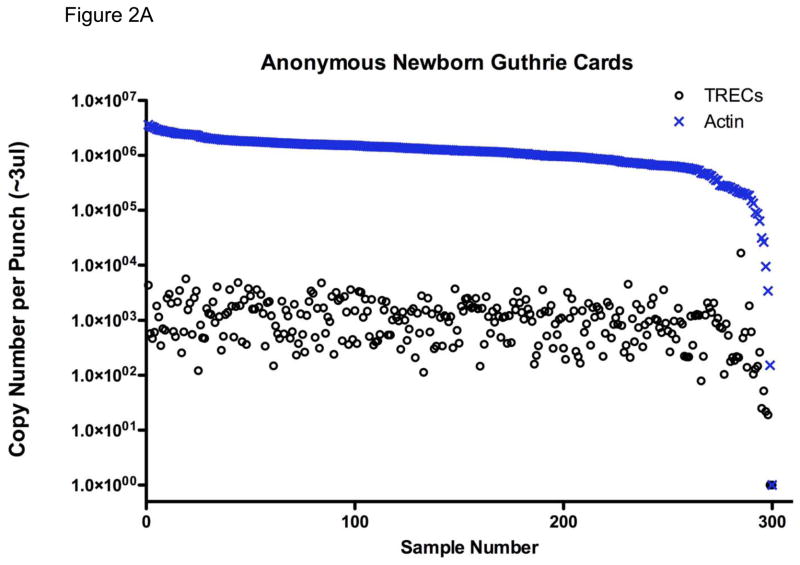
In contrast to its shortcomings as a clinical test, TREC determination, of all the approaches considered to date, is the only one that has proven clinical utility for SCID newborn screening. In 2005, Chan and Puck published the first method for DBS screening for SCID using TRECs as the analyte.50 DNA was extracted from DBS and used for quantitative PCR (qPCR) for TRECs. A β-actin gene segment could be amplified from the same DNA as a control to distinguish samples with low TRECs due to lack of naïve T cells vs. samples with poor DNA yield or quality. As shown in Figure 2A, anonymous DBS samples obtained for routine newborn screening had a mean of 1000 TRECs per punch in the assay as performed in the Puck laboratory. Occasional DBS failed to amplify TREC DNA adequately; such samples would have repeat determinations made and β-actin gene amplification performed, and if both TREC and β-actin PCR failed, as shown in the lower right of the graph in Figure 2A, a new blood spot would be needed from the infant. The initial publication of Chan and Puck had 1.4% of samples with failed PCR, an unacceptably high rate for large scale screening; however, subsequent refinements reduced the false positive rate more than 10-fold (discussed below). Eleven states routinely obtain a second heelstick from all infants at around two weeks of age, making a repeat sample readily available if needed.46
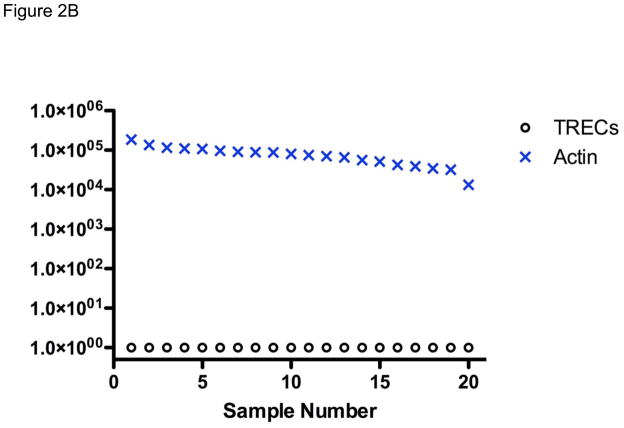
Figure 2.
TREC (○) and actin (×) copy number measured in DNA isolated from (A) 300 anonymous dried blood spots (DBS) obtained in newborn nurseries, showing one sample, far right, with failure of DNA amplification; and (B) 20 newborn nursery DBS recovered from storage from infants later found to have SCID.
From states such as California that archive residual dried blood spots from all infants for appropriate research and quality control purposes, we were able to obtain the original nursery DBS samples from 20 infants who were eventually diagnosed with SCID of diverse genotypes. As shown in Figure 2B, TRECs were uniformly undetectable in these samples even though positive amplification of the genomic β actin DNA was confirmed in each case.50,51 Morinishi et al. obtained similar results from newborn blood spots recovered from 5 SCID infants in Japan.52
Advantages of TRECs as a newborn screening analyte include ability to use dried blood spots, low cost, high throughput, and high sensitivity—that is, avoidance of false negative results from infants with SCID who have high numbers of B lymphocytes, maternal T cell engraftment52,53 or oligoclonally expanded T cells. Furthermore, additional conditions in which T cell numbers are low, such as complete DiGeorge syndrome, would also be predicted to be found. Complete DiGeorge syndrome is potentially treatable by thymus transplantation.54
The TREC method was first adapted to high throughput use in a public health setting by the Wisconsin State Newborn Screening Program, which started a state-wide pilot SCID screening program in 2008.55,56 Massachusetts,57,58 California, New York and other states have followed. In addition to maintaining high sensitivity for detecting SCID cases, TREC test specificity has been excellent with automated methods that have reduced the number of DNA amplification failures to 0.1% or lower. This is as good or better than the false positive rates of many tests currently in use for newborn screening for other conditions.
