Abstract
The present manuscript extends our de novo peptide design approach to the synthesis and evaluation of a new generation of reversed-phase HPLC peptide standards with the same composition and minimal sequence variation (SCMSV). Thus, we have designed and synthesized four series of peptide standards with the sequences Gly-X-Leu-Gly-Leu-Ala-Leu-Gly-Gly-Leu-Lys-Lys-amide, where the N-terminal is either Nα-acetylated (Series 1) or contains a free α-amino group (Series 3); and Gly-Gly-Leu-Gly-Gly-Ala-Leu-Gly-X-Leu-Lys-Lys-amide, where the N-terminal is either Nα-acetylated (Series 2) or contains a free α-amino group (Series 4). In this initial study, the single substitution position, X, was substituted with alkyl side-chains (Ala<Val<Ile, in order of increasing hydrophobicity) or aromatic side-chains (Phe, Tyr). Peptide series pairs 1/2 and 3/4 thus represent SCMSV peptides, with the substitution site, X, being towards the N- or C-terminal, albeit with identical adjacent residues (Gly-X-Leu) to maintain the same environment around position X. In addition, peptide series pairs 1/3 and 2/4 enable an examination of the effect of a free, positively charged α-amino group on peptide retention behaviour relative to a blocked N-terminus. Peptide mixtures were run at pH 2 on columns with a variety of stationary phase selectivity (C8, C18, polar endcapped, polar embedded, ether-linked phenyl and phenyl-hexyl) under linear gradient conditions with acetonitrile or methanol as organic modifier. It was interesting to note that the addition of the hydroxyl group to the aromatic ring in a 12-residue Tyr SCMSV peptide pair had a dramatic effect on resolution compared to the Phe peptide pair. In addition, SCMSV peptide pairs with the β-branched Val and Ile side-chains at position X were the most difficult to separate compared to SCMSV peptides containing the aromatic side-chains Tyr and Phe. In this initial study, SCMSV peptide pairs proved to be a potent test of the selectivity of reversed-phase packing materials. In addition, mixtures of SCMSV peptide standards to assess overall capabilities of stationary phases to resolve complex peptide mixtures underlined the useful complementarity of combinations of different columns and elution conditions to maximize flexibility in peptide applications. Finally, our controlled, de-novo designed peptide approach should spur the development of more quantitative selectivity parameters for peptide separations, such as those already available for small molecules, enhancing further the universal value of utilizing peptide standards to compare column performances in the separation of peptide mixtures.
I. Introduction
Our laboratory has been active for over 25 years in the de novo design and synthesis of peptide standards for monitoring high-performance liquid chromatography (HPLC) column performance in several modes of HPLC: size-exclusion chromatography (SEC) [1–5]; cation-exchange chromatography (CEX) [2,3,5,6]; hydrophilic interaction/cation-exchange chromatography (HILIC/CEX) [7]; and reversed-phase HPLC (RP-HPLC) [2,3,5,8–15]. Our extensive work in this area is a result of our premise that peptide solutes are best suited for monitoring peptide behaviour during HPLC, since it is preferable to use standards that are structurally similar to the sample of interest. This is particularly pertinent to RP-HPLC when one considers the significant partitioning of small molecules between the mobile phase and stationary phase, with concomitant employment of isocratic elution for effective separation of such small solutes [16]. In contrast, peptides and proteins exhibit relatively limited partitioning between the mobile phase and stationary phases [16,17], requiring gradients of organic modifier for their elution and separation.
As we noted in a previous paper [15], compared to a wealth of information published over the past three decades on the assessment of RP-HPLC stationary phases for small molecule separations (references 18–22 and references therein represent excellent reviews), the lack of data derived from RP-HPLC retention behaviour of peptides or proteins, as opposed to the mixtures of small molecules almost invariably employed, is striking. Indeed, the requirement of peptide standards for assessing column selectivity for peptidic solutes, as opposed to small molecule separations, was clearly demonstrated by our previous study [15], where the retention behaviour of mixtures of peptide standards on different RP-HPLC stationary phases was compared to the PQRI database of several hundred different stationary phases [www.usp.org/USPNF/columns.html]. Briefly, thousands of small molecule separations on many silica-based columns generated a column selectivity factor (denoted as F) arising from five column properties: hydrophobicity, steric interaction, H-bond acidity, H-bond basicity and ion-exchange capacity), with reference 19 a useful review. Columns with F values differing by ≤ 3 are generally viewed as being essentially equivalent. To summarize, there was no correlation whatsoever between the results we observed utilizing peptide standards and the relative equivalency of the same columns generated by the PQRI calculator. Thus, our continuing efforts to make up for this deficiency in assessing the suitability of different phases for peptide separations involve utilizing peptide standards, specifically de novo designed and synthesized peptide standards. Such designed standards avoid the disadvantages of the use of random mixtures of native peptides which is generally not informative, since if an improvement in selectivity is observed, the reason may be unclear due to this random nature where size, amino acid composition and sequence of the peptides in the mixture may vary considerably. Hence, our preferred approach has always been the design and synthesis of peptide standards with minimal sequence variation to probe column selectivity for the separation of peptide mixtures.
Our previous approach to de novo design of RP-HPLC peptide standards has been to ensure only subtle changes in overall hydrophilicity/hydrophobicity between peptides in a peptide mixture (i.e., single substitution of an amino acid residue from one peptide to the next), in order to test RP-HPLC column selectivity. We now wished to design a new generation of peptide standards for an even more rigorous assessment of RP-HPLC stationary phase selectivity for peptide separations: peptide standards with same amino acid composition and minimal sequence variation (SCMSV) (Table 1). That is, in the absence of any variation in side-chain nearest-neighbour and/or peptide conformation effects, can a reversed-phase packing separate peptide pairs where the only difference between the two peptides is the location of a single amino acid? In addition, does the nature of the substituted amino acid (e.g., alkyl, aromatic) affect the degree of resolution of the peptide pairs?
Table 1.
Sequences of synthetic model peptides used in this study
| Peptide Sequence | Name | |||||||
|---|---|---|---|---|---|---|---|---|
| Series 1: | Ac- | G | X | LGLALG | G | LKK | -amide | AcN-X |
| Series 2: | Ac- | G | G | LGLALG | X | LKK | -amide | AcC-X |
| Series 3: | +NH3- | G | X | LGLALG | G | LKK | -amide | AmN-X |
| Series 4: | +NH3- | G | G | LGLALG | X | LKK | -amide | AmC-X |
Residues/residue sequences identical in all for peptide series are boxed. The substitution site, X, was substituted with Ala (A), Val (V), Ile (I), Tyr (Y)or Phe (F). Ac- denotes Nα -acetyl and -amide denotes Cα -amide. Series 1 and 2 and Series 3 and 4 are SCMSV peptides. The substitution site, X, has identical adjacent residues (G-X-L) in all peptides.
Clearly, concomitant with the design of new and novel peptide standards is evidence of their value for assessment of the effectiveness of RP-HPLC packings with defined and varying selectivities. Developments in surface modification of silica-based packings over the past 25 years have included polar-embedded phases [15,18,19,23–55], polar-endcapped phases [15,19,35,40,42,45,47,50–52] and phases containing phenyl groups [15,19,45,56–65], all with the purpose of providing alternative selectivity compared to traditional alkyl (generally C8, C18 groups [3,5]) functional groups. Thus, the present paper extends our peptide design approach to SCMSV peptide standards and their effectiveness in monitoring the resolving capabilities of stationary phases of widely differing properties in the presence of acetonitrile or methanol as organic modifier. Two approaches are being used to evaluate the potential of the SCMSV peptide standards: (i) application of specific SCMSV peptide pairs (e.g., alkyl or aromatic) to RP-HPLC on stationary phases of varying selectivity; (ii) application of mixtures of SCMSV peptide standards to assess overall ability of stationary phases of varying selectivity to resolve more complex SCMSV peptide mixtures.
2. Experimental
2.1 Materials
HPLC-grade water was prepared by an E-pure water purification system from Barnstead International (Dubuque, IA, USA). Trifluoroacetic acid (TFA) was obtained from Pierce (Rockford, IL, USA). HPLC-grade acetonitrile and methanol were obtained from Fisher Scientific (Fairlawn, NJ, USA).
