Abstract
Sumoylation is a posttranslational modification implicated in a variety of cellular activities, and its role in a number of human pathogeneses such as cleft lip/palate has been well documented. However, the importance of the SUMO conjugation pathway in cardiac development and functional disorders is newly emerging. We previously reported that knockout of SUMO-1 in mice led to congenital heart diseases (CHDs). To further investigate the effects of imbalanced SUMO conjugation on heart development and function and its underlying mechanisms, we generated transgenic (Tg) mice with cardiac-specific expression of SENP2, a SUMO-specific protease that deconjugates sumoylated proteins, to evaluate the impact of desumoylation on heart development and function. Overexpression of SENP2 resulted in premature death of mice with CHDs—atrial septal defects (ASDs) and/or ventricular septal defects (VSDs). Immunobiochemistry revealed diminished cardiomyocyte proliferation in SENP2-Tg mouse hearts compared with that in wild type (WT) hearts. Surviving SENP2-Tg mice showed growth retardation, and developed cardiomyopathy with impaired cardiac function with aging. Cardiac-specific overexpression of the SUMO-1 transgene reduced the incidence of cardiac structural phenotypes in the sumoylation defective mice. Moreover, cardiac overexpression of SENP2 in the mice with Nkx2.5 haploinsufficiency promoted embryonic lethality and severity of CHDs, indicating the functional interaction between SENP2 and Nkx2.5 in vivo. Our findings indicate the indispensability of a balanced SUMO pathway for proper cardiac development and function.
Keywords: SUMO, SENP2, congenital heart defects, cardiomyopathy, Nkx2.5
1. Introduction
Normal cardiac development is a complex process that requires sophisticated cooperation between a variety of transcription factors, co-factors, and signal transduction pathways. Interference with the function of any of these factors and/or signal transduction pathways may cause cardiac malformation, i.e. congenital heart diseases (CHDs) and/or cardiac dysfunction. The most common form of CHDs is abnormal septation (atrial septal defects or ASDs, and ventricular septal defects or VSDs) (1–6). To date, chromosomal and Mendelian syndromes accounted for only 20% of CHDs (7). Thus, the unifying molecular mechanisms responsible for the vast majority of ASDs/VSDs remain to be elucidated.
Recent studies suggested that SUMO (small ubiquitin-related modifier) modification plays a role in cardiac development and function (8, 9). SUMO belongs to the superfamily of ubiquitin-like proteins (ULPs). SUMO modification, which appears to be involved in regulation of many cellular events including cell cycle progression and chromatin remodeling (10–12), is a process in which SUMO proteins are covalently and reversibly attached to targets via a series of enzymatic reactions, requiring heterodimeric E1 (SAE1/SAE2), lone E2 (Ubc9) and a number of E3 ligases (13). So far three conjugatable SUMO proteins, SUMO-1, -2 and -3, have been identified in vertebrates, and they exhibit sequence identity to various degrees with each other (~50% homology between SUMO-1 and SUMO-2/3, but ~97% identity between the active forms of SUMO-2 and SUMO-3). SUMO-1 and SUMO-2/3 shared overlapped targets but also had distinct substrates (14), suggesting that they may play different roles in cellular processes.
One defining characteristic of SUMO conjugation is its reversibility. SUMO conjugates can be readily deconjugated by a class of enzymes named SENPs (sentrin-specific proteases). So far six SENP family members (SENP1, SENP2, SENP3, SENP5, SENP6, SENP7) with desumoylation activity have been identified in human (15), among which SENP1, SENP2, and SENP5 exhibited both endopeptidase and isopeptidase activities (16–18). However, the isopeptidase activity of SENP2 and SENP5 prevailed over the endopeptidase activity in vivo (18), suggesting that both may serve as sumoylation suppressors. SENPs activity was relatively specific for distinct SUMO family members (19, 20). For instance, SENP1 and SENP2 generally targeted all SUMO isoforms for deconjugation (16), whereas SENP3, 5, 6 and 7 preferentially targeted SUMO-2/3 conjugates (21–24). Knockout of SENP1 or SENP2 in mice led to embryonic lethality (25–27), suggesting that the SUMO pathway components played an essential role in normal murine embryogenesis. There is evidence that the SUMO pathway was involved in a number of human pathogeneses such as neurodegeneration (28), diabetes (29), prostate and breast cancer (30), and cleft lip/palate (31). More recently, examination of two naturally occurring mutants of the nuclear structural protein, lamin A, showed reduced sumoylation, which may be associated with human familial cardiomyopathy (32). Our demonstration that cardiac transcription factors such as Nkx2.5, GATA4, and myocardin were SUMO targets (33–35) pointed to a role for sumoylation in cardiovascular development. Indeed, SUMO-1 knockout mice exhibited CHDs, although the penetrance of cardiac phenotypes was potentially affected by genetic background (36). To further understand the contribution of the SUMO pathway to cardiac development and function, we overexpressed human SUMO-specific protease SENP2 in murine cardiomyocytes under the control of murine α-myosin heavy chain (α-MHC) promoter. As hypothesized, in the heart, SENP2 overexpression promoted desumoylation, leading to congenital heart malformations and cardiac dysfunction.
