Abstract
DNA methylation is commonly thought of as a "molecular lock" that leads to permanent gene silencing. To investigate this notion, we tested 24 different HDAC inhibitors (HDACi) on colon cancer cells that harbor a GFP locus stably integrated and silenced by a hypermethylated CMV promoter. We found that HDACi efficiently reactivated expression of GFP and many other endogenous genes silenced by DNA hypermethylation. After treatment, all promoters were marked with active chromatin, yet DNA hypermethylation did not change. Thus, DNA methylation could not prevent gene reactivation by drug-induced resetting of the chromatin state. In evaluating the relative contribution of DNA methylation and histone modifications to stable gene silencing, we followed expression levels of GFP and other genes silenced by DNA hypermethylation over time after treatment with HDACi or DNA demethylating drugs. Reactivation of methylated loci by HDACi was detectable for only 2 weeks, whereas DNA demethylating drugs induced permanent epigenetic reprogramming. Therefore, DNA methylation cannot be considered as a lock for gene expression, but rather as a memory signal for long-term maintenance of gene silencing. These findings define chromatin as an important druggable target for cancer epigenetic therapy and suggest that removal of DNA methylation signals is required to achieve long-term gene reactivation.
Introduction
Epigenetic marks such as histone modifications and DNA methylation are involved in cell memory expression patterns which are transmitted through cell division (1). Chromatin modifications are required in all organisms while DNA methylation is not present in some lower organisms like worms and flies, suggesting that chromatin has a broader epigenetic function in gene regulation (1). Histone acetylation is associated with open chromatin and gene expression while removal of acetyl groups by histone deacetylases is observed in inactive chromatin. In higher organisms, DNA methylation plays an important role in several physiological processes, including X chromosome inactivation, genomic imprinting, silencing of germ cell specific genes and repetitive elements (2). In cancer, tumor suppressor genes (TSG) are silenced by both DNA hypermethylation and chromatin repressive marks (2, 3).
A common hypothesis is that DNA methylation serves as a «molecular lock» that prevents reprogramming and is responsible for stable gene silencing (1, 4, 5). This concept was built on indirect observations whereby hypermethylated genes in cancer cells could be reactivated only after removal of promoter DNA hypermethylation using hypomethylating drugs such as decitabine (5-aza-2’-deoxycytidine, 5-AZA-CdR). By contrast chromatin acetylation induced by histone deacetylase inhibitors (HDACi) such as Trichostatin A (TSA) could not reactivate hypermethylated genes in cancer cells (6–8). However, more recently some reports have shown that HDACi such as TSA and depsipeptide (Depsi) produce gene reactivation from hypermethylated promoters without any changes in DNA methylation at the promoter level (9–12). Since these reports were against the current paradigm, a more detailed look at this issue was needed.
One of the problems in studying DNA methylation-associated silencing of TSG is that reactivation of these genes can impair cellular growth and be difficult to detect and quantitate. A selectable system was recently described to overcome this issue. YB5 cells are derived from the SW48 colon cancer cell line transfected with a green fluorescent protein (GFP) driven by a hypermethylated cytomegalovirus (CMV) promoter packaged into inactive chromatin. YB5 carries a single copy of CMV/GFP stably integrated in chromosome 1. GFP can be reactivated in YB5 cells by treatment with 5-AZA-CdR when its promoter region is demethylated and also marked by active chromatin signals characterized by H3K9 acetylation, low level of H3K27 trimethylation and low nucleosome density (13).
In this paper, we use YB5 cells (and 5 other cancer cell lines) to show that the vast majority of HDACi tested can reactivate genes silenced by promoter hypermethylation without detectable changes in DNA methylation. We further show that while DNA methylation cannot prevent gene activation by chromatin reprogramming, it is essential for long-term gene silencing.
Materials and Methods
Cell culture and drug treatments
All cell lines were obtained from American Type Culture Collection. Cell lines were authenticated at MD Anderson Cancer Center with short tandem repeat PCR method. YB5 cell line is a colon cancer cell line generated from SW48 as previously described (13). YB5 cell line was cultured in L-15 medium while MCF-7, K562, MDA-MD-231, and PC-3 cells were cultured in RPMI 1640. Both cell culture media contained 10% fetal bovine serum and 100 µg/mL Penicillin-Streptomycin solution. Cells growing in log phase were treated with decitabine (5-AZA-CdR) at 50 nM for 72h. Drug and medium were replaced every day. Cells were cultured an additional 24h without drug prior to analysis. HDAC inhibitors (HDACi) were dissolved either in DMSO, ethanol or PBS according the manufacturers’ recommendations. HDACi were added for 24h at various concentrations prior to analysis.
Histone extraction and Western blots
Total protein extracts were prepared using RIPA buffer as described previously (14) in the presence of sodium butyrate (5 mM) to avoid in vitro histone deacetylation and resolved on 15% SDS-polyacrylamide gels. Antibodies used were H3K9-acetylation (07–352 Millipore), H4K16-acetylation (39168, Active motif) and total H3 (ab1791 Abcam).
Fluorescence-activated cell sorting analysis and cell sorting
YB5 cells were trypsinized and stained with propidium iodide (PI). GFP and PI fluorescence were measured by Gallios flow cytometer (Beckman Coulter). Data were analyzed using Kaluza® software. GFP cell sorting was performed using BD FACSAriaII. GFP fluorescence of samples was analyzed post-sorting to assess the purity of the sorted cells.
RNA extraction, cDNA synthesis, quantitative real-time PCR
Total RNA (2 µg) was extracted using Trizol (Invitrogen) and cDNA was synthesized using High-Capacity cDNA Kit (Applied Biosystems). Quantification of cDNA was done by qPCR with the Universal PCR Master Mix (Bio-Rad) using ABI Prism 7900HT. Results were obtained from at least three independent experiments where each sample was analyzed in duplicate. 18S was used as a reference gene. cDNA synthesis used the same amount of RNA after treatment with different drugs. All primers, except GFP primers that were described previously (13), are listed in supplemental Table S1. 5′ Rapid amplification of cDNA ends (5′-RACE) was done as previously described (13).
DNA extraction and DNA methylation analysis
DNA extraction and bisulfite conversion, pyrosequencing and bisulfite cloning/sequencing were carried out as previously described (13). All primers are listed in supplemental Table S1, except for all GFP primers that were described previously (13).
Chromatin immunoprecipitation (ChIP)
ChIP was performed as described previously (14). Antibodies used were anti–histone H3 (ab1791, Abcam), anti–histone H3K9 acetylation (07–352, Millipore), anti–histone H3K27 trimethylation (07–449, Millipore), anti-histone H3K4 trimethylation (17–614, Millipore), anti-histone H3K36 trimethylation (ab9050, Abcam) and anti-IgG (ab46540, Abcam). Quantification of ChIP DNA was done by qPCR, and primers/probes are listed in supplemental Table S1. Each ChIP assay was validated using targets for the various modifications (ACTB for active histone mark and LINE-1 for inactive histone mark). The value of each histone modification was determined by H3 and IgG normalization using the equation: Fold enrichment = 2^-[Ct(Ab) − Ct(H3)] – 2^-[Ct(IgG) − Ct(H3)].
