Abstract
Only two genome-wide association studies (GWAS) have been conducted to date to identify potential markers for total mortality after diagnosis of breast cancer. Here we report the identification of two SNPs associated with total mortality from a two-stage GWAS conducted among 6,110 Shanghai-resident Chinese women with TNM stage I-IV breast cancer. The discovery stage included 1,950 patients and evaluated 613,031 common SNPs. The top 49 associations were evaluated in an independent replication stage of 4,160 Shanghai breast cancer patients. A consistent and highly significant association with total mortality was documented for SNPs rs3784099 and rs9934948. SNP rs3784099, located in the RAD51L1 gene, was associated with total morality in both the discovery stage (P=1.44×10−8) and replication stage (P=0.06; P-combined=1.17×10−7). Adjusted hazard ratios (HR) for total mortality were 1.41 (95%CI=1.18–1.68) for the AG genotype and 2.64 (95%CI=1.74–4.03) for the AA genotype, when compared with the GG genotype. The variant C allele of rs9934948, located on chromosome 16, was associated with a similarly elevated risk of total mortality (P-combined: 5.75×10−6). We also observed this association among 1,145 breast cancer patients of European-ancestry from the Nurses’ Health Study (NHS; P=0.006); the association was highly significant in a combined analysis of NHS and Chinese data (P=1.39×10−7). Similar associations were observed for these two SNPs with breast cancer-specific mortality. This study provides strong evidence suggesting that the RAD51L1 gene and a chromosome 16 locus influence breast cancer prognosis.
Keywords: breast cancer, survival, genome-wide association study, Asian population, RAD51L1 gene
INTRODUCTION
Breast cancer is one of the most common malignancies among women in many countries, including China. Despite generally good prognosis for breast cancer patients, wide variation exists in survival, even after accounting for clinical prognostic factors, suggesting that genetic susceptibility may influence breast cancer outcomes. Over the past 10 years, candidate-gene studies, including our own (1–5), have found several genetic variants to be related to breast cancer prognosis. These genetic variants are found primarily in breast cancer susceptibility genes (e.g., BRCA1, BRCA2, TP53) (6–8) or genes involved in drug metabolism (e.g., CYP2D6, NQO1) (9, 10) and tumor microenvironment regulation (e.g., TGFβ1, VEGF, CCND1, PAI1, MMP7) (1–5). However, very few of these associations have been confirmed. Given that almost all previous studies used the candidate-gene approach, in which only a limited number of genetic variants are investigated and the choice of variant is based on our limited knowledge of the underlying biology of cancer, more comprehensive genomic investigations of breast cancer prognosis are urgently needed. Recent genome-wide association studies (GWAS) have identified genetic variants related to breast cancer risk that have been robustly replicated across populations (11–18). Many GWAS-identified genetic markers are located in regions that had never been suspected of being related to cancer susceptibility. To our knowledge, only two studies have evaluated genetic factors in relation to breast cancer survival using the GWAS approach (19, 20) and both were conducted among women of European ancestry. The first study reported a single nucleotide polymorphism (SNP) in the OCA2 gene (rs4778137) associated with total mortality among women of European ancestry with estrogen receptor (ER)-negative tumors at P=5×10−4 (19). However, the second study, conducted as part of the Cancer Genetic Markers of Susceptibility (CGEMS) study, found no SNPs with a statistically significant association with breast cancer survival (20).
Over the past 15 years, we have conducted multiple, large-scale, population-based studies of breast cancer among Chinese women in Shanghai (14, 21, 22). Using the data collected from these studies, we evaluated lifestyle determinants of breast cancer survival (21, 23, 24). In addition to the candidate-gene studies reported previously (1–5), we recently conducted a two-stage GWAS among 6,110 patients (719 deaths) with stage I to IV breast cancer recruited in the Shanghai studies to identify novel genetic variants associated with breast cancer survival. To evaluate the generalizability of our findings to other ethnic groups, we investigated these GWAS-identified SNPs in CGEMS data from 1,145 breast cancer patients (229 deaths) of European ancestry who participated in the Nurses’ Health Study (NHS).
METHODS
Overall Study Design and Study Populations
Samples included in this GWAS came from participants of the Shanghai Breast Cancer Study (SBCS) and Shanghai Breast Cancer Survival Study (SBCSS). Details on the methodology of the parent studies have been described previously (14, 21). Briefly, the SBCS is a population-based, case-control study that recruited incident breast cancer patients and controls in urban Shanghai between August 1996 and March 1998 and again between April 2002 and February 2005 (14). A total of 3,448 patients were recruited (participation rate: 86.7%); 90.6% of participants provided a blood or exfoliated buccal cell sample. The SBCSS also was conducted in urban Shanghai and recruited 5,042 breast cancer patients between March 2002 and April 2006 (participation rate: 80.1%); 98% of patients provided an exfoliated buccal cell sample (14, 21). All participants of both studies provided written informed consent before participating in the study and the Institutional Review Boards of all institutes involved approved the study protocols. Medical charts for breast cancer patients were reviewed to verify cancer diagnosis and obtain treatment information. Cancer patients have been followed for survival status and breast cancer recurrence through a combination of record linkages with the Shanghai Vital Statistics Registry and in-person surveys. Due to a time overlap during recruitment, 1,469 women participated in both the SBCS and SBCSS. After taking these overlaps into consideration and excluding patients with stage 0 disease (n=190), those for whom we had no information on survival status (n=185), and genotyping failures due to limited DNA (n=17), a total of 6,110 participants remained in the present study. The discovery stage of this study included 1,950 participants. Genomic DNA samples were genotyped primarily using the Affymetrix Genome-Wide Human SNP Array 6.0. From the discovery stage, two batches of SNPs were selected for replication in an independent set of samples from the Shanghai studies. Criteria used to select SNPs for validation were: 1) P<=0.001 under the additive model for either the total mortality (overall survival) or breast cancer recurrence (disease-free survival) analysis; for SNPs that are on or close to metastasis genes or genes previous indicated in breast cancer prognosis, the P-value was relaxed to <=0.01; 2) Minor allele frequency >10%; 3) Exhibited high quality genotype cluster plots; 4) In regions where multiple SNPs in linkage disequilibrium (LD) (r2>=0.6) met the above criteria, the SNP with the lowest P value was chosen. The first batch (the top 30 SNPs not in LD) was selected when GWAS data were available for 1,436 participants. Twenty-nine of these SNPs were successfully genotyped in an independent set of 3,881 Shanghai study participants. The second batch (the top 20 SNPs not in LD and not overlapping with those included in the first batch) was selected after an additional 514 breast cancer patients (for a total of 1,950 participants included in the discovery stage) were scanned. These 20 SNPs were genotyped in an independent set of 4,160 Shanghai study participants. Among women who participated in the two batches of validation studies, 3,522 overlapped.
