Abstract
We review studies of subunit III-depleted cytochrome c oxidase (CcO III (−)) that elucidate the structural basis of steady-state proton uptake from solvent into an internal proton transfer pathway. The removal of subunit III from R. sphaeroides CcO makes proton uptake into the D pathway a rate-determining step, such that measurements of the pH dependence of steady-state O2 consumption can be used to compare the rate and functional pKa of proton uptake by D pathways containing different initial proton acceptors. The removal of subunit III also promotes spontaneous suicide inactivation by CcO, greatly shortening its catalytic lifespan. Because the probability of suicide inactivation is controlled by the rate at which the D pathway delivers protons to the active site, measurements of catalytic lifespan provide a second method to compare the relative efficacy of proton uptake by engineered CcO III (−) forms. These simple experimental systems have been used to explore general questions of proton uptake by proteins, such as the functional value of an initial proton acceptor, whether an initial acceptor must be surface-exposed, which side chains will function as initial proton acceptors and whether multiple acceptors can speed proton uptake.
Keywords: Cytochrome c oxidase, Proton uptake, Proton transfer, D pathway, Proton acceptor
A reasonably large number of enzymes and pumps carry out long-range proton transfer (i.e. >10 Å) [1, 2]. Pathways leading to an internal proton sink often begin with an initial proton acceptor, a protein side chain that accepts a proton from solvent and transfers it into the pathway. The proton transfer pathway leading from the initial acceptor to the proton sink is composed of hydrogen-bonded waters, hydrogen-bonded amino acid side chains with exchangeable protons, or (most often) a combination of these two components. With some exceptions, protons are thought to move rapidly along these hydrogen-bonded chains of proton carriers by non-ionic Grotthuss-type proton transfer, in which a carrier simultaneously acquires a proton from the preceding carrier while releasing a proton to the next [3].
For each O2 consumed, the aa3-type cytochrome c oxidases (CcO) of mitochondria and α proteobacteria transfer four protons from the inner surface of the complex to the heme a3-CuB O2 reduction site, located within the transmembrane region of subunit I, and four protons to a nearby site for proton pumping. Two long proton transfer pathways, termed the K and D pathways, carry out this function [2–5]. The K pathway takes up two substrate protons (those protons used in the reduction of O2 to H2O) during the reduction of heme a3 and CuB prior to O2 binding [6–8]. The initial proton acceptor of the K pathway is Glu-101 of subunit II [9, 10]. (Residue numbers of the aa3-type CcO of R. sphaeroides are used throughout.) From Glu-101 a pathway composed of amino acid side chains and internal waters transfers the protons to the active site [4, 5].
The D pathway takes up the remaining two substrate protons of the catalytic cycle (CC) plus all four of the pump protons (those destined to be pumped through the protein, across the membrane) [3–6]. The D-pathway begins at Asp-132 of subunit I and extends approximately 26 Å to Glu-286, close to the active site [3, 4] (Figure 1). From Glu-286 protons are directed, alternately, to the heme a3-CuB center for O2 reduction chemistry or to the central proton pumping element, likely in the region of the D propionate of heme a3 [11, 12]. The pathway for protons between Asp-132 and Glu-286 appears to consist entirely of a series of waters whose positions in CcO structures are highly conserved [12–15]. In the crystal structures of bacterial and mitochondrial CcO, the amide side chain of a conserved asparagine (Asn-139) interrupts the water chain, creating what should be a block for proton transfer [14–16]. However, molecular dynamics simulations show that the amide side chain and the internal waters of the D pathway are in motion, which allows formation of a continuous water path for proton conduction [15].
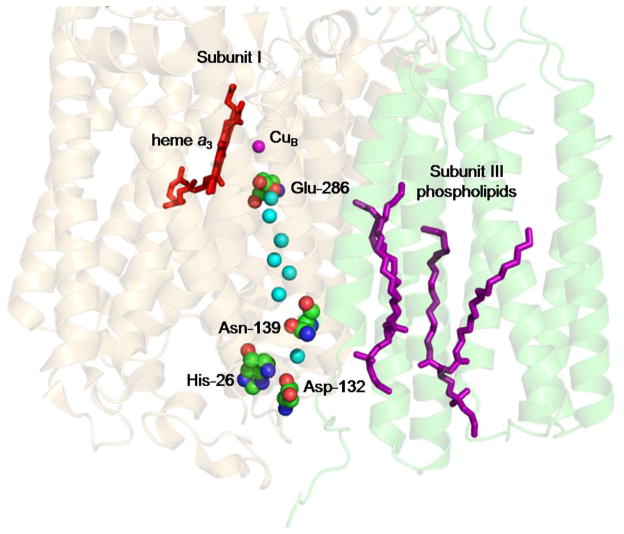
Figure 1.
The D pathway of the aa3-type CcO of Rhodobacter sphaeroides. Subunit I is shadowed in brown, subunit III in green. The two phospholipids bound within the cleft of subunit III are in purple. The structure shown is PDB 1M56 [17] since this is the only structure to date of wild-type R. sphaeroides CcO that resolves subunit III to high resolution. Seven waters of the D pathway are shown in cyan.
Asp-132, the initial proton acceptor of the D pathway, is located at an interface of subunits I and III [17, 18]. Only one oxygen of its carboxyl group is exposed at the bottom of a well-like depression in the protein surface, a structure that limits the exposure of Asp-132 to bulk solvent. Half of the residues lining the well are from subunit I and half from subunit III. Subunit III is one of the three proteins, along with subunits I and II, that make up the catalytic core of mitochondrial CcO and the aa3-type CcOs of α proteobacteria [19, 20]. Like subunit I, subunit III is almost entirely transmembrane, with seven helices compared to the twelve of subunit I, and it binds to a face of subunit I opposite from the face that binds the two transmembrane helices of subunit II [17, 21]. Subunit III contains no redox centers but it does bind two phospholipids in highly conserved binding sites located in a deep cleft [17, 22, 23]. Interactions of subunit I with these lipids provides much of the binding interaction between the two subunits [23]. The relatively weak association of subunit III with subunit I allows its removal from CcO by relatively mild procedures with the retention of O2 reduction activity by the remaining complex [24–27]. The removal of subunit III has been found to lower the stoichiometry of proton pumping of bacterial and mitochondrial CcO [26, 28–30] but not always [31]. The reason for the observed changes in proton pumping remains to be elucidated. It should be noted that this review focuses on the uptake of substrate protons; the topic of proton pumping is not further addressed.
Two of the effects of removing subunit III from CcO are particularly relevant to the study of proton uptake. First, the absence of subunit III increases the exposure of Asp-132 to bulk solvent, lowers its pKa and makes proton uptake into the D pathway a rate determining step at pH values >6.5 [20, 28, 32, 33]. Thus, the rate and pH dependence of O2 reduction activity by WT III (−) reflects the rate and pH dependence of proton uptake into the D pathway [27]. The evidence for this has been detailed in previous studies and is further reviewed below.