Gene expression and DNA methylation microarrays
Gene expression analysis was performed using the Agilent whole genome array (G4112F) that was scanned using the Agilent G2505B scanner. Data represents the average expression level of two independent experiments. DNA methylation analysis using high-throughput methylation profiling by MCA coupled to CpG island microarray (MCAM) was performed as described previously (15). After analysis, genes with M values more than 1.3 were considered methylated. Microarray data sets were deposited in the Gene Expression Omnibus (GEO) database with the accession number GSE34077.
Statistical analysis
Differences between groups were assessed using a t-test. A 2-sided P value of 0.05 was considered statistically significant.
Results
HDAC inhibitors induce GFP reactivation from a DNA hypermethylated promoter
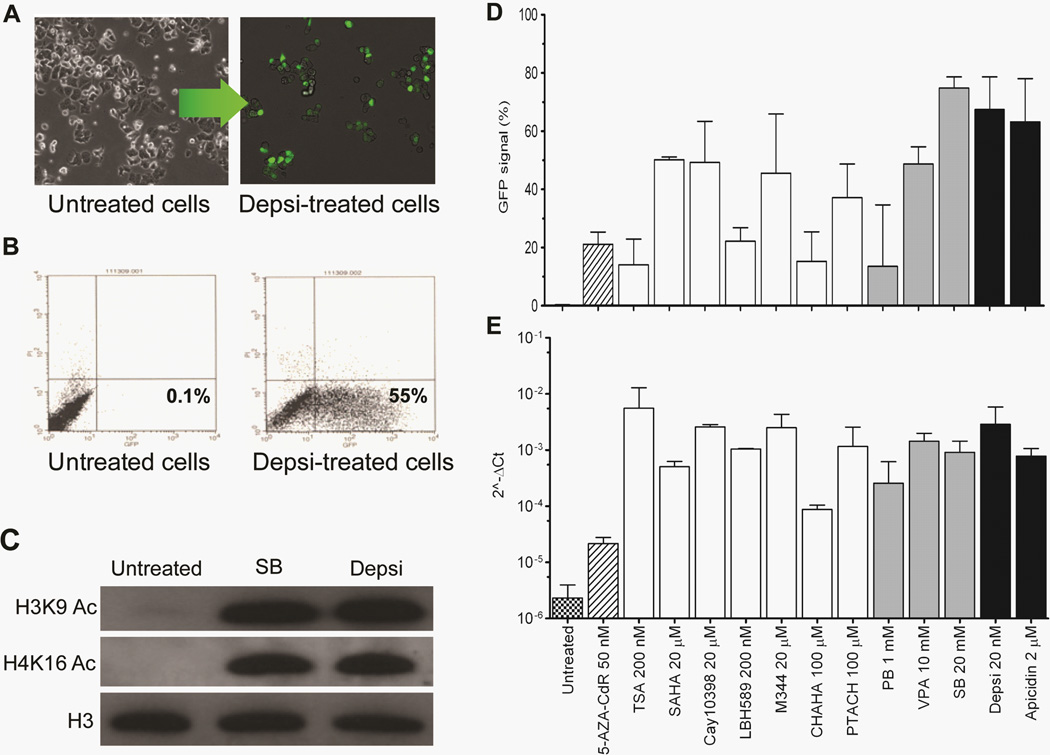
To examine the effects of HDACi on gene silencing by DNA methylation, YB5 cells were treated with 24 different HDACi that belong to 8 different chemical classes in a wide range of concentrations (Table 1) and 17 of them reactivated GFP. Although GFP reactivation was detected in a wide range of concentrations (even at high doses, Table 1), the subsequent experiments were performed at doses where the percentage of dead cells after treatment was less than 30% and where GFP reactivation was the highest as detected by FACS analysis. Following treatment with depsipeptide (Depsi, for 24h at 20 nM), GFP expression was detectable in ~50% of cells as seen by fluorescent microscopy (Fig. 1A), quantified by FACS analysis (Fig. 1B), and associated with extensive global histone acetylation (Fig. 1C). HDACi produced GFP mRNA and GFP fluorescence as early as 12h after treatment (Figure S1 A–B). GFP reactivation was not specific to a molecular structure or chemical class of these epigenetic drugs since the vast majority of HDACi tested activated this hypermethylated locus (Fig. 1D and Table 1). Moreover, GFP fluorescence and mRNA levels were stronger after a 24h treatment with HDACi than after a 72h treatment with 5-AZA-CdR (Fig. 1D–E). We confirmed by 5’RACE experiments that GFP mRNA originated from its own promoter and not from an alternative transcription start site (data not shown).
Table 1.
Effects of various HDAC inhibitors (HDACi) on GFP expression in YB5 cells detected by FACS analysis after a 24h treatment. HDACi targets (specific isoforms or entire HDAC class) are shown in the table. Concentration (Conc.) range tested on the YB5 system is indicated in the table.
| Drugs | Structural class | Target HDAC | Conc. tested | GFP expression |
|---|---|---|---|---|
| SAHA | Hydroxamic acid | I, IIa, IIb, IV | 0.3–20 µM | Yes |
| TSA | Hydroxamic acid | I, II | 50–200 nM | Yes |
| Oxamiflatin | Hydroxamic acid | HDAC 3, 6 | 100–400 nM | Yes |
| APHA8 | Hydroxamic acid | HDAC 1 | 1–25 µM | Yes |
| Cay 10398 | Hydroxamic acid | HDAC 1 | 10–40 µM | Yes |
| M344 | Hydroxamic acid | HDAC 1, 6 | 1–4 µM | Yes |
| CHAHA | Hydromaxic acid | HDAC 6 | 0.2–1.5 µM | Yes |
| LBH-589 | Hydromaxic acid | Pan-HDACi | 0.02–20 µM | Yes |
| Depudicin | TSA-like but non hydroxamate | HDAC 1 | 5–25 µM | Yes |
| PTACH | TSA-like but non hydroxamate | --- | 1–10 µM | Yes |
| Valproic acid | Short-chain fatty acid | HDAC 2 | 0,25–10 mM | Yes |
| Phenylbutyrate | Short-chain fatty acid | I | 1 mM | Yes |
| Sodium butyrate | Short-chain fatty acid | I, IIa | 5–20 mM | Yes |
| Depsipeptide | Cyclic peptide | HDAC 1, 2 | 1 nM-2 µM | Yes |
| Apicidin | Cyclic peptide | I, II | 0.4–40 µM | Yes |
| HC-Toxin | Cyclic peptide | HDAC 1, 2, 3, 8 | 25–100 nM | Yes |
| Sirtinol | Hydroxy-naphthaldehyde | Sirt 2 | 1.25–100 µM | Yes |
| Splitomicin | Naphthalenes | Sirt 2 | 0.1–75 µM | No |
| Ex527 | Tetrahydrocarbazoles | Sirt 1 | 20–100 µM | No |
| CHIC-35 | ND | Sirt 1 | 0.1–1 µM | No |
| Cambinol | β-naphthol | Sirt1, 2 | 2.5–25 µM | No |
| PCI-34051 | Hydroxamic acid | HDAC 8 | 1–10 µM | No |
| BATCP | ND | HDAC 6 | 1–10 µM | No |
| Scriptaid | Hydroxamic acid | HDAC 1, 2, 8 | 0.05– 1 µM | No |
ND: not determined.
Figure 1.