2. Materials and Methods
2.1 Materials
Dulbecco's modified Eagle medium (DMEM) was obtained from Thermo Scientific. Lipofectamine 2000, cloned reverse transcriptase, NuPAGE SDS gels and all restriction enzymes used for subcloning were purchased from Invitrogen. All animals were obtained from Harlan (Houston). Antibodies against SUMO-1, SUMO-2/3, SENP2, caveolin 3 (Cav3), GAPDH and agarose-conjugated SUMO-1 antibody were purchased from Santa Cruz. Anti-flag antibody (M2) and WGA (wheat germ agglutinin)-TRITC, and neutral red were from Sigma. Anti-cleaved Caspase 3 (Asp175) antibody was from Cell Signaling, and anti-Ki67 antibody from Abcam. Biotinylated anti-rabbit secondary antibody, Vectastain ABC Kit and DAB kit were purchased from Vector Laboratories. 5-Bromo-2'-dexoyuridine (BrdU) and BrdU in situ detection kit were from BD. ApopTag® Plus Peroxidase In Situ Apoptosis Detection Kit for TUNEL assay was from Chemicon. The expression vectors encoding flag-tagged human SENP2 wild type (wt) and catalytic mutant were gifts from Dr. Edward Yeh (University of Texas MD Anderson Cancer Center, Houston). ANF luciferase reporter (ANF-Luc) was provided by Dr. Mona Nemer (University of Ottawa, Canada). Nkx2.5 expression vector was detailed previously (37, 38).
2.2 Generation of transgenic and knockout mice
The α-MHC-flag-SENP2 transgene was constructed as described (39) under the control of mouse α-MHC promoter (provided by Dr. J. Robbins, University of Cincinnati). The orientation of inserted transgene was confirmed by DNA sequencing. The construct was microinjected into the pronucleus of fertilized eggs from FVB mice to generate founder (F0) SENP2 transgenic (SENP2-Tg) mice, which were crossed back to C57BL/6 mice for over one to five generations. Genomic DNA was isolated from tail biopsies performed on weaned animals (approximately 3-week old pups) and screened by PCR. The expression of transgene in SENP2-Tg mouse hearts was verified by Western blot and/or RT-PCR. The approach used to generate α-MHC-flag-SUMO-1 transgenic mice (SUMO-1-Tg) was previously described (36). The heterozygous Nkx2.5 knockout (Nkx2.5cre/+) mouse line was generated previously (40) and designated as Nkx2.5+/− in the present study. The SUMO-1 knockout mouse line was provided by Dr. Michael Kuehn at the National Cancer Institute (41). The crossbreeding among various knockout and/or transgenic mice was performed as needed. Animals were handled in accordance with institutional guidelines with IACUC approval.