A second consequence of the removal of subunit III is a dramatic increase in the probability of spontaneous mechanism-based inactivation (suicide inactivation) during catalytic turnover [24, 34]. In other words, normally subunit III lowers the probability of suicide inactivation and thereby extends the catalytic lifespan (the number of catalytic cycles until irreversible inactivation) of CcO by 600-fold or more [24, 35]. The source of suicide inactivation has been traced to a catastrophic event at the heme a3-CuB active site [24, 35] that results in the loss of CuB. The phenomenon of suicide inactivation is not restricted to bacterial oxidases; the removal of subunit III also leads to the inactivation of rat liver and bovine heart CcO during turnover (Thompson & Ferguson-Miller; Hill & Hosler; Prochaska; unpublished data). The ability of subunit III to prevent the premature loss of this major energy conserving machine of the cell may explain why it is as well conserved as subunit I, the core catalytic subunit [20]. The effect of subunit III on proton uptake into the D pathway and the effect of subunit III on catalytic lifespan are linked: slow proton uptake promotes suicide inactivation [35]. Because of this, measurements of catalytic lifespan (detailed below) can also be used to compare the relative proton uptake capabilities of D pathways of different CcO forms. (It is useful to note that due to the weak binding of subunit III, most preparations of bacterial aa3-type CcOs contain substoichiometric amounts of the subunit. Although the subunit III content of CcO preparations is rarely measured, it can be a source of experimental variability, especially in mutant CcO forms that further weaken the subunit I–III interaction.)
The processes by which protons are transferred from solvent into the protein, crossing the barrier from outside to inside, can be viewed as a process distinct from that of proton transfer within the protein. Subunit III-depleted CcO forms are useful tools for the study of proton uptake because 1) proton uptake into the D pathway becomes rate-determining in the absence of subunit III, and 2) the entry region of the D pathway is more exposed to bulk solvent, which allows the introduction of alternative initial proton acceptors. This review focuses on the insights gained from analyses of proton uptake by CcO III (−) in WT and mutant forms during continuous catalytic turnover. Measurements of the pH dependence of O2 reduction activity and catalytic lifespan are compared with results obtained from single catalytic cycle (CC) experiments [5] yielding further insight. The enzyme used is the aa3-type CcO of the bacterium Rhodobacter sphaeroides, one of the well-studied bacterial models of mitochondrial CcO [19, 20].
1. Using steady-state activity to study proton uptake
1.1. In the absence of subunit III, steady-state CcO activity can be a measure of the rate of steady-state proton uptake by the D pathway
In the absence of subunit III proton uptake into the D pathway has been shown to be a rate-determining step, as explained below. During the four electron reduction of one O2 to two waters (a single catalytic cycle, CC) following the photo-dissociation of CO from fully-reduced CcO, two substrate protons are taken up via the D pathway, one during the P→F transition and one during the final F→O transition [5]. In single CC experiments where proton uptake was measured directly using a pH-sensitive dye, the removal of subunit III from WT CcO strongly inhibited the rate of proton uptake into the D pathway at pH 8, but the rate of proton uptake returned to normal at pH 5.5 [32]. This indicated that the removal of subunit III alters the pH dependence of proton uptake, which was further examined by determining the pH dependence of the F→O transition, since this partial reaction includes the transfer of a proton from bulk solvent to the active site via the D pathway [5]. As expected from the direct measurements of proton uptake, the rate of the F→O transition was low at pH 8 but normal at lower pH [32]. The pH dependence of the F→O transition for WT III (−) showed an apparent pKa ~7, significantly shifted from the apparent pKa of 8.6 measured for the F→O transition of WT III (+). In further experiments, the rate and pH dependence of the F→O transition was used to compare proton uptake by D pathways of CcO III (−) forms with widely variant situations for proton capture at the protein surface [33]. These experiments showed that the F→O transition was rate limited by the proton uptake event, i.e. by the transfer of a proton into the D pathway. Moreover, it was determined that the pH profile of the F→O transition primarily or solely reflects the proton affinity of the initial acceptor. For example, the apparent pKa ~7 measured for the F→O transition in WT III (−) reflects the affinity of Asp-132 for protons in the absence of subunit III [32, 33] (Table 1).
Table 1.
Proton uptake and activity of purified CcO III (−) forms with different initial proton acceptors for the D pathway
| CcO form | Initial proton acceptor of D pathway | Apparent pKa of single proton uptake during the F→O transition | Functional pKa of steady-state proton uptakea | TNmax of cytochrome c-driven O2 consumption at pH 6.5 (% of WT III (+)) |
|---|---|---|---|---|
| WT III (+) | Asp-132 | 8.6 [32] | 8.4 (8.3–8.5)b [20, 27, 28] | 2062 (100) |
| WT III (−) | Asp-132 | ~7 [32, 33] | 7.0–7.3 [20, 27, 28] | 1835 (89) |
| D132A/H III (−) | None | >10 [33] | >9c [33] | ~400 (19) |
| D132A-H26A III (−) | None | ND | >9c [33] | 793 (40) |
| WT III (−) + Aa | Asp-132 +Aa? | 7.6 [33] | 7.8 (7.7–7.9) [28] | 1479 (72) |
| N139D-D132N III(−) | Asp-139 | ND | 7.7 (7.5–7.9) [27] | 1799 (87) |
| N139C-D132N III(−) | Cys-139 | ND | 8.0 (7.9–8.1) [27] | 1587 (77) |
| N139D III(−) | Asp-139 & Asp-132 | ND | 8.3 (8.2–8.4) [27] | 2505 (121) |
| N139C III(−) | Cys-139 & Asp-132 | ND | 8.2 (8.2–8.3) [27] | 2256 (109) |
| H26D-D132A III(−) | Asp-26 | ND | 7.0 (6.9–7.1) [this report] | 1568 (76) |
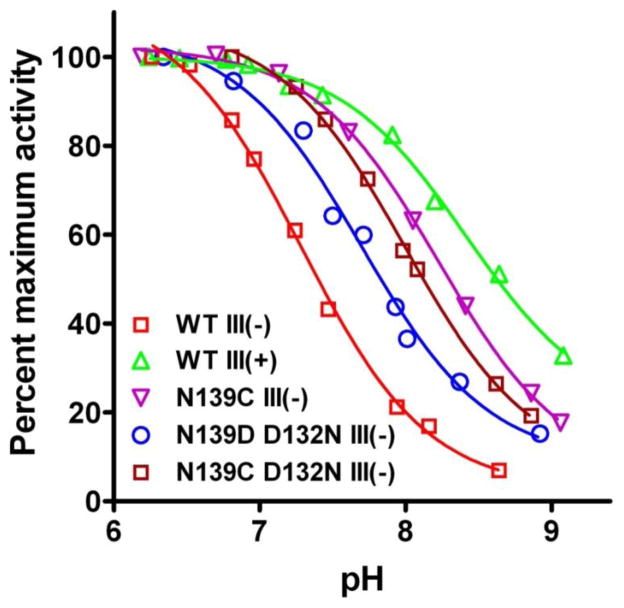
It was also discovered that the pH profile of steady-state O2 reduction by WT III (−) (Figure 2) mirrors that of the pH profile for the F→O transition [27, 28, 32, 33]. Moreover, the rates of steady-state O2 reduction [27] match those of the F→O transition [32]. These findings indicate that proton uptake into the D pathway limits continuous, steady-state O2 reduction of WT III (−) in the same way that it limits the rate of the F→O step during a single catalytic cycle of the enzyme. Therefore, the apparent pKa of steady-state O2 reduction (7–7.3) also reports the proton affinity of Asp-132 (Table 1). Importantly, the apparent pKa of steady-state O2 reduction tracks with the steady-state pKa of the F→O transition when the initial proton acceptor of the D pathway is altered. For example, the carboxyl group of the fatty acid arachidonic acid (Aa) can substitute for the carboxyl of Asp-132 if Asp-132 is missing; when Asp-132 is present the addition of Aa raises the apparent pKa of proton uptake, presumably via electrostatic interaction of the carboxyl groups [33]. As the apparent pKa of the F→O transition of WT III (−) rises from 7 to 7.6 upon the addition of Aa [33], the apparent pKa of steady-state O2 reduction also rises, from 7–7.3 to 7.8 [28](Table 1).