Various HDAC inhibitors (HDACi) induce significant expression of genes silenced by promoter DNA hypermethylation. A, Representative pictures by fluorescent microscopy and B, FACS analysis of untreated YB5 cells and treated with the HDACi depsipeptide (Depsi) at 20 nM for 24h. On the FACS scatter plot, the x-axis and the y-axis represent respectively GFP and propidium iodide (PI) fluorescence. C, Western blots show H3K9 and H4K16 acetylation after a 24h treatment with sodium butyrate (SB) at 20 mM and Depsi at 20 nM. D, GFP protein expression detected by FACS analysis and E, GFP mRNA expression detected by qPCR after a 24h treatment using 12 different HDACi (n>3). HDACi that belong to the hydroxamic acid group are shown in white bars whereas the short-chain fatty acid and the cyclic peptide HDACi are respectively shown in grey and black bars. GFP expression is also obtained after a treatment with 5-AZA-CdR, at 50 nM daily for 72h followed by 24h without drug, as a positive control. Each experiment was done three separate times: shown is the mean ± SEM.
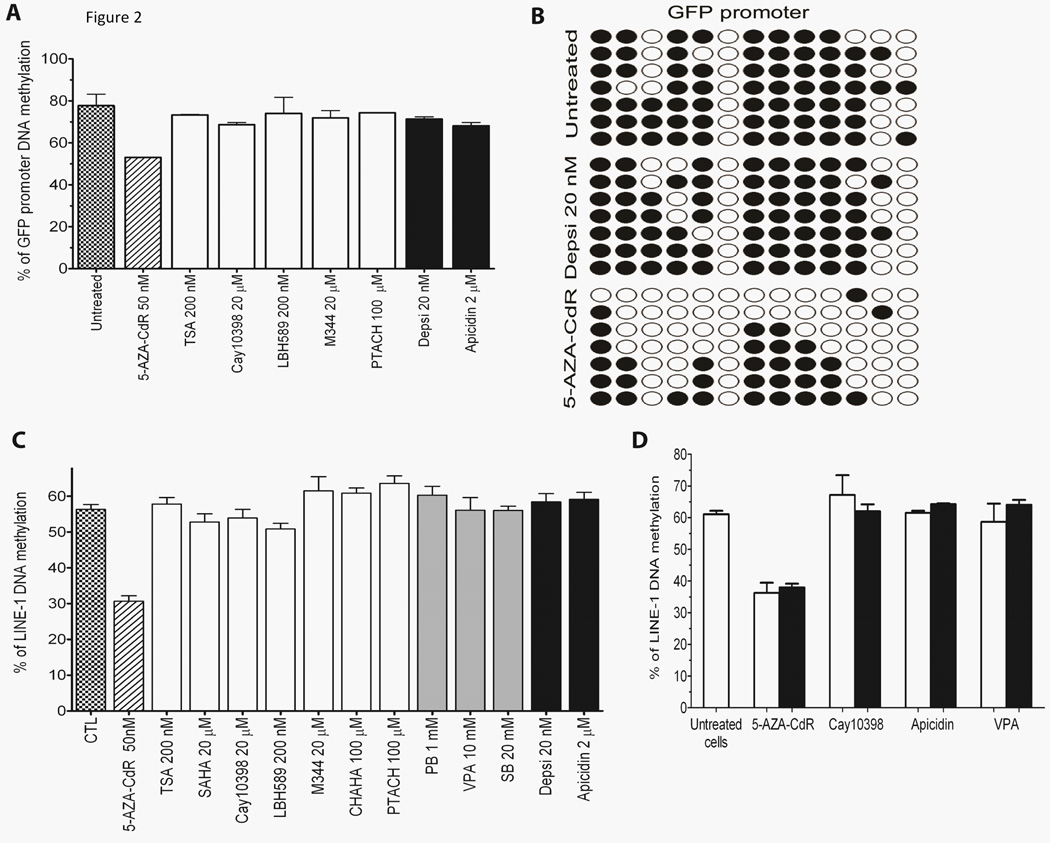
It has previously been suggested that HDACi can lead to DNA demethylation (16–18). To test this, DNA methylation levels were measured after treatment with 7 different HDACi and 5-AZA-CdR was used as a control for DNA hypomethylation. Analyses were performed by bisulfite pyrosequencing and cloning/sequencing at the GFP promoter (respectively Fig. 2A–B). No changes were detected after treatment with any of the HDACi tested after 24h treatment. Similarly, there were no effects on global DNA methylation assessed by bisulfite pyrosequencing of LINE-1 methylation after 24h treatment (Fig. 2C) or 10 days following treatment (Fig. 2D). Only treatment with 5-AZA-CdR reduced DNA methylation levels. These results and others (19) clearly show that HDACi do not alter DNA methylation levels of cancer cells. Thus, HDACi can induce gene reactivation through a DNA hypermethylated promoter without any change in DNA methylation levels. These results do not support the lock hypothesis and are in agreement with more recent findings demonstrating that HDACi can reactivate hypermethylated genes.
Figure 2.
HDAC inhibitors do not alter DNA methylation at promoter levels. A, DNA methylation analysis of the GFP promoter region by pyrosequencing. YB5 cells were treated for 24h with different HDACi at the doses indicated in the graph (n>3). B, DNA methylation analysis of GFP promoter CGI by bisulfite cloning/sequencing in YB5 cells untreated, and treated with Depsi (20 nM for 24h) or 5-AZA-CdR (50 nM for 72h). White and black circles indicate unmethylated and methylated cytosine within a CpG site, respectively. C, DNA methylation analysis of LINE-1 promoter analyzed by bisulfite pyrosequencing after 24h treatment with different HDACi at the doses indicated in the graph (n>3). HDACi that belong to the hydroxamic acid group are shown in white bars whereas the short-chain fatty acid and the cyclic peptide HDACi are shown in grey and black bars, respectively. D, DNA methylation analysis of LINE-1 promoter analyzed by bisulfite pyrosequencing immediately after treatment (white bars) or after 10 days of culture without any drugs (black bars; n=2). YB5 cells were treated with different HDACi (Cay10398 at 20 µM, Apicidin at 2 µM and VPA at 10 mM) for 24h. In all experiments, 5-AZA-CdR treatment at 50 nM for 72h was used as a positive control for DNA demethylation.