To explore the generalizability of the study findings to other ethnic groups, we selected eight SNPs with possible associations with breast cancer survival in the Shanghai studies and evaluated their associations with breast cancer survival in European-ancestry Americans using data from the CGEMS project. These SNPs were chosen based on the significance level of the association found in the discovery stage and/or the consistency of the associations observed in both discovery and replication stages. The CGEMS project included 1,145 post-menopausal breast cancer cases from the NHS whose DNA samples were scanned using the Illumina HumanHap500 array (12, 16). The NHS is a prospective cohort of 121,700 registered nurses who resided in 11 US states and enrolled in the study in 1976. Follow-up was conducted by personal mailings and searches of the National Death Index (20).
Genotyping and Quality Control Procedures
We included three positive quality control (QC) samples purchased from Coriell Cell Repositories (Coriell Institute, Camden, NJ, USA) and a negative QC sample (water) in each of the 96-well test plates. The average concordance among the QC samples was 99.85% (median: 100%). The gender of all scanned samples was confirmed. Genetically identical, unexpected duplicated samples were excluded, as were close relatives with a pairwise proportion of IBD estimate >0.25. Multidimensional scaling (MDS) analyses of pooled data including 210 unrelated HapMap subjects, and our study data showed that all our study participants clustered closely with HapMap Asians. All samples with a call rate <95% were excluded. In addition, each SNP met following inclusion criteria: 1) Minor allele frequency (MAF) ≥0.05; 2) Call rate ≥95%; 3) P for Hardy-Weinberg equilibrium (HWE) ≥0.000001; 4) concordance ≥95% among duplicated QC samples. After exclusions, 607,728 SNPs from batch 1 and 613,031 SNPs from batch 2 were available for the statistical analyses used to select promising SNPs for replication. The genotyping and QC protocols for the CGEMS project are described in detail elsewhere (12, 16).
Genotyping for the replication stage was performed on the iPLEX™ Sequenom MassARRAY® platform. Polymerase chain reaction (PCR) and extension primers were designed by using the MassARRAY Assay Design 3.0 software (Sequenom, Inc). PCR and extension reactions were performed according to the manufacturer’s instructions, and extension product sizes were determined by mass spectrometry using the Sequenom iPLEX system. In each 96-well plate, two negative controls (water), two blinded duplicates, and two samples from the HapMap project were included. The concordance was 100% for all SNPs for both the blinded duplicates and the HapMap samples. We also included 65 participants who were genotyped by using the Affymetrix 6.0 array on the Sequenom genotyping platform and found 100% consistency for data generated on these two platforms. All SNPs showed high call rates (>95%).
Statistical Analyses
A set of 4,305 SNPs (not in LD) with a MAF>0.35 and at least 100kb apart was selected to evaluate the population structure. The inflation factor λ was estimated to be 1.045, suggesting that population substructure, if present, should not have any appreciable effect on the results.
Outcomes of the study were total mortality (for the overall survival analysis) and breast cancer recurrence (for the disease-free analysis). For the overall survival analysis, follow-up time was calculated as the number of days between the date of cancer diagnosis and the date of death or date of last record linkage for survivors. For the disease-free survival analysis, follow-up time was calculated as the number of days between the date of cancer diagnosis and disease recurrence or date of last survey for women who did not have disease recurrence or die of breast cancer. For 62 women who died of breast cancer but were missing information on disease recurrence, we imputed the date for recurrence based on the TNM stage-specific recurrence rate estimated for the current study. The delayed-entry Cox proportional hazards regression models were used to derive hazard ratios (HR) for total mortality and breast cancer recurrence in association with each SNP with adjustment for age. Additional adjustments for known clinical predictors for breast cancer prognosis, including breast cancer stage (TNM), estrogen/progesterone receptor (ER/PR) status, ever use of chemotherapy, radiotherapy, and tamoxifen did not materially change the results. We also examined the influence of population substructure by adjusting for the first 5 principal components derived based on 196,471 SNPs with a pairwise LD of r2<0.2 that were selected using PLINK (25, 26). We observed no appreciable changes in study results (data not shown). Thus, the results presented in this article were not adjusted for population substructure.
RESULTS
Clinical characteristics of study participants are presented in Table 1. Patients included in the discovery stage were younger, more like to have late-stage disease, lower 5-year survival rates, and to have received chemotherapy, radiotherapy, or tamoxifen than those included in the replication stage. These differences reflect differences in study enrollment criteria (the SBCS, contributed the majority of cases to the discovery stage and disproportionally recruited younger women with breast cancer) and, possibly, temporal changes in breast cancer treatment protocols and outcomes.
Table 1.
