Abstract
The expression of proinflammatory cytokines and chemokines in response to T cell receptor (TCR) agonists is regulated by the CARMA1 signalosome through the coordinated assembly of complexes containing the BCL10 adaptor protein. We describe a novel mechanism to negatively regulate the CARMA1 signalosome by the “death” adaptor protein, CRADD/RAIDD. We show that CRADD interacts with BCL10 through its caspase recruitment domain (CARD) and suppresses interactions between BCL10 and CARMA1. TCR agonist-induced interaction between CRADD and BCL10 coincides with reduction of its complex formation with CARMA1 in wild-type, as compared to Cradd-deficient, primary cells. Finally, Cradd-deficient spleen cells, CD4+ T cells, and mice respond to T cell agonists with strikingly higher production of proinflammatory mediators, including γ-interferon, IL-2, TNF-α, and IL-17. These results define a novel role for CRADD as a negative regulator of the CARMA1 signalosome and suppressor of T helper (Th)1- and Th17-mediated inflammatory responses.
INTRODUCTION
Proinflammatory pathways are maintained through the assembly of oligomeric signaling complexes, known as “signalosomes” (1). One of them is the CARMA1 [caspase-recruitment domain (CARD) membrane-associated guanylate kinase (MAGUK)] signalosome employed by multiple immunoreceptors, including the T and B cell antigen receptors (2, 3).
In T cells, antigen receptor (TCR) engagement results in phosphorylation-induced conformational changes in CARMA1 necessary for the recruitment of BCL10, together with the MALT1 paracaspase protein (4–6). These CARD domain-mediated interactions between BCL10 and CARMA1 (7, 8) are required for TCR-coupled expression of cytokines under NF-κB control. While searching in silico for additional proteins bearing CARD and DEATH domains, a novel adaptor protein, designated CRADD [caspase and receptor interacting protein (RIP) adaptor with death domain; Mouse Genome Informatics (MGI) 1336168], or RAIDD (RIP-associated ICH-1/CED-3 homologous protein with a death domain), was discovered and characterized for its potential role in apoptosis (9, 10). CRADD was subsequently linked to DNA damage based on its inclusion in a complex with PIDD (p53-induced death domain protein) and caspase-2, termed the PIDDosome (11, 12). However, further studies provided little support for this concept and described other functions for PIDD and caspase-2 in cell cycle control (13–17) and for CRADD/RAIDD, PIDD and caspase-2 in the cellular responses to lethal heat shock (18). As an alternative model of CRADD function, we hypothesized that CRADD may play a role in TCR signal transduction as an adaptor protein through its CARD domain. Herein, we establish a novel immunoregulatory function for CRADD/RAIDD as a suppressor of the CARMA1 signalosome and as a negative regulator of the T cell-mediated proinflammatory response.
MATERIALS AND METHODS
Mice
Animal work was done in accordance with the Animal Care and Use Committee guidelines for Vanderbilt University. Animals were housed in specific pathogen-free conditions. The Cradd gene [Mouse Genome Informatics (MGI), 1336168] was disrupted in ES cells, introduced into the germ line (19) (Supplemental Figure 1), and maintained in a 129SVJ-C57BL/6 background.
Cell culture
RBC-lysed spleen cells from wild-type and Cradd−/− mice were cultured in RPMI medium 1640 with 10% FBS at 37°C in a CO2 incubator. Other cell types were cultured in DMEM medium with 10% FBS.
MACS
CD4+ T cells were purified with the CD4+ Isolation Kit II and naïve T cells were purified through the naïve cell purification kit according to the manufacturer’s instructions (Miltenyi). Isolated CD4+ T cells were >95% pure; naïve T cells were >92% pure as determined by identifying CD4+CD44loCD62Lhi cells.
Immunoprecipitation
Tissues from wild-type and Cradd−/− mice were rinsed in PBS, frozen in liquid N2, resuspended in 10 ml hypotonic buffer (20 mM HEPES-KOH, 10 mM KCl, 1 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, pH 7.5) and subjected to three freeze/thaw cycles. Splenocytes and cultured cells were lysed in hypotonic buffer + 0.1% NP-40. Lysates were pre-cleared with 50 μl protein G Sepharose beads (Sigma) at 4°C for one hour and incubated with 5 μg of the appropriate antibodies overnight with protein G beads at 4°C. The beads were washed three times before the proteins were separated by SDS/PAGE.