Figure 2.
pH dependence of steady-state activity by wild-type CcO and CcO III (−) forms with different initial proton acceptors for the D pathway (see Table 1). The data indicate the distribution of functional pKas of proton uptake and the extension of the pH range of enzyme activity afforded by manipulation of the proton uptake step. Each data set, taken from Varanasi & Hosler [27], is normalized by setting the highest measured activity as 100%; fits are to the Henderson-Hasselbalch function. The functional pKa values are presented in Table 1.
These experiments show that measurements of the pH dependence of steady-state O2 reduction in CcO lacking subunit III can be used, with some limitations, to compare the ability of different groups to transfer solvent protons into the D pathway. Because proton uptake from solvent is rate-determining in the III (−) forms, the pH dependence of steady-state activity reflects the proton affinity of the initial proton acceptor. The pH vs. activity curves do not yield a true, equilibrium pKa for the initial acceptor, but rather its ‘functional’ pKa which depends upon the inherent pKa of the group and the rates at which it is protonated and deprotonated during steady-state O2 reduction. The normal unaltered K pathway of CcO appears to be able to deliver protons to the active site, in steps where it is required, more rapidly than the D pathway [27]. Therefore, proton uptake by the K pathway is not reflected in the pH dependence of steady-state activity by the subunit III-depleted CcO forms. (It should be noted that an exception is D132A III (−). Here, the extremely high functional pKa of the D pathway (>10) [33] allows a different pH-dependent process, likely the K pathway, to control steady-state O2 reduction at physiologic pH values [28].)
We present below a brief set of empirical conclusions about the process of proton uptake gleaned from analyses of steady-state O2 reduction by subunit III-depleted CcO forms. Along with knowledge obtained from other enzymes and from computational approaches, such information may be useful for understanding proton pathway design and the physiological regulation of proton pathways in proteins.
1.2. Is a protein-supplied proton acceptor at the beginning of the pathway required for physiologic rates of proton uptake?
Not necessarily. D132A III (−) appears to lack a protein group that functions as an initial proton acceptor [33]. His-26, the neighbor of Asp-132 (Figure 1), does not substitute as an initial proton acceptor in the absence of Asp-132 [33]. The functional pKa of proton uptake by D132A III (−) is high (>10) indicating that proton uptake is driven by the consumption of protons by the proton sink of the pathway, i.e. Glu-286 and the high pKa groups that form during O2 reduction at the active site [33]. Apparently the removal of subunit III, along with the alteration of Asp-132 to alanine, allows sufficient space for a short file of waters to transfer protons from solvent to the first crystal water of the D pathway, seen below Asn-139 (Figure 1). Therefore the D pathway of D132A III (−) is likely composed of only water from solvent to Glu-286. Nonetheless, the pathway achieves a rate of proton uptake of ~400 s−1 (Table 1) [33]. Moreover, when both His-26 and Asp-132 are altered to alanine, the rate of proton uptake doubles (D132A-H26A III (−); Table 1), suggesting that a water file that transfers protons from solvent forms more easily in the additional space created by the double mutant. While D132A III (−) and D132A-H26A III (−) are engineered proteins, one of the pathways proposed for proton transfer to the quinone binding site of E. coli succinate dehydrogenase is composed solely of coordinated waters [36, 37]. Given the rates of catalytic turnover reported for this enzyme, a ‘water-only’ pathway cannot be discounted on the basis of kinetics.
1.3. Must the initial protein-supplied proton acceptor be exposed to bulk solvent in order to facilitate efficient proton uptake?
No. Asp-139 of N139D-D132N III (−) lies 6Å inside CcO, within the D pathway, where it functions as an efficient initial proton acceptor [27]. Protons are likely transferred to Asp-139 from solvent via a short chain of two to three waters. (Because protons are transferred from solvent to Asp-139 via water it might be questioned why Asp-139 is assigned as the “initial” proton acceptor rather than water. The assignment is based on three characteristics of Asp-139 [27]. One, the carboxyl of Asp-139 is the group that controls the rate and functional pKa of proton uptake. Two, Asp-139 is the first protein group of this D pathway. Three, Asp-139 is deprotonated to its anionic form as it transfers a proton. This distinguishes it from a Grotthuss-type proton transfer element, such as the waters leading to solvent.) The functional pKa of Asp-139 is greater than that of Asp-132 (Table 1; Figure 2), indicating that shielding the initial proton acceptor from bulk solvent increases its affinity for protons. This strategy likely elevates the pKa of Asp-132 in normal CcO [38], where subunit III shields the carboxyl group from bulk solvent.
1.4. What amino acid side chains can function as initial proton acceptors?
A survey of known and proposed proton transfer pathways indicates a preference for carboxyl side chains. Several characteristics of carboxyl side chains seem ideal for this role. Once protonated, the hydroxyl of a carboxyl group can rapidly move from an ‘outside’, solvent-exposed position to an ‘inside’ position close to the next element of the proton transfer chain by simple rotation of the carboxyl group about the terminal C-C bond of the side chain [39]. Native CcO exposes one oxygen of the Asp-132 carboxyl to solvent while the other oxygen remains internal, within hydrogen bond distance of the first water of the D pathway. Free rotation about the terminal C-C bond allows the two oxygens to rapidly trade places without disrupting the integrity of the protein surface [39]. An obvious advantage is that the negative charge of the deprotonated carboxyl group attracts protons. Moreover, molecular dynamics simulations show that carboxyl groups organize short water files within solvent that facilitate proton transfer to the carboxylate [40].