Demographic characteristics of participants of the Shanghai Breast Cancer Study (1996–2005) and Shanghai Breast Cancer Survival Study (2002–2006)
| Discovery Stage | Replication Stage | |
|---|---|---|
| No. of cases | 1,950 | 4,160 |
| Total no. of deaths | 331 | 388 |
| No. of recurrences | 411 | 504 |
| No. of breast cancer deaths | 299 | 331 |
| Age at diagnosis (median, range) | 48.3 (22.6–69.9) | 51.2 (20.4–75.0) |
| Year of follow-up (median) | 6.1 (0.3–8.7) | 3.7 (0.5–8.2) |
| TNM (%) | ||
| I | 29.88 | 35.83 |
| II | 59.07 | 53.39 |
| III | 10.03 | 10.24 |
| IV | 1.02 | 0.54 |
| ER status (%) | ||
| Positive | 63.71 | 64.34 |
| Negative | 36.29 | 35.66 |
| PR status(%) | ||
| Positive | 63.24 | 58.03 |
| Negative | 36.76 | 41.97 |
| Chemotherapy (%) | ||
| Yes | 94.2 | 91.69 |
| No | 5.8 | 8.31 |
| Radiotherapy (%) | ||
| Yes | 36.58 | 32.23 |
| No | 63.42 | 67.77 |
| Tamoxifen use (%) | ||
| Yes | 67.09 | 51.27 |
| No | 32.91 | 48.73 |
| Mastectomy (%) | ||
| Yes | 99.9 | 99.73 |
| No | 0.1 | 0.27 |
| 5-year survival rate (%) | 85.85 | 89.12 |
Patients with missing data were excluded.
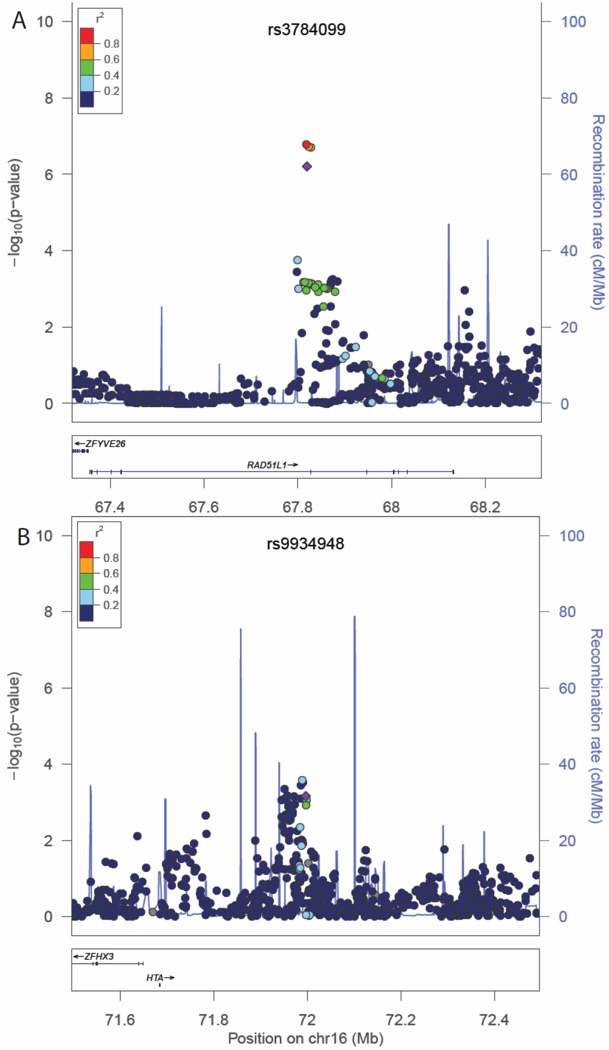
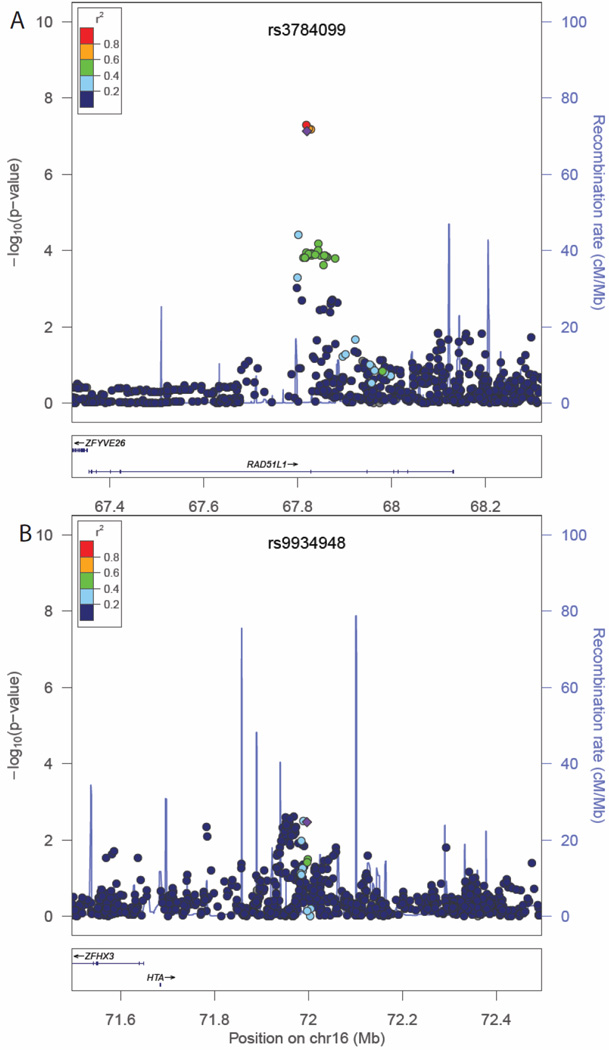
Of the top 50 SNPs chosen for replication, 49 SNPs were successfully genotyped. Associations for all SNPs with breast cancer outcomes and P-values for HWE tests are presented in Supplementary Table S1. A nominally statistically significant (P≤0.05) or marginally significant (P<0.06) association with total mortality was observed for four SNPs: rs3784099, rs9934948, rs729438, and rs1769441, and the directions of the associations were consistent in both the discovery and replication stages (Supplementary Table 1). For two SNPs, rs3784099 on chromosome 14 and rs9934948 on chromosome 16, the P-value for the combined analysis reached 1.17×10−7 and 5.75×10−6, respectively (Table 2). SNP rs3784099 was associated with total mortality with a per-allele HR of 1.79 (95% confidence interval [CI]=1.46–2.19, Ptrend=1.44×10−8) in the discovery stage, 1.22 (95%CI=0.99–1.52, Ptrend=0.06) in the replication stage, and 1.49 (95%CI=1.28–1.72, Ptrend=1.17×10−7) for all samples combined. In the recurrence analyses, the Ptrend for rs3784099 was of marginal statistical significance in the replication stage (P=0.07), and the direction of the association was consistent with the discovery stage. In the combined analysis, the per-allele HR for recurrence for rs3784099 was 1.43 (95%CI=1.25–1.64, Ptrend=2.83×10−7). SNP rs9934948 showed a statistically significant association with total mortality (P=0.03) but was not significantly associated with recurrence (P= 0.32) in the replication stage. In combined analyses, per-allele HRs were 1.29 (95%CI=1.16–1.44, Ptrend=5.75×10−6) for total mortality and 1.19 (95%CI=1.08–1.31, Ptrend=7.32×10−4) for recurrence. Regional association plots for these 2 SNPs are presented in Figures 1 and 2. The associations of these two SNPs with total mortality and recurrence did not vary by ER or menopausal status (Table 3). The vast majority of deaths among study participants were due to breast cancer (88%). In analyses of breast cancer-specific mortality, associations similar to those for total mortality were observed (HR=1.45, 95%CI=1.24–1.70, Ptrend=3.8×10−6 for rs3784099 and HR= 1.27, 95% CI=1.13–1.43, Ptrend=6.0×10−5 for rs9934948), although the P-values were increased due to decreased number of events (data not shown in tables).