2.3 Cell culture, transfection, Western blot, immunoprecipitation and in vivo sumoylation assay
HeLa cells were cultured in DMEM supplemented with 10% FBS and Gentamicin (10 μg/ml). Transfections were performed on HeLa cells cultured on either 6 cm plates for Western blot analysis or 12 well plates for luciferase activity assays using Lipofectamine 2000 based on the protocol provided by the supplier. Luciferase activity assays were performed as previously described (34, 38). Promoter activity was expressed as the ratio of luciferase activity induced by the presence of specific factor(s) to the control group (empty vector alone). Data shown were expressed as mean ± standard error of the mean (SEM) from at least two independent assays, with each carried out in duplicate. Western blot and in vivo sumoylation assays were detailed previously (33, 38, 42). Briefly, 100 μg of protein lysates from mouse hearts or 40 μg of HeLa cell lysates containing overexpressed proteins were purified in the presence of 25 mM isopeptidase inhibitor N-ethylmaleimide (NEM), which prevents desumoylation of SUMO-conjugated substrates. For immunoprecipitation, protein lysates were prepared from frozen mouse hearts and for each sample, 400 μg of total protein was diluted to a final concentration of 0.4 mg/ml in lysis buffer. An appropriate amount of bead-conjugated anti-SUMO-1 antibody was added to the sample and incubated for 4 hours at 4°C on a rotary platform. The beads were subsequently pelleted by centrifugation, washed five times with lysis buffer, and resuspended in 4x NuPAGE sample buffer. These protein lysates were subsequently subjected to 4–12% NuPAGE, transferred to PVDF membrane, detected with the desired antibody, which was then visualized with chemiluminescence.
2.4 Histopathology
Mouse embryos at various developmental stages (E14.5, E15.5, E16.5 or E17.5, as needed), neonatal hearts were dissected and fixed overnight in 4% paraformaldehyde (PFA). Adult mouse hearts were fixed in 10% formaldehyde. Hematoxylin and eosin (H&E) or Masson's trichrome staining was performed on heart sections (10 μm) according to standard protocols.
2.5 Immunostaining
For cleaved caspase 3 immunostaining, antigen retrieval was performed by boiling slides in 10 mM sodium citrate (pH 6.0, 15 min) followed by treatment with 3% H2O2 to quench endogenous peroxidase. Specimens were blocked in 2% normal goat serum (NGS) for 1 h at room temperature (RT) and incubated for 1 hr at RT with anti-cleaved caspase 3 (Asp175) antibody (1:200), followed by incubation in biotinylated anti-rabbit secondary antibody (1:200) for 30 min at RT. Sections were processed by immunoperoxidase labeling using the Vectastain ABC Kit and visualized with a DAB kit. Cells were counterstained with the neutral red.
To detect DNA synthesis in cardiomyocytes, BrdU was injected intraperitoneally into pregnant female mice bearing E14.5 or 15.5 embryos at a dose of 100 μg/g body weight. Sixteen hours after injection, embryonic hearts were dissected, fixed in 4% PFA, dehydrated in increasing concentrations of ethanol (up to final of 100%), and sectioned (5 μm). BrdU staining was conducted according to the protocol provided by the manufacturer. Ki67 staining was conducted with anti-Ki67 antibody (1:200). Anti-caveolin 3 antibody (1:8000) was used as a cardiomyocyte marker. Sections were then incubated with the following secondary antibodies: anti-rabbit TRITC antibody or anti-mouse FITC antibody (Jackson ImmunoResearch). Sections were mounted with Vectashield with DAPI (Vector Laboratory). BrdU and Ki67 labeling indices were calculated based on scores obtained from 6 different embryonic or adult hearts (3 for each group), with each at least 100 cells (embryonic hearts) or 5 fields (adult hearts) randomly selected for evaluation. To assess the apoptosis, TUNEL staining was performed based on the standard protocol provided by the manufacturer.
2.6 Cardiomyocyte size measurement
WGA-TRITC staining was performed on heart sections of E14.5 or age-matched adult WT and SENP2-Tg mice according to the manufacturer's protocol. Surface areas of cardiomyocytes were measured in 10–20 randomly selected fields from each individual heart sample using Software ImageJ. The average surface area calculated for WT cardiomyocytes was taken as 100%.
2.7 Echocardiography
Mice of interest with various ages as designed were anesthetized by inhalation of 1% isofluorane and fixed to a warm pad for transthoracic measurements of cardiac function using two dimensional M-mode of a Vevo 770 in vivo micro-imaging system (Visual Sonics, Toronto, Canada). Chest hair was removed and contacted with the probe to record cardiac function indices. An expert analyst who was blinded to the genotype of animals evaluated cardiac function and heart dimensions.