HDACi induce gene reactivation of endogenous genes silenced by promoter DNA hypermethylation
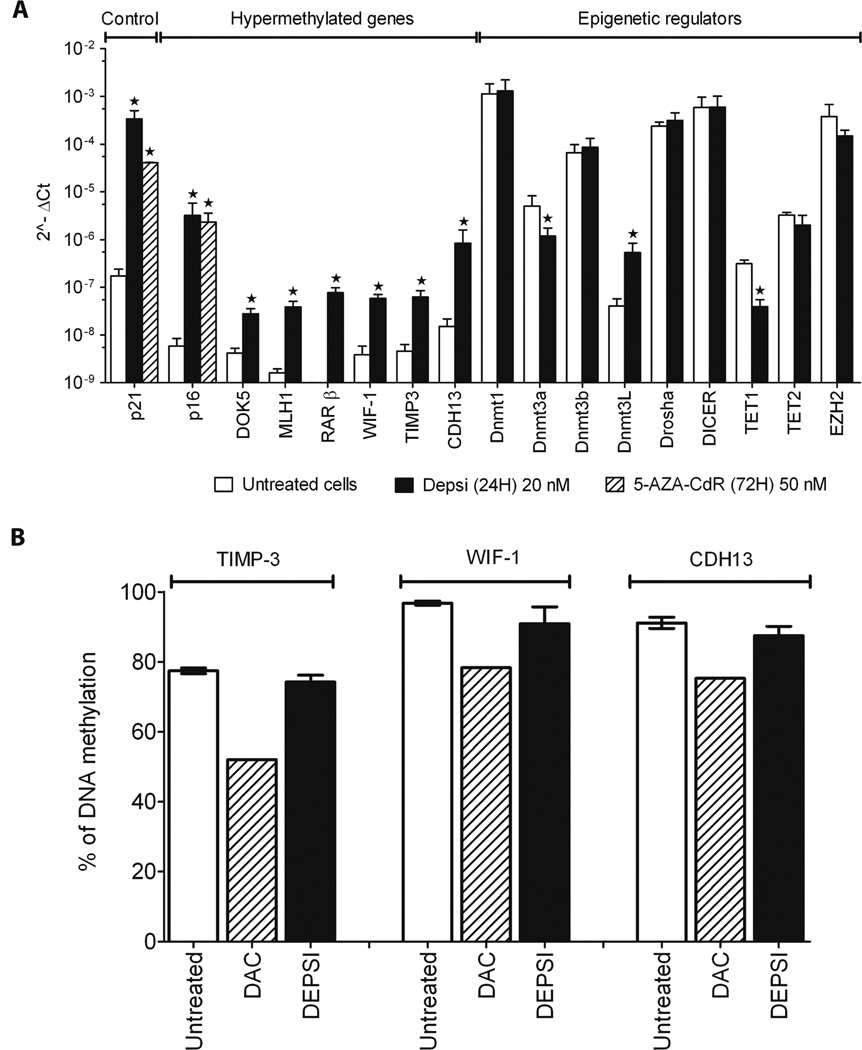
Since these data are not in agreement with other studies on gene reactivation induced by HDACi (6, 7), we asked whether this effect was specific to the GFP locus or could be observed in other methylated genes in various cancer cell lines. First, we analyzed in YB5 cells gene reactivation of other hypermethylated genes in response to Depsi (Fig. 3A) and other HDACi (Fig. S2A–B). For this, we selected 7 TSG silenced by DNA hypermethylation in YB5 cells (85–100% methylation as detected by bisulfite pyrosequencing). These play roles in mediating cell adhesion (CDH13), metastasis (TIMP-3), cell cycle (P16), DNA mismatch repair (MLH1), Wnt pathway signaling (WIF-1), MAP kinase signaling (DOK5), and cell differentiation (RAR-β). Among these, all but one (RAR-β) are driven by promoter CpG Islands (CGI) (20). These genes are epigenetically inactivated in many cancers. 24h treatment with Depsi and other HDACi reactivated all these hypermethylated genes as detected by qPCR (Fig. 3A and Fig. S2A–B), while DNA methylation levels did not change as compared to untreated cells (Fig. 3B). These results were extended to four other cancer cell lines (K562, a CML cell line; MCF-7 and MDA-MB-231, breast cancer cell lines; and PC-3, a prostate cancer cell line) with six different genes (CDH1, MGMT, NPM2, RASSF1A, DOK5, and PDLIM4; Fig. S3 A–D) whose promoter methylation levels vary between 65 and 100% methylation as detected by pyrosequencing. Most of them showed reactivation after HDACi treatment. The induction of p21, a cyclin-dependent kinase inhibitor, by HDACi was used as a positive control for HDACi activity since it is considered as a hallmark of the effect of HDACi on gene expression. Our data are in agreement with previous reports on single genes that HDACi such as TSA, phenylbutyrate and LBH589 (9, 11, 12, 21, 22) or SIRT1 inhibition (10) can induce gene reactivation of hypermethylated genes without alteration in DNA methylation. Our findings on numerous genes and in different cancer cell lines demonstrate that chromatin acetylation induced by HDACi overcomes DNA hypermethylation silencing and induces gene reactivation. All together, these data demonstrate that chromatin remodeling allows a subset of TSG silenced by DNA hypermethylation to be reactivated in response to HDACi.
Figure 3.
HDACi reactivates endogenous hypermethylated genes in YB5 cell line. A, Gene expression was detected by qPCR of untreated (white bars) and treated YB5 cells with Depsi at 20 nM for 24h (black bars). Tested genes were divided in three categories: control genes, hypermethylated genes and epigenetic regulators. A star indicates a significant difference between untreated and treated cells (p<0.05). Each experiment was done three separate times: shown is the mean ± SEM. B, DNA methylation analysis of TIMP-3, WIF-1 and CDH13 promoters analyzed by pyrosequencing. YB5 cells were treated with hypomethylating drug 5-AZA-CdR or with HDACi Depsi (n>3).
Chromatin remodeling despite DNA hypermethylation
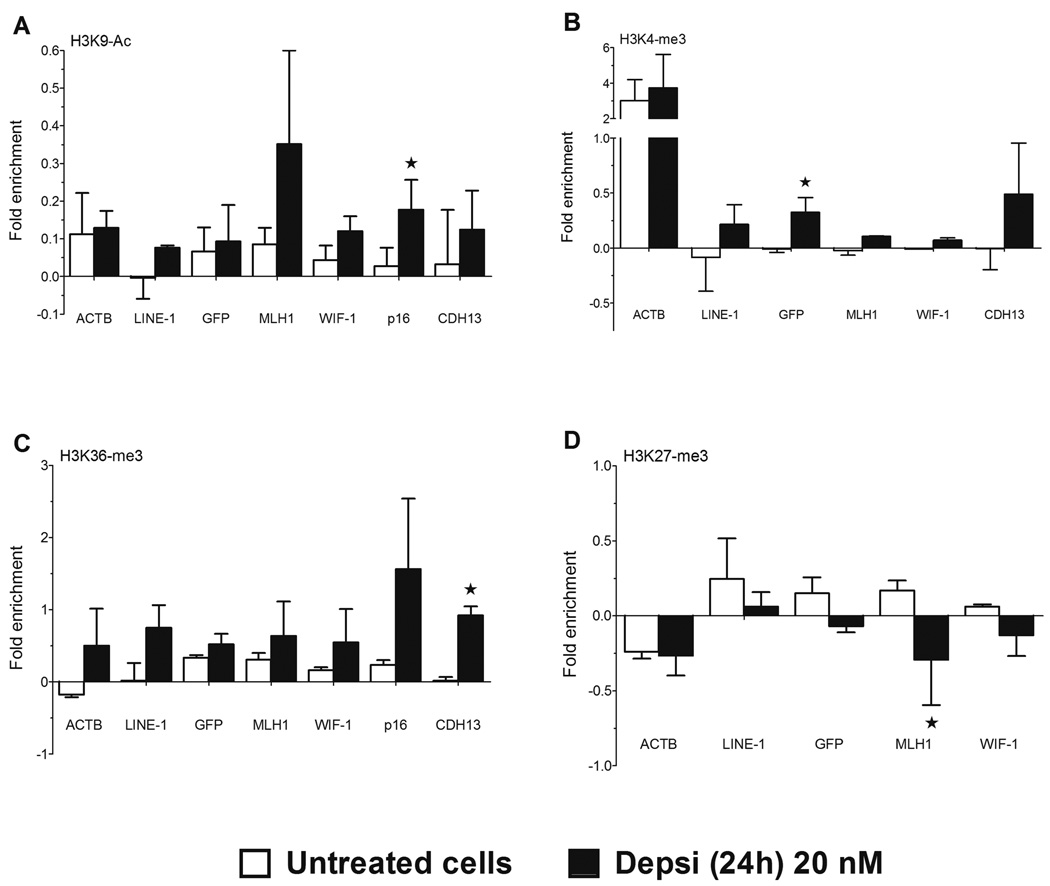
Since HDACi-induced gene reactivation was not associated with DNA demethylation, we investigated the effect of the treatment on chromatin modifications at the promoter regions of these hypermethylated genes. We performed chromatin immunoprecipitation assays (ChIP) coupled with qPCR analysis for H3K9-Ac, H3K4-me3, H3K36-me3 (marks associated with gene activity), and H3K27-me3 (modification associated with gene repression) in YB5 cells untreated and treated with Depsi for 24h at 20 nM (Fig. 4A–D). Interestingly, we recently reported that regardless of DNA methylation status, gene reactivation is associated with a promoter region marked by active chromatin marks and low nucleosome density (13). Following Depsi treatment, all promoter regions of GFP, CDH13, MLH1, and WIF-1 showed an increase in activating marks (1.5 to 24.5 fold) such as H3K9-Ac (Fig. 4A), H3K4-me3 (Fig. 4B), and H3K36-me3 (Fig. 4C). By contrast, H3K27-me3, a surrogate for chromatin inactivated by polycomb, was reduced after Depsi treatment by 1.4 to 4 fold (Fig. 4D). Interestingly, Depsi treatment did not seem to have modified nucleosome density (measured by ChIP-PCR of H3/input) on the promoter region of these genes (data not shown). To rule out indirect effects through other chromatin regulators, we measured the expression of DNMT1, DNMT3a, DNMT3b, DNMT3L, DROSHA, DICER, TET1, TET2, and EZH2. We found that most did not change significantly after Depsi treatment (Fig. 3A). These data show that HDACi directly induced chromatin remodeling and this was associated with gene reactivation from DNA hypermethylated promoters.