Table 2.
Associations of breast cancer outcomes with the top two SNPs a
| SNP |
No. of Events |
All participants |
HR (95% CI) (per allele) |
HR (95% CI) (heterozygous) |
HR (95% CI) (homozygous) |
P trend | |
|---|---|---|---|---|---|---|---|
| stage | |||||||
| rs3784099 | (A/G, 0.13) b | Total Mortality | |||||
| Discovery | 276 | 1436 | 1.79 ( 1.46 – 2.19 ) | 1.77 ( 1.37 – 2.28 ) | 3.31 ( 1.92 – 5.71 ) | 1.44E-08 | |
| Replication | 338 | 3809 | 1.22 ( 0.99 – 1.52 ) | 1.14 ( 0.89 – 1.47 ) | 2.05 ( 1.05 – 4.00 ) | 6.44E-02 | |
| Combined | 614 | 5245 | 1.49 ( 1.28 – 1.72 ) | 1.41 ( 1.18 – 1.68 ) | 2.64 ( 1.74 – 4.03 ) | 1.17E-07 | |
| Recurrence | |||||||
| Discovery | 343 | 1405 | 1.73 ( 1.43 – 2.09 ) | 1.69 ( 1.34 – 2.13 ) | 3.20 ( 1.86 – 5.50 ) | 1.75E-08 | |
| Replication | 456 | 3809 | 1.20 ( 0.99 – 1.46 ) | 1.06 ( 0.84 – 1.34 ) | 2.32 ( 1.33 – 4.05 ) | 7.05E-02 | |
| Combined | 799 | 5214 | 1.43 ( 1.25 – 1.64 ) | 1.32 ( 1.12 – 1.55 ) | 2.69 ( 1.82 – 3.96 ) | 2.83E-07 | |
| rs9934948 | (C/T, 0.46) b | Total Mortality | |||||
| Discovery | 276 | 1436 | 1.45 ( 1.23 – 1.71 ) | 1.37 ( 1.01 – 1.85 ) | 2.09 ( 1.51 – 2.90 ) | 9.71E-06 | |
| Replication | 338 | 3807 | 1.18 ( 1.02 – 1.37 ) | 1.29 ( 0.99 – 1.68 ) | 1.40 ( 1.03 – 1.90 ) | 2.76E-02 | |
| Combined | 614 | 5243 | 1.29 ( 1.16 – 1.44 ) | 1.31 ( 1.08 – 1.60 ) | 1.67 ( 1.34 – 2.09 ) | 5.75E-06 | |
| Recurrence | |||||||
| Discovery | 343 | 1405 | 1.37 ( 1.18 – 1.59 ) | 1.26 ( 0.96 – 1.65 ) | 1.86 ( 1.38 – 2.49 ) | 4.20E-05 | |
| Replication | 457 | 3807 | 1.07 ( 0.93 – 1.23 ) | 1.11 ( 0.88 – 1.40 ) | 1.14 ( 0.87 – 1.50 ) | 3.24E-01 | |
| Combined | 800 | 5212 | 1.19 ( 1.08 – 1.31 ) | 1.16 ( 0.98 – 1.39 ) | 1.41 ( 1.16 – 1.73 ) | 7.32E-04 | |
Adjusted for age at diagnosis.
Effect allele/reference allele, effect allele frequency, all SNPs were coded on the forward strand.
Figure 1.
Regional association plots for total mortality in association with rs3784099 (panel A) and rs9934948 (panel B). Regional association plots for total mortality in association with rs3784099 (panel A) and rs9934948 (panel B). Chromosomal position is given on the X axis and the association P-values (−log10 P) on the Y axis. Referent SNPs are identified with purple diamonds and pairwise linkage disequilibrium with adjacent SNPs as measured by r2 values (according to the HapMap 1000 Genomes June 2010 CHB+JPT data) is indicated with the color of each circle. Refseq genes are shown beneath each plot.
Figure 2.
Regional association plots for recurrence/breast cancer-specific mortality in association with rs3784099 (panel A) and rs9934948 (panel B). Regional association plots for recurrence/breast cancer-specific mortality in association with rs3784099 (panel A) and rs9934948 (panel B). Chromosomal position is given on the X axis and the association P-values (−log10 P) on the Y axis. Referent SNPs are identified with purple diamonds and pairwise linkage disequilibrium with adjacent SNPs as measured by r2 values (according to the HapMap 1000 Genomes June 2010 CHB+JPT data) is indicated with the color of each circle. Refseq genes are shown beneath each plot.
Table 3.