The imidazole side chain of surface-exposed histidine residues is another proton-carrying group often present at the sites of proton uptake. The imidazole group is argued to be well-suited for proton capture with a pKa near that of the pH of solvent [41]. In addition to rotation of the imidazole ring around the Cβ-Cγ bond, histidines can swing the entire imidazole function about Cβ. Such motion has been well-documented for carbonic anhydrase, where a histidine functions as the proton exit group by rotating 100 degrees about Cβ in order to transfer protons from an internal water network to bulk solvent [42]. Two histidines transfer protons into the pathway leading to QB of the bacterial photosynthetic reaction center [43]. Their side chains extend into bulk solvent where they are free to move. The alteration of both histidines to alanine inhibits proton uptake but leaves more residual proton uptake activity (as percent of normal) than does the removal of Asp-132 from the D pathway. Perhaps a surface-exposed aspartic acid, the next side chain of the QB pathway [44], provides an alternative entry point in the absence of the histidines.
The experimental data to date indicates that histidine does not readily substitute for the carboxyl proton acceptors of the D and K pathways of CcO. His-26 near Asp-132 does not become the initial proton acceptor in the absence of Asp-132 (e.g. in D132A III (−); [33], even though a carboxyl side chain at this position appears to function as an alternative proton acceptor (H26D-D132A; Table 1). Computer modeling indicates that movement of His-26 of D132A III (−) will be more restricted than that of Asp-126 in H26D-D132A III (−), which could prevent His-26 from participating in proton uptake. In another example, histidine cannot substitute for Asp-132 at position 132; D132H shows the same properties as D132A (unpublished data). Computer modeling shows energetically favorable rotamer positions for His-132 in ‘inside’ and ‘outside’ orientations that could cycle to carry out proton uptake, but the transition between these two rotamers is inhibited by surrounding protein. At the entry to the K pathway of CcO, His-96(II) fails to substitute for Glu-101(II) when the latter is altered to alanine [45, 46] or when Glu-101 itself is altered to histidine [45]. Steric restriction of histidine movement is a less obvious reason for the failure of histidine at these sites to facilitate proton uptake. It may be that the anionic nature of the Glu-101(II) is required to coordinate a crystal water at the entrance of the pathway because the carboxyl functions of arachidonic acid, cholate and deoxycholate will restore K pathway function in the absence of Glu-101 [46, 47].
Thus far, a naturally occurring proton transfer pathway using cysteine as the initial acceptor has not been identified, perhaps because the reactivity of the sulfhydryl group disfavors its use. Nevertheless, cysteine shares many of the useful characteristics listed above for carboxyl proton acceptors, e.g. the ability to reversibly protonate in the physiologic pH range, a negative charge in its deprotonated state that may help organize local water files and the ability to transfer a proton from ‘outside’ to ‘inside’ by rotation of the sulfhydryl about the terminal C-S bond. In fact, we have shown that a cysteine at position 139 of the D pathway will function as an initial acceptor [27]. In light of this, it is curious that cysteine at position 132 is completely nonfunctional (unpublished data).
1.5. Do multiple proton entry paths enhance proton uptake?
They can. The presence of Asp-132 at the entrance of the D pathway, plus either Asp-139 or Cys-139, creates two overlapping pathways for proton entry as long as subunit III is removed [27]. One pathway can be written as “solvent→Asp-132→H20→Asp-139 (Cys-139)→” and the other as “solvent→2–3 H2O→Asp-139 (Cys-139)→”. The presence of these two entry pathways speeds the uptake of substrate protons to rates greater than that of wild-type CcO (Table 1). At low pH (e.g. 6.5), where both acceptors are largely protonated, the two pathways are incompletely additive (i.e. the rate of proton uptake does not double). At high pH, however, the rate of proton uptake by the CcO III (−) forms with two entry pathways is five-fold greater than that of normal CcO [27]. Apparently, electrostatic interaction between the two deprotonated acceptors (Asp-139/Asp-132 or Cys-139/Asp-132) promotes the protonation of each, and thereby increases the rate of proton uptake. Such electrostatic control of proton entry does not appear to occur in the D pathway of normal CcO. However, in the proton transfer pathway leading from the outer surface of nitric oxide reductase, electrostatically interacting glutamates control the pKa of a coordinated water proposed to be the initial proton acceptor, thus setting the functional pKa of proton uptake [48]. Similar chemistry has been proposed for the proton release group of bacteriorhodopsin [49].
2. Using catalytic lifespan to study D pathway function
2.1. What is catalytic lifespan?
In what may be its most obvious function, subunit III greatly extends the catalytic lifespan of CcO by decreasing the probability of spontaneous suicide inactivation during turnover [24, 35]. Inactivation results from a catastrophic event at the heme a3-CuB center during turnover that include an apparent loss of CuB [24], although the details of the inactivation mechanism are not yet clarified. Our measure to compare the propensity of different CcO forms to suicide inactivate is the CC50, defined as the number of catalytic cycles (O2→2H2O) required for 50% of the original population to become inactive. The CC50 value is also the average lifespan of any of the CcO molecules in the population, measured in terms of catalytic cycles, hence the term ‘catalytic lifespan’. Lifespan is not defined in units of time because the rate of turnover has no effect on the probability of inactivation, only the number of turnovers. A greater CC50 indicates a longer catalytic lifespan as well as a lower probability of inactivation during any catalytic cycle.
Numerous experiments show that slowing proton uptake into the D pathway increases the probability of inactivation and shortens catalytic lifespan [35]. In contrast, slowing proton transfer by the K pathway has no effect on the probability of suicide inactivation [35]. Neither does slowing electron input into the active site [35]. Thus, the strong effect of subunit III on catalytic lifespan is directly linked to the ability of subunit III to facilitate proton uptake into the D pathway. This relationship means that measurements of catalytic lifespan provide a measure, independent of O2 reduction activity, with which to compare steady-state proton uptake by the D pathways of different CcO forms.