Immunoblot Analysis
Total cell lysates or immunoprecipitated proteins were separated by 10–12% SDS/PAGE, transferred onto nitrocellulose membranes (Amersham Biosciences), and incubated with the appropriate antibodies reactive with CRADD (ProteinTech Group, Inc); MYC, BCL10, and PIDD (Santa Cruz Biotechnology); AU1 (QED Bioscience); Caspase-2, TBP (Abcam); SLP76 (Novus Biologicals); ZAP70, CD3 (BD Pharmingen); CARMA1 (Ana-Spec); GAL-4 (Millipore); LAT, MALT1, NFkB p65 (Cell Signaling); and β-actin (Sigma). Immunoblots were scanned using Li-COR’s Odyssey infrared imaging system with appropriate secondary antibodies. Relative protein levels were determined based on two color infrared fluorescence with normalizing cellular proteins.
Preparation of Nuclear Extracts
Cytoplasmic and nuclear extracts were prepared as described with minor modifications (20). Cells were washed with ice-cold PBS and then lysed by incubation in 200 μl of lysis buffer (10 mM HEPES, pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 0.4% Nonidet P-40, 1 mM DTT, and protease inhibitor cocktail (Sigma) on ice for 5 min. Nuclei were pelleted by centrifugation, supernatant saved as the cytoplasmic extract, and nuclear pellet washed in 1 ml lysis buffer. Nuclear extracts were prepared by resuspension in salt extraction buffer (20 mM HEPES, pH 7.9, 400 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, and protease inhibitor cocktail), followed by shaking for 30 mins, centrifugation and removal of genomic DNA.
Mammalian two-hybrid assay
293T cells were transfected with plasmid vectors expressing GAL4 DNA binding domain and VP16 trans-activation domain fusion proteins according to the Matchmaker two-hybrid system (Clontech). Chloramphenicol acetyltransferase activities indicative of protein interactions were measured by the FAST CAT assay (Molecular Probes).
Cytokine/chemokine assays
Cytokines and chemokines in plasma or tissue culture media were assayed by cytometric bead array (BD Biosciences, San Diego, CA) and analyzed on a FACS Calibur flow cytometer. Standard curves were determined for each cytokine.
RESULTS AND DISCUSSION
CRADD interacts with BCL10 through CARD domain engagement
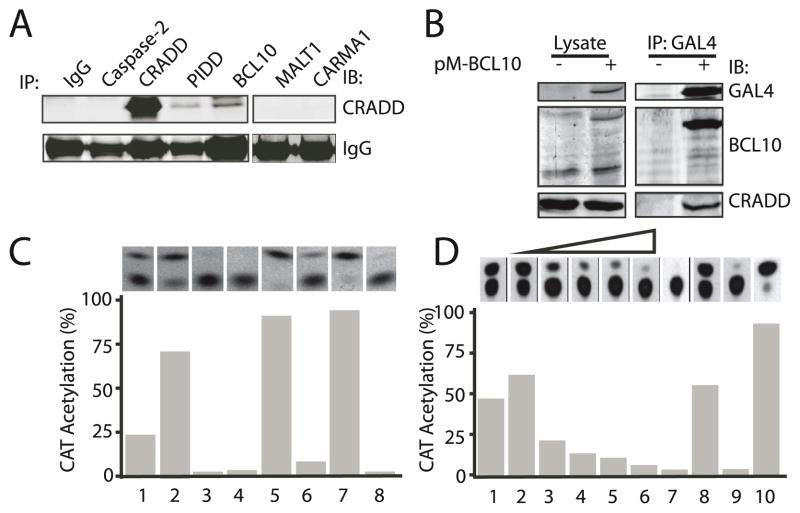
To test our hypothesis that CRADD interacts with the CARMA1 signalosome, we performed Western blot analysis using extracts prepared from primary spleen cells. As shown in Figure 1A, CRADD specifically co-precipitated with BCL10 and with PIDD, which served as a positive control, but not with MALT1, CARMA1 or caspase 2. Endogenous cellular CRADD protein was also recovered in human embryonic kidney 293T cells by using an epitope-tagged BCL10 protein (Figure 1B), suggesting that the two proteins interact directly. Moreover, in a mammalian two-hybrid assay, CRADD specifically interacted with BCL10 through the CARD domain (Figure 1C, column 2) and not the DEATH domain (Figure 1C, column 3). CRADD did not interact with MALT1 (Figure 1C, column 4), consistent with co-immunoprecipitation data (Figure 1A).