Figure 4.
Depsipeptide induces chromatin modifications on promoters of DNA hypermethylated genes. Chromatin-immunoprecipitation analysis of A, H3K9-Ac; B, H3K4-me3; C, H3K36-me3, and D, H3K27-me3 were performed in YB5 cells untreated (white bars) and treated with Depsi at 20 nM for 24h (black bars). Chromatin status at the promoter of GFP, CDH13, MLH1, P16 and WIF-1 was quantified by qPCR and expressed as fold enrichment, using the following equation 2^-[Ct(Ab) – Ct(H3)] – 2^-[Ct(IgG) – Ct(H3)]. Histone marks at ACTB and LINE-1 promoters were used as controls for active and inactive genes, respectively. Each experiment was done three separate times: shown is the mean ± SEM. A star indicates a significant difference between untreated and treated cells (p<0.05).
Genome wide effects of Depsipeptide on hypermethylated genes
We next investigated on a genomic scale the effect of Depsi on gene reactivation of hypermethylated genes. We combined the data of two independent gene expression microarrays of untreated and Depsi-treated YB5 cells (Fig. S4A) with whole genome DNA methylation data using Methylation CpG island microarray (MCAM, Fig. S4B) (15). As previously reported by other groups (23, 24), we found that HDACi increased the expression of 11% of the genes with the same amount of genes being repressed (11%; Fig. S4D). Whole genome methylation data showed that this colon cancer cell line has more than 330 detectable hypermethylated promoter CGIs (i.e, 6% of the gene promoters on the array; Fig. S4B). When combining the data of both gene expression and DNA methylation microarrays, we could analyze gene expression and DNA methylation of more than 4,300 genes (Fig. S4C). In this gene list, 258 genes showed promoter DNA hypermethylation and 8% (21 genes) of these were reactivated following HDACi treatment (Fig. S4E–F). Considering that 80–90% of hypermethylated genes in cancer are not expressed in normal tissues and thus lack the appropriate transcription factor for activation, our data suggest that the majority of ‘inducible’ genes are actually induced by HDACi (25). By contrast 14% of the unmethylated genes could be reactivated by treatment with Depsi.
DNA methylation is a long-term memory signal for gene silencing
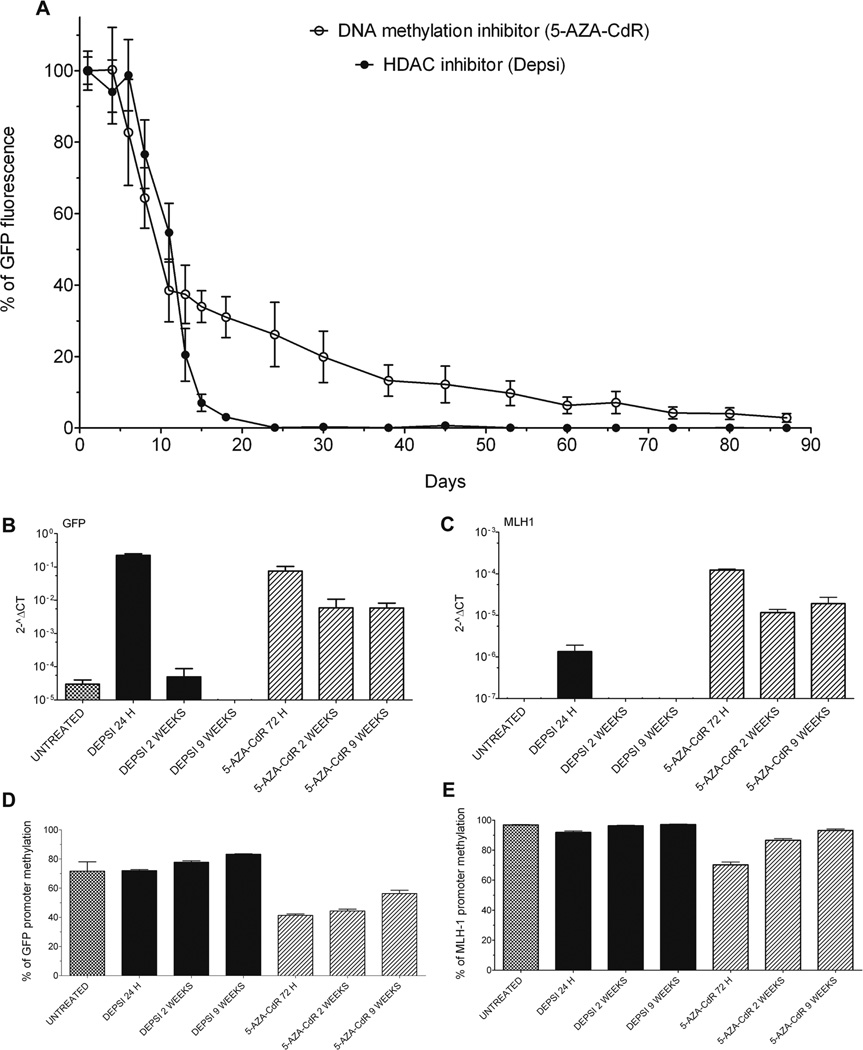
The data described so far demonstrate that rapid activation of a DNA hypermethylated promoter is possible with strong drug-induced chromatin acetylation. These results raise the question of the importance of DNA methylation in gene silencing. To study the relative contribution of DNA methylation versus chromatin modifications in gene silencing, we compared the long-term effects of Depsi and 5-AZA-CdR treatment on gene expression and DNA methylation. YB5 cells were treated with Depsi (24h) or 5-AZA-CdR (72h) and were then subjected to cell sorting to obtain enriched GFP+ cell populations (purity ~70% of GFP+ cells for each treatment conditions). GFP positive cells were cultured post-sorting without drugs and GFP expression was followed for more than 3 months by FACS analysis (Fig. 5A). During the first week post-sorting, the population of Depsi-treated YB5 cells was mostly GFP+. Ten days post-sorting, approximately 60% of the cells treated with Depsi lost GFP expression and 2 weeks post-treatment, GFP expression was rare among these cells. These data were confirmed with other HDACi such as VPA, Apicidin, Cay 10398, and TSA (data not shown). GFP expression was undetectable 25 days following Depsi-treatment and was similar to untreated cells for the rest of the experiment.