2.2 Instrumentation
All analytical runs were carried out on an Agilent 1100 Series liquid chromatograph (Agilent Technologies, Foster City, CA, USA).
2.3 RP-HPLC columns
(i) Luna C8 (5 μm particle size, 100 Å pore size); (ii) Luna C18 (5 μm, 100 Å); (iii) Synergi Hydro-RP (4 μm, 80 Å); (iv) Synergi Fusion-RP (4 μm, 80 Å); (v) Synergi Polar-RP (4 μm, 80 Å); (vi) Luna Phenyl-Hexyl (5 μm, 100 Å). All columns are silica-based with dimensions of 250 × 3 mm I.D. The columns were donated by Phenomenex (Torrance, CA, USA).
2.3 RP-HPLC run conditions
Runs were carried out using linear AB gradient elution (0.25% acetonitrile/min or 0.5% methanol/min) at a flow-rate of 0.5 ml/min at 25°C, where eluent A is 0.2% aq. TFA and eluent B is 0.18% TFA in acetonitrile or methanol. The shallow gradient rates were designed to help maximize observed differences in peptide separations on different columns.
2.4 Model synthetic SCMSV peptides
The peptide standards were synthesized by solid-phase synthesis methodology using conventional Boc- and Fmoc-chemistry, as described previously [66,67] and purified and characterized as described previously [68].
3. Results
3.1 Design of peptide standards
The sequences of the four series of 12-residue peptide standards are shown in Table 1, where the N-terminal is either Nα-acetylated (Series 1 and 2) or contains a free α-amino group (Series 3 and 4). The single substitution site, X, is either towards the N-terminus (Series 1 and 3) or towards the C-terminus (Series 2 and 4). Thus, peptide series pairs 1/2 and 3/4 represent peptides with the same composition but different sequences. The length of the peptides was designed to approximate the average size of cleavage fragments from proteolytic digests of proteins. The presence of only basic amino acid residues (lysine), i.e., no acidic, potentially negatively charged residues, the blocked C-terminus for all peptides, and the use of mobile phases at pH values (~pH 2) far below the pKa values of the N-terminal amino group and the lysine side-chain amino group, ensures a constant net positive charge on the peptides of either +2 (Series 1 and 2) or +3 (Series 3 and 4). The presence of the lysine residues also ensures good peptide solubility. In addition, peptide series pairs 1/3 and 2/4 enable an examination of the effect of a free, positively charged α-amino group relative to a blocked N-terminus on peptides of identical sequence.
In a previous paper [69], we categorized two types of sequence-dependent effects that may affect peptide retention behaviour: (1) conformational effects resulting in an apparent reduction or enhancement of the overall hydrophobicity of a peptide as a result of its adopting an unique conformation on interacting with the hydrophobic stationary phase, compared to the hydrophobicity of the peptide if it existed as a random coil; (2) nearest-neighbour effects resulting in sequence-dependent variability of peptide retention behaviour but independent of a defined secondary structure. Houghten and DeGraw [70] demonstrated how peptides with the same amino acid composition but different sequences (SCDS) can have very different RP-HPLC retention times. However, these peptides were glycine insertion analogues, in which one glycine was inserted into the original 13-residue sequence between each position, i.e., the environment around the substitution site varied, often considerably, from analogue to analogue. Thus, the positional effect, if any, on peptide retention behaviour could not be delineated from potential nearest-neighbour and/or conformational effects. Induction of secondary, specifically α-helical, structure by the hydrophobic environment of RP-HPLC [69,71–73] may also produce significant differences in retention behaviour of SCDS peptides [69,74], Thus, a key design parameter of our new standards was to eliminate any nearest-neighbour and/or conformational differences between pairs of peptides identical except for the substitution position of a particular amino acid residue. To this end, a random coil nature of the peptides is ensured by the inclusion of four helix-disrupting glycine residues [75,76] throughout the peptide sequences [Table 1). In addition, whether substitution position X is towards the N-terminus (Series 1 and 3) or towards the C-terminus (Series 2 and 4), the adjacent residues are identical (i.e., G-X-L) (Table 1), ensuring the same nearest-neighbour environment surrounding the substitution site in all four peptide series. It should also be noted that a considerable majority (10 out of 12 residues) of the amino acid sequence of all four peptide series, as denoted by boxed residues in Table 1, is identical. Thus, the single substitution site in the sequences, coupled with this minimal sequence variation, prompted the term “same composition and minimal sequence variation” (SCMSV) to describe these peptide standards.
For this initial study, position X of the peptide sequence is substituted by Ala, Val, Ile, Phe or Ty217 r. Thus, these peptides represent series of peptides with single substitutions of alkyl side-chains (Ala < Val < Ile) in order of increasing hydrophobicity [77,78] alkyl versus aromatic side-chains (Phe, Tyr) and a side-chain also with polar character (hydroxyl group of Tyr). Finally, the presence of an N-terminal positive charge on peptide series 3 and 4 increases their overall polarity compared with their Nα-acetyl counterparts, Series 1 and 2, respectively. The denotions of the peptides identify the relative substitution position within the sequence, as well as whether the peptide has a free or blocked N-terminus. For example, AcC-I denotes the peptide analogue with Ile substituted towards the C-terminus and a blocked (Nα-acetyl) N-terminus; AmN-A denotes the peptide analogue with Ala substituted towards the N-terminus and a free α-amino group at the N-terminal.
It is important to note that the Series 2 and 4 peptides were previously reported by us to be useful in assessing column and solvent selectivity on standard, polar-embedded and polar endcapped columns [15]. However, it should be stressed that this earlier work examined selectivity differences in resolving mixtures of peptides of varying hydrophobicity. The present study, in contrast, presents a more potent challenge of resolving peptide pairs varying in only positional differences of identical amino acids, i.e., “Same Composition Minimal Sequence Variation” (SCMSV) peptides.
Series 1/2 and Series 3/4 peptide pairs were examined by circular dichroism (CD) spectroscopy in the presence of 0.2% aq. TFA, 0.2% aq. TFA containing 50% (v/v) acetonitrile and 0.2% aq. TFA containing 50% (v/v) trifluoroethanol (TFE). As expected, all peptides exhibited negligible, random coil structure (i.e., no higher orders of structure such as secondary α-helix or β-sheet) in 0.2% aq. TFA in the presence or absence of acetonitrile, as well as identical spectral profiles in the presence of the helix-inducing solvent TFE. The significance of these observations is that no structure would be induced on binding to the reversed-phase columns that would account for resolution of SCMSV peptide pairs.
3.2 Stationary Phases
In order to test the general utility of the peptide standards, a range of packing materials with different selectivities was selected. Concomitantly, of course, this approach serves to examine the ability of stationary phases with purposely designed selectivities to effect difficult peptide separations. Employing multiple stationary phases from the same manufacturer had the advantage of ruling out silica source as a variable when assessing the influence of different stationary phases on peptide resolution.
The Luna C8 and C18 columns represent traditionally employed stationary phases containing alkyl functional groups [3,5]. It should be noted that the following two phases (polar endcapped and polar embedded) are modified C18 phases.
Polar-endcapped phases are designed to enable the retention of polar analytes on phases containing long alkyl chains, e.g., C18, under highly aqueous conditions [15,39,40,42], hence the term “Aqua” phase [45] and the polar endcapping group of the Synergi Hydro-RP phase being denoted as “Aq” for “aqueous”. The value of these groups (the nature of which is proprietary for the Synergi Hydro-RP phase) lies in easy wetting of the silica surface and, thus, allowing full interaction of solutes with the long C18 alkyl chains.
Polar-embedded phases are produced by insertion of a polar functional group (e.g., urea, carbamate, amide, ether) within the alkyl chain. The Synergi Fusion-RP is actually a mixed phase, containing ODS chains and shorter polar-embedded chains. However, for ease of comparison, it is referred to simply as a polar-embedded phase in the present study. Such polar embedded phases are reported to reduce retention of polar and basic small molecule solutes, with non-polar analytes less affected. On the other hand, enhanced retention can occur via hydrogen bonding to the polar-embedded groups relative to non-polar-embedded phases.