2.2. Catalytic lifespan of CcO III (−) forms with different initial proton acceptors
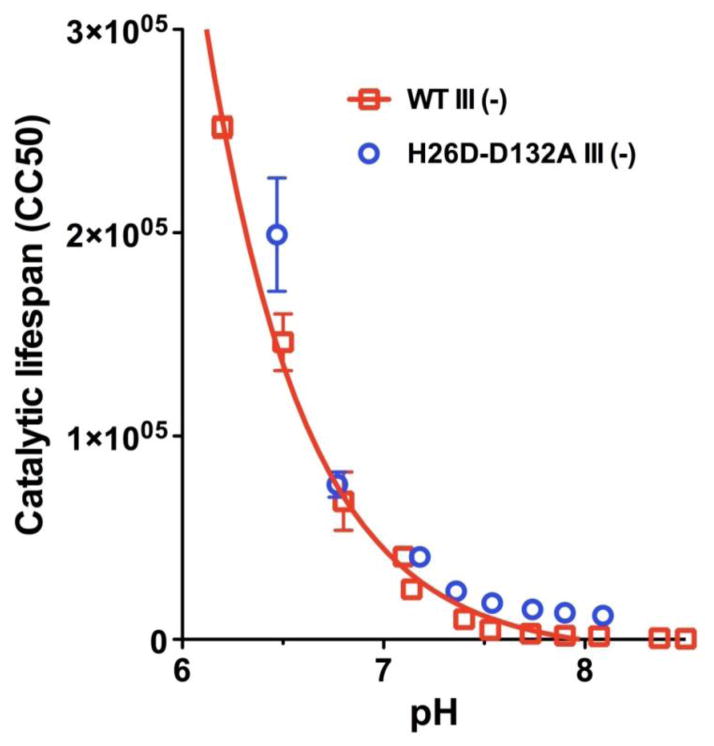
Figures 3 and 4 show how measurements of catalytic lifespan can be applied to compare the D pathways of five CcO forms with different initial proton acceptors. The first observation is that the catalytic lifespan of WT III (−) declines with pH (Figure 3), as does its steady state activity (Figure 2), because both depend upon the rate of proton uptake into the D pathway via Asp-132. However, the pH dependence of lifespan shows no maximum at low pH; the data can be fit to a single exponential decay function but not the Henderson-Hasselbalch relationship. The mechanistic implications of this are discussed below. Figure 3 also compares the catalytic lifespans of CcO III (−) with Asp-26 or Asp-132 as initial proton acceptors of the D pathway (H26D-D132A III (−) vs. WT III (−)). The overlap of the data across a range of pH values indicates that Asp-26 facilitates rates of proton uptake similar to Asp-132, consistent with the similar steady-state activities of these two CcO forms (Table 1).
Figure 3.
pH dependence of the catalytic lifespan of WT III (−), with Asp-132 as the initial proton acceptor of the D pathway, and H26D-D132A III (−), with Asp-26 as the initial acceptor. Catalytic lifespans were measured for the purified CcO forms in dodecyl maltoside solution supplemented with exogenous soybean phospholipid as detailed in Mills and Hosler [35] using the buffer systems of Varanasi and Hosler [27]. The data for WT III (−) is fit to a single exponential decay function using GraphPad Prism.
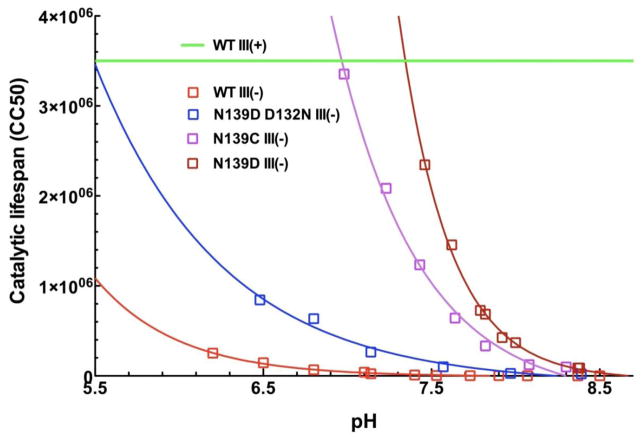
Figure 4.
pH dependence of the catalytic lifespan of CcO III (−) forms with Asp-132, Asp-139, Asp-132 plus Cys 139 or Asp-132 plus Asp-139 as the initial proton acceptors of the D pathway, compared to normal CcO containing subunit III (WT III (+). The green line represents the minimum catalytic lifespan of WT III (+) across this pH range; the long lifespan of WT III (+) is difficult to measure with accuracy. Catalytic lifespan measurements and exponential fits were performed as in Figure 3.
Figure 4 compares the catalytic lifespans afforded by four previously characterized D pathway mutants with different initial proton acceptors: Asp-132 (WT III (−)), Asp-139 (N139D-D132N III (−), Asp-132 plus Cys-139 (N139C III (−) and Asp-132 plus Asp-139 (N139D III (−). Steady-state O2 reduction measurements have shown that Asp-139 facilitates a rate of steady-state proton uptake similar to that of Asp-132 [27] (Table 1), but the functional pKa of proton uptake via Asp-139 is shifted to a higher value (Table 1; Figure 2). These findings are corroborated by the catalytic lifespan measurements since the lifespans of N139D-D132N III (−) and WT III (−) are similar but the pH dependence of catalytic lifespan for N139D-D132N III (−) is shifted toward higher pH values (Figure 4). Steady-state O2 reduction analyses indicate that the two proton acceptors present in N139C III (−) and N139D III (−) interact to increase the functional pKas of proton uptake to 8.2 and 8.3, respectively [27] (Table 1; Figure 2). Again, the catalytic lifespan data are consistent with these findings in that both mutant CcOs exhibit pH dependence curves shifted to even higher pH values (Figure 4).
Below pH 8, the rates of steady-state proton uptake by N139D III (−) and N139C III (−) exceed that of (WT III (+)) [27]. This unusual situation allows a test of whether catalytic lifespan will continue to increase along with proton uptake, as predicted by extrapolation of the exponential relationship between pH and lifespan in WT III (−). The data indicate that it does. The pH profiles of Figure 4 show that by pH 7 the catalytic lifespans of these highly active subunit III-depleted CcOs are within the range of lifespans measured for normal CcO containing subunit III. In terms of protection from suicide inactivation, rapid proton uptake by the D pathway can compensate for the absence of subunit III.
2.3. Relating catalytic lifespan to the rate of proton uptake
Another way to examine lifespan data is to plot catalytic lifespan as a function of the rate of proton uptake (Figure 5). In the lifespan measurement, the initial rate of CcO activity (as TN) is also the rate of proton uptake of every active CcO molecule in the population. The data of Figure 5 indicate that lifespan increases exponentially with the rate of proton uptake. Once rapid proton uptake is achieved, small increases in the rate of proton uptake have an increasingly greater effect in lowering the probability of suicide inactivation. This suggests that the mechanism of suicide inactivation includes a fast, reversible proton-dependent alteration(s) that precedes a final, irreversible step.
Figure 5.