2.8 Microarray assay, reverse transcription and semi-quantitative/quantitative PCR (qPCR)
RNA was extracted from age-matched WT or SENP2-Tg mouse hearts using Trizol according to the manufacturer's protocol. Microarray service was provided by Phalanx Biotech (OneArray Express). Reverse transcription reaction was performed using 1 μg total RNA per reaction and cloned reverse transcriptase, followed by either semi-quantitative PCR or qPCR (MX3000, Stratagene) using the following gene-specific primers: SENP2: forward, 5' GCTAAGGTTCTCGGCACCATT 3'; reverse, 5' ATTACAAGCAGAAGACACCATG 3'. Nkx2.5 (43): forward, 5' TCTCCGATCCATCCCACTTTATTG 3'; reverse, 5' TTGCGTTACGCACTCACTTTAATG 3'. GATA4 (44): forward, 5' GCCTGTATGTAATGCCTGCG 3'; reverse, 5' CCGAGCAGGAATTTGAAGAGG 3'. SRF (45): forward, 5' CCAGCGCTGTCAGCAGTGCCAAC 3'; reverse, 5' GCTGCTCCCAGCTTGCTGCCCTATC 3'. SUMO-1: forward, 5' TCTGACCAGGAGGCAAAACC 3'; reverse, 5' CTAAACCGTCGAGTGACCCC 3'. SUMO-2: forward, 5' GACGAGAAACCCAAGGAAGG 3'; reverse, 5' CTCCAGTCTGCTGCTGGAAC 3'. SUMO-3: forward, 5' CCAAGGAGGGTGTGAAGACA 3'; reverse, 5' TCAATAGCACAGGTCAGGACA 3'. Mlc2-a (46): forward, 5' AGGCACAACGTGGCTCTTCT 3'; reverse, 5' AGCTGGGAATAGGTCTCCTTCA 3'. MLC2-v (47): forward, 5' GCCAAGAAGCGGATAGAAGG 3'; reverse, 5' CTGTGGTTCAGGGCTCAGTC 3'. skeletal α-actin (46) : forward, 5' AGACACCATGTGCGACGAAGA 3'; reverse, 5' CCGTCCCCAGAATCCAACACGA 3'. GAPDH: forward, 5' ATGTTCCAGTATGACTCCACTCAC 3'; reverse, 5' GAAGACACCAGTAGACTCCACGA 3'.
2.9 Statistics
The unpaired Student's t test, Fisher's Exact Probability Test or Chi-square Test was used to determine statistical significance between groups, as applicable, and results were shown in the respective Figure legend. P<0.05 was defined as significant and p<0.01 as highly significant.
3 Results
3.1 SENP2 was expressed in the developing heart during mouse embryogenesis
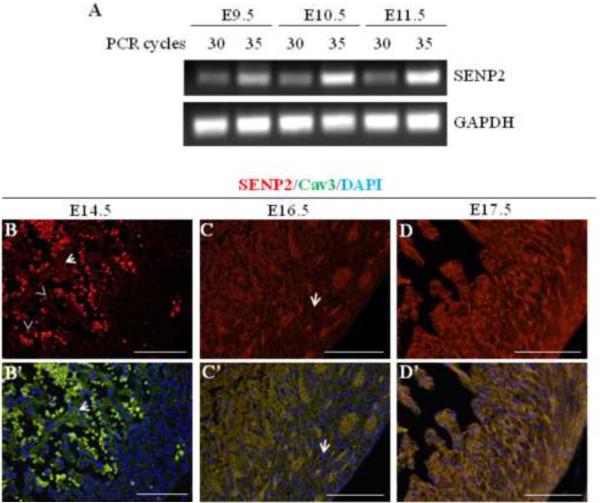
To assess the presence of endogenous SENP2 transcripts in the developing murine heart, semi-quantitative RT-PCR was performed using RNA samples purified from embryonic hearts at three different developmental stages (E9.5, E10.5 and E11.5); SENP2-specific primers were used under two PCR conditions, one at 30 and one at 35 cycles, for each RT sample. SENP2 transcripts were detected as early as E9.5 (Fig. 1A), and appeared to be upregulated during this developmental period (from E9.5 to E11.5). To investigate the expression of SENP2 in heart at later embryonic stages, immunostaining was performed on developing hearts at E14.5, E16.5 and E17.5. SENP2 was detected at all stages examined (Fig. 1B–D', arrows); however, SENP2 expression appeared to be more restricted to the trabecular myocardium at E14.5 (Fig. 1B & B', as exemplified by arrows) and was barely seen in the compact zone at this stage. With the progress of development, however, SENP2 expression was weakly detected in the compact myocardium at E16.5 (Fig. 1C & C', as exemplified by arrows) and became abundant in this region at E17.5 (Fig. 1D & D'). Also, SENP2 staining overlapped with Cav3 staining, but not with DAPI (as exemplified by arrows in Fig. 1B–C'), suggesting that under physiological conditions SENP2 was not localized inside the nucleus, an observation that was consistent with a previous report (48). These observations indicate developmental and spatial regulation of SENP2 expression in the developing heart during embryogenesis.