Figure 1. CRADD interacts with BCL10 through the CARD domain and prevents CARMA1 binding to BCL10.
(A) CRADD and BCL10 co-immunoprecipitate. Protein complexes precipitated from primary spleen cells with antibodies to the indicated proteins (IP) were immunoblotted (IB) with anti-CRADD antibody.
(B) Association between epitope-tagged BCL10 and cellular CRADD protein. 293T cells were transfected with pM-BCL10, which expresses a Gal4BCL10 fusion protein. Lysates (left panel) or immune complexes precipitated with anti-Gal4 antibodies (right panel) from control (−) or transfected (+) cells were subjected to western blot analysis (IB) using antibodies to the indicated proteins.
(C) CRADD interacts with BCL10 via the CARD domain. Mammalian two-hybrid assay in 293 T cells expressing proteins with DNA binding (DB) and trans-activation (AD) domains: DB-CRADD + AD-BCL10 (1); DB-CRADDCARD + AD-BCL10 (2); DB-CRADDDEATH + AD-BCL10 (3); DB-CRADD + AD-MALT1 (4); DB-p53 + AD-T antigen (5); DB-p53 + AD-Polyoma coat protein (6); DB-AD-M3-vp16 (7) and chloramphenicol acetyl transferase (CAT) reporter plasmid only (8). CAT activity indicative of protein interaction was assayed by thin layer chromatography (upper panel) and quantified by densitometry (lower panel).
(D) CRADD competitively interferes with interactions between BCL10 and the CARMA1 CARD domain. Two-hybrid assay of protein interactions in 293T cells expressing: DB-CRADD + AD-BCL10 (1); DB-CARMA1CARD + AD-BCL10 + increasing amounts (0– 40 μg) of Myc-CRADD (2–6); 10 μg Myc-CRADD (7); DB-p53 + AD-T antigen (8); CAT reporter only (9); and DB-AD-M3-vp16 (10).
We then confirmed that the CARD domains of BCL10 and CARMA1 interact in the mammalian two-hybrid assay (Figure 1D, column 1) and showed that CRADD blocks this interaction in a dose-dependent manner (Figure 1D, columns 2–6). Thus, we document a novel interaction of CRADD with BCL10 through CARD domain engagement and exclusion of CARMA1 from its union with BCL10.
Agonist-induced CRADD-BCL10 interaction coincides with reduced BCL10-CARMA1 assembly and NF-κB activation in primary cells
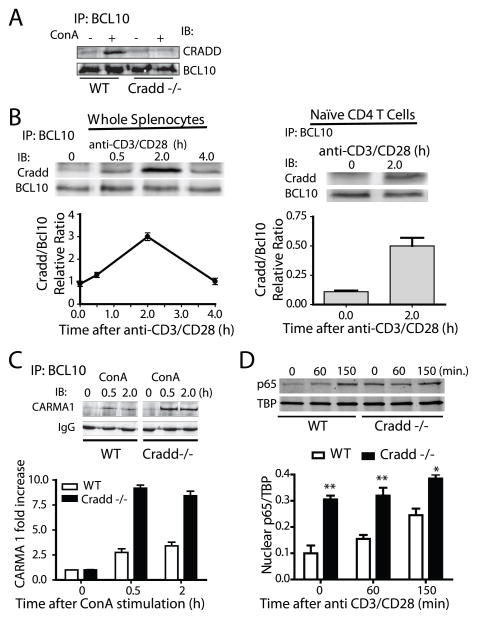
We determined whether Cradd deficiency had a measurable impact on CARMA1 signalosome formation. To generate Cradd-deficient mice, we employed ES cells containing a gene trap mutation (19) (Fig. S1 A–D). Homozygous mutant animals displayed no obvious developmental abnormalities or altered lymphocyte profiles in the spleen or thymus, as previously reported (13, 14). We then tested the effect of Cradd deficiency on agonist-induced BCL10-CARMA1 interactions. In Cradd-sufficient mouse spleen cells, the amount of CRADD that co-precipitated with BCL10 was enhanced by concanavalin A (ConA), a broadly acting T cell agonist and potent inducer of systemic inflammation (Fig. 2A), and was also induced by the T cell specific stimulus anti-CD3/CD28 in a time-dependent manner (Fig. 2B). Moreover, we analyzed purified naïve (CD44loCD62Lhi) CD4+ T cells following anti-CD3/CD28 –evoked TCR activation. Similar to whole splenocytes, an interaction between CRADD and BCL10 was induced at 2h after stimulation (Fig. 2B). Cradd deficiency had a striking impact on CARMA1 signalosome formation by enhancing BCL10 and CARMA1 interaction induced by ConA (Figure 2C). Notably, the amount of CARMA1 that co-precipitated with BCL10 after ConA treatment was 5-fold (at 30 min) and 3.5-fold (at 2h) higher in Cradd-deficient as compared to Cradd-sufficient spleen cells. The enhancement of CARMA1 signalosome assembly in CRADD-deficient cells resulted in predicted downstream activation events as demonstrated by elevated levels of nuclear RelA/p65 (p65) in Cradd-deficient purified CD4+ T cells (Fig. 2D).