For small molecules, polar-endcapped stationary phases have generally shown less change in RP-HPLC selectivity than polar-embedded phases compared with corresponding alkyl-silica columns, possibly due to a lesser accessibility of endcapping groups compared to embedded groups [19]. In addition, the overall hydrophobicity of polar-embedded phases is generally less than corresponding alkyl-silica phases with ligands of identical chain length [15].
The Synergi Polar-RP phase contains an ether-linked group with polar endcapping. In a similar manner to the polar-endcapped column, the “Aq” denotions on the Polar-RP phase refers to “aqueous”. Such a phase, in a similar manner to polar-embedded phases, is designed for enhanced polar selectivity relative to non-modified alkyl phases. In addition, the presence of phenyl groups may be expected to enhance aromatic solute selection (see Phenyl-Hexyl phase below), with either groups having been used effectively as polar-enhancing agents [40].
The Luna Phenyl-Hexyl phase employs a hexyl alkyl linker to the silica support as opposed to the traditional propyl chain in other phenyl phases. Phenyl groups have frequently been shown to exhibit preferential retention of aromatic solutes compared to alkyl phases [15,56–64], this preferred retention generally being attributed to π-π interactions between such solutes and the phenyl groups of the stationary phase [19,63,64].
3.3 RP-HPLC of SCMSV peptide pairs on stationary phases of varying selectivity
Mixtures of sets of SCMSV peptide pairs were applied to the six columns under gradient elution conditions of 0.25% acetonitrile/min or 0.5% methanol/min, such shallow gradient conditions being employed to maximize selectivity differences. Acetonitrile is, of course, the most widely used organic modifier for peptide separations [3,5]. The more polar methanol has been used as an alternate organic modifier, often for more hydrophilic peptide solutes [3,5,15] or for areas where methanol is more preferable to acetonitrile, e.g., peptide drug purification. Doubling the gradient rate when using methanol (0.5%/min compared with 0.25% acetonitrile/min) was required for elution of peptides within a reasonable time, methanol being a weaker eluting solvent than acetonitrile. In addition, doubling of the gradient rate of methanol relative to acetonitrile resulted in similar peptide elution times in both solvents, allowing a valid comparison of the relative selectivity of peptide separations between the two mobile phases on the range of stationary phases.
For comparison purposes, the six columns were arranged in tables and figures to reflect a progression of phase type: the C8 was followed by the longer alkyl chain C18, these more traditionally employed phases being followed by two columns which are essentially modified C18 phases, either polar endcapped (Hydro) or polar embedded (Fusion); the final two columns both contain phenyl-modified supports, one with an ether linkage between the aromatic ring and the alkyl chain (Polar) and the other being the Phenyl-Hexyl phase.
Table 2 reports retention time differences between SCMSV peptide pairs, with calculated resolution of peptide pairs shown in Table 3.
Table 2.
RP-HPLC Retention Time Differences (ΔtR) Between Peptide Pairs of the Same Composition and Minimal Sequence Variation
| Peptide Pairsa | C8b | C18 | Hydro | Fusion | Polar | PH | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CH3CNc | MeOH | CH3CN | MeOH | CH3CN | MeOH | CH3CN | MeOH | CH3CN | MeOH | CH3CN | MeOH | |
| AmN-Y/AmC-Y | 4.3d | 3.1 | 3.9 | 2.9 | 4.2 | 3.5 | 6.1* | 5.1* | 3.1 | 4.0 | 2.8 | 3.1 |
| AmN-F/AmC-F | 2.6 | 1.0 | 2.4 | 1.8 | 2.8 | 2.5 | 5.4* | 4.4* | 2.1 | 2.0 | 1.5 | 1.3 |
| AcN-Y/AcC-Y | 4. 6 | 4.0 | 3.7 | 3.7 | 4.0 | 4.7 | 6.8* | 6.3* | 2.2 | 4.5 | 2.1 | 3.6 |
| AcN-F/AcC-F | 1.9 | 2.1 | 1.5 | 2.4 | 1.9 | 3.1 | 5.2* | 5.0* | 0.4 | 2.1 | 0.3 | 1.7 |
| AmN-A/AmC-A | 3.4 | 2.4 | 3.2 | 2.3 | 3.5 | 2.7 | 4.9* | 4.3* | 3.5 | 3.5 | 3.0 | 2.6 |
| AmN-V/AmC-V | 0.5 | 0 | 0 | 0 | 0 | 0 | 1.8* | 1.5* | 1.0 | 1.5 | 0.5 | 0.8 |
| AcN-A/AcC-A | 2.7 | 2.8 | 2.6 | 2.8 | 2.8 | 2.9 | 4.1* | 4.0* | 1.7 | 2.7 | 1.6 | 2.2 |
| AcN-V/AcC-V | 0.6 | 0 | 1.4 | 0 | 1.2 | 0 | 1.7* | 2.3* | 0.8 | 1.8 | 1.3 | 0.9 |
| AmN-I/AmC-I | 0.7 | 0.3 | 0 | 0.7 | 0 | 0 | 2.7* | 2.0* | 1.3 | 1.9 | 0.6 | 1.0 |
| AcN-I/AcC-I | 0.4 | 0 | 1.6 | 0 | 1.4 | 0 | 1.8* | 2.4* | 0.9 | 1.8 | 1.6 | 0.9 |
Sequences and denotions of peptides are shown in Table 1.
Descriptions of columns shown in Section 3.2.
RP-HPLC conditions for both CH3CN and MeOH mobile phases are shown in Section 2.3.
Retention time differences in min.
e For the majority of peptide pairs, analogues with the amino acid substitution towards the N-terminal were eluted prior to those substituted towards the C-terminal (Table 1); the bold, italicized values represent values where this situation was reversed.
Denotes best ΔtR values on the six columns tested.
Table 3.
RP-HPLC Resolutiona of Peptide Pairs of the Same Composition and Minimal Sequence Variation
| Peptide Pairsb | C8c | C18 | Hydro | Fusion | Polar | PH | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CH3CNd | MeOH | CH3CN | MeOH | CH3CN | MeOH | CH3CN | MeOH | CH3CN | MeOH | CH3CN | MeOH | |
| AmN-Y/AmC-Y | 5.8 | 4.3 | 4.7 | 3.6 | 5.0 | 3.6 | 5.9* | 4.6* | 3.1 | 3.4 | 3.3 | 3.5 |
| AmN-F/AmC-F | 2.8 | 1.2 | 2.4 | 1.8 | 2.7 | 1.8 | 4.5* | 3.6* | 1.9 | 1.5 | 1.8 | 1.7 |
| AcN-Y/AcC-Y | 5.2 | 4.3 | 3.7 | 3.9 | 4.3 | 4.7 | 6.1* | 5.2* | 2.3 | 3.4 | 2.0 | 3.7 |
| AcN-F/AcC-F | 2.2 | 2.4 | 1.6 | 2.7 | 1.8 | 2.7 | 4.2* | 4.0* | n/c | 1.7 | n/c | 1.8 |
| AmN-A/AmC-A | 4.2 | 2.4 | 4.3 | 2.7 | 3.6 | 3.9* | 4.6* | 3.3 | 3.6 | 3.1 | 3.4 | 2.7 |
| AmN-V/AmC-V | n/ce | 0 | 0 | 0 | 0 | 0 | 1.3* | 1.0 | 0.8 | 1.1* | n/c | 0.7 |
| AcN-A/AcC-A | 2.8 | 2.7 | 2.7 | 2.5 | 2.7 | 2.1 | 3.3* | 2.8* | 1.5 | 2.0 | 1.5 | 2.1 |
| AcN-V/AcC-V | n/c | 0 | 1.4* | 0 | 1.1 | 0 | 1.3 | 1.7* | 0.7 | 1.3 | 1.2 | 0.9 |
| AmN-I/AmC-I | 1.0 | n/c | 0 | 0.9 | 0 | 0 | 2.5* | 1.8 | 1.5 | 1.9* | 0.8 | 1.2 |
| AcN-I/AcC-I | n/c | 0 | 2.0* | 0 | 1.3 | 0 | 1.6 | 2.0* | 0.9 | 1.8 | 1.7 | 1.0 |
Resolution (Rs) values were calculated according to the relationship Rs = 1.176Δt/w1+w2, where Δt is the difference in RP-HPLC retention times between peptide pairs and w1 and w2 are the peak width, at half height of the peptides (< 1% variation from multiple runs of all peptide pairs).