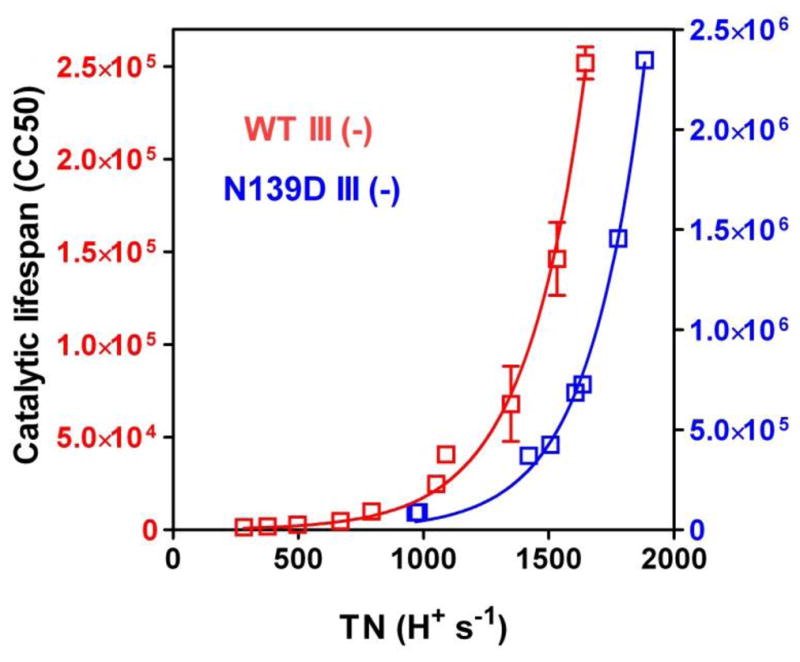
The exponential relationship of catalytic lifespan to the rate of steady-state proton uptake for CcO III (−) forms with Asp-132 (WT III (−) or Asp-132 plus Asp-139 (N139D III (−) as initial proton acceptors of the D pathway. The rate of steady-state proton uptake of substrate protons is the turnover number, expressed as H+ consumption (H+ s−1). The Y axis on the left shows the CC50 values measured for WT III (−) while the ten-fold higher CC50 values of the Y axis on the right are those measured for N139D III (−). The data are fit to a single exponential growth functions using GraphPad Prism.
The D pathway carries out six proton uptake events during each catalytic cycle. Four pump protons are taken up along with two substrate protons, one during the PM→F transition and one during the F→O transition [5]. Catalytic lifespan depends upon the rate of uptake of one or more of these protons, but which? Thus far, there exists no correlation between the ability of a CcO form to pump protons and its catalytic lifespan. If pump protons are not directly involved in the mechanism of suicide inactivation, then comparisons of catalytic lifespan are reporting on the ability of different D pathways to deliver substrate protons during the PM→F or the F→O steps, or both. Such a comparison is presented in Figure 5, which shows that at any given rate of steady-state proton uptake N139D III (−) has a greater lifespan that WT III (−). (Note that the CC50 values on the right Y axis of Figure 5, corresponding to N139D III (−), are an order of magnitude greater than the CC50 values on the left Y axis, corresponding to WT III (−).) These data suggest that N139D III (−) takes up a proton during the PM→F or the F→O steps of the catalytic cycle at a greater rate than does WT III (−); this would identify the step involved in suicide inactivation. Proton uptake by N139D III (−) during these two transitions is being examined, but it is already documented that the rate of the F→O transition of N139D III (+) is twice as fast as that of WT III (+) [50].
Conclusions
Relatively simple steady-state assays for O2 reduction rates and catalytic lifespan can be applied to CcO III (−) forms to compare the efficiency of steady-state proton uptake by altered D pathways as well as to compare the proton affinity of the initial proton acceptors of altered pathways. The addition of such information to that obtained from single CC experiments yields new insight into the process of proton uptake and suicide inactivation.
Highlights.
Subunit III-depleted cytochrome c oxidase provides information about proton uptake.
Steady-state activity compares proton uptake via different initial proton acceptors.
Catalytic lifespan is another method to compare different proton uptake strategies.
Acknowledgments
Supported by National Institutes of Health Grant GM 56824
The authors thank Shelagh Ferguson-Miller for valuable discussion and critical reading of the manuscript.
Abbreviations
- Aa
arachidonic acid
- CC
catalytic cycle
- CcO III (−)
cytochrome c oxidase lacking subunit III
- TN
turnover number, as e− or H+ consumed per second per CcO
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Decoursey TE. Voltage-gated proton channels and other proton transfer pathways. Physiol Rev. 2003;83:475–579. doi: 10.1152/physrev.00028.2002. [DOI] [PubMed] [Google Scholar]
- 2.Wraight CA. Chance and design--proton transfer in water, channels and bioenergetic proteins. Biochim Biophys Acta. 2006;1757:886–912. doi: 10.1016/j.bbabio.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 3.Brzezinski P, Ädelroth P. Design principles of proton-pumping haem-copper oxidases. Curr Opin Struct Biol. 2006;16:465–472. doi: 10.1016/j.sbi.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 4.Hosler JP, Ferguson-Miller S, Mills DA. Energy transduction: Proton transfer through the respiratory complexes. Annu Rev Biochem. 2006;75:165–187. doi: 10.1146/annurev.biochem.75.062003.101730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brändén G, Gennis RB, Brzezinski P. Transmembrane proton translocation by cytochrome c oxidase. Biochim Biophys Acta. 2006;1757:1052–1063. doi: 10.1016/j.bbabio.2006.05.020. [DOI] [PubMed] [Google Scholar]
- 6.Forte E, Scandurra FM, Richter OM, D’Itri E, Sarti P, Brunori M, Ludwig B, Giuffre A. Proton uptake upon anaerobic reduction of the Paracoccus denitrificans cytochrome c oxidase: a kinetic investigation of the K354M and D124N mutants. Biochemistry. 2004;43:2957–2963. doi: 10.1021/bi035863u. [DOI] [PubMed] [Google Scholar]
- 7.Vygodina TV, Pecoraro C, Mitchell D, Gennis R, Konstantinov AA. Mechanism of inhibition of electron transfer by amino acid replacement K362M in a proton channel of Rhodobacter sphaeroides cytochrome c oxidase. Biochemistry. 1998;37:3053–3061. doi: 10.1021/bi971876u. [DOI] [PubMed] [Google Scholar]
- 8.Ganesan K, Gennis RB. Blocking the K-pathway still allows rapid one-electron reduction of the binuclear center during the anaerobic reduction of the aa(3)-type cytochrome c oxidase from Rhodobacter sphaeroides. Biochim Biophys Acta. 2010 doi: 10.1016/j.bbabio.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brändén M, Tomson F, Gennis RB, Brzezinski P. The Entry Point of the K-Proton-Transfer Pathway in Cytochrome c Oxidase. Biochemistry. 2002;41:10794–10798. doi: 10.1021/bi026093+. [DOI] [PubMed] [Google Scholar]
- 10.Tomson FL, Morgan J, Gu G, Barquera B, Vygodina TV, Gennis RB. Substitutions for Glutamate 101 in Subunit II of Cytochrome c Oxidase from Rhodobacter sphaeroides Result in Blocking the Proton-Conducting K-Channel. Biochemistry. 2003;42:1711–1717. doi: 10.1021/bi026750y. [DOI] [PubMed] [Google Scholar]