Figure 1. SENP2 gene was expressed in mouse heart during embryogenesis.
A. Semi-quantitative RT-PCR was performed on RNA samples purified from embryonic hearts at various developmental stages (E9.5, E10.5 and E11.5) to detect SENP2 mRNA. PCR for each sample was carried out with two cycles (30 and 35, respectively). GAPDH served as a loading control. B–D'. Immunostaining shows SENP2 expression in hearts at later developmental stages. Hearts of three embryonic stages, E14.5 (B–B'), E16.5 (C–C'), and E17.5 (D–D') were examined. Panel B, C & D show single SENP2 staining, while B', C' & D' display the merged images of three stainings: SENP2 (red), Cav3 (green, positive for cardiomyocyte), and DAPI (blue, for nucleus). The arrows in B & B' show examples of non-nuclear staining of SENP2, and the dotted staining (arrowheads) indicates blood cells. Note that in the compact zone, SENP2 was barely detected at E14.5, but was detectable at E16.5 (C & C', arrows) and became abundant at E17.5, indicating developmental and spatial regulation of SENP2 expression during cardiogenesis.
3.2 Generation of transgenic (Tg) mice overexpressing human SENP2 in heart
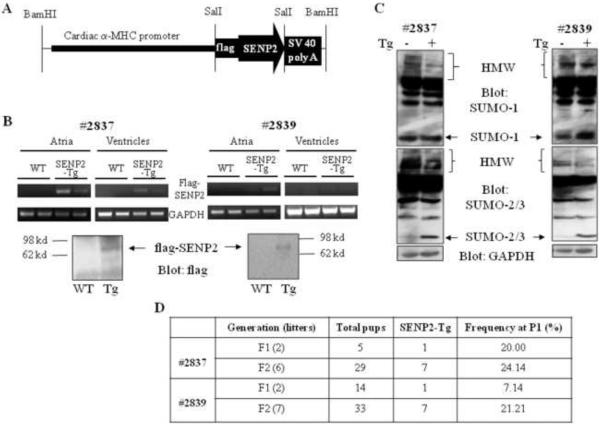
The flag-tagged human SENP2 transgene was designed to be expressed under the control of α-MHC promoter in murine cardiomyocytes (SENP2-Tg) (Fig. 2A). Two lines of SENP-Tg mice, #2837 and #2839, expressed the flag-tagged SENP2 transgene in murine hearts (Fig. 2B). Atria and ventricles were dissected from E16.5 mouse hearts from the flag-SENP2 transgenic lines and RNA was purified. Semi-quantitative RT-PCR indicated that the flag-SENP2 transgene was expressed in the atria of the mice from both SENP2-Tg lines (#2837 and #2839); however, it was only clearly seen in the ventricles of the mice from line #2837 but not from line #2839 (Fig. 2B, upper panels). Variability in expression of the transgene in different transgenic mouse lines was expected and was probably associated with different copy numbers of transgene. We also detected flag-SENP2 via Western blot in the two-week postnatal (P14) mouse hearts from both lines (Fig. 2B, bottom panels). Subsequent sumoylation assays revealed decreased SUMO-1, -2 and -3 conjugates in the SENP2-Tg hearts (Fig. 2C). Thus, in line with the previous report (18), SENP2 served as a sumoylation pathway repressor in vivo. The P1 frequency in F1 and F2 generations of both SENP2-Tg lines was much lower than the expected 50% Mendelian rate (Fig. 2D), indicating prenatal loss of SENP2-Tg mice.
Figure 2. Generation of SENP2 transgenic (Tg) mice.