Sequences and denotions of peptides are shown in Table 1.
Descriptions of columns shown in Section 3.2.
RP-HPLC conditions for both CH3CN and MeOH mobile phases are shown in Section 2.3
n/c denotes non-calculable.
Denotes best resolution on the six columns tested.
From Table 2, the great majority of elution orders of the SCMSV peptide pairs showed analogues with the substitution towards the N-terminus being eluted prior to those substituted towards the C-terminus. Exceptions were mainly seen with the acetylated Val- and Ile-substituted analogues in the presence of acetonitrile on five of the six columns, together with the acetylated Phe-substituted analogues on the Polar and Phenyl-Hexyl columns. Note that, due to the extra positive charge on the non-acetylated peptides, they were eluted prior to their acetylated counterparts due to their subsequent lesser hydrophobicity.
From Tables 2 and 3, it is interesting to note that, of the alkyl side-chains, the Ala-substituted analogues showed significantly more separation than analogues substituted with the bulkier Val and Ile side-chains. In fact, the Ala-substituted analogues were resolved to a similar magnitude to the aromatic amino acid-substituted peptides. It can also be seen that the Tyr-substituted peptide pairs were consistently resolved better than the Phe-substituted pairs.
Finally, with a few exceptions involving mainly Val- and Ile-substituted analogues, the presence or absence of an N-terminal α-amino group or the use of acetonitrile versus methanol generally had little effect on the resolution of a SCMSV peptide pair. Such exceptions, together with column specific-effects, are discussed below.
3.3.1 Resolution of SCMSV peptides with aromatic side-chain substitutions: Phe (F), Tyr (Y)
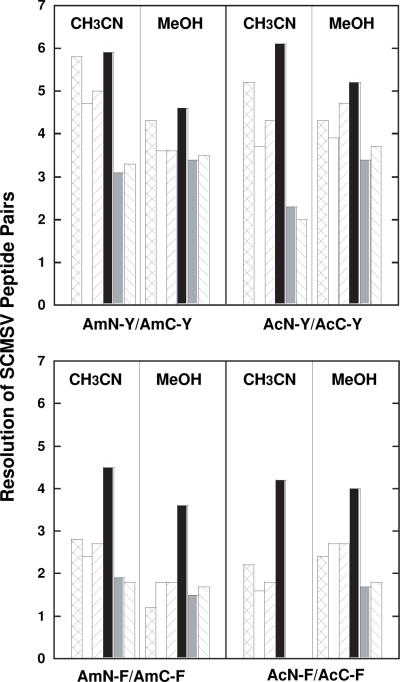
Resolution results of SCMSV peptide pairs substituted with Tyr (top) or Phe (bottom) are shown in Fig. 1. The Tyr analogues were always eluted prior to corresponding Phe analogues, due to the greater hydrophobicity of Phe compared to Tyr [77]. From Fig.1, the superior resolution of all Tyr-substituted SCMSV peptide pairs compared to the Phe-substituted peptides is apparent. The standout observation here is the superior resolution of both the Phe- and Tyr-substituted peptide pairs by the Fusion (polar embedded) column compared to all other phases, this superiority being particularly clear for the Phe-substituted pairs. The overall similarity in resolution when comparing the non-acetylated analogues to those containing a free N-terminal α-amino group, in the presence of acetonitrile or methanol, is also apparent.
Fig. 1.
Effect of stationary phase or organic modifier on resolution of SCMSV peptide pairs substituted with aromatic tyrosine (Tyr, Y) and phenylalanine (Phe, F) amino acids at position X (Table 1). Column denotions: C8, cross-hatched; C18, white; Hydro, upward slanted line; Fusion, black; Polar, grey; Phenyl-Hexyl, downward slanted line. Sequences of AmN, AcN, AmC and AcC peptides shown in Table 1. HPLC conditions shown in Section 2.3. Data shown in Table 3.
Concomitant with the ability of a particular column/organic modifier combination to resolve SCMSV peptide pairs, interesting selectivity differences between the columns were apparent. Thus, the Fusion column was able to resolve to baseline all eight peptides in an eight-peptide mixture (AmN-Y/AmC-Y, AmN-F/AmC-F, AcN-Y/AcC-Y, AcN-F/AcC-F), an achievement not observed with the other columns. Also, only one example of co-elution of a peptide pair was apparent, that of AcN-F/AcC-F on the Polar and Phenyl-Hexyl columns column in the presence of acetonitrile; however, the peptide pair was resolved on the Polar and Phenyl-Hexyl columns in the presence of methanol. Clearly, serendipity was involved in that no SCMSV peptides resolved on the Fusion column were co-eluted with peptides from other pairs, unlike the other columns where, even if resolution of all SCMSV peptide pairs was achieved, such co-elution with other SCMSV peptides occurred. However, it was interesting to note that, as has been observed in a previous paper from this laboratory [15], the polar-embedded Fusion phase achieved the best separation of a peptide mixture over a shorter run time compared to other phases. Such useful selectivity differences between columns are illustrated later in this paper. Finally, it should be stressed that these peptide pairs containing Tyr or Phe substitutions are the most significant standards for evaluating packing materials for researchers utilizing peptides varying in aromaticity/types of aromatic residue substitutions.
3.3.2 Resolution of SCMSV peptides with alkyl side-chain substitutions: Ala (A), Val (V), Ile(I)
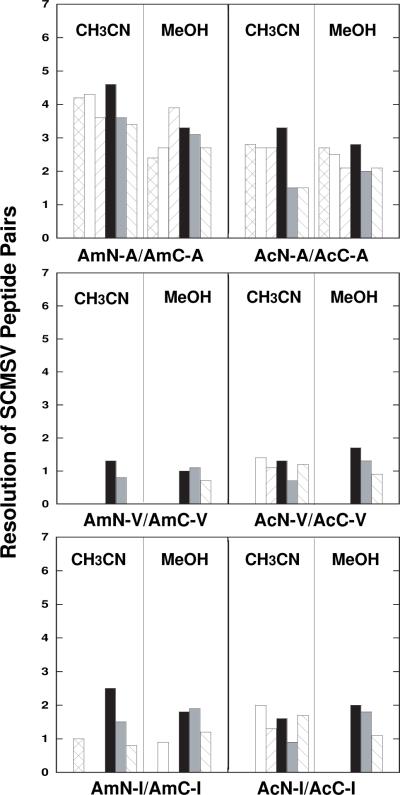
Resolution results of SCMSV peptide pairs substituted with Ala (top), Val (middle) or Ile (bottom) are shown in Fig. 2. Note that the Val-substituted analogues were always eluted later than their less hydrophobic [77] Ala-substituted counterparts. From Fig. 2, the superior resolution of the Ala-substituted SCMSV pairs compared to the more bulky β-branched Val- and Ile-substituted analogues is clear. Indeed, the Val- and Ile-substituted SCMSV pairs proved to be the most difficult of all analogues tested to be resolved under all column/mobile phase combinations. Overall, there was no particular standout column for these alkyl-substituted SCMSV separations, although the Fusion column was generally at least as good, usually superior, to the other phases for all peptide pairs and run conditions over a shorter run time. However, interesting selectivity differences between the columns for peptide mixtures containing Ala- and Val-substituted or Ile-substituted SCMSV peptide analogues were still observed. Thus, concerning the Ala- and Val-substituted analogues, the Fusion column was again able to resolve all peptides in a peptide mixture, this time a six-peptide mixture (AmN-V/AmC-V, AcN-A/AcC-A, AcN-V/AcC-V), in the presence of acetonitrile or methanol, as was the Polar column in the presence of methanol. Unlike the separations of the Tyr- and Phe-substituted peptide pairs where, with one exception, peptide co-elution was not due to inability to resolve SCMSV peptide pairs, the Val-substituted SCMSV analogues often proved difficult to resolve. Thus, the AmN-V/AmC-V pair was co-eluted on the C18 column in the presence of acetonitrile and methanol; in addition, the AcN-V/AcC-V pair was co-eluted on the C18 column in the presence of methanol but resolved in the presence of acetonitrile (Table 3). On the Polar column, the AmN-V/AmC-V and AcN-V/AcC-V peptide pairs were both resolved on the Polar column in the presence of methanol but both pairs showed doublets in the presence of acetonitrile.