- 11.Kaila VR, Sharma V, Wikstrom M. The identity of the transient proton loading site of the proton-pumping mechanism of cytochrome c oxidase. Biochim Biophys Acta. 2011;1807:80–84. doi: 10.1016/j.bbabio.2010.08.014. [DOI] [PubMed] [Google Scholar]
- 12.Zhu J, Han H, Pawate A, Gennis RB. Decoupling mutations in the D-channel of the aa3-type cytochrome c oxidase from Rhodobacter sphaeroides suggest that a continuous hydrogen-bonded chain of waters is essential for proton pumping. Biochemistry. 2010;49:4476–4482. doi: 10.1021/bi100344x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qin L, Liu J, Mills DA, Proshlyakov DA, Hiser C, Ferguson-Miller S. Redox-dependent conformational changes in cytochrome c oxidase suggest a gating mechanism for proton uptake. Biochemistry. 2009;48:5121–5130. doi: 10.1021/bi9001387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koepke J, Olkhova E, Angerer H, Muller H, Peng G, Michel H. High resolution crystal structure of Paracoccus denitrificans cytochrome c oxidase: new insights into the active site and the proton transfer pathways. Biochim Biophys Acta. 2009;1787:635–645. doi: 10.1016/j.bbabio.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 15.Henry RM, Yu CH, Rodinger T, Pomes R. Functional hydration and conformational gating of proton uptake in cytochrome c oxidase. J Mol Biol. 2009;387:1165–1185. doi: 10.1016/j.jmb.2009.02.042. [DOI] [PubMed] [Google Scholar]
- 16.Cukier RI. Quantum molecular dynamics simulation of proton transfer in cytochrome c oxidase. Biochim Biophys Acta. 2004;1656:189–202. doi: 10.1016/j.bbabio.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 17.Svensson-Ek M, Abramson J, Larsson G, Tornroth S, Brzezinski P, Iwata S. The X-ray crystal structures of wild-type and EQ(I-286) mutant cytochrome c oxidases from Rhodobacter sphaeroides. J Mol Biol. 2002;321:329–339. doi: 10.1016/s0022-2836(02)00619-8. [DOI] [PubMed] [Google Scholar]
- 18.Muramoto K, Hirata K, Shinzawa-Itoh K, Yoko-o S, Yamashita E, Aoyama H, Tsukihara T, Yoshikawa S. A histidine residue acting as a controlling site for dioxygen reduction and proton pumping by cytochrome c oxidase. Proc Natl Acad Sci USA. 2007;104:7881–7886. doi: 10.1073/pnas.0610031104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richter OM, Ludwig B. Electron transfer and energy transduction in the terminal part of the respiratory chain - lessons from bacterial model systems. Biochim Biophys Acta. 2009;1787:626–634. doi: 10.1016/j.bbabio.2009.02.020. [DOI] [PubMed] [Google Scholar]
- 20.Hosler JP. The influence of subunit III of cytochrome c oxidase on the D pathway, the proton exit pathway and mechanism-based inactivation in subunit I. Biochim Biophys Acta. 2004;1655:332–339. doi: 10.1016/j.bbabio.2003.06.009. [DOI] [PubMed] [Google Scholar]
- 21.Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 Å. Science. 1996;272:1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- 22.Shinzawa-Itoh K, Aoyama H, Muramoto K, Terada H, Kurauchi T, Tadehara Y, Yamasaki A, Sugimura T, Kurono S, Tsujimoto K, Mizushima T, Yamashita E, Tsukihara T, Yoshikawa S. Structures and physiological roles of 13 integral lipids of bovine heart cytochrome c oxidase. EMBO J. 2007;26:1713–1725. doi: 10.1038/sj.emboj.7601618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Varanasi L, Mills D, Murphree A, Gray J, Purser C, Baker R, Hosler J. Altering conserved lipid binding sites in cytochrome c oxidase of Rhodobacter sphaeroides perturbs the interaction between subunits I and III and promotes suicide inactivation of the enzyme. Biochemistry. 2006;45:14896–14907. doi: 10.1021/bi061390q. [DOI] [PubMed] [Google Scholar]
- 24.Bratton MR, Pressler MA, Hosler JP. Suicide inactivation of cytochrome c oxidase: catalytic turnover in the absence of subunit III alters the active site. Biochemistry. 1999;38:16236–16245. doi: 10.1021/bi9914107. [DOI] [PubMed] [Google Scholar]
- 25.Thompson DA, Gregory L, Ferguson-Miller S. Cytochrome c Oxidase Depleted of Subunit III: Proton-Pumping, Respiratory Control, and pH Dependence of the Midpoint Potential of Cytochrome a. J Inorg Biochem. 1985;23:357–364. doi: 10.1016/0162-0134(85)85046-7. [DOI] [PubMed] [Google Scholar]
- 26.Wilson KS, Prochaska LJ. Phospholipid vesicles containing bovine heart mitochondrial cytochrome c oxidase and subunit III-deficient enzyme: analysis of respiratory control and proton translocating activities. Arch Biochem Biophys. 1990;282:413–420. doi: 10.1016/0003-9861(90)90137-n. [DOI] [PubMed] [Google Scholar]
- 27.Varanasi L, Hosler J. Alternative initial proton acceptors for the D pathway of Rhodobacter sphaeroides cytochrome c oxidase. Biochemistry. 2011;50:2820–2828. doi: 10.1021/bi102002v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mills DA, Tan Z, Ferguson-Miller S, Hosler J. A role for subunit III in proton uptake into the D pathway and a possible proton exit pathway in Rhodobacter sphaeroides cytochrome c oxidase. Biochemistry. 2003;42:7410–7417. doi: 10.1021/bi0341307. [DOI] [PubMed] [Google Scholar]
- 29.Thompson DA, Gregory L, Ferguson-Miller S. Cytochrome c oxidase depleted of subunit III: proton-pumping, respiratory control, and pH dependence of the midpoint potential of cytochrome a. J Inorg Biochem. 1985;23:357–364. doi: 10.1016/0162-0134(85)85046-7. [DOI] [PubMed] [Google Scholar]