Performance of the TREC test in state-wide newborn screening
Results of initial state screening programs in Wisconsin and Massachusetts have been published and are summarized by Routes et al. (see also in this issue).55,59 California began TREC screening in August, 2010, within three months of the recommendation by the U. S. Department of Health and Human Services Secretary to add SCID to the nationally reviewed uniform panel of conditions subject to newborn screening. A collaboration was formed between the California State Genetic Disease Laboratory and PerkinElmer, Inc., to adapt and implement the TREC and β-actin detection protocols of Chan and Puck and the Wisconsin pilot program. Table 3 summarizes the results of the first year of screening for TRECs in California, the state with the largest annual number of births in the U. S. and a population that is ethnically very diverse. In a single year over 500,000 infants were screened for TRECs. Test specificity was excellent, with only 8 per 10,000 infants requiring a second heelstick sample; as with all other newborn screening tests, the great majority of the incomplete results from the initial sample were from preterm and ill infants in neonatal intensive care nurseries, who make up about 11% of all infants. Of the initial samples with failure to amplify a normal copy number of both TREC and β-actin DNA, designated DNA amplification failures (DAF), the majority were from infants whose birth weight was under 1500 grams; and nearly half of the failed samples were obtained from indwelling vascular lines rather than the preferred heelsticks. It is hypothesized that heparin anticoagulant may contribute to inhibition of PCR when samples are obtained from indwelling lines.
Table 3. Summary of California newborn screening with TRECs in the first year.
| Over 500,000 births screened. |
| DNA amplification failures (DAF), <0.08%, requiring second heelstick.* |
| 84% from neonatal intensive care units. |
| 56% from infants who were <1500 g at birth. |
| 44% had the failed sample obtained from an intravenous line. |
| 50 infants had two DAF results or a positive result (0.01% of births), requiring CBC and lymphocyte analysis by flow cytometry. |
| 20 of these (40%) had low T cells confirmed. |
| Diagnoses among infants with low T cells: |
| 6 SCID**: |
| 2 IL-7 recptor defects |
| 2 RAG1 defects |
| 2 Common γchain defects |
| 1 Omenn syndrome*** with RAG2 defect |
| 3 SCID variant with no known gene defect |
| 4 Syndromes associated with T lymphocytopenia: |
| 3 DiGeorge syndrome (1 complete) |
| 1 Trisomy 21 |
| 6 Secondary T lymphocytopenia |
| 2 Gastroschesis |
| 1 Gastrointestinal atresia |
| 3 Prematurity |
Since all screening tests other than TRECs are performed in regional laboratories throughout California, with samples then forwarded to a central laboratory for TREC testing, most newborns were 2 weeks old when the TREC result was available. When a SCID-specific second heelstick was needed, older age usually resulted in a normal TREC value.
Only one SCID case had a positive family history leading to testing at birth.
Signs of Omenn syndrome in the first weeks of life had led to the diagnosis just before the TREC test was reported.
Fifty infants (one per 10,000 births) had either 2 DAF samples or an initial positive sample, with low or absent TRECs, but a normal β-actin copy number. These infants were had venous blood obtained for complete blood count, differential white blood count and lymphocyte subset enumeration by flow cytometry. An important feature of screening for SCID and related disorders in California has been inclusion within the newborn screening program of not only TREC testing, but also the follow-up lymphocyte subset enumeration, which is performed in a single reference laboratory (Quest Nichols Institute, San Juan Capistrano, CA). All samples are thus stained for a uniform set of markers—T cells (CD3), helper (CD4) and cytotoxic (CD8) cells, naïve and memory phenotype cells (CD45RA/RO), B cells (CD19) and NK cells (CD16/CD56)—and analyzed with consistent methodology. In addition, all results are interpreted by two designated immunology experts for the screening program, who also oversee referral for treatment of patients with low T cells.
Conditions detected in California by screening with TRECs are also shown in Table 3. Diagnoses have included six cases of typical SCID, two each with mutations in IL2RG, IL7R and RAG1/2. One case of Omenn syndrome had mutation of RAG2. In each of these cases TRECs were undetectable and β-actin normal in the initial DBS sample. The variant SCID cases have been healthy, but are being followed by immunologists for abnormal T cell proliferation in vitro and T lymphocytopenia persisting for at least 6 months. In only one instance, an infant with an X-linked IL2RG mutation, was there a positive family history of an affected older brother that would have prompted newborn detection at birth in the absence of screening. The infant with Omenn syndrome had developed erythroderma and been diagnosed just before the positive TREC test was reported.
A range of T lymphocytopenic conditions other than SCID has been found by TREC screening in California (Table 3), as well as other states. Some syndromes, including DiGeorge syndrome and CHARGE syndrome (ocular coloboma, heart malformation, choanal atresia, retardation of growth and development, genital and ear anomalies) are well recognized to have variable degrees of T cell impairment; in others such as trisomy 21 a spectum of defects in adaptive and innate immunity has been observed, but not well characterized.60 Secondary T lymphocytopenia has also been found through TREC screening, with California cases including gastrointestinal malformations and extreme prematurity. Population based forward screening with TRECs thus identifies known and previously unrecognized associations with low T cells. Regardless of the cause, infants who have significantly low T cells (<1,500/uL) should not be given live vaccines and should be referred to immunology centers for assessment.