A. Schematic representation of the construct for generation of SENP2-Tg mouse lines. B. Transcripts and protein level of transgenic flag-tagged human SENP2 were detected in the SENP2-Tg mouse hearts but not in the WT hearts. Upper panel: Reverse transcription followed by PCR was performed on total RNA purified from atria and ventricles of E16.5 WT and SENP2-Tg mouse hearts (lines #2837 and #2839) to detect mRNA of flag-tagged SENP2 and GAPDH (as a control) using specific oligos. Lower panel: Western blot was performed on lysates extracted from P14 WT and SENP2-Tg mouse hearts from both lines to detect flag-tagged SENP2 in SENP2-Tg hearts. Arrows indicate flag-tagged SENP2. C. Suppressed conjugation of SUMO-1 (upper panel) and SUMO-2/3 (middle panel) in SENP2-Tg mouse hearts by overexpression of SENP2. GAPDH served as a loading control (lower panel). HMW, high molecular weight SUMO conjugates. D. Summary of transmission at F1 and F2 generations of both SENP2-Tg lines. Note that the frequency at P1 of F1 and F2 from both lines was lower than the expected Mendelian rate of 50%.
3.3 SENP2-Tg mice exhibited CHDs
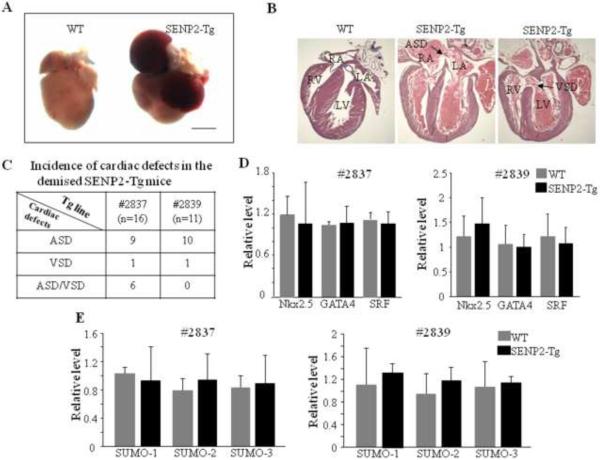
Starting from F3, ~50% and ~80% of the transgenic mice from lines #2839 and #2837 died in the first week after birth, respectively. The mutant hearts obtained from the demised SENP2-Tg mice had atria that were engorged with blood and were enlarged compared with its WT littermate's hearts (Fig. 3A, representative data were shown). Necropsies on those demised SENP2-Tg newborn pups revealed a high preponderance of either ASDs or VSDs or both (Fig. 3B & C). Since a number of transcription factors are critically involved in normal cardiac structural formation, as a first step to elucidate the potential molecular basis underlying those CHDs observed in the SENP2-Tg mice, we carried out RT-qPCR on RNA samples from WT and SENP2-Tg hearts from both lines at E16.5 using specific oligos against GATA4, SRF, Nkx2.5, but detected no significant decrease in the transcription levels of these factors for either line compared to WT (Fig. 3D). Our microarray analysis on the same RNA samples mentioned above also revealed no significant changes in the transcription levels of notch receptors, prox1, smad4, Tbx1, Tbx5 or myocardin (Supplemental Table 1), factors which were shown to be important for proper cardiac morphogenesis, although up- and down- regulation of the transcripts of a number of genes were observed (Supplemental Table 2 & 3). The transcription levels of SUMO-1, -2 and -3 in the SENP2-Tg hearts were not significantly affected by overexpressed SENP2 (Fig. 3E). Collectively, these data indicate that SENP2 overexpression caused ASDs/VSDs via a novel mechanism, one which may involve regulation of the activity, but not the level, of factors that are critical for normal structural morphogenesis.
Figure 3. SENP2-Tg mice exhibited congenital heart diseases.
A. Global morphological view of hearts from one of the demised SENP2-Tg mice and a WT littermate control at P2. Scale bar = 1 mm. B. Histological evaluations revealed ASDs/VSDs in SENP2-Tg mice (middle and right panels) in comparison to littermate control (left panel) at P4. Arrows indicate location of ASD (middle panel) and VSD (right panel), respectively, with the following abbreviations: RA, right atrium; RV, right ventricle; LA, left atrium; LV, left ventricle; ASD, atrial septal defect; VSD, ventricular septal defect. C. Summary of incidence of cardiac defects in the demised SENP2-Tg mice. D. RT-qPCR analysis of RNA samples isolated from E16.5 WT and SENP2-Tg hearts revealed no significant changes in the transcription levels of genes indicated between WT and SENP2-Tg mice. E. RT-qPCR revealed no significant changes in the transcription levels of SUMO factors in E16.5 hearts between the WT and SENP2-Tg mice. n=4 for each group.