Figure 5.
Chromatin remodeling reactivated transiently expression of hypermethylated genes while DNA hypomethylation could stably reactivate gene expression. A, YB5 cells were either treated 24h with Depsi at 20 nM or 72h with 5-AZA-CdR at 50 nM. GFP+ cells were enriched by cell sorting (purity ~70%). Time course of GFP fluorescence was measured by FACS and expressed as a percentage of GFP fluorescence measured immediately post-sorting (n=3). B, GFP and C, MLH1 expression levels were measured by qPCR in YB5 untreated cells or sorted cells after the treatment with Depsi or 5-AZA-CdR at the time indicated on the graph (n=3). DNA methylation analysis of D, GFP and E, MLH1 promoters by pyrosequencing of YB5 untreated cells or sorted cells treated with Depsi or 5-AZA-CdR at the time indicated on the graph (n=3).
Results obtained with YB5 cells treated with 5-AZA-CdR exhibited a different pattern (Fig. 5A). For the first week, the vast majority of the cells exhibited GFP fluorescence. Then, the percentage of cells showing GFP fluorescence decreased to 50% and 35% after 10 days and 25 days post-treatment, respectively. The number of GFP+ decreased slowly thereafter and after 3 months, 3% of YB5 cells treated with 5-AZA-CdR still exhibited GFP fluorescence (Fig. 5A). We similarly investigated the RNA expression of several hypermethylated genes including GFP (Fig. 5B), MLH1 (Fig. 5C), CDH13 (Fig. S5A), WIF-1 (Fig. S5B), and TIMP-3 (Fig. S5C) and found that gene expression was activated immediately by treatment with either epigenetic drug, but expression was silenced 2 weeks after Depsi treatment while it was maintained following treatment with 5-AZA-CdR for up to 9 weeks. Interestingly, other chromatin modifiers such as histone methyltransferase inhibitors were previously shown to induce transient gene activation which returned to the original state upon drug removal (26). As previously mentioned, after Depsi treatment, DNA methylation in the promoter regions of these hypermethylated genes did not change. By contrast, after 5-AZA-CdR treatment, methylation levels decreased significantly at GFP (Fig. 5D), MLH1 (Fig. 5E), CDH13 (Fig. S5D), WIF-1 (Fig. S5E), TIMP-3 (Fig. S5F), and LINE-1 (Fig. S5G).
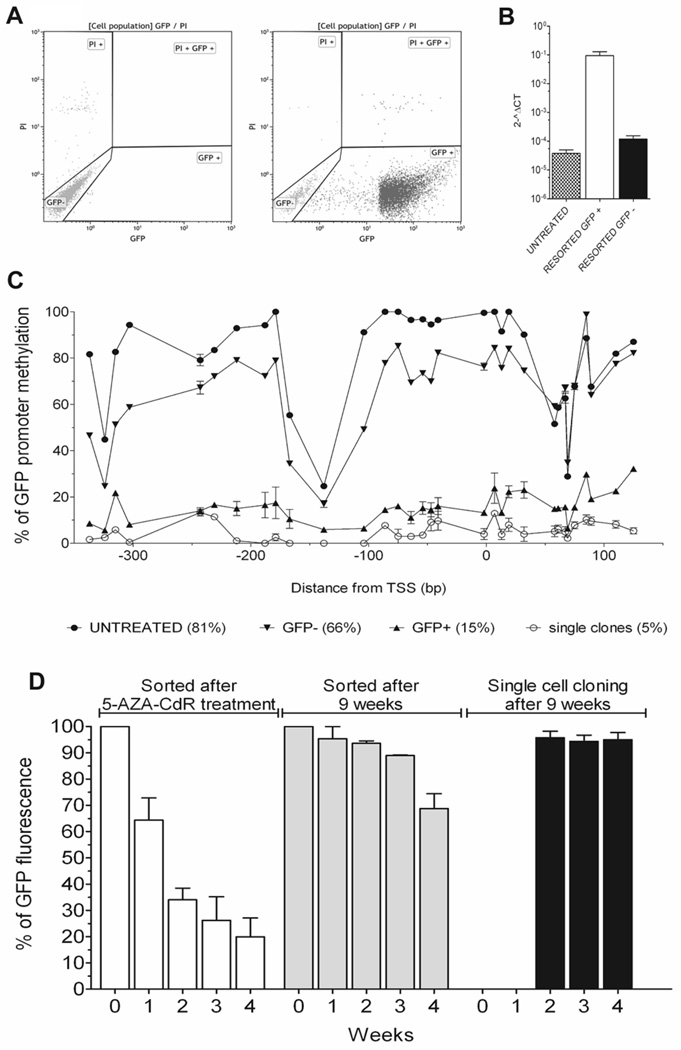
The gradual loss of GFP expression after 5-AZA-CdR withdrawal was coincided with gradual remethylation of the CMV promoter. Although it has been reported that the p16CDKN2A/INK4 locus was remethylated after 5-AZA-CdR removal, it has been suggested that the apparent remethylation was due to clonal replacement by a subset of cancer cells that were not affected by 5-AZA-CdR (19, 27). Cells treated with hypomethylating agents tend to have a longer cell cycle because of the reexpression of growth regulatory signals. Therefore, hypomethylated cell populations can be easily replaced by more rapidly growing methylated populations, which can bias the measurement of DNA methylation (19). To address this issue, we performed two series of cell sorting experiments using YB5 cells cultured 9 weeks without drug following initial 5-AZA-CdR treatment (Fig. 6A). The purity of the sorted GFP+ cells exceeded 90% while the GFP− cells contained only 0.2% GFP positive cells. Gene expression and DNA methylation analysis of GFP+ and GFP− cells at 9 weeks post-treatment revealed that gene reactivation in GFP+ (Fig. 6B) was at this late time point associated with an unmethylated promoter region with an average methylation of 15% (Fig. 6C). GFP− cells exhibited 1000 times less GFP mRNA and a promoter closer to the methylation level of untreated cells with an average methylation of 66% (Fig. 6B–C). This gene expression pattern was also observed for other TSG such as TIMP-3, CDH13, and MLH1 while their DNA methylation level was reduced in the GFP+ and GFP− cells (Figure S6A–F). Global DNA methylation measured by the LINE-1 assay did not change significantly between untreated cells, GFP+ and GFP− cells (Figure S6G).
Figure 6.