Associations of breast cancer outcomes with the two newly identified SNPs and one GWAS-identified SNP by tumor and participant characteristics a
| SNP b | No. of Events |
All participants |
HR (95% CI) per allele |
HR (95% CI) heterozygous |
HR (95% CI) homozygous |
P trend |
|---|---|---|---|---|---|---|
| Total Mortality | ||||||
| ER-positive breast cancer | ||||||
| rs3784099 (A/G, 0.13) | 282 | 3097 | 1.71 ( 1.39 – 2.11 ) | 1.67 ( 1.29 – 2.15 ) | 3.19 ( 1.73 – 5.86 ) | 5.11 × 10−07 |
| rs9934948 (C/T, 0.46) | 283 | 3100 | 1.23 ( 1.05 – 1.45 ) | 1.24 ( 0.93 – 1.66 ) | 1.52 ( 1.10 – 2.10 ) | 0.012 |
| rs4778137 (C/G, 0.19) | 150 | 1252 | 1.27 ( 0.96 – 1.68 ) | 1.10 ( 0.78 – 1.56 ) | 2.29 ( 1.16 – 4.55 ) | 0.091 |
| ER-negative breast cancer | ||||||
| rs3784099 (A/G, 0.13) | 226 | 1726 | 1.13 ( 0.87 – 1.47 ) | 0.94 ( 0.68 – 1.29 ) | 2.41 ( 1.23 – 4.72 ) | 0.354 |
| rs9934948 (C/T, 0.46) | 225 | 1721 | 1.26 ( 1.05 – 1.52 ) | 1.29 ( 0.93 – 1.79 ) | 1.60 ( 1.10 – 2.33 ) | 0.013 |
| rs4778137 (C/G, 0.19) | 90 | 691 | 0.95 ( 0.64 – 1.41 ) | 0.86 ( 0.54 – 1.36 ) | 1.37 ( 0.43 – 4.38 ) | 0.792 |
| Pre-menopausal women | ||||||
| rs3784099 (A/G, 0.13) | 305 | 2725 | 1.52 ( 1.25 – 1.86 ) | 1.47 ( 1.14 – 1.88 ) | 2.58 ( 1.47 – 4.52 ) | 4.16 × 10−05 |
| rs9934948 (C/T, 0.46) | 305 | 2720 | 1.29 ( 1.10 – 1.51 ) | 1.10 ( 0.83 – 1.45 ) | 1.64 ( 1.21 – 2.22 ) | 0.002 |
| rs4778137 (C/G, 0.19) | 197 | 1354 | 1.29 ( 1.02 – 1.64 ) | 1.11 ( 0.81 – 1.50 ) | 2.34 ( 1.32 – 4.15 ) | 0.037 |
| Post-menopausal women | ||||||
| rs3784099 (A/G, 0.13) | 309 | 2520 | 1.45 ( 1.17 – 1.80 ) | 1.35 ( 1.05 – 1.74 ) | 2.83 ( 1.50 – 5.35 ) | 6.23 × 10−04 |
| rs9934948 (C/T, 0.46) | 309 | 2523 | 1.30 ( 1.11 – 1.52 ) | 1.58 ( 1.19 – 2.09 ) | 1.71 ( 1.23 – 2.37 ) | 0.001 |
| rs4778137 (C/G, 0.19) | 134 | 955 | 1.18 ( 0.87 – 1.60 ) | 1.29 ( 0.91 – 1.83 ) | 0.93 ( 0.29 – 2.93 ) | 0.278 |
| Recurrence | ||||||
| ER-positive breast cancer | ||||||
| rs3784099 (A/G, 0.13) | 383 | 3084 | 1.50 ( 1.23 – 1.82 ) | 1.38 ( 1.09 – 1.75 ) | 2.95 ( 1.69 – 5.16 ) | 5.47 × 10−05 |
| rs9934948 (C/T, 0.46) | 384 | 3087 | 1.17 ( 1.02 – 1.36 ) | 1.18 ( 0.91 – 1.51 ) | 1.38 ( 1.03 – 1.84 ) | 0.029 |
| rs4778137 (C/G, 0.19) | 213 | 1239 | 1.15 ( 0.91 – 1.47 ) | 1.09 ( 0.82 – 1.47 ) | 1.58 ( 0.80 – 3.10 ) | 0.246 |
| ER-negative breast cancer | ||||||
| rs3784099 (A/G, 0.13) | 298 | 1717 | 1.19 ( 0.94 – 1.50 ) | 0.95 ( 0.71 – 1.27 ) | 2.83 ( 1.58 – 5.07 ) | 0.153 |
| rs9934948 (C/T, 0.46) | 298 | 1712 | 1.11 ( 0.94 – 1.31 ) | 1.09 ( 0.82 – 1.46 ) | 1.23 ( 0.88 – 1.73 ) | 0.225 |
| rs4778137 (C/G, 0.19) | 121 | 682 | 0.90 ( 0.63 – 1.28 ) | 0.91 ( 0.62 – 1.35 ) | 0.73 ( 0.18 – 2.95 ) | 0.548 |
| Pre-menopausal women | ||||||
| rs3784099 (A/G, 0.13) | 420 | 2702 | 1.39 ( 1.16 – 1.68 ) | 1.32 ( 1.05 – 1.64 ) | 2.37 ( 1.38 – 4.05 ) | 4.48 × 10−04 |
| rs9934948 (C/T, 0.46) | 420 | 2697 | 1.21 ( 1.05 – 1.39 ) | 1.11 ( 0.87 – 1.41 ) | 1.45 ( 1.11 – 1.91 ) | 0.007 |
| rs4778137 (C/G, 0.19) | 262 | 1331 | 1.16 ( 0.94 – 1.44 ) | 1.05 ( 0.80 – 1.38 ) | 1.76 ( 1.00 – 3.10 ) | 0.175 |
| Post-menopausal women | ||||||
| rs3784099 (A/G, 0.13) | 379 | 2512 | 1.48 ( 1.21 – 1.81 ) | 1.32 ( 1.04 – 1.68 ) | 3.26 ( 1.86 – 5.70 ) | 1.14 × 10−04 |
| rs9934948 (C/T, 0.46) | 380 | 2515 | 1.17 ( 1.01 – 1.36 ) | 1.24 ( 0.96 – 1.60 ) | 1.37 ( 1.02 – 1.84 ) | 0.035 |
| rs4778137 (C/G, 0.19) | 175 | 947 | 1.03 ( 0.78 – 1.35 ) | 1.16 ( 0.85 – 1.57 ) | 0.45 ( 0.11 – 1.83 ) | 0.846 |
Combined data from all study stages; adjusted for age at diagnosis
Effect allele/reference allele, effect allele frequency; all SNPs were coded on the forward strand.