3.4 SENP2-Tg mice exhibited growth retardation and cardiac dysfunction
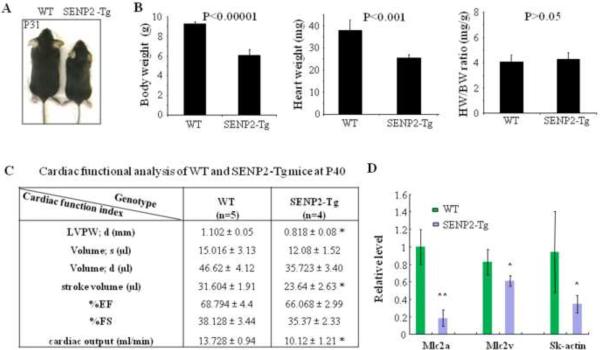
Although immediate postnatal death was observed in both SENP2-Tg mouse lines, a proportion of SENP2-Tg mice did survive to adulthood. However, those surviving SENP2-Tg mice exhibited growth retardation with no detectable cardiac defects (Fig. 4A). Cardiac index heart weight/body weight ratio (HW/BW) showed no significant increase between WT (n=5) and SENP2-Tg (n=4) mice of either line at P18 (Fig. 4B, right panel; only data from line #2839 were shown), suggesting no obvious dilated and/or hypertrophic cardiomyopathy in the SENP2-Tg mice at early stage. However, the body and heart weights in SENP2-Tg mice were significantly lower than those of WT littermates (Fig. 4B, left and middle panels, respectively). To gain insight into the cardiac function of SENP2-Tg mice, we chose line #2839 for detailed non-invasive cardiac functional analysis. At P40, the mutant mice exhibited significantly reduced cardiac output, stroke volume and thinner diastolic left ventricular posterior wall (LVPW; d) compared with those of WT littermates (Fig. 4C), while no significant difference in the percentage (%) of ejection fraction (EF) and the percentage (%) of fractional shortening (FS) between WT and mutant line #2839 was observed. Poor growth of the SENP2-Tg mice was associated with reduced heart size and functional impairment (decreased cardiac output and stroke volume) at this developmental stage. In addition, a number of contractile proteins such as Mlc2a, Mlc2v and skeletal α-actin (SK-actin) were significantly downregulated (Fig. 4D).
Figure 4. SENP2-Tg mice exhibited growth retardation.
A. Comparative body size of a SENP2-Tg mouse (line #2839) and a WT littermate at P31. B. Heart weight (HW) and body weight (BW) of SENP2-Tg mice (n=4) at P18 were significantly lower than those of littermates (n=5, P<0.001 and 0.00001, respectively), but cardiac index HW/BW ratio showed no significant difference (P=0.66). C. Non-invasive cardiac functional analysis of WT and SENP2-Tg mice at P40 using echocardiography showed thinner LVPW in diastole, reduced stroke volume and cardiac output in SENP2-Tg mice. Abbreviations: LVPW, left ventricular posterior wall; d, diastole; s, systole. D. RT-qPCR on RNA samples purified from both WT and SENP2-Tg mouse hearts at P18 revealed down-regulation of transcripts of a number of contractile proteins. *, P<0.05; **, P<0.01. The unpaired Student's t test was applied to determine statistical significance between groups.
To further characterize cardiac phenotype(s) present in the adult SENP2-Tg mice, we performed cardiac functional analysis on mice at 3 and 12 months (mo) of age. Intriguingly, while at P40 SENP2-Tg mice had significantly diminished stroke volume and cardiac output (Fig. 4C), at 3 and 12 mo, SENP2-Tg mice exhibited no significant differences in these two indices compared with WT mice (Table 1), an indication that cardiac functional recovery occurred. At 3 mo, the diastolic LVPW was still thinner in the SENP2-Tg mice than in WT, but the LV mass/body weight index was significantly higher (SENP2-Tg vs. WT: 4.911 ± 0.28 vs. 3.603 ± 0.27 mg/g, p<0.05). In contrast, at 12 mo, diastolic LVPW was no longer thinner in SENP2-Tg mice relative to WT. In addition, at 12 mo, the increased LV mass/BW ratio observed in the SENP2-Tg mice at 3 mo persisted, but this was now accompanied by a decrease in %EF (WT vs. SENP2-Tg: 71.90 ± 3.75 vs. 57.90 ± 4.74, p<0.05) and in %FS (WT vs. SENP2-Tg: 40.76 ± 3.11 vs. 29.97 ± 3.05, p<0.05).