Fig. 2.
Effect of stationary phase or organic modifier on resolution of SCMSV peptide pairs substituted with alanine (Ala, A), valine (Val, V) or isoleucine (Ile, I) at position X (Table 1). Column denotions: C8, cross-hatched; C18, white; Hydro, upward slanted line; Fusion, black; Polar, grey; Phenyl-Hexyl, downward slanted line. Sequences of AmN, AcN, AmC and AcC peptides shown in Table 1. HPLC conditions shown in Section 2.3. Data shown in Table 3.
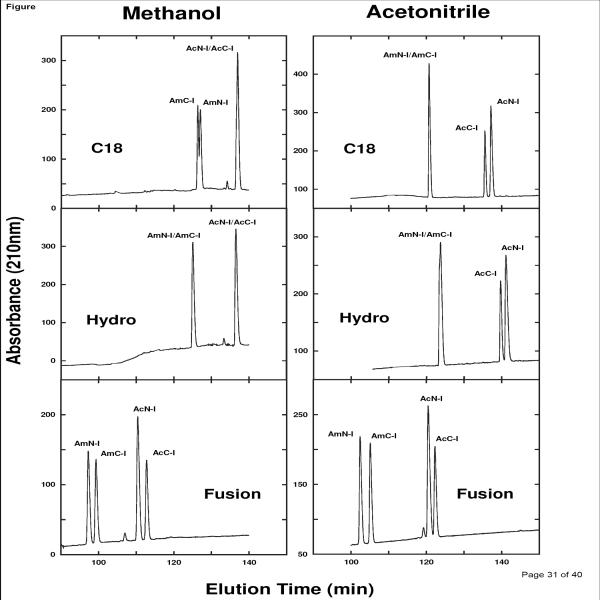
Fig. 3 clearly shows the variation in stationary phase ability to separate the difficult Ile-substituted SCMSV pairs, in addition to some interesting selectivity differences between the two organic modifiers. Thus, on the C18 column, the AmN-I/AmC-I pair formed a doublet in the presence of methanol, with the AcN-I/AcC-I pair being completely co-eluted; in contrast, the AmN-I/AmC-I pair was co-eluted in the presence of acetonitrile, with the acetylated pair being resolved to baseline. Both peptide pairs were co-eluted on the Hydro column in the presence of methanol, while the AcN-I/AcC-I pair was resolved to baseline in the presence of acetonitrile. Finally, the Fusion column was able to resolve both SCMSV peptide pairs to baseline in the presence of either organic modifier, again within a shorter separation time than all other columns.
Fig. 3.
Effect of stationary phase (C18, Hydro, Fusion) or organic modifier on resolution of SCMSV peptides substituted with isoleucine (IleI) at position X (Table 1). HPLC conditions shown in Section 2.3.
Finally, it is clear that the Ile- and Val-substituted peptide pairs have potentially excellent value in assessing which RP-HPLC packings are most suitable to solve very difficult separation problems.
3.4 RP-HPLC of mixtures of SCMSV peptide standards
The peptide standards described in the present study serve a dual role of not only assessing the ability of reversed-phase stationary phases of varying properties in the presence of acetonitrile or methanol to resolve specific SCMSV peptide pairs, as described above, but also to monitor the effectiveness generally of such columns to resolve more complex peptide mixtures. Thus, the advantages of selectivity differences between columns were now examined further with various mixtures of peptide standards, with representative peptide elution profiles highlighting the value of column and/or mobile phase variation. In order to underline this role of monitoring column selectivity, additional SCMSV peptides substituted with glycine (Gly, G), aspartic acid (D) or glutamic acid (Glu, E) were included in peptide mixtures: AmN-G, AcN-G, AmN-D, AcN-D, AmN-E and AcN-E.
3.4.1 Effect of varying stationary phase on separation of peptide standards
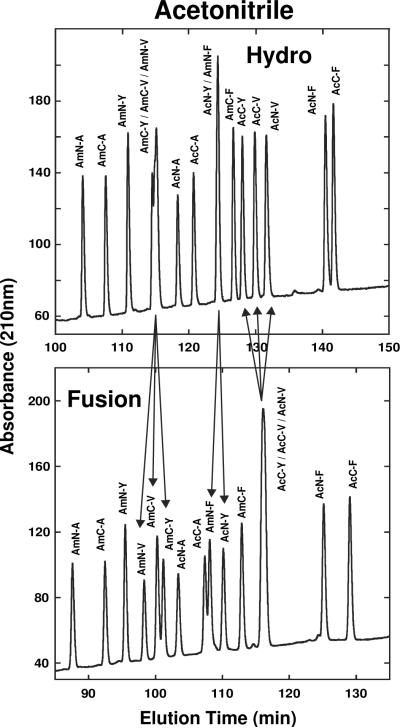
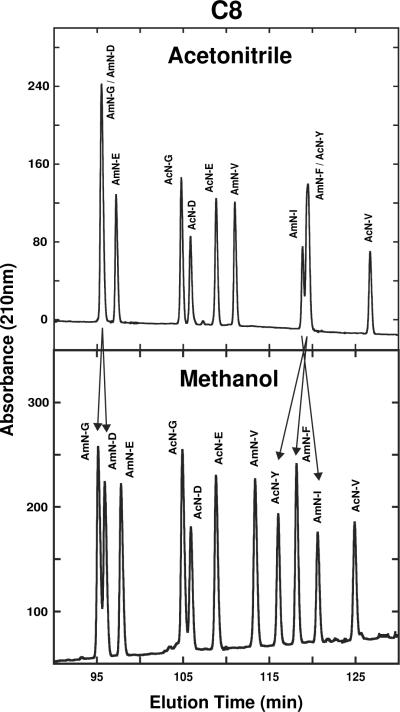
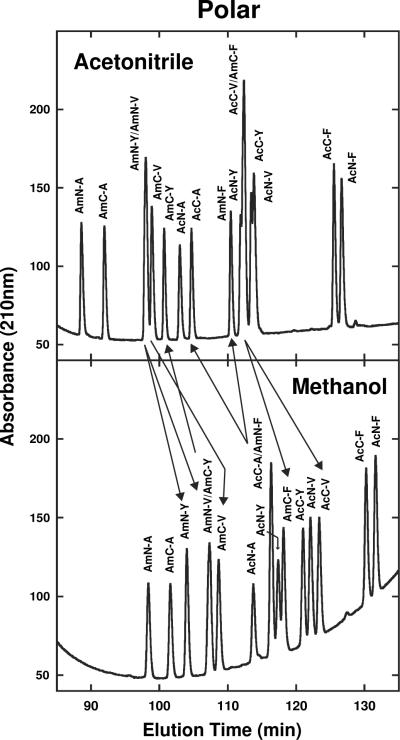
Fig. 4 compares elution profiles of a 16-peptide mixture of SCMSV peptide standards on the Hydro (top) and Fusion (bottom) columns with acetonitrile as organic modifier. This is an excellent example of column complementarity being able to separate all peptides in a peptide mixture, when the individual columns are unable to achieve resolution of all peptides. Thus, peptides co-eluted on the polar-endcapped Hydro column (AmC-Y/AmC-V/AmN-V and AcN-Y/AmN) were resolved on the polar-embedded Fusion column (follow arrows in Fig. 4); in contrast, peptides AcC-Y/AcC-V/ AcN-V which were co-eluted on the Fusion column were completely resolved on the Hydro column (follow arrows in Fig. 4).
Fig. 4.
Effect of stationary phase (Hydro, Fusion) on SCMSV peptide elution profiles with acetonitrile as organic modifier. Peptide sequences shown in Table 1. HPLC conditions shown in Section 2.3.