DNA methylation serves as a mechanism for gene expression memory. Cell sorting of GFP+ cells was performed 9 weeks following 5-AZA-CdR treatment. A, YB5 cells were sorted twice for maximum of purity. GFP− cells (Left panel; 0.2% of GFP+ cells) and GFP+ cells (Right panel; 90% of GFP+ cells) were expanded prior to molecular analysis of gene expression and DNA methylation. B, GFP expression measured by qPCR of YB5 cells untreated or sorted GFP+ and GFP−, 9 weeks post-5-AZA-CdR treatment. C, DNA methylation analysis performed by pyrosequencing of the entire GFP promoter region of these sorted YB5 cells. Interestingly, average DNA methylation levels at this promoter were 81% and 66 % in untreated cells and GFP− cells, respectively. Average DNA methylation levels in this locus was only 15 % in GFP+ cells. Moreover, methylation level of single clones isolated by cell sorting, 9 weeks following 5-AZA-CdR treatment, showed 5 % methylation levels which is similar to the background of the assay. D, Time-dependent GFP expression measured by FACS analysis on YB5 cells sorted immediately following 5-AZA-CdR treatment at 70 % purity of GFP+ cells (left), 9 weeks post-5-AZA-CdR treatment after 2 additional rounds of cell sorting, at 90 % purity of GFP+ cells (middle), and after single cell cloning for GFP+ cells performed 9 weeks after the initial 5-AZA-CdR treatment (right).
In order to eliminate the effect of clonal replacement, we performed cell sorting and single cloning experiments (Fig. 6D). After clonal expansion of these single clones to obtain enough cells (~2 weeks), we monitored their GFP fluorescence over time (black bars, Fig. 6D) as compared to sorted cells obtained after 5-AZA-CdR treatment when the purity was ~70% (white bars, Fig. 6D) and 9 weeks after treatment when the purity exceeded 90% (grey bars, Fig. 6D). Single cell clones of these GFP+ YB5 cells obtained 9 weeks after 5-AZA-CdR without drugs revealed that 92–97% stably express GFP for up to 6 months post-treatment demonstrating stable epigenetic reprogramming (Fig. 6D). Interestingly, the GFP promoter region was completely demethylated in these GFP+ clones (Fig. 6C). These results clearly show that DNA methylation is the molecular mechanism responsible for long-term gene silencing. Thus, full epigenetic reprogramming and switching from the silent to the expressed state can be accomplished by complete promoter demethylation which is correlated with RNA pol II occupancy (28).
Discussion
Early studies have reported that TSG silenced by promoter DNA hypermethylation could be reactivated only after the removal of methylation marks. In these studies, treatment with TSA, an HDACi, could not produce gene reactivation of genes silenced by promoter DNA hypermethylation (6, 7). Based on these indirect observations, the function of promoter DNA methylation, as a signal for gene silencing, has been considered as a ‘lock’ for gene expression. However, other studies have reported that hypermethylated genes can be reactivated by TSA and other HDACi without any loss in promoter DNA methylation (9–12). These reports put in jeopardy the lock hypothesis which has been the paradigm for more than a decade. Hence, we chose to investigate this issue by looking at the effects of more than 20 different HDACi on reactivation of genes silenced by promoter DNA methylation.
Using the well-characterized YB5 system and other cancer cell lines, we discovered that most of the HDACi-tested could reactivate hypermethylated genes in a dose-dependent pattern regarding their chemical class and HDAC affinity. DNA methylation analysis revealed that gene reactivation was generated without any loss of promoter DNA methylation. Methylation levels were carefully assessed before and after treatment by pyrosequencing and bisulfite cloning sequencing since it was reported that HDACi could potentially reduce DNA methylation levels by non-specific mechanisms (16–18). However, our study, as well as others (19) show that methylation levels did not change 24h after HDACi exposure or several days post-treatment. Therefore, these data confirm that HDACi can reactivate gene expression through hypermethylated promoters, which demonstrates that DNA methylation does not lock gene expression in that it does not prevent reactivation by chromatin remodeling. It is not clear why previous studies reported that HDACi do not reactivate the expression of hypermethylated genes, though it may relate to the use of low doses of HDACi for short periods of time, and the use of insensitive methods for gene expression analysis. Alternately, it is possible that some genes/cell lines are resistant to this effect, though we observed it for most genes and most cell lines tested.
The fact that DNA methylation does not lock gene expression raises the question of the relative contribution of DNA methylation and chromatin modifications to gene silencing. The YB5 system was particularly suitable to investigate this issue since after treatment with either Depsi or with 5-AZA-CdR, we were able to sort the GFP-expressing cells and monitor GFP fluorescence for several months. We discovered that a treatment with HDACi can transiently reactivate hypermethylated genes (GFP and other TSG) for up to 2 weeks without any changes in DNA methylation level in their promoter regions. On the other hand, treatment with 5-AZA-CdR leads to gene reactivation of GFP and other TSG for several months. Moreover, the decline in GFP-expressing cells after 5-AZA-CdR, thought largely to be due to remethylation, is in fact attributable in part to clonal replacement by YB5 cells that are methylated and do not express GFP. Indeed, cell sorting and single cell cloning 9 weeks after drug removal led to clones where the promoter region was completely demethylated, and expression permanently on. Thus, efficient demethylation leads to permanent gene reactivation, showing that DNA methylation provides a memory signal for the silent state.
Our data show that the respective roles of DNA methylation and chromatin remodeling can be completely separated using the YB5 selectable system. The chromatin state determines the immediate gene expression potential, while DNA methylation provides a long-term memory for gene silencing. Thus, DNA methylation does not provide a ‘lock’ function as previously thought, because gene expression can be restored by drug-induced chromatin modifications without any DNA demethylation (i.e. without breaking the lock). Rather, DNA methylation provides a ‘spring’ function, which does not suppress gene expression but brings back silencing, presumably through the previously defined order of events: methyl-binding protein recruitment, histone deacetylation, histone methylation, HP1 binding and so on (3). This explains why physiologically, DNA methylation at promoter CGIs is only involved when very long-term silencing is required, and why it provides such a selective advantage to cancer cells when TSG are silenced by this mechanism (3). Interestingly, after treatment with HDACi, gene expression is not sufficient to lead to permanent expression and DNA demethylation. It is possible that gene reactivation induced by HDACi may be caused by either i) bypassing transcription factors, whereby histone acetylation will directly trigger RNA pol II activation leading to reactivation or ii) transient binding of transcription factors to promoter regions, with gene silencing rapidly restored by repressive signals arising from DNA methylation. Restoring a silenced state is likely when histones are replaced during cell divisions in the face of persistent DNA methylation. Importantly after the treatment with hypomethylating drugs, it has been previously demonstrated that removal of DNA methylation marks will allow the binding of transcription factors leading to permanent epigenetic resetting promoting the emergence of stably reactivated clones. This was shown by 5-AZA-CdR-induced DNA demethylation in YB5 cells where the CREB transcription factor bound only the CMV promoter only when it was hypomethylated (13).
These data have implications for therapeutic intervention. We show that genes silenced by DNA hypermethylation in cancer can be significantly but transiently reactivated through chromatin remodeling without any changes in DNA methylation, and this may be part of the clinical mechanisms of action of HDACi. Moreover, our results provide a molecular explanation for the synergy between decitabine and HDACi (6, 29) in which the combination induces more complete epigenetic reprogramming. Finally, while these findings validate chromatin as a key target for therapeutic intervention in cancer, they also suggest that stable reprogramming may require the removal of DNA methylation signals.