Figure 2. CRADD-BCL10 interaction is induced by agonist stimulation and coincides with suppression of BCL10-CARMA1 interaction whereas NF-κB activation is attenuated in Cradd-sufficient as compared to Cradd-deficient spleen cells.
(A) Agonist-induced association between cellular BCL10 and CRADD proteins in spleen cells. Lysates from Cradd-sufficient wild-type (WT) and Cradd-deficient spleen cells, before (−) and 2 h after (+) treatment with ConA (1 μg/ml), were precipitated with anti-BCL10 antibody and probed with anti-CRADD and anti-BCL10 antibodies. CRADD signal was not detected in anti-BCL10 immunoprecipitate analyzed in cells derived from Cradd-deficient mice indicating the specificity of the anti-CRADD antibody.
(B) TCR- mediated association between BCL10 and CRADD in whole splenocytes and naïve CD4+ T cells. Lysates were obtained from Cradd-sufficient splenocytes following stimulation with 1μg/ml of anti-CD3/CD28 antibodies at the indicated intervals. The association between CRADD and BCL10 was inducible, reached a maximum at 2h, and returned to baseline by 4h post-activation. CD44hiCD62Llo naïve CD4+ T cells were purified by MACS and stimulated with plate bound anti-CD3/CD28 antibodies for 2 h at which time lysates were obtained and the interaction between BCL10 and CRADD determined by co-immunoprecipitation. CRADD interaction with BCL10 was significantly increased at 2 h (p<0.05, t-test)
(C) CRADD suppresses CARMA1-BCL10 interactions. Proteins precipitated with anti-BCL10 antibody from Cradd-sufficient and Cradd-deficient spleen cells after treatment with ConA (1 μg/ml) were immunoblotted using anti-CARMA1 antibody. CARMA1 signal in Cradd-sufficient (WT) cells was reduced as compared to cells obtained from Cradd-deficient mice at all times after stimulation; the relative increase in CARMA1 expression as compared to time 0 is shown (p<0.0001, ANOVA).
(D) Nuclear NF-κB p65 from Cradd-sufficient or Cradd-deficient purified CD4+ T cells stimulated for the indicated times (minutes) with plate bound anti-CD3 (5 μg/ml) plus anti-CD28 (2.5 μg/ml) was detected by Western blot analysis of nuclear extracts (*, <0.05; **, <0.01; Cradd-sufficient vs. Cradd-deficient, repeated measures ANOVA with Bonferroni post-test). TATA-binding protein (TBP) serves as a loading control for nuclear proteins.