SNP rs9934948 was associated with total mortality in European-ancestry Americans and the direction of association was the same as that observed in the Shanghai studies (Supplementary Table S2). The age-adjusted HRs were 3.27 (95%CI=0.75–14.19) for the CT genotype and 4.70 (95%CI=1.11–19.97) for the CC genotype compared with the TT genotype (Ptrend=0.006). Meta-analyses combining Shanghai samples and CGEMS data showed a combined P-value of 1.39×10−7. Data on recurrence were not available for CGEMS participants.
DISCUSSION
In this two-stage GWAS of breast cancer survival conducted among Chinese women, we found strong evidence for an association of SNP rs3784099 with total mortality and with recurrence and breast cancer-specific mortality. This SNP is located on chromosome 14 in intron 7 of the RAD51L1 gene, an established cancer susceptibility gene (27). The RAD51L1 gene encodes a protein that is part of the RAD51 family, which is essential for DNA repair by homologous recombination. Over-expression of this gene has been shown to cause cell cycle delay and apoptosis (27, 28). The RAD51L1 gene is not ubiquitously expressed, but it is significantly expressed in breast cancer-derived MCF7 cells (29). A recent GWAS identified a SNP in this gene, rs999737, to be associated with breast cancer risk (16). SNP rs999737, however, was not related to breast cancer survival in our study (data not presented), nor is it in LD with SNP rs3784099 (r2=0 in Asians and r2=0.032 in Europeans based on HapMap data).
SNP rs3784099 is also associated with differential expression of two other genes involved in cancer, SNCG and CTF1, according to the SCAN database (30), which uses HapMap human lymphoblastoid cell lines to identify expression quantitative trait loci (eQTL) (31). Both genes yielded a P value of 0.0001 in cell lines of European ancestry (CEU), though the specific allele of rs3784099 responsible for increased/decreased expression is not apparent in this resource. The SNCG gene encodes synuclein gamma, also known as breast cancer-specific protein 1 (32). Up-regulation of the SNCG gene has been shown to enhance cancer cell motility and contributes to cancer cell survival (32). There are indications that the SNCG gene may be involved in late-stage breast and ovarian cancer metastasis by enhancing cell motility through activation of RHO-family small GTPases and extracellular signal-regulated kinases (ERK) (32, 33). Over-expression of the SNCG gene is a marker for breast cancer progression and a potential target for breast cancer treatment (32, 34). The CTF1 gene is a transcription factor that can delimit chromatin boundaries and thereby block the propagation of silent chromatin (35). These data provide additional support for the association between rs3784099 and breast cancer outcomes observed in our study.
SNP rs9934948 resides on chromosome 16, in the middle of a gene desert with its nearest neighboring genes, ZFHX3 and PSMD7, 346kb and 891kb away, respectively. ZFHX3 is one of the homeobox genes that are often located in gene deserts. PSMD7 is a proteasome component (36) and has been previously shown to be one of the genes most impacted by siRNA knock down of the ER in MCF cells (37). Proteasome activity is increased in tumor cells, resulting in increased turnover rates for signaling molecules that are involved in the regulation of cell growth and apoptosis (38). These biological links and the strong association of this SNP with total mortality observed among breast cancer survivors of European ancestry in CGEMS data support a possible role for rs9934948 in breast cancer prognosis.
To date, only one GWAS-identified SNP, rs4778137, has been associated with breast cancer survival, although the association for this SNP did not reach the conventional genome-wide significance level of 5×10−8 (only 5×10−4) (19). We evaluated this SNP using the scanned data from our discovery stage and found that rs4778137 was significantly associated with total mortality (per allele HR=1.25, 95%CI=1.03–1.51, Ptrend= 0.02; data not shown in tables). The association was observed predominantly among pre-menopausal women (per allele HR=1.29, 95%CI=1.02–1.64) and women with ER-positive breast cancer (per allele HR=1.27, 95%CI=0.96–1.68). Thus, our results provide some support for the association identified by the previous GWAS conducted among women of European ancestry.
Given the difference in genetic architecture across ethnic groups, disease-associated SNPs identified in one population are often not replicated directly in another population. In a recent study conducted among approximately 6,000 female Chinese cancer patients and controls in Shanghai, only 8 of the 12 breast cancer risk SNPs identified in women of European ancestry could be directly replicated (39). Therefore, it is not surprising that the top SNP identified by our study, rs3784099, was not directly replicated in the CGEMS data. Differences in study eligibility could also have contributed to the lack of replication. For example, CGEMS only included post-menopausal women, while the SBCS over-sampled younger breast cancer patients. However, we did not find the association of rs3784099 to be modified by menopausal status. On the other hand, replication of an association in other ethnic groups, as is the case with rs9934948, provides additional evidence for a true association.
In summary, we found that genetic variants in the RAD51L1 gene and chromosome 16 were associated with survival among breast cancer patients. Additional research on the genetic regions and genes identified by our study would lead to a better understanding of the biological mechanisms responsible for breast cancer progression and survival.
Supplementary Material
Acknowledgement
Financial Support: This work was supported by the US National Institutes of Health/National Cancer Institute (grant numbers: R01CA118229 to X.O.S. and R01CA124558, R01CA064277, and R01CA090899 to W.Z.), as well as the US Department of Defense (DOD) Breast Cancer Research Program (Idea Awards BC011118 to X.O.S. and BC050791 to Q.C.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or Department of Defense. Sample preparation and discovery stage genotyping were conducted at the Survey and Biospecimen and Microarray Shared Resources, which are supported in part by the Vanderbilt-Ingram Cancer Center (P30 CA68485).