Outcome tracking
TREC newborn screening followed by lymphocyte subset enumeration has now been proven to have clinical utility, with several states having implemented their own TREC test protocols that have acceptable false positive rates, no reported false negatives, and successful implementation into each state’s work flow. Many infants with otherwise unsuspected SCID or related T cell disorders have been referred for prompt evaluation and treatment, and anecdotal reports of successful outcomes are already emerging. As more experience accumulates and more states wish to add newborn TREC screening, it will be important to document outcomes of the current programs. Not only the total incidence but also the severity spectrum and relative incidence of these rare conditions in different population subgroups remains to be defined. We have worked with the Newborn Screening Translational Research Network (NBSTRN) to establish categories for tracking and reporting all of the cases of T lymphocytopenia that have been found by prospective screening so that diagnoses and screening test performance can be compared and analyzed.61
While the full range of diagnoses has yet to become established, Table 4 summarizes currently recognized categories that are also consistent with the definitions put forward by the Primary Immune Deficiency Treatment Consortium (PIDTC).62,63 This organization, part of the NIH-funded Rare Diseases Clinical Research Network, was established to conduct multicenter studies of SCID and other diseases treated by cellular therapies. Even though the range for T cells in normal infants born at full term is >2,500/uL,64 an overall definition of clinically significant T lymphocytopenia in newborns for the California screening program (including term and preterm births) is taken to be fewer than 1,500 T cells/uL. The conditions with low or absent TRECs and this degree of T lymphocytopenia fall into five categories.61–63 Typical SCID (Table 4.I) is defined by the PIDTC as fewer than 300 autologous T cells/uL of peripheral blood and less than 10% of normal T cell proliferation to the mitogen PHA. Causes include null mutations of the genes listed in Table 2, and may also include other genes preventing T cell development that have not yet been discovered. Leaky SCID (Table 4.II) has no maternal engraftment and T cell counts ranging from 300 to 1500/uL; infants may have a later age of onset of clinical symptoms (in the absence of screening) or may present in infancy as Omenn syndrome, defined by erythroderma rash, adenopathy and oligoclonal, poorly functioning T cells accompanied by hypomorphic mutations in RAG1, RAG2 or other known SCID genes in Table 2. Variant SCID (Table 4.III) is defined as absence of a known SCID gene defect and 300–1500 autologous T cells/uL with impaired responses to mitogens. Further research is expected to define the molecular defects underlying the T lymphocytopenia in these infants.
Table 4. Conditions with low or absent TRECs and clinically significant T lymphocytopenia (<1,500 T cells/uL).
| I. Typical SCID (see genotypes in Table 2), defined as <300 autologous T cells/uL and <10% of normal proliferation to the mitogen PHA. |
| II. Leaky SCID, due to incomplete (hypomorphic) mutation(s) in a typical SCID gene, with 300–1,500 T cells/uL and impaired, but not absent (10–30% of normal) proliferation to PHA. |
| III. Variant SCID, with no defect in a known SCID gene and 300–1,500 T cells/uL that demonstrate impaired function. |
| IV. Syndromes with variably affected cellular immunity that may be severe: |
| Complete DiGeorge syndrome* |
| Partial DiGeorge syndrome with low T lymphocytes* |
| CHARGE syndrome* |
| Jacobsen syndrome* |
| Trisomy 21* |
| RAC2 dominant interfering mutation* |
| DOCK8 deficient hyper-IgE syndrome** |
| Cartilage hair hypoplasia |
| V. Secondary T lymphocytopenia: |
| Neonatal cardiac surgery with thymectomy* |
| Neonatal leukemia* |
| Gastroschesis* |
| Third spacing* |
| Extreme prematurity (resolves to normal with time)* |
| Possibly severe prenatal HIV disease (hypothesized, but not observed to date) |
, observed to have low or absent TRECs upon newborn screening in one or more cases to date in US pilot programs or published reports.
observed to have low or absent TRECs in one or more cases after diagnosis; newborn samples not available.
An important category of T lymphocytopenia is multi-system syndromes with T cell defects (Table 4.IV). Patients with these syndromes have variable expression of congenital abnormalities and a wide spectrum of T cell dysgenesis, from virtually normal immunity to defects as profound as seen in typical SCID. Infants who have severely affected T cell production are expected to have positive TREC screens, while others with the same syndrome but more normal T cells will not be detected. DiGeorge syndrome, usually associated with chromosome 22.11 deletion; trisomy 21; and CHARGE syndrome (with ocular coloboma, heart defect, atresia of nasal choanae, retardation of growth and development, genitourinary abnormality, and ear abnormality) can all present with life-threatening infections in infancy due to T cell deficiency and have all been identified by neonatal TREC screening.52,55,61 In addition, RAC2 deficiency, previously known only as a granulocyte disorder, has been diagnosed following newborn screening with low TRECs and T lymphocytopenia,55 as has Jacobsen syndrome associated with terminal deletions of chromosome 11q24. Siblings affected with DOCK8 deficient hyper-IgE syndrome with lymphocytopenia had undetectable TRECs during childhood, though newborn samples have not yet been available.65
Finally, secondary T cell defects (Table 4.V) are characterized by acquired conditions with no intrinsic thymic defect, but increased T cell loss. These include congenital heart defects, neonatal leukemia, lymphocyte extravasation or third spacing, lymphangiectasia, and possibly severe congenital HIV infection, though this has not been confirmed to date. Some secondary defects, such as lymphocytopenia associated with extreme low birthweight are not yet fully understood, but may resolve over time. Note that all of these conditions are important to bring to clinical attention to avoid infectious diarrhea following rotavirus vaccine66 or complications following inappropriate transfusions, as well as to monitor for development of infections.