Cradd deficiency enhances agonist-induced cytokine and chemokine expression in mice, isolated spleen cells, and CD4+ T cells and is reversed by retroviral CRADD gene transfer
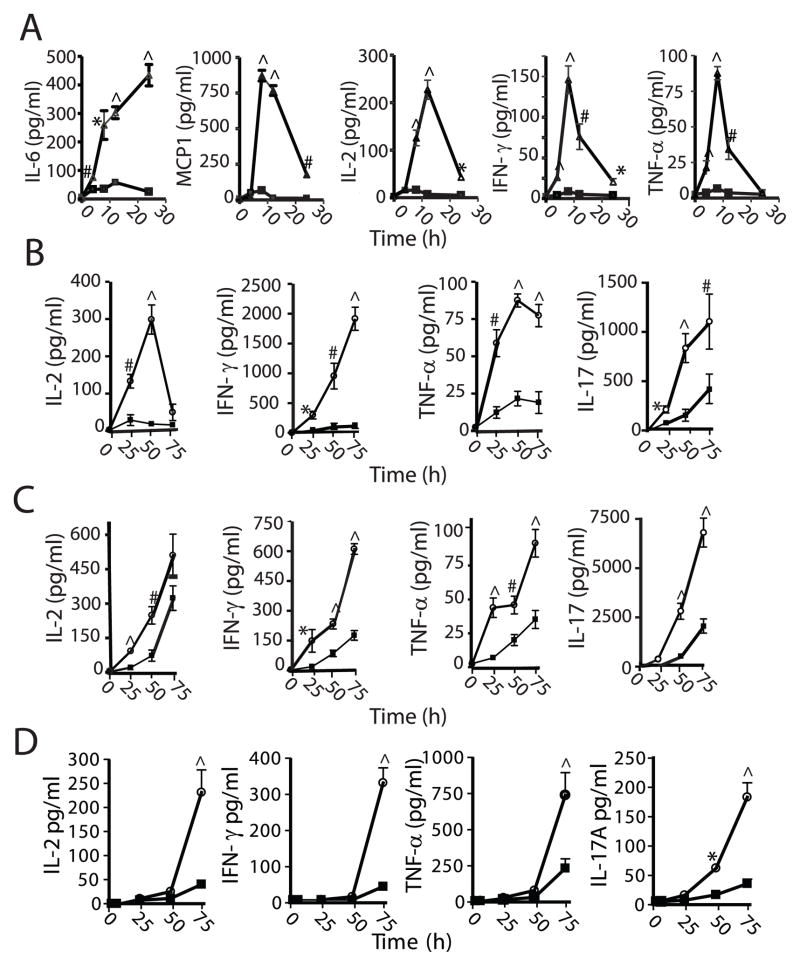
Heightened agonist-induced interactions between CARMA1 and BCL10 and enhanced NF-κB activation in Cradd-deficient T cells suggested that CRADD suppresses signaling pathways involved in agonist-induced cytokine and chemokine expression. Confirming this hypothesis, cytokine and chemokine expression was remarkably enhanced in Cradd-deficient mice following I.V. administration of ConA (Fig 3A). Plasma levels of TNF-α, INF-γ, IL-2, IL-6, and monocyte chemotactic protein-1 (MCP-1), which peaked 8 to 12h after ConA injection, were 8 to 20-fold higher in Cradd-deficient as compared to wild-type animals. Of note, extracts of spleen cells isolated from these animals at 6 h after ConA injection immunoprecipitated with anti-CD3 antibody demonstrated association of CRADD with components of the TCR signaling complex ZAP70, SLP-76, and LAT (Fig. S2 A and B).
Figure 3. Enhanced cytokine expression in Cradd-deficient mice, isolated spleen cells, and CD4+T cells.
Cytokines produced in mice following IV injection of ConA (A) or by spleen cells following stimulation with 1 μg/ml Con A (B) or with plate-immobilized anti-CD3 (2 μg/ml) and anti-CD28 (1 μg/ml) antibodies (C), or purified CD4+ T cells stimulated with anti-CD3/CD28 antibodies (D) were determined from wild-type (solid squares) and homozygous mutant mice (open circles). The levels of IL-6, MCP-1, IL-2, IFN-γ and TNF-α in plasma, and IL-2, IFN-γ, IL-17 and TNF-α in cell culture supernatants were measured by CBA assay. Mean values (± S.D.) for 9 independent experiments (A–C) or 3 experiments (D) are plotted. (*, <0.05; #, <0.005; and ^, <0.0005, repeated measures ANOVA with Bonferroni post-test).
Enhanced responses reflected by higher levels of IL-2, IFN-γ and TNF-α, characteristic of a T helper 1 (Th1) response, were also found in purified spleen cells from Cradd-knockout animals after treatment with ConA (Fig 3B), TCR agonistic anti-CD3/CD28 antibodies (Fig 3C), and superantigen staphylococcal enterotoxin B (SEB) (not shown). Likewise, higher levels of IL-17, a hallmark of Th17 cells, were induced in splenocytes derived from Cradd-deficient mice. We verified this remarkable enhancement of Th1- and Th17-type cytokine responses in purified CD4+ T cells stimulated with anti- CD3/CD28 (Fig 3D). By contrast, levels of IL4 and IL13, characteristic of T helper 2 (Th2) responses, remained below the threshold of detection under the same experimental conditions (not shown).