The authors wish to thank study participants and research staff for their contributions and commitment to this project, Ms. Regina Courtney for DNA preparation, and Ms. Bethanie Rammer for editorial support in the preparation of this manuscript.
Abbreviations
- CGEMS
Cancer Genetic Markers of Susceptibility
- CI
confidence interval
- eQTL
expression quantitative trait loci
- ER
estrogen receptor
- GRS
genetic risk score
- GWAS
genome-wide association study
- HR
hazard ratio
- HWE
Hardy-Weinberg equilibrium
- LD
linkage disequilibrium
- MAF
minor allele frequency
- MDS
multidimensional scaling
- NHS
Nurses’ Health Study
- PCR
polymerase chain reaction
- PR
progesterone receptor
- QC
quality control
- SBCS
Shanghai Breast Cancer Study
- SBCSS
Shanghai Breast Cancer Survival Study
- SNP
single nucleotide polymorphism
- TNM
tumor-node-metastasis
- UTR
untranslated region
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
Author Contributions: Xiao Ou Shu conceived, designed, and implemented the study; analyzed and interpreted the data; drafted the manuscript.; and obtained funding for the study. Jirong Long analyzed data and contributed to writing the manuscript. Wei Lu designed and implemented the study. Chun Li contributed to data analysis and data interpretation. Wendy Y. Chen contributed to the data analysis and data interpretation and provided critical review of the manuscript. Ryan Delahanty contributed to the writing of the manuscript. Jiarong Cheng contributed to study implementation. Hui Cai contributed to the data analysis. Ying Zheng contributed to the study design and implementation. Jiajun Shi performed the genotyping. Kai Gu contributed to study implementation. Wen-Jing Wang contributed to study implementation. Peter Kraft contributed to data interpretation and provided critical review of the paper. Yu-Tang Gao designed and implemented the study. Qiuyin Cai contributed to the design and implementation of the study, interpretation of the data, and funding of the study. Wei Zheng conceived, designed, and implemented the study; contributed to interpretation of the data; provided critical review of the paper; and contributed to the funding of the study.
Xiao Ou Shu confirms that she had full access to all of the study data and bears final responsibility for the decision to submit the paper.
Reference List
- 1.Shu XO, Gao YT, Cai Q, Pierce L, Cai H, Ruan ZX, et al. Genetic polymorphisms in the TGF-beta 1 gene and breast cancer survival: a report from the Shanghai Breast Cancer Study. Cancer Res. 2004;64:836–839. doi: 10.1158/0008-5472.can-03-3492. [DOI] [PubMed] [Google Scholar]
- 2.Lu H, Shu XO, Cui Y, Kataoka N, Wen W, Cai Q, et al. Association of genetic polymorphisms in the VEGF gene with breast cancer survival. Cancer Res. 2005;65:5015–5019. doi: 10.1158/0008-5472.CAN-04-2786. [DOI] [PubMed] [Google Scholar]
- 3.Shu XO, Moore DB, Cai Q, Cheng J, Wen W, Pierce L, et al. Association of cyclin D1 genotype with breast cancer risk and survival. Cancer Epidemiol Biomarkers Prev. 2005;14:91–97. [PubMed] [Google Scholar]
- 4.Zhang X, Shu XO, Cai Q, Ruan Z, Gao YT, Zheng W. Functional plasminogen activator inhibitor-1 gene variants and breast cancer survival. Clin Cancer Res. 2006;12:6037–6042. doi: 10.1158/1078-0432.CCR-05-2851. [DOI] [PubMed] [Google Scholar]
- 5.Beeghly-Fadiel A, Shu XO, Long J, Li C, Cai Q, Cai H, et al. Genetic polymorphisms in the MMP-7 gene and breast cancer survival. Int J Cancer. 2009;124:208–214. doi: 10.1002/ijc.23859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chappuis PO, Kapusta L, Begin LR, Wong N, Brunet JS, Narod SA, et al. Germline BRCA1/2 mutations and p27(Kip1) protein levels independently predict outcome after breast cancer. J Clin Oncol. 2000;18:4045–4052. doi: 10.1200/JCO.2000.18.24.4045. [DOI] [PubMed] [Google Scholar]
- 7.Kriege M, Seynaeve C, Meijers-Heijboer H, Collee JM, Menke-Pluymers MB, Bartels CC, et al. Sensitivity to first-line chemotherapy for metastatic breast cancer in BRCA1 and BRCA2 mutation carriers. J Clin Oncol. 2009;27:3764–3771. doi: 10.1200/JCO.2008.19.9067. [DOI] [PubMed] [Google Scholar]
- 8.Xu Y, Yao L, Ouyang T, Li J, Wang T, Fan Z, et al. p53 Codon 72 polymorphism predicts the pathologic response to neoadjuvant chemotherapy in patients with breast cancer. Clin Cancer Res. 2005;11:7328–7333. doi: 10.1158/1078-0432.CCR-05-0507. [DOI] [PubMed] [Google Scholar]
- 9.Fagerholm R, Hofstetter B, Tommiska J, Aaltonen K, Vrtel R, Syrjakoski K, et al. NAD(P)H:quinone oxidoreductase 1 NQO1*2 genotype (P187S) is a strong prognostic and predictive factor in breast cancer. Nat Genet. 2008;40:844–853. doi: 10.1038/ng.155. [DOI] [PubMed] [Google Scholar]
- 10.Hoskins JM, Carey LA, McLeod HL. CYP2D6 and tamoxifen: DNA matters in breast cancer. Nat Rev Cancer. 2009;9:576–586. doi: 10.1038/nrc2683. [DOI] [PubMed] [Google Scholar]