We expect that previously unrecognized conditions with low T cells will come to light as TREC screening continues and expands. Furthermore, many primary immunodeficiency diseases are not detectable by TREC screening. Diseases in which T cells develop in the thymus to the point of production of the δRec-ΨJα TREC (Figure 1), but do not have full functional maturation will be missed. For example, newborns with Zap70 deficiency, MHC Class II deficiency and NF-kappa-b essential modulator (NEMO) deficiency have had normal TRECs. (ref 50 and unpublished observations of J Puck). It is thus important to educate providers to remain alert for signs of cellular immune defects in all infants, including those who have had newborn TREC screening.
Future prospects and challenges
Now that TREC screening has become available and its effectiveness has been shown, spreading its implementation to all states is important. With increasing limitations in health care budgets, states considering adding TREC screening must be provided with data on costs and outcomes of treatments for patients diagnosed with SCID and related disorders without vs. with screening. While improved survival for SCID patients diagnosed early vs. late has been well demonstrated,35–37 rigorous cost effectiveness analysis of SCID newborn screening is not yet available. Ongoing efforts to regionalize testing and treatment centers of excellence for these rare conditions may be more effective than having each state develop its own.
As screening becomes widespread it will be possible to define the true incidence and proportions of each genotype of SCID in unbiased populations, rather than relying on data from HCT centers that may not include all infants without a family history or access to expert medical care.37
Optimal treatments for SCID remain controversial and need to be established by multicenter studies. The PIDTC has taken the important step of prospectively enrolling patients treated for SCID in a large number of participating centers; a consistent protocol of outcome measures collected over time will help to identify best practices for managing these rare diseases.
As extraordinary advances in molecular technology enter the clinical arena, deep sequencing, such as whole exome or whole genome sequencing, may soon become routine. Increasing numbers of medically actionable genotypes, DNA variants that confer risk and are treatable, are driving the trend toward “personalized medicine.” Improvement and public acceptance of this new technology will facilitate identification of rare genotypes with clinical consequences. It is possible that future newborns will have the DNA sequence of their entire genome determinated at birth, from which a blueprint of their risks for a great variety of conditions affecting health can be ascertained, including SCID and other defects of the immune system. Even predisposition to the more common multifactorial immune disorders with later onset may become possible through such sequence analysis of DNA from newborns. However, since genotypes do not fully predict phenotype for these conditions, much more needs to be learned about the true predictive value of deep sequencing.
Table 1. Test Methods Proposed to Screen for Severe Comlbined Immunodeficiency.
| Analyte | Measurement | Platform | Uses dried blood spot | Comment |
|---|---|---|---|---|
| Lymphocytes | CBC and differential white blood count to determine lymphocyes/uL | Hospital clinical lab test on liquid cord blood or postnatal blood (heelstick or venous) | No | Costly, decentralized; false negatives for SCID with oligoclonal T cells, elevated B cells or maternal cells |
| Interleukin-7 | IL-7 immuno-assay | ELISA or other method | Maybe | Stability, purification and assay problems; possible 2nd tier test with TRECs |
| T cell proteins CD3, CD45 | Immuno-assay for low CD3 (multiplex) | Automated fluorescent bead capture | Yes | Limited availability; false negatives for maternal or oligoclonal T cells; possible 2nd tier test with TRECs |
| Mutation in SCID gene | DNA sequence determination | Custom resequencing array | Yes | False negatives; possible 2nd tier test or future whole genome sequencing |
| T cell receptor excision circle | Copy number of circular DNA joints; comparison with control genomic DNA segment | Quantitative PCR on DNA | Yes | Biomarker for newly produced T cells; now used by several states; samples yielding poor DNA are inadequate |
Acknowledgments
Sources of Funding: This work was made possible by funding from NIH (NHGRI Division of Intramural Research, NIAID for RO1 AI078248 and PIDTC U54 AI082973, NICHD for RO3 HD 060311 and NCRR UCSF CTSI UL1 RR024131) and from The Jeffrey Modell Foundation.
Thanks to Dr. Fred Lorey and colleagues at the California Dept. of Public Health and Genetic Disease Laboratory, and to Perkin Elmer and Quest Nichols Institute for design and conduct of testing. Drs. Joseph Church, Mort Cowan, Christopher Dvorak, Nina Kapoor, David Lewis, Sean McGhee, Ted Moore, E. R. Stiehm, Chu Ri Shin and Ken Weinberg provided helpful discussions.