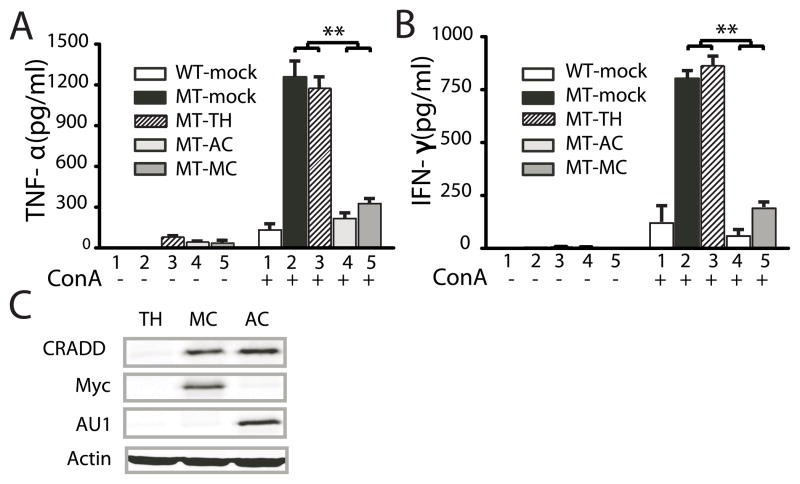
As a final demonstration of the inhibitory function of CRADD in T cell signaling, CRADD expression was restored in Cradd-deficient spleen cells by retroviral gene transfer. Spleen cells were infected with retroviruses containing the CRADD gene together with the Thy1.1 gene that encodes a cell surface protein used to immunoselect retrovirally infected cells, and epitope tags, c-Myc (MC) and AU1 (AC). As expected for the complementation of phenotypes caused by loss of Cradd function, TNF-α (Fig. 4A) and IFN-γ (Fig. 4B) were suppressed to nearly wild-type levels in mutant cells infected with CRADD expression vectors but not in cells infected with an empty vector (TH), consistent with the expression of epitope-tagged CRADD protein in virally-infected spleen cells (Fig. 4C). Thus, we showed that the cytokine suppressing activity of CRADD can be restored by its gene transfer in mutant spleen cells and established that the cytokine-enhancing mutant phenotype was caused by Cradd deficiency.
Figure 4. Restoration of cytokine suppression by retroviral Cradd gene transfer to Cradd−/− spleen cells.
(A) ConA-induced TNF-α expression in spleen cells infected with Cradd-expression vectors. TNF-α or IFN-γ expression was measured in 106 Thy1.1+ primary spleen cells (isolated 24 h post infection) without/with ConA treatment (1 μg/ml) for 8 h: wild-type spleen cells (1,WT-mock), Cradd-deficient spleen cells (2, MT-mock); and Cradd-deficient spleen cells infected with empty (3, MT-TH), AU1-Cradd (4, MT-AC) and Myc-Cradd (5, MT-MC) expression vectors. IL-2 expression was not measured, since the cytokine was added to the media to enhance T cell viability. Mean values (± S.D.) for three experiments using fresh spleen cells are plotted. P values were determined by one-way ANOVA (**, <0.005).
(B) ConA-induced IFN-γ expression in spleen cells infected with Cradd-expression vectors as described above.
(C) Expression of epitope-tagged CRADD proteins in spleen cells that were infected with empty (TH), or with c-Myc- (MC) or AU1- (AC) tagged Cradd MSCV retroviral vectors encoding additional surface marker Thy1.1. After 24 h, cell lysates from magnetically separated Thy1.1+ cells were immunoblotted using the indicated antibodies.