- 11.Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, Hankinson SE, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stacey SN, Manolescu A, Sulem P, Rafnar T, Gudmundsson J, Gudjonsson SA, et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2007;39:865–869. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 14.Zheng W, Long J, Gao YT, Li C, Zheng Y, Xiang YB, et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat Genet. 2009;41:324–328. doi: 10.1038/ng.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Long J, Cai Q, Shu XO, Qu S, Li C, Zheng Y, et al. Identification of a functional genetic variant at 16q12.1 for breast cancer risk: results from the Asia Breast Cancer Consortium. PLoS Genet. 2010;6:e1001002. doi: 10.1371/journal.pgen.1001002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomas G, Jacobs KB, Kraft P, Yeager M, Wacholder S, Cox DG, et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1) Nat Genet. 2009;41:579–584. doi: 10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turnbull C, Ahmed S, Morrison J, Pernet D, Renwick A, Maranian M, et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet. 2010;42:504–507. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Antoniou AC, Wang X, Fredericksen ZS, McGuffog L, Tarrell R, Sinilnikova OM, et al. A locus on 19p13 modifies risk of breast cancer in BRCA1 mutation carriers and is associated with hormone receptor-negative breast cancer in the general population. Nat Genet. 2010;42:885–892. doi: 10.1038/ng.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Azzato EM, Tyrer J, Fasching PA, Beckmann MW, Ekici AB, Schulz-Wendtland R, et al. Association between a germline OCA2 polymorphism at chromosome 15q13.1 and estrogen receptor-negative breast cancer survival. J Natl Cancer Inst. 2010;102:650–662. doi: 10.1093/jnci/djq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Azzato EM, Pharoah PD, Harrington P, Easton DF, Greenberg D, Caporaso NE, et al. A genome-wide association study of prognosis in breast cancer. Cancer Epidemiol Biomarkers Prev. 2010;19:1140–1143. doi: 10.1158/1055-9965.EPI-10-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shu XO, Zheng Y, Cai H, Gu K, Chen Z, Zheng W, et al. Soy food intake and breast cancer survival. JAMA. 2009;302:2437–2443. doi: 10.1001/jama.2009.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng W, Chow WH, Yang G, Jin F, Rothman N, Blair A, et al. The Shanghai Women's Health Study: rationale, study design, and baseline characteristics. Am J Epidemiol. 2005;162:1123–1131. doi: 10.1093/aje/kwi322. [DOI] [PubMed] [Google Scholar]
- 23.Chen X, Lu W, Zheng Y, Gu K, Chen Z, Zheng W, et al. Exercise, tea consumption, and depression among breast cancer survivors. J Clin Oncol. 2010;28:991–998. doi: 10.1200/JCO.2009.23.0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Epplein M, Zheng Y, Zheng W, Chen Z, Gu K, Penson D, et al. Quality of life after breast cancer diagnosis and survival. J Clin Oncol. 2011;29:406–412. doi: 10.1200/JCO.2010.30.6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Purcell S. PLINK. http://pngu mgh harvard edu/~purcell/plink/ 2012 Available from: URL: http://pngu.mgh.harvard.edu/~purcell/plink/ [Google Scholar]
- 26.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thacker J. The RAD51 gene family, genetic instability and cancer. Cancer Lett. 2005;219:125–135. doi: 10.1016/j.canlet.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 28.Yoo S, McKee BD. Overexpression of Drosophila Rad51 protein (DmRad51) disrupts cell cycle progression and leads to apoptosis. Chromosoma. 2004;113:92–101. doi: 10.1007/s00412-004-0300-x. [DOI] [PubMed] [Google Scholar]
- 29.Takaku M, Machida S, Hosoya N, Nakayama S, Takizawa Y, Sakane I, et al. Recombination activator function of the novel R. J Biol Chem. 2009;284:14326–14336. doi: 10.1074/jbc.M807715200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cox N, Nicolae DL, Dolan ME, Gamazon ER. SCAN: SNP and CNV Annotation Database. http://www scandb org/newinterface/about html. 2012 Available from: URL: http://www.scandb.org/newinterface/about.html. [Google Scholar]
- 31.Gamazon ER, Zhang W, Konkashbaev A, Duan S, Kistner EO, Nicolae DL, et al. SCAN: SNP and copy number annotation. Bioinformatics. 2010;26:259. doi: 10.1093/bioinformatics/btp644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jia T, Liu YE, Liu J, Shi YE. Stimulation of breast cancer invasion and metastasis by synuclein gamma. Cancer Res. 1999;59:742–747. [PubMed] [Google Scholar]
- 33.Wu K, Weng Z, Tao Q, Lin G, Wu X, Qian H, et al. Stage-specific expression of breast cancer-specific gene gamma-synuclein. Cancer Epidemiol Biomarkers Prev. 2003;12:920–925. [PubMed] [Google Scholar]
- 34.Liu J, Spence MJ, Zhang YL, Jiang Y, Liu YE, Shi YE. Transcriptional suppression of synuclein gamma (SNCG) expression in human breast cancer cells by the growth inhibitory cytokine oncostatin M. Breast Cancer Res Treat. 2000;62:99–107. doi: 10.1023/a:1006418219012. [DOI] [PubMed] [Google Scholar]
- 35.Singh H, Sekinger EA, Gross DS. Chromatin and cancer: causes and consequences. J Cell Biochem Suppl. 2000 Suppl 35:61–68. [PubMed] [Google Scholar]
- 36.Tanaka K, Tsurumi C. The 26S proteasome: subunits and functions. Mol Biol Rep. 1997;24:3–11. doi: 10.1023/a:1006876904158. [DOI] [PubMed] [Google Scholar]
- 37.Luqmani YA, Al AA, Al BM, Abraham G, El ZM. Modification of gene expression induced by siRNA targeting of estrogen receptor alpha in MCF7 human breast cancer cells. Int J Oncol. 2009;34:231–242. [PubMed] [Google Scholar]
- 38.Almond JB, Cohen GM. The proteasome: a novel target for cancer chemotherapy. Leukemia. 2002;16:433–443. doi: 10.1038/sj.leu.2402417. [DOI] [PubMed] [Google Scholar]
- 39.Zheng W, Wen W, Gao YT, Shyr Y, Zheng Y, Long J, et al. Genetic and clinical predictors for breast cancer risk assessment and stratification among Chinese women. J Natl Cancer Inst. 2010;102:972–981. doi: 10.1093/jnci/djq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.