Abbreviations
- ADA
Adenosine deaminase
- SCID
Severe combined immunodeficiency
- ALC
Absolute lymphocyte count
- DAF
DNA amplification failure
- DBS
Dried blood spot
- HCT
Hematopoietic cell transplantation
- NBS
Newborn screening
- PITC
Primary Immune Deficiency Treatment Consortium
- qPCR
Quantitative polymerase chain reaction
- RAG1
RAG2, Recombinase activating genes 1 and 2
- TCR
T cell receptor
- TREC
T cell receptor excision circle
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Puck JM. X-linked severe combined immunodeficiency. In: Ochs H, Smith CIE, Puck JM, editors. Primary Immunodeficiency Diseases: A Molecular and Genetic Approach. 2. NY (NY): Oxford Univ Press; 2007. pp. 123–36. [Google Scholar]
- 2.Gatti RA, Meuwissen HJ, Allen Hd, Hong R, Good RA. Immunological reconstitution of sex-linked lymphopenic immunological deficiency. Lancet. 1968;2(7583):1366–9. doi: 10.1016/s0140-6736(68)92673-1. [DOI] [PubMed] [Google Scholar]
- 3.O’Reilly RJ, Friedrich W, Small TN. Transplantation approaches for severe combined immunodeficiency disease, Wiskott-Aldrich syndrome, and other lethal genetic combined immunodeficiency disorders. In: Forman SJ, Blume KG, Thomas ED, editors. Bone marrow transplantation. Boston (MA): Blackwell Scientific Publications; 1994. pp. 849–67. [Google Scholar]
- 4.Buckley RH, Schiff RI, Schiff SE, Markert LM, Williams LW, Harville TO, et al. Human severe combined immunodeficiency: genetic phenotypic, and functional diversity in one hundred eight infants. J Pediatr. 1997;130:378–87. doi: 10.1016/s0022-3476(97)70199-9. [DOI] [PubMed] [Google Scholar]
- 5.Knutsen AP, Wall DA. Umbilical cord blood transplantation in severe T-cell immunodeficiency disorders: two-year experience. J Clin Immunol. 2000;20:466–76. doi: 10.1023/a:1026463900925. [DOI] [PubMed] [Google Scholar]
- 6.Smogorzewska EM, Brooks J, Annett G, Kapoor N, Crooks GM, Kohn DB, et al. T cell depleted haploidentical bone marrow transplantation for the treatment of children with severe combined immunodeficiency. Arch Immunol Ther Exp (Warsz) 2000;48:111–8. [PubMed] [Google Scholar]
- 7.Buckley RH. Molecular defects in human severe combined immunodeficiency and approaches to immune reconstitution. Ann Rev Immunol. 2004;22:625–55. doi: 10.1146/annurev.immunol.22.012703.104614. [DOI] [PubMed] [Google Scholar]
- 8.Grunebaum E, Mazzolari E, Porta F, Dallera D, Atkinson A, Reid B, et al. Bone marrow transplantation for severe combined immune deficiency. JAMA. 2006;295:508–18. doi: 10.1001/jama.295.5.508. [DOI] [PubMed] [Google Scholar]
- 9.Buckley RH, Fischer A. Bone marrow transplantation for primary immunodeficiency diseases. In: Ochs H, Smith CIE, Puck JM, editors. Primary immunodeficiency diseases: a molecular and genetic approach. 2. NY (NY): Oxford Univ Press; 2007. pp. 669–87. [Google Scholar]
- 10.Cavazzana-Calvo M, Carlier F, Le Deist F, Morillon E, Taupin P, Gautier D, et al. Long-term T-cell reconstitution after hematopoietic stem-cell transplantation in primary T-cell-immunodeficient patients is associated with myeloid chimerism and possibly the primary disease phenotype. Blood. 2007;109:4575–81. doi: 10.1182/blood-2006-07-029090. [DOI] [PubMed] [Google Scholar]
- 11.Buckley RH. Transplantation of hematopoietic stem cells in human severe combined immunodeficiency: longterm outcomes. Immunol Res. 2011;49:25–43. doi: 10.1007/s12026-010-8191-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaspar HB, Aiuti A, Porta F, Candotti F, Hershfield MS, Notarangelo LD. How I treat ADA deficiency. Blood. 2009;114:3524–32. doi: 10.1182/blood-2009-06-189209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischer A, Hacein-Bey-Abina S, Cavazzana-Calvo M. Gene therapy for primary adaptive immune deficiencies. J Allergy Clin Immunol. 2011;127:1356–9. doi: 10.1016/j.jaci.2011.04.030. [DOI] [PubMed] [Google Scholar]
- 14.Aiuti A, Cattaneo F, Galimberti S, Benninghoff U, Cassani B, Callegaro L, et al. Gene therapy for immunodeficiency due to adenosine deaminase deficiency. New Engl J Med. 2009;360:447–58. doi: 10.1056/NEJMoa0805817. [DOI] [PubMed] [Google Scholar]
- 15.Noguchi M, Yi H, Rosenblatt HM, Filipovich AH, Adelstein S, Modi WS, McBride OW, Leonard WJ. Interleukin-2 receptor gamma chain mutation results in X-linked severe combined immunodeficiency in humans. Cell. 1993;73:147–57. doi: 10.1016/0092-8674(93)90167-o. [DOI] [PubMed] [Google Scholar]