Cumulatively, our results identify a previously unrecognized immunoregulatory function of the CRADD adaptor protein as a suppressor of TCR agonist-evoked signaling. This function is dependent on the CARD domain of CRADD, which binds to BCL10 and interferes with signal-induced targeting of BCL10 to CARMA1, an essential step in formation of the CBM (CARMA1, BCL10, MALT) complex required for activation of the NF-κB signaling pathway via TRAF6 and NEMO/IKK gamma (21, 22). This new suppressor function of CRADD operates in primary T cells, as judged by (i) associations between CRADD and BCL10 in spleen cells and CD44hiCD62Llo naïve CD4+ T cells stimulated with T cell-specific agonist anti-CD3/CD28 (Fig. 2B), (ii) strikingly enhanced cytokine expression in Cradd-deficient spleen cells and purified CD4+ T cells treated with the T cell-specific agonists, anti-CD3/CD28 and SEB and displaying Th1 and Th17 cytokine profiles (Fig. 3), and (iii) enhanced recovery of activated T cells from the spleens of Con A-treated Cradd-deficient mice as assessed by CD69 expression (not shown). Given the widespread involvement of BCL10 and CARMA proteins as regulators of NEMO/IKK gamma in other immunoreceptor-activated signaling pathways (2, 3), CRADD may also suppress immunoreceptor signaling in other cell types. Moreover, CRADD may also function as a regulatory adaptor in other signaling pathways, which awaits future analysis.
We propose that CRADD provides the first example of a TCR-proximal adaptor function that negatively regulates the CARMA1 signalosome thereby limiting potentially harmful proinflammatory cytokine and chemokine expression under conditions of excessive or inappropriate T cell stimulation. Further understanding of CRADD regulation will inform therapeutic strategies to control the inflammatory responses in human disease.
Supplementary Material
Acknowledgments
This work was supported by the US Public Health Service NIH Grants: HL687440 to HER and JH, HL069452, HL085833 and AA015752 to JH, 1KO8 DK090146 to DJM, and by a Kleberg Family Grant to HER. Additional support was provided by the Department of Microbiology and Immunology and by a Cancer Center Support grant (P30CA68485).
We wish to thank Robert Collins for histological analysis, Luc Van Kaer for the initial screen of immune system defects in Cradd-deficient mice, Dean Ballard and Eric Sebzda for helpful discussions, Abudi Nashabi for technical assistance, and Susanna Richards for editorial assistance.
References
- 1.Park HH, Lo YC, Lin SC, Wang L, Yang JK, Wu H. The death domain superfamily in intracellular signaling of apoptosis and inflammation. Annu Rev Immunol. 2007;25:561–586. doi: 10.1146/annurev.immunol.25.022106.141656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rawlings DJ, Sommer K, Moreno-Garcia ME. The CARMA1 signalosome links the signalling machinery of adaptive and innate immunity in lymphocytes. Nat Rev Immunol. 2006;6:799–812. doi: 10.1038/nri1944. [DOI] [PubMed] [Google Scholar]
- 3.Thome M. Multifunctional roles for MALT1 in T-cell activation. Nat Rev Immunol. 2008;8:495–500. doi: 10.1038/nri2338. [DOI] [PubMed] [Google Scholar]
- 4.Matsumoto R, Wang D, Blonska M, Li H, Kobayashi M, Pappu B, Chen Y, Wang D, Lin X. Phosphorylation of CARMA1 plays a critical role in T Cell receptor-mediated NF-kappaB activation. Immunity. 2005;23:575–585. doi: 10.1016/j.immuni.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 5.Wang D, Matsumoto R, You Y, Che T, Lin XY, Gaffen SL, Lin X. CD3/CD28 costimulation-induced NF-kappaB activation is mediated by recruitment of protein kinase C-theta, Bcl10, and IkappaB kinase beta to the immunological synapse through CARMA1. Mol Cell Biol. 2004;24:164–171. doi: 10.1128/MCB.24.1.164-171.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun Z, Arendt CW, Ellmeier W, Schaeffer EM, Sunshine MJ, Gandhi L, Annes J, Petrzilka D, Kupfer A, Schwartzberg PL, Littman DR. PKC-theta is required for TCR-induced NF-kappaB activation in mature but not immature T lymphocytes. Nature. 2000;404:402–407. doi: 10.1038/35006090. [DOI] [PubMed] [Google Scholar]