- 30.Sarti P, Jones MG, Antonini G, Malatesta F, Colosimo A, Wilson MT, Brunori M. Kinetics of Redox-Linked Proton Pumping Activity of Native and Subunit III-Depleted Cytochrome c Oxidase: A Stopped-Flow Investigation. Proc Natl Acad Sci USA. 1985;82:4876–4880. doi: 10.1073/pnas.82.15.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hendler RW, Pardhasaradhi K, Reynafarje B, Ludwig B. Comparison of Energy-transducing Capabilities of the Two- and Three-subunit Cytochromes aa3 from Paracoccus denitrificans and the 13-subunit Beef Heart Enzyme. Biophys J. 1991;60:415–423. doi: 10.1016/S0006-3495(91)82067-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gilderson G, Salomonsson L, Aagaard A, Gray J, Brzezinski P, Hosler J. Subunit III of cytochrome c oxidase of Rhodobacter sphaeroides is required to maintain rapid proton uptake through the D pathway at physiologic pH. Biochemistry. 2003;42:7400–7409. doi: 10.1021/bi0341298. [DOI] [PubMed] [Google Scholar]
- 33.Ädelroth P, Hosler J. Surface proton donors for the D-pathway of cytochrome c oxidase in the absence of subunit III. Biochemistry. 2006;45:8308–8318. doi: 10.1021/bi0605843. [DOI] [PubMed] [Google Scholar]
- 34.Haltia T, Semo N, Arrondo JL, Goni FM, Freire E. Thermodynamic and structural stability of cytochrome c oxidase from Paracoccus denitrificans. Biochemistry. 1994;33:9731–9740. doi: 10.1021/bi00198a044. [DOI] [PubMed] [Google Scholar]
- 35.Mills DA, Hosler JP. Slow proton transfer through the pathways for pumped protons in cytochrome c oxidase induces suicide inactivation of the enzyme. Biochemistry. 2005;44:4656–4566. doi: 10.1021/bi0475774. [DOI] [PubMed] [Google Scholar]
- 36.Cheng VWT, Johnson A, Rothery RA, Weiner JH. Alternative sites for proton entry from the cytoplasm to the quinone binding site in Escherichia coli succinate dehydrogenase. Biochemistry. 2008;47:9107–9116. doi: 10.1021/bi801008e. [DOI] [PubMed] [Google Scholar]
- 37.Horsefield R, Yankovskaya V, Sexton G, Whittingham W, Shiomi K, Omura S, Byrne B, Cecchini G, Iwata S. Structural and computational analysis of the quinone-binding site of complex II (succinate-ubiquinone oxidoreductase): a mechanism of electron transfer and proton conduction during ubiquinone reduction. J Biol Chem. 2006;281:7309–7316. doi: 10.1074/jbc.M508173200. [DOI] [PubMed] [Google Scholar]
- 38.Wikström M, Verkhovsky MI. The D-channel of cytochrome oxidase: An alternative view. Biochim Biophys Acta. 2011 doi: 10.1016/j.bbabio.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 39.Chen K, Hirst J, Camba R, Bonagura CA, Stout CD, Burgess BK, Armstrong FA. Atomically defined mechanism for proton transfer to a buried redox centre in a protein. Nature. 2000;405:814–817. doi: 10.1038/35015610. [DOI] [PubMed] [Google Scholar]
- 40.Gu W, Helms V. Tightly connected water wires facilitate fast proton uptake at the proton entrance of proton pumping proteins. J Am Chem Soc. 2009;131:2080–2081. doi: 10.1021/ja809301w. [DOI] [PubMed] [Google Scholar]
- 41.Marantz Y, Einarsdottir OO, Nachliel E, Gutman M. Proton-collecting properties of bovine heart cytochrome c oxidase: kinetic and electrostatic analysis. Biochemistry. 2001;40:15086–15097. doi: 10.1021/bi010453w. [DOI] [PubMed] [Google Scholar]
- 42.Domsic JF, Williams W, Fisher SZ, Tu C, Agbandje-McKenna M, Silverman DN, McKenna R. Structural and kinetic study of the extended active site for proton transfer in human carbonic anhydrase II. Biochemistry. 2010;49:6394–6399. doi: 10.1021/bi1007645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ädelroth P, Paddock ML, Tehrani A, Beatty JT, Feher G, Okamura MY. Identification of the proton pathway in bacterial reaction centers: decrease of proton transfer rate by mutation of surface histidines at H126 and H128 and chemical rescue by imidazole identifies the initial proton donors. Biochemistry. 2001;40:14538–14546. doi: 10.1021/bi011585s. [DOI] [PubMed] [Google Scholar]
- 44.Xu Q, Axelrod HL, Abresch EC, Paddock ML, Okamura MY, Feher G. X-Ray structure determination of three mutants of the bacterial photosynthetic reaction centers from Rb. sphaeroides; altered proton transfer pathways. Structure. 2004;12:703–715. doi: 10.1016/j.str.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 45.Tomson FL, Morgan JE, Gu G, Barquera B, Vygodina TV, Gennis RB. Substitutions for glutamate 101 in subunit II of cytochrome c oxidase from Rhodobacter sphaeroides result in blocking the proton-conducting K-channel. Biochemistry. 2003;42:1711–1717. doi: 10.1021/bi026750y. [DOI] [PubMed] [Google Scholar]
- 46.Qin L, Mills DA, Hiser C, Murphree A, Garavito RM, Ferguson-Miller S, Hosler J. Crystallographic location and mutational analysis of Zn and Cd inhibitory sites and role of lipidic carboxylates in rescuing proton path mutants in cytochrome c oxidase. Biochemistry. 2007;46:6239–6248. doi: 10.1021/bi700173w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qin L, Mills DA, Buhrow L, Hiser C, Ferguson-Miller S. A conserved steroid binding site in cytochrome c oxidase. Biochemistry. 2008;47:9931–9933. doi: 10.1021/bi8013483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Flock U, Thorndycroft FH, Matorin AD, Richardson DJ, Watmough NJ, Ädelroth P. Defining the proton entry point in the bacterial respiratory nitric-oxide reductase. J Biol Chem. 2008;283:3839–3845. doi: 10.1074/jbc.M704615200. [DOI] [PubMed] [Google Scholar]
- 49.Phatak P, Ghosh N, Yu H, Cui Q, Elstner M. Amino acids with an intermolecular proton bond as proton storage site in bacteriorhodopsin. Proc Natl Acad Sci USA. 2008;105:19672–19677. doi: 10.1073/pnas.0810712105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pawate AS, Morgan J, Namslauer A, Mills D, Brzezinski P, Ferguson-Miller S, Gennis RB. A mutation in subunit I of cytochrome oxidase from Rhodobacter sphaeroides results in an increase in steady-state activity but completely eliminates proton pumping. Biochemistry. 2002;41:13417–13423. doi: 10.1021/bi026582+. [DOI] [PubMed] [Google Scholar]