Supplementary Material
Acknowledgements
This work was supported by National Institutes of Health grants CA100632, CA121104, and a grant from the Stand Up to Cancer foundation. JPI is an American Cancer Society Clinical Research professor supported by a generous gift from the F. M. Kirby Foundation. DNA sequencing and Flow Cytometry in the respective cores at the M. D. Anderson Cancer Center is supported by Core Grant CA16672 from the National Institutes of Health.
References
- 1.Bird A. DNA methylation patterns and epigenetic memory. Genes & evelopment. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 2.Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taby R, Issa JP. Cancer Epigenetics. CA: a cancer journal for clinicians. 2010 doi: 10.3322/caac.20085. [DOI] [PubMed] [Google Scholar]
- 4.Siegfried Z, Cedar H. DNA methylation: a molecular lock. Curr Biol. 1997;7:R305–R307. doi: 10.1016/s0960-9822(06)00144-8. [DOI] [PubMed] [Google Scholar]
- 5.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 6.Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nature genetics. 1999;21:103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 7.Suzuki H, Gabrielson E, Chen W, et al. A genomic screen for genes upregulated by demethylation and histone deacetylase inhibition in human colorectal cancer. Nature genetics. 2002;31:141–149. doi: 10.1038/ng892. [DOI] [PubMed] [Google Scholar]
- 8.Meng CF, Zhu XJ, Peng G, Dai DQ. Promoter histone H3 lysine 9 di-methylation is associated with DNA methylation and aberrant expression of p16 in gastric cancer cells. Oncology reports. 2009;22:1221–1227. doi: 10.3892/or_00000558. [DOI] [PubMed] [Google Scholar]
- 9.Fujii S, Luo RZ, Yuan J, et al. Reactivation of the silenced and imprinted alleles of ARHI is associated with increased histone H3 acetylation and decreased histone H3 lysine 9 methylation. Hum Mol Genet. 2003;12:1791–1800. doi: 10.1093/hmg/ddg204. [DOI] [PubMed] [Google Scholar]
- 10.Pruitt K, Zinn RL, Ohm JE, et al. Inhibition of SIRT1 reactivates silenced cancer genes without loss of promoter DNA hypermethylation. PLoS genetics. 2006;2:e40. doi: 10.1371/journal.pgen.0020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bovenzi V, Momparler RL. Antineoplastic action of 5-aza-2'-deoxycytidine and histone deacetylase inhibitor and their effect on the expression of retinoic acid receptor beta and estrogen receptor alpha genes in breast carcinoma cells. Cancer chemotherapy and pharmacology. 2001;48:71–76. doi: 10.1007/s002800100294. [DOI] [PubMed] [Google Scholar]
- 12.Yang X, Ferguson AT, Nass SJ, et al. Transcriptional activation of estrogen receptor alpha in human breast cancer cells by histone deacetylase inhibition. Cancer research. 2000;60:6890–6894. [PubMed] [Google Scholar]
- 13.Si J, Boumber YA, Shu J, et al. Chromatin remodeling is required for gene reactivation after decitabine-mediated DNA hypomethylation. Cancer research. 2010;70:6968–6977. doi: 10.1158/0008-5472.CAN-09-4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kondo Y, Shen L, Cheng AS, et al. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nature genetics. 2008;40:741–750. doi: 10.1038/ng.159. [DOI] [PubMed] [Google Scholar]
- 15.Estecio MR, Yan PS, Ibrahim AE, et al. High-throughput methylation profiling by MCA coupled to CpG island microarray. Genome research. 2007;17:1529–1536. doi: 10.1101/gr.6417007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiong Y, Dowdy SC, Podratz KC, et al. Histone deacetylase inhibitors decrease DNA methyltransferase-3B messenger RNA stability and down-regulate de novo DNA methyltransferase activity in human endometrial cells. Cancer research. 2005;65:2684–2689. doi: 10.1158/0008-5472.CAN-04-2843. [DOI] [PubMed] [Google Scholar]
- 17.Wu LP, Wang X, Li L, et al. Histone deacetylase inhibitor depsipeptide activates silenced genes through decreasing both CpG and H3K9 methylation on the promoter. Molecular and cellular biology. 2008;28:3219–3235. doi: 10.1128/MCB.01516-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.You JS, Kang JK, Lee EK, et al. Histone deacetylase inhibitor apicidin downregulates DNA methyltransferase 1 expression and induces repressive histone modifications via recruitment of corepressor complex to promoter region in human cervix cancer cells. Oncogene. 2008;27:1376–1386. doi: 10.1038/sj.onc.1210776. [DOI] [PubMed] [Google Scholar]
- 19.Bender CM, Pao MM, Jones PA. Inhibition of DNA methylation by 5-aza-2'-deoxycytidine suppresses the growth of human tumor cell lines. Cancer research. 1998;58:95–101. [PubMed] [Google Scholar]
- 20.Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sirchia SM, Ren M, Pili R, et al. Endogenous reactivation of the RARbeta2 tumor suppressor gene epigenetically silenced in breast cancer. Cancer research. 2002;62:2455–2461. [PubMed] [Google Scholar]
- 22.Zhou Q, Atadja P, Davidson NE. Histone deacetylase inhibitor LBH589 reactivates silenced estrogen receptor alpha (ER) gene expression without loss of DNA hypermethylation. Cancer biology & herapy. 2007;6:64–69. doi: 10.4161/cbt.6.1.3549. [DOI] [PubMed] [Google Scholar]
- 23.Drummond DC, Noble CO, Kirpotin DB, Guo Z, Scott GK, Benz CC. Clinical development of histone deacetylase inhibitors as anticancer agents. Annual review of pharmacology and toxicology. 2005;45:495–528. doi: 10.1146/annurev.pharmtox.45.120403.095825. [DOI] [PubMed] [Google Scholar]
- 24.Peart MJ, Smyth GK, van Laar RK, et al. Identification and functional significance of genes regulated by structurally different histone deacetylase inhibitors. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:3697–3702. doi: 10.1073/pnas.0500369102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sproul D, Nestor C, Culley J, et al. Transcriptionally repressed genes become aberrantly methylated and distinguish tumors of different lineages in breast cancer. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:4364–4369. doi: 10.1073/pnas.1013224108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miranda TB, Cortez CC, Yoo CB, et al. DZNep is a global histone methylation inhibitor that reactivates developmental genes not silenced by DNA methylation. Molecular cancer therapeutics. 2009;8:1579–1588. doi: 10.1158/1535-7163.MCT-09-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Egger G, Aparicio AM, Escobar SG, Jones PA. Inhibition of histone deacetylation does not block resilencing of p16 after 5-aza-2'-deoxycytidine treatment. Cancer research. 2007;67:346–353. doi: 10.1158/0008-5472.CAN-06-2845. [DOI] [PubMed] [Google Scholar]
- 28.Kagey JD, Kapoor-Vazirani P, McCabe MT, Powell DR, Vertino PM. Long-term stability of demethylation after transient exposure to 5-aza-2'-deoxycytidine correlates with sustained RNA polymerase II occupancy. Mol Cancer Res. 2010;8:1048–1059. doi: 10.1158/1541-7786.MCR-10-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Belinsky SA, Klinge DM, Stidley CA, et al. Inhibition of DNA methylation and histone deacetylation prevents murine lung cancer. Cancer research. 2003;63:7089–7093. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.