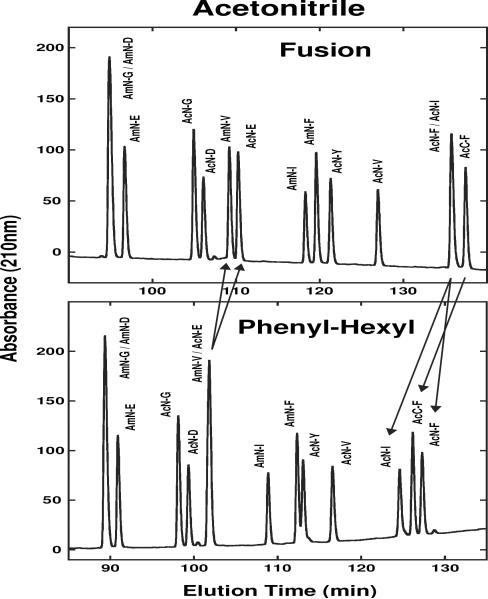
Fig. 5 compares elution profiles of a 14-peptide mixture of SCMSV peptides on the Fusion (top), and Phenyl-Hexyl (bottom) columns with acetonitrile as organic modifier. Again, individual columns were not able to resolve completely all peptides in the peptide mixture but, when employed independently, the two columns would enable the identification/separation of all peptides due to selectivity differences for closely-eluted peptides. Thus, peptides AmN-V/AcN-E, co-eluted on the Phenyl-Hexyl column, are resolved to baseline on the Fusion column; conversely, peptides AcN-F/AcN-I, co-eluted on the Fusion column, are resolved on the Phenyl-Hexyl column. Note also the reversal in elution order of AcN-F and AcC-F between the two columns. The greater retention of the aromatic residue-substituted peptides (Phe, Tyr) compared to the alkyl-substituted peptides on the Phenyl-Hexyl column relative to the Fusion column is likely due to π-π interactions of the aromatic side-chains with the phenyl groups on the stationary phase, as has been observed previously [15]. This would lead to the separation of AcC-F from AcN-I on the Phenyl-Hexyl column, as well as the movement of AmN-F and AcN-Y relative to AmN-I and AcN-V on this column (AmN-F and AcN-Y elutions are delayed on the Phenyl-Hexyl column compared relative to AmN-I compared to the Fusion column).
Fig. 5.
Effect of stationary phase (Fusion, Phenyl-Hexyl) on SCMSV peptide elution profile with acetonitrile as organic modifier. Peptide sequences shown in Table 1. HPLC conditions shown in Section 2.3.
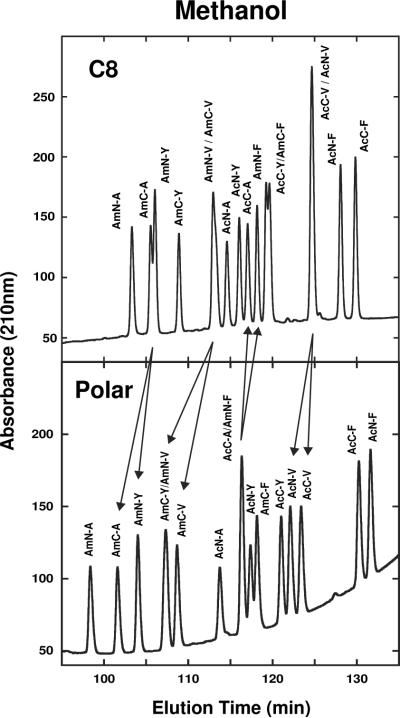
Fig. 6 compares elution profiles of the same 16-peptide mixture seen in Fig. 4 on the C8 (top) and Polar (bottom) columns in the presence of methanol. Again, selectivity differences between the two stationary phases allows manipulation of the elution profile of the peptide standards. Examples include the poor separation/co-elution of peptides AmC-A/AmN-Y, AmN-V/AmC-V and AcN-V/AcC-V on the C8 column but their resolution from each other on the Polar column (follow arrows in Fig. 6); conversely, co-eluted AcC-A/AmN-F on the Polar column were resolved to baseline on the C8 column. Note again the reversal in elution order of AcN-F and AcC-F between the two columns.
Fig. 6.
Effect of stationary phase (C8, Polar) on SCMSV peptide elution profiles with methanol as organic modifier. Peptide sequences shown in Table 1. HPLC conditions shown in Section 2.3.
3.4.2 Effect of varying organic modifier on separation of peptide standards
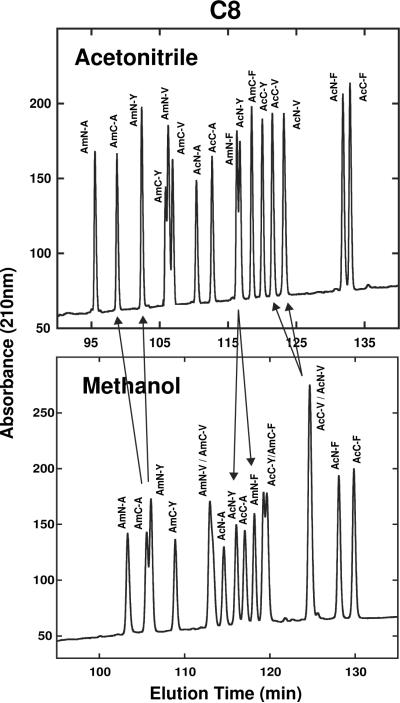
Fig. 7 compares elution profiles of the same 16-peptide mixture seen in Figs. 4 and 6 in the presence of acetonitrile (top) or methanol (bottom) on the C8 column. Again, useful complementarity between the profiles is apparent. Representative selectivity differences between the two systems include the co-elution of peptides AmC-A/AmN-Y and AcN-V/AcC-V in methanol and their separation in acetonitrile (follow arrows in Fig. 7); and the poor resolution of AmN-F/AcN-Y in acetonitrile with complete separation in methanol (follow arrows).
Fig. 7.
Effect of organic modifier on SCMSV peptide elution profiles on the C8 column. Peptide sequences shown in Table 1. HPLC conditions shown in Section 2.3.
An interesting observation from Fig. 8 is the resolution of peptide AmN-G from AmN-D on the C8 column in methanol. Although not separated completely to baseline, this particular pair of peptides proved to be extremely difficult to resolve. Indeed between the six different columns and the two organic modifiers, the profile shown in Fig. 8 (bottom) was the sole occasion that this separation was achieved. Also note the excellent resolution in methanol of AcN-Y/AmN-F/AmN-I compared to the poor resolution/co-elution of these three peptides in acetonitrile.
Fig. 8.
Effect of organic modifier on SCMSV peptide elution profiles on the C8 column. Peptide sequences shown in Table 1. HPLC conditions shown in Section 2.3.
Finally, Fig. 9 shows complementary resolution of the 16-peptide mixture on the Polar column in the presence of acetonitrile (top) and methanol (bottom). There are some particularly large shifts in relative retention times between the two conditions (examples denoted by arrows for AmC-V/AmC-F, which are coeluted in acetonitrile but widely separated in methanol; and AcC-A/AmN-F, which are coeluted in methanol but widely separated in acetonitrile), together with switches in elution order (e.g., AmC-Y/AmC-V), leading to considerable, and useful, changes in the elution profiles.
Fig. 9.
Effect of organic modifier on SCMSV peptide elution profiles on the Polar column. Peptide sequences shown in Table 1. HPLC conditions shown in Section 2.3.