- 16.Puck JM, Deschênes SM, Porter JC, Dutra AS, Brown CJ, Willard HF, Henthorn PS. The interleukin-2 receptor gamma chain maps to Xq13. 1 and is mutated in X-linked severe combined immunodeficiency, SCIDX1. Hum Molecular Genet. 1993;2:1099–104. doi: 10.1093/hmg/2.8.1099. [DOI] [PubMed] [Google Scholar]
- 17.Hershfield MS. PEG-ADA replacement therapy for adenosine deaminase deficiency: an update after 8. 5 years. Clin Immunol Immunopathol. 1995;76:S228–32. doi: 10.1016/s0090-1229(95)90306-2. [DOI] [PubMed] [Google Scholar]
- 18.Puel A, Ziegler SF, Buckley RH, Leonard WJ. Defective IL7R expression in T(−) B(+)NK(+) severe combined immunodeficiency. Nat Genet. 1998;20:394–7. doi: 10.1038/3877. [DOI] [PubMed] [Google Scholar]
- 19.Roberts JL, Lengi A, Brown SM, Chen M, Zhou YJ, O’Shea JJ, Buckley RH. Janus kinase 3 (JAK3) deficiency: clinical, immunologic, and molecular analyses of 10 patients and outcomes of stem cell transplantation. Blood. 2004;103:2009–18. doi: 10.1182/blood-2003-06-2104. [DOI] [PubMed] [Google Scholar]
- 20.Moshous D, Callebaut I, de Chasseval R, Poinsignon C, Villey I, Fischer A, de Villartay JP. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell. 2001;105:177–186. doi: 10.1016/s0092-8674(01)00309-9. [DOI] [PubMed] [Google Scholar]
- 21.Li L, Moshous D, Zhou Y, Wang J, Xie G, Salido E, et al. A founder mutation in Artemis, an SNM1-like protein, causes SCID in Athabascan-speaking Native Americans. J Immunol. 2002;168:6323–9. doi: 10.4049/jimmunol.168.12.6323. [DOI] [PubMed] [Google Scholar]
- 22.de Villartay J-P, Schwarz K, Villa A. VDJ recombination defects. In: Ochs H, Smith CIE, Puck JM, editors. Primary Immunodeficiency Diseases, a Molecular and Genetic Approach. 2. NY (NY): Oxford Univ Press; 2007. pp. 153–68. [Google Scholar]
- 23.van der Burg M, van Veelen LR, Verkaik NS, Wiegant WW, Hartwig NG, Barendregt BH, et al. A new type of radiosensitive T-B-NK+ severe combined immunodeficiency caused by a LIG4 mutation. J Clin Invest. 2006;116:137–45. doi: 10.1172/JCI26121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kung C, Pingel JT, Heikinheimo M, Klemola T, Varkila K, Yoo LI, et al. Mutations in the tyrosine phosphastase CD45 gene in a child with severe combined immunodeficiency disease. Nature Med. 2000;6:343–5. doi: 10.1038/73208. [DOI] [PubMed] [Google Scholar]
- 25.Tchilian EZ, Wallace DL, Wells RS, Flower DR, Morgan G, Beverley PC. A deletion in the gene encoding the CD45 antigen in a patient with SCID. J Immunol. 2001;166:1308–13. doi: 10.4049/jimmunol.166.2.1308. [DOI] [PubMed] [Google Scholar]
- 26.Dadi HK, Simon AJ, Roifman CM. Effect of CD3δ deficiency on maturation of α/β and γ/δ T-cell lineages in severe combined immunodeficiency. New Engl J Med. 2003;349:1821–8. doi: 10.1056/NEJMoa031178. [DOI] [PubMed] [Google Scholar]
- 27.Regueiro JR, Español T. CD3 and CD8 deficiencies. In: Ochs H, Smith CIE, Puck JM, editors. Primary Immunodeficiency Diseases, a Molecular and Genetic Approach. 2. NY (NY): Oxford Univ Press; 2007. pp. 216–26. [Google Scholar]
- 28.Recio MJ, et al. Differential biological role of CD3 chains revealed by human mmunodeficiencies. J Immunol. 2007;178:2556–64. doi: 10.4049/jimmunol.178.4.2556. [DOI] [PubMed] [Google Scholar]
- 29.Goldman FD, Ballas ZK, Schutte BC, Kemp J, Hollenback C, Noraz N, Taylor N. Defective expression of p56lck in an infant with severe combined immunodeficiency. J Clin Invest. 1998;102:421–9. doi: 10.1172/JCI3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pignata C, Fiore M, Guzzetta V, Castaldo A, Sebastio G, Porta F, Guarino A. Congenital Alopecia and nail dystrophy associated with severe functional T-cell immunodeficiency in two sibs. Am J Med Genet. 1996;65:167–70. doi: 10.1002/(SICI)1096-8628(19961016)65:2<167::AID-AJMG17>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 31.Bertrand Y, Muller SM, Casanova JL, Morgan G, Fischer A, Friedrich W, et al. Reticular dysgenesis: HLA non-identical bone marrow transplants in a series of 10 patients. Bone Marrow Transplantation. 2002;29:759–62. doi: 10.1038/sj.bmt.1703531. [DOI] [PubMed] [Google Scholar]
- 32.Tangsinmankong N, Day NK, Nelson RP, Jr, Puck JM, Good RA. Severe combined immunodeficiency in an infant with multiple congenital abnormalities. J Allergy Clin Immunol. 1999;103:1222–3. doi: 10.1016/s0091-6749(99)70207-1. [DOI] [PubMed] [Google Scholar]