- 7.Gaide O, Martinon F, Micheau O, Bonnet D, Thome M, Tschopp J. Carma1, a CARD-containing binding partner of Bcl10, induces Bcl10 phosphorylation and NF-kappaB activation. FEBS Lett. 2001;496:121–127. doi: 10.1016/s0014-5793(01)02414-0. [DOI] [PubMed] [Google Scholar]
- 8.Bertin J, Wang L, Guo Y, Jacobson MD, Poyet JL, Srinivasula SM, Merriam S, DiStefano PS, Alnemri ES. CARD11 and CARD14 are novel caspase recruitment domain (CARD)/membrane-associated guanylate kinase (MAGUK) family members that interact with BCL10 and activate NF-kappa B. J Biol Chem. 2001;276:11877–11882. doi: 10.1074/jbc.M010512200. [DOI] [PubMed] [Google Scholar]
- 9.Ahmad M, Srinivasula SM, Wang L, Talanian RV, Litwack G, Fernandes-Alnemri T, Alnemri ES. CRADD, a novel human apoptotic adaptor molecule for caspase-2, and FasL/tumor necrosis factor receptor-interacting protein RIP. Cancer Res. 1997;57:615–619. [PubMed] [Google Scholar]
- 10.Duan H, V, Dixit M. RAIDD is a new ‘death’ adaptor molecule. Nature. 1997;385:86–89. doi: 10.1038/385086a0. [DOI] [PubMed] [Google Scholar]
- 11.Tinel A, Tschopp J. The PIDDosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress. Science. 2004;304:843–846. doi: 10.1126/science.1095432. [DOI] [PubMed] [Google Scholar]
- 12.Park HH, Logette E, Raunser S, Cuenin S, Walz T, Tschopp J, Wu H. Death domain assembly mechanism revealed by crystal structure of the oligomeric PIDDosome core complex. Cell. 2007;128:533–546. doi: 10.1016/j.cell.2007.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berube C, Boucher LM, Ma W, Wakeham A, Salmena L, Hakem R, Yeh WC, Mak TW, Benchimol S. Apoptosis caused by p53-induced protein with death domain (PIDD) depends on the death adapter protein RAIDD. Proc Natl Acad Sci U S A. 2005;102:14314–14320. doi: 10.1073/pnas.0506475102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horvat S, Medrano JF. Lack of Socs2 expression causes the high-growth phenotype in mice. Genomics. 2001;72:209–212. doi: 10.1006/geno.2000.6441. [DOI] [PubMed] [Google Scholar]
- 15.Manzl C, Krumschnabel G, Bock F, Sohm B, Labi V, Baumgartner F, Logette E, Tschopp J, Villunger A. Caspase-2 activation in the absence of PIDDosome formation. J Cell Biol. 2009;185:291–303. doi: 10.1083/jcb.200811105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Read SH, Baliga BC, Ekert PG, Vaux DL, Kumar S. A novel Apaf-1-independent putative caspase-2 activation complex. J Cell Biol. 2002;159:739–745. doi: 10.1083/jcb.200209004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cuenin S, Tinel A, Janssens S, Tschopp J. p53-induced protein with a death domain (PIDD) isoforms differentially activate nuclear factor-kappaB and caspase-2 in response to genotoxic stress. Oncogene. 2008;27:387–396. doi: 10.1038/sj.onc.1210635. [DOI] [PubMed] [Google Scholar]
- 18.Milleron RS, Bratton SB. Heat shock induces apoptosis independently of any known initiator caspase-activating complex. J Biol Chem. 2006;281:16991–17000. doi: 10.1074/jbc.M512754200. [DOI] [PubMed] [Google Scholar]
- 19.Lin Q, Donahue SL, Moore-Jarrett T, Cao S, Osipovich AB, Ruley HE. Mutagenesis of diploid mammalian genes by gene entrapment. Nucleic Acids Res. 2006;34:e139. doi: 10.1093/nar/gkl728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Torgerson TR, Colosia AD, Donahue JP, Lin YZ, Hawiger J. Regulation of NF-kappa B, AP-1, NFAT, and STAT1 nuclear import in T lymphocytes by noninvasive delivery of peptide carrying the nuclear localization sequence of NF-kappa B p50. J Immunol. 1998;161:6084–6092. [PubMed] [Google Scholar]
- 21.Blonska M, Lin X. CARMA1-mediated NF-kappaB and JNK activation in lymphocytes. Immunol Rev. 2009;228:199–211. doi: 10.1111/j.1600-065X.2008.00749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lobry C, Lopez T, Israel A, Weil R. Negative feedback loop in T cell activation through IkappaB kinase-induced phosphorylation and degradation of Bcl10. Proc Natl Acad Sci U S A. 2007;104:908–913. doi: 10.1073/pnas.0606982104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.