4. Conclusions
The present study reports our initial results of our de novo design of synthetic peptide standards with the same (amino acid) composition and minimal sequence variation (SCMSV). Such peptides, which may be predicted to be difficult to separate, were designed to be a potent test of the selectivity of reversed-phase packing materials. SCMSV peptide pairs were thus applied to RP-HPLC on a range of stationary phases (C8 and C18 alkyl, polar embedded, polar endcapped, ether-linked phenyl and phenyl-hexyl) in the presence of acetonitrile or methanol as organic modifier. In addition, mixtures of SCMSV peptide standards to assess overall capabilities of stationary phases to resolve complex peptide mixtures were also examined under various combinations of column and organic modifier. In general, SCMSV peptide pairs with the β-branched Val and Ile side-chains at position X were the most difficult to separate compared to SCMSV peptides containing the aromatic side-chains Tyr and Phe (compare Figs. 1 and 2). For example, the AmN-V/AmC-V peptide pair was co-eluted on the C18 and Hydro columns in acetonitrile and methanol but could be resolved on the Fusion column with a resolution of 1.0 or greater in either solvent (Table 3). The column containing aromatic groups (Phenyl-Hexyl and Polar) might be expected to resolve peptide pairs containing single aromatic residues (Tyr or Phe). In our SCMSV peptide pairs because of preferential retention of aromatic solutes. However, this was not the case. For example, the AcN-Y/AcC-Y pair was resolved on all six columns with the poorest resolution of 2.0 and 2.3 obtained on the Phenyl-Hexyl and Polar columns, respectively, in acetonitrile compared to a resolution of 6.1 on the Fusion column. In addition, the poorest resolution values of 3.7 and 3.4 for this peptide pair in methanol were obtained on the Phenyl-Hexyl and Polar columns, respectively, compared to a resolution of 5.2 on the Fusion column (Table 3). Similar results were obtained when the substituted aromatic residue was Phe instead of Tyr (AcN-F/AcC-F), albeit the resolution values were always significantly greater for the Tyr-substitutes peptide pair compared to the Phe-substituted peptide pair (Table 3). It is interesting that the addition of the hydroxyl group to the aromatic ring in the 12-residue Tyr peptide pair would have such a dramatic effect on resolution compared to the Phe peptide pair. In general, the Fusion column provided the best resolution of the ten SCMSV peptide pairs on the six columns tested in both acetonitrile and methanol. However, the C18 (in acetonitrile), Hydro (in methanol) and Polar (in methanol) columns did provide the best resolution for specific peptide pairs (Table 3).
The potential of these novel standards and the complementarity of different columns and mobile phase conditions was made clear as was the value to the researcher of having access to more than one stationary phase type, coupled with changes in organic modifier, to maximize flexibility in analytical and preparative peptide RP-HPLC applications. Finally, although the results of this early study were essentially qualitative in nature, our controlled, de-novo designed peptide approach may also spur the development of more quantitative selectivity parameters for peptide separations, such as those already available for small molecules, enhancing further the universal value of utilizing peptide standards to compare column performances in the separation of peptide mixtures.
Highlights
-
>
This paper introduces a new generation of reversed-phase HPLC peptide standards
-
>
These new standards vary only subtly in amino acid sequence
-
>
The standards allow a potent test of selectivity of reversed-phase packings
Acknowledgements
This work was supported by an NIH grant (RO1 GM61855) to Robert. S. Hodges and the John Stewart Chair in Peptide Chemistry to Robert S. Hodges. We are grateful to Tivadar Farkas of Phenomenex for donating the reversed-phase columns.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Mant CT, Parker JMR, Hodges RS. J. Chromatogr. 1987;397:99. doi: 10.1016/s0021-9673(01)84993-1. [DOI] [PubMed] [Google Scholar]
- [2].Hodges RS, Parker JMR, Mant CT, Sharma RR. J. Chromatogr. 1988;458:147. doi: 10.1016/s0021-9673(00)90560-0. [DOI] [PubMed] [Google Scholar]
- [3].Mant CT, Hodges RS, editors. HPLC of Peptides and Proteins: Separation Analysis and Conformation. CRC Press, Boca Ration; FL: 1991. [Google Scholar]
- [4].Mant CT, Chao H, Hodges RS. J. Chromatogr. A. 1997;791:85. doi: 10.1016/s0021-9673(97)00767-x. [DOI] [PubMed] [Google Scholar]
- [5].Mant CT, Hodges RS. In: HPLC of Biological Macromolecules. Gooding KM, Regnier FE, editors. Marcel Dekker; New York: 2002. p. 433. [Google Scholar]
- [6].Burke TWL, Mant CT, Black JA, Hodges RS. J. Chromatogr. 1989;476:377. doi: 10.1016/s0021-9673(01)93883-x. [DOI] [PubMed] [Google Scholar]
- [7].Mant CT, Hodges RS. J. Sep. Sci. 2008;31:1573. doi: 10.1002/jssc.200700619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Mant CT, Hodges RS, Magazine LC. Liq. Chromatogr. HPLC. 1986;4:250. [Google Scholar]
- [9].Mant CT, Hodges RS. Chromatographia. 1987;24:805. [Google Scholar]
- [10].Mant CT, Burke TWL, Black JA, Hodges RS. J. Chromatogr. 1988;458:193. doi: 10.1016/s0021-9673(00)90564-8. [DOI] [PubMed] [Google Scholar]
- [11].Sereda TJ, Mant CT, Quinn AM, Hodges RS. J. Chromatogr. 1993;646:17. doi: 10.1016/s0021-9673(99)87003-4. [DOI] [PubMed] [Google Scholar]
- [12].Shibue M, Mant CT, Hodges RS. J. Chromatogr. A. 2005;1080:49. doi: 10.1016/j.chroma.2005.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Shibue M, Mant CT, Hodges RS. J. Chromatogr. A. 2005;1080:58. doi: 10.1016/j.chroma.2005.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Shibue M, Mant CT, Hodges RS. J. Chromatogr. A. 2006;1080:68. doi: 10.1016/j.chroma.2005.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Mant CT, Cepeniene D, Hodges RS. J. Sep. Sci. 2010;33:3005. doi: 10.1002/jssc.201000518. [DOI] [PubMed] [Google Scholar]
- [16].Mant CT, Burke TWL, Hodges RS. Chromatographia. 1987;24:565. [Google Scholar]
- [17].Mahoney WC. Biochim. Biophys. Acta. 1982;704:284. doi: 10.1016/0167-4838(82)90158-3. [DOI] [PubMed] [Google Scholar]
- [18].Vervoort RJM, Debets AJJ, Claessens HA, Cramers CA, de Jong GJ. J. Chromatogr. A. 2000;897:1. doi: 10.1016/s0021-9673(00)00811-6. [DOI] [PubMed] [Google Scholar]
- [19].Snyder LR, Dolan JW, Carr PW. J. Chromatogr. A. 2004;1060:77. [PubMed] [Google Scholar]
- [20].Stella C, Rudaz S, Veuthey J-L, Tchalpa A. Chromatographia. 2001;53:S132. [Google Scholar]
- [21].Claessens HA. TRAC. 2001;20:563. [Google Scholar]
- [22].Neue UD. J. Sep. Sci. 2007;30:1611. doi: 10.1002/jssc.200700082. [DOI] [PubMed] [Google Scholar]
- [23].Nomura A, Yamada J, Tsunoda K. Anal. Sci. 1987;3:209. [Google Scholar]