- 33.Gilroy RK, Coccia PF, Talmadge JE, Hatcher LI, Pirruccello SJ, Shaw BW, Jr, et al. Donor immune reconstitution after liver-small bowel transplantation for multiple intestinal atresia with immunodeficiency. Blood. 2004;103:1171–4. doi: 10.1182/blood-2003-04-1187. [DOI] [PubMed] [Google Scholar]
- 34.Puck JM, Middelton LA, Pepper AE. Carrier and prenatal diagnosis of X-linked severe combined immunodeficiency: mutation detection methods and utilization. Hum Genet. 1997;99:628–33. doi: 10.1007/s004390050418. [DOI] [PubMed] [Google Scholar]
- 35.Myers LA, Patel DD, Puck JM, Buckley RH. Hematopoietic stem cell transplantation for SCID in the neonatal period leads to superior thymic output and improved survival. Blood. 2002;99:872–8. doi: 10.1182/blood.v99.3.872. [DOI] [PubMed] [Google Scholar]
- 36.Brown L, Xu-Bayford J, Allwood Z, Slatter M, Cant A, Davies EG, et al. Neonatal diagnosis of severe combined immunodeficiency leads to significantly improved survival outcome: the case for newborn screening. Blood. 2011;117:3243–6. doi: 10.1182/blood-2010-08-300384. [DOI] [PubMed] [Google Scholar]
- 37.Chan A, Scalchunes C, Boyle M, Puck JM. Early vs. Delayed Diagnosis of Severe Combined Immunodeficiency: A Family Perspective Survey. Clin Immunol. 2011;138:3–8. doi: 10.1016/j.clim.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Griffith LM, Cowan MJ, Notarangelo LD, Puck JM, Buckley RH, Candotti F, et al. Allogeneic hematopoietic cell transplantation for primary immune deficiency diseases: current status and critical needs. J Allergy Clin Immunol. 2008;122:1087–96. doi: 10.1016/j.jaci.2008.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lindegren ML, Kobrynski L, Rasmussen SA, Moore CA, Grosse SD, Vanderford ML, et al. Applying public health strategies to primary immunodeficiency diseases: a potential approach to genetic disorders. MMWR Recomm Rep. 2004;53(RR-1):1–29. [PubMed] [Google Scholar]
- 40.Guthrie R, Susi I. A simple phenylalanine method for detecting phenylketonuria in large populations of newborn infants. Pediatrics. 1963;32:318–43. [PubMed] [Google Scholar]
- 41.Meryash DL, Levy HL, Guthrie R, Warner R, Bloom S, Carr JR. Prospective study of early neonatal screening for phenylketonuria. N Engl J Med. 1981;304:294–6. doi: 10.1056/NEJM198101293040510. [DOI] [PubMed] [Google Scholar]
- 42.Puck JM SCID Newborn Screening Working Group. Population-based newborn screening for severe combined immunodeficiency: steps toward implementation. J Allergy Clin Immunol. 2007;120:760–768. doi: 10.1016/j.jaci.2007.08.043. [DOI] [PubMed] [Google Scholar]
- 43.Tarini BA, Clark SJ, Pilli S, Dombkowski KJ, Korzeniewski SJ, Gebremariam A, et al. False-positive newborn screening result and future health care use in a state medicaid cohort. Pediatrics. 2011;128:715–22. doi: 10.1542/peds.2010-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McGhee SA, Stiehm ER, Cowan M, Krogstad P, McCabe ER. Two-tiered universal newborn screening strategy for severe combined immunodeficiency. Mol Genet Metab. 2005;86:427–30. doi: 10.1016/j.ymgme.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 45.Janik DK, Lindau-Shepard B, Comeau AM, Pass KA. A multiplex immunoassay using the Guthrie specimen to detect T-cell deficiencies including severe combined immunodeficiency disease. Clin Chem. 2010;56:1460–5. doi: 10.1373/clinchem.2010.144329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Newborn Screening Status Report (updated 11/21/11) National Newborn Screening and Genetics Resource Center; [Cited 2011 December 12.] Available from http://genes-r-us.uthscsa.edu/nbsdisorders.pdf. [Google Scholar]
- 47.Lebet T, Chiles R, Hsu AP, Mansfield E, Warrington JA, Puck JM. Mutations causing severe combined immunodeficiency: detection with a custom resequencing microarray. Genet in Med. 2008;10:575–85. doi: 10.1097/gim.0b013e31818063bc. [DOI] [PubMed] [Google Scholar]
- 48.Hazenberg MD, Verschuren MC, Hamann D, Miedema F, van Dongen JJ. T cell receptor excision circles as markers for recent thymic emigrants: basic aspects, technical approach, and guidelines for interpretation. J Mol Med. 2001;79:631–40. doi: 10.1007/s001090100271. [DOI] [PubMed] [Google Scholar]
- 49.Douek DC, McFarland RD, Keiser PH, Gage EA, Massey JM, Haynes BF, et al. Changes in thymic function with age and during the treatment of HIV infection. Nature. 1998;396:690–5. doi: 10.1038/25374. [DOI] [PubMed] [Google Scholar]
- 50.Chan K, Puck J. Development of population–based newborn screening for severe combined immunodeficiency. J Allergy Clin Immunol. 2005;115:391–8. doi: 10.1016/j.jaci.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 51.Puck JM. Neonatal screening for severe combined immunodeficiency. Curr Opin Pediatr. 2011;23:667–73. doi: 10.1097/MOP.0b013e32834cb9b0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morinishi Y, Imai K, Nakagawa N, et al. Identification of severe combined immunodeficiency by T-cell receptor excision circles quantification using neonatal Guthrie cards. J Pediatr. 2009;155:829–833. doi: 10.1016/j.jpeds.2009.05.026. [DOI] [PubMed] [Google Scholar]
- 53.Patel DD, Gooding ME, Parrott RE, Curtis KM, Haynes BF, Buckley RH. Thymic function after hematopoietic stem-cell transplantation for the treatment of severe combined immunodeficiency. N Engl J Med. 2000;342:1325–32. doi: 10.1056/NEJM200005043421804. [DOI] [PubMed] [Google Scholar]
- 54.Markert ML, Devlin BH, Chinn IK, McCarthy EA. Thymus transplantation in complete DiGeorge anomaly. Immunol Res. 2009;44:61–70. doi: 10.1007/s12026-008-8082-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Routes JM, Grossman WJ, Verbsky J, Laessig RH, Hoffman GL, Brokopp CD, Baker MW. Statewide newborn screening for severe T-cell lymphopenia. JAMA. 2009;302:2465–70. doi: 10.1001/jama.2009.1806. [DOI] [PubMed] [Google Scholar]
- 56.Baker MW, Grossman WJ, Laessig RH, Hoffman GL, Brokopp CD, Kurtycz DF, et al. Development of a routine newborn screening protocol for severe combined immunodeficiency. J Allergy Clin Immunol. 2009;124:522–7. doi: 10.1016/j.jaci.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 57.Gerstel-Thompson JL, Wilkey JF, Baptiste JC, Navas JS, Pai SY, Pass KA, et al. High-throughput multiplexed T-cell-receptor excision circle quantitative PCR assay with internal controls for detection of severe combined immunodeficiency in population-based newborn screening. Clin Chem. 2010;56:1466–74. doi: 10.1373/clinchem.2010.144915. [DOI] [PubMed] [Google Scholar]
- 58.Hale JE, Bonilla FA, Pai SY, Gerstel-Thompson JL, Notarangelo LD, Eaton RB, Comeau AM. Identification of an infant with severe combined immunodeficiency by newborn screening. J Allergy Clin Immunol. 2010;126:1073–4. doi: 10.1016/j.jaci.2010.08.043. [DOI] [PubMed] [Google Scholar]
- 59.Chase NM, Verbsky JW, Routes JM. Newborn screening for T-cell deficiency. Curr Opin Allergy Clin Immunol. 2010;10:521–5. doi: 10.1097/ACI.0b013e32833fd6fe. [DOI] [PubMed] [Google Scholar]
- 60.Ram G, Chinen J. Infections and immunodeficiency in Down syndrome. Clin Exp Immunol. 2011;164:9–16. doi: 10.1111/j.1365-2249.2011.04335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Newborn Screening Translational Research Network, NBSTRN. doi: 10.32481/djph.2021.12.010. [Cited 2011 Dec. 12] Available from http://www.nbstrn.org/supported-projects/severe-combined-immunodeficiencies. [DOI] [PMC free article] [PubMed]
- 62.Griffith LM, Cowan MJ, Notarangelo LD, Puck JM, Buckley RH, Candotti F, et al. Improving cellular therapy for primary immune deficiency diseases: recognition, diagnosis, and management. Allergy Clin Immunol. 2009;124:1152–60. doi: 10.1016/j.jaci.2009.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Primary Immune Deficiency Treatment Consortium. Rare Diseases Clinical Research Network. 2011 [Cited 2011 Dec. 12.] http://rarediseasesnetwork.epi.usf.edu/PIDTC/SCID/index.htm.
- 64.Shearer WT, Rosenblatt HM, Gelman RS, Oyomopito R, Plaeger S, Stiehm ER, et al. Lymphocyte subsets in healthy children from birth through 18 years of age: the Pediatric AIDS Clinical Trials Group P1009 study. J Allergy Clin Immunol. 2003;112:973–80. doi: 10.1016/j.jaci.2003.07.003. [DOI] [PubMed] [Google Scholar]
- 65.Dasouki M, Okonkwo KC, Ray A, Folmsbeel CK, Gozales D, Keles S, Puck JM, Chatila T. Deficient T Cell Receptor Excision Circles (TRECs) in autosomal recessive hyper IgE syndrome caused by DOCK8 mutation: Implications for pathogenesis and potential detection by newborn screening. Clin Immunol. 2011;141:128–32. doi: 10.1016/j.clim.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Patel NC, Hertel PM, Estes MK, de la Morena M, Petru AM, Noroski LM. Vaccine-acquired rotavirus in infants with severe combined immunodeficiency. N Engl J Med. 2010;362:314–9. doi: 10.1056/NEJMoa0904485. [DOI] [PMC free article] [PubMed] [Google Scholar]