- [24].Ascah TL, Feibush B. J. Chromatogr. 1990;506:357. [Google Scholar]
- [25].Buszewski B, Schmid J, Albert K, Bayer E. J. Chromatogr. 1991;552:415. doi: 10.1016/s0021-9673(01)95957-6. [DOI] [PubMed] [Google Scholar]
- [26].Jaroniec M. J. Chromatogr. A. 1993;656:37–50. [Google Scholar]
- [27].Buszewski B, Jaroniec M, Gilpin RK. J. Chromatogr. A. 1994;668:293. [Google Scholar]
- [28].O'Gara JE, Alden BA, Walter TH, Petersen JS, Niederländer CL, Neue UD. Anal. Chem. 1995;67:3809. [Google Scholar]
- [29].Ascah TL, Kallury KML, Szafranski CA, Corman SD, Lui F. J. Liq. Chromatogr. Relat. Technol. 1996;19:3049. [Google Scholar]
- [30].Jaroniec CP, Gilpin RK, Jaroniec M. J. Chromatogr. A. 1998;797:103. [Google Scholar]
- [31].Sándi A, Szepesy L. J. Chromatogr. A. 1998;818:1. [Google Scholar]
- [32].McCalley DV. J. Chromatogr. A. 1999;844:23. [Google Scholar]
- [33].O'Gara JE, Walsh DP, Alden BA, Casellini P, Walter TH. Anal. Chem. 1999;71:2992. doi: 10.1021/ac9900331. [DOI] [PubMed] [Google Scholar]
- [34].Neue UD, Serowik E, Iraneta P, Alden BA, Walter TH. J. Chromatogr. A. 1999;849:87. [Google Scholar]
- [35].Neue UD, Alden BA, Walter TH. J. Chromatogr. A. 1999;849:101. [Google Scholar]
- [36].Morrison RD, Dolan JW. LC.GC. 2000;9:936. [Google Scholar]
- [37].Neue UD, Cheng Y-F, Lu Z, Alden BA, Iraneta PC, Phoebe CH, Van Tran K. Chromatographia. 2001;54:169. [Google Scholar]
- [38].Engelhardt H, Grüner R, Scherer M. Chromatographia. 2001;5:S154. [Google Scholar]
- [39].O'Gara JE, Walsh DP, Phoebe CH, Jr., Alden BA, Bouvier ESP, Iraneta PC, Capparella M, Walter TH. LC.GC. 2001;19:632. [Google Scholar]
- [40].Majors RE, Przybyciel M. LC.GC. 2002;7:584. [Google Scholar]
- [41].Silva CT, Bachmann S, Schefer RR, Albert K, Jardim ICSF, Airoldi C. J. Chromatogr. A. 2002;948:85. doi: 10.1016/s0021-9673(01)01263-8. [DOI] [PubMed] [Google Scholar]
- [42].Layne J. J. Chromatogr. A. 2002;957:149. doi: 10.1016/s0021-9673(02)00193-0. [DOI] [PubMed] [Google Scholar]
- [43].Silva CR, Jardim ICSF, Airoldi C. J. Chromatogr. A. 2003;987:127. doi: 10.1016/s0021-9673(02)01807-1. [DOI] [PubMed] [Google Scholar]
- [44].Silva CR, Jardim ICSF, Airoldi C. J. Chromatogr. A. 2003;987:139. doi: 10.1016/s0021-9673(02)01947-7. [DOI] [PubMed] [Google Scholar]
- [45].Euerby MR, Petersson P. J. Chromatogr. A. 2003;994:13. doi: 10.1016/s0021-9673(03)00393-5. [DOI] [PubMed] [Google Scholar]
- [46].McCalley DV. J. Sep. Sci. 2003;26:187. [Google Scholar]
- [47].Wilson NS, Gilroy J, Dolan JW, Snyder LR. J. Chromatogr. A. 2004;1026:91. doi: 10.1016/j.chroma.2003.11.041. [DOI] [PubMed] [Google Scholar]
- [48].Silva CR, Collins CH, Jardin ICSF, Airoldi C. J. Chromatogr. A. 2004;1030:157. doi: 10.1016/j.chroma.2003.11.016. [DOI] [PubMed] [Google Scholar]
- [49].Hamada T, Tanaka H, Izumine H, Ohira M. J. Chromatogr. A. 2004;1043:27. doi: 10.1016/j.chroma.2004.01.018. [DOI] [PubMed] [Google Scholar]
- [50].Euerby MR, Petersson P. J. Chromatogr. A. 2005;1088:1. doi: 10.1016/j.chroma.2004.10.027. [DOI] [PubMed] [Google Scholar]
- [51].Le Mapihan K, Vial J, Jardy A. J. Chromatogr. A. 2005;1088:16. doi: 10.1016/j.chroma.2005.02.023. [DOI] [PubMed] [Google Scholar]
- [52].Lesellier E, West C, Tchapla A. J. Chromatogr. A. 2006;1111:62. doi: 10.1016/j.chroma.2006.01.107. [DOI] [PubMed] [Google Scholar]
- [53].Liu X, Bordunov A, Tracy M, Slingsby R, Avdalovic N, Pohl C. J. Chromatogr. A. 2006;1119:120. doi: 10.1016/j.chroma.2005.12.097. [DOI] [PubMed] [Google Scholar]
- [54].Neu UD, O'Gara JE, Méndez A. J. Chromatogr. A. 2006;1127:161. doi: 10.1016/j.chroma.2006.06.006. [DOI] [PubMed] [Google Scholar]
- [55].Jing L-L, Jiang R, Liu P, Wang P-A, Shi T-Y, Sun X-L. J. Sep. Sci. 2009;32:212. doi: 10.1002/jssc.200800468. [DOI] [PubMed] [Google Scholar]
- [56].Tanaka N, Tokuda Y, Iwaguchi K, Araki M. J. Chromatogr. 1982;239:761. [Google Scholar]
- [57].Hanai T, Hubert J. J. Chromatogr. 1984;291:81. [Google Scholar]
- [58].Antle PE, Goldberg AP, Snyder LR. J. Chromatogr. 1985;321:1. [Google Scholar]
- [59].Glajch JL, Gluckman JC, Charikofsky JG, Minor JJ, Kirkland JJ. J. Chromatogr. 1985;318:23. [Google Scholar]
- [60].Mant CT, Zhou NE, Hodges RS. J. Chromatogr. 1989;476:363. doi: 10.1016/s0021-9673(01)93882-8. [DOI] [PubMed] [Google Scholar]
- [61].Thevenon-Emeric G, Tchalpa A, Martin M. J. Chromatogr. 1991;550:267. [Google Scholar]
- [62].Lean J, Reubsaet E, Vieskar R. J. Chromatogr. A. 1999;841:147. [Google Scholar]
- [63].Croes K, Steffens A, Marchand DH, Snyder LR. J. Chromatogr. A. 2005;1098:123. doi: 10.1016/j.chroma.2005.08.090. [DOI] [PubMed] [Google Scholar]
- [64].Marchand DH, Croes K, Dolan JW, Snyder LR, Henry RA, Kallury KMR, Waite S, Carr PW. J. Chromatogr. A. 2005;1062:65. doi: 10.1016/j.chroma.2004.11.014. [DOI] [PubMed] [Google Scholar]
- [65].Jardim ICSF, Maldaner L, Lourenço J, Fioravanti LMA, Collins CH. J. Sep. Sci. 2010;33:2917. doi: 10.1002/jssc.201000313. [DOI] [PubMed] [Google Scholar]
- [66].Tripet B, Wagschal K, Lavigne P, Mant CT, Hodges RS. J. Mol. Biol. 2000;300:377. doi: 10.1006/jmbi.2000.3866. [DOI] [PubMed] [Google Scholar]
- [67].Tripet B, Cepeniene D, Kovacs JM, Mant CT, Krokhin OV, Hodges RS. J. Chromatogr. A. 2007;1141:212. doi: 10.1016/j.chroma.2006.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Chen Y, Mant CT, Hodges RS. J. Chromatogr. A. 2007;1140:112. doi: 10.1016/j.chroma.2006.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Zhou NE, Mant CT, Hodges RS. Peptide Res. 1990;3:8. [PubMed] [Google Scholar]
- [70].Houghten RA, DeGraw ST. J. Chromatogr. 1987;385:223. doi: 10.1016/s0021-9673(01)94599-6. [DOI] [PubMed] [Google Scholar]
- [71].Blondelle SE, Ostresh JM, Houghten RA, Pérez-Payá E. Biophys, J. 1995;68:351. doi: 10.1016/S0006-3495(95)80194-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Purcell AW, Aguilar MI, Wettenhall REW, Hearn MTW. Pept. Res. 1995;8:160. [PubMed] [Google Scholar]
- [73].Steer DL, Thompson PE, Blondelle SE, Houghten RA, Aguilar MI. J. Pept. Res. 1998;51:401. doi: 10.1111/j.1399-3011.1998.tb00638.x. [DOI] [PubMed] [Google Scholar]
- [74].Ostresh JM, Büttner K, Houghten RA. In: HPLC of Peptides and Proteins: Separation Analysis and Conformation. Mant CT, Hodges RS, editors. CRC Press, Boca Raton; FL; 1991. p. 633. [Google Scholar]
- [75].Zhou NE, Monera OD, Kay CM, Hodges RS. Protein Pept. Lett. 1994;1:114. [Google Scholar]
- [76].Monera OD, Sereda TJ, Zhou NE, Kay CM, Hodges RS. J. Pept. Sci. 1995;1:319. doi: 10.1002/psc.310010507. [DOI] [PubMed] [Google Scholar]
- [77].Kovacs JM, Mant CT, Hodges RS. Biopolymers (Pept. Sci.) 2006;84:298. doi: 10.1002/bip.20417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Mant CT, Kovacs JM, Kim H-M, Pollock DD, Hodges RS. Biopolymers (Pept. Sci.) 2009;92:573. doi: 10.1002/bip.21316. [DOI] [PMC free article] [PubMed] [Google Scholar]