Table 1.
Cardiac functional analysis of WT and SENP2-Tg mice at ages of three and twelve months
| Age | Three months | Twelve Months | ||
|---|---|---|---|---|
| Genotype | WT | SENP2-Tg | WT | SENP2-Tg |
| n=5 | n=4 | n=6 | n=4 | |
| LVPW; d (mm) | 1.20 ± 0.08 | 0.803 ± 0.06** | 1.28 ± 0.20 | 1.33 ± 0.22 |
| Volume; s (μl) | 21.83 ± 7.14 | 16.92 ± 1.10 | 16.41 ± 5.11 | 22.95 ± 7.08 |
| Volume; d (μl) | 63.52 ± 9.25 | 52.69 ± 2.98 | 52.41 ± 9.98 | 50.95 ± 11.18 |
| stroke volume (μl) | 41.69 ± 5.08 | 35.77 ± 2.93 | 36.00 ± 4.94 | 28.01 ± 4.10 |
| %EF | 68.51 ± 7.12 | 67.65 ± 2.36 | 71.90 ± 3.75 | 57.90 ± 4.74+ |
| %FS | 39.00 ± 5.37 | 37.02 ± 1.96 | 40.76 ± 3.11 | 29.97 ± 3.05+ |
| cardiac output (ml/min) | 14.69 ± 1.93 | 14.66 ± 2.88 | 17.35 ± 2.32 | 11.44 ± 1.49 |
| LV mass (mg) | 127.37 ± 10.38 | 97.73 ± 9.61 | 117.84 ± 17.49 | 132.77 ± 8.79 |
| BW (g) | 35.56 ± 1.92 | 19.80 ± 1.11** | 41.58 ± 3.04 | 29.30 ± 1.84+ |
| LV mass/BW | 3.60 ± 0.27 | 4.91 ± 0.28* | 2.87 ± 0.38 | 4.56 ± 0.32+ |
Cardiac functions of WT and SENP2 mice at indicated ages were analyzed by Vevo 770 as detailed in Materials and Methods. The total animal number (n) of each genotype group analyzed was shown. The unpaired Student's t test was used for statistical significance determination between groups of the same age.
p<0.05
p<0.01, compared with the WT group at three months age.
p<0.05, compared with the WT group at twelve months age.
EF, left ventricular ejection faction; FS, left ventricular fractional shortening. Note that the thinner LVPW in diastole in SENP2-Tg mice measured at age of three months was completely recovered in SENP2-Tg mice at the age of twelve months, and the compromised contractility indicated by reduced %EF and %FS was present only in SENP2-Tg mice at twelve months age.
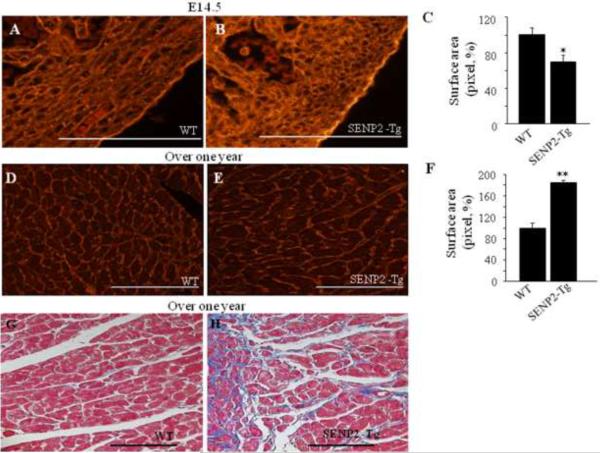
To further understand the cellular basis underlying the cardiac phenotypes in the SENP2-Tg animals at an early developmental stage as well as in aging mice, we evaluated the cardiomyocyte sizes using quantitative WGA-TRITC staining. At E14.5, the average of cardiomyocyte areas in SENP2-Tg embryos was ~30% smaller than that of WT (Fig. 5A–C, p<0.05), indicating impaired cardiomyocyte maturation. However, the cardiomyocyte size of the SENP2-Tg mice at age of over one year was significantly increased compared with WT (Fig. 5D–F, p<0.001), and was accompanied by fibrosis (Fig. 5G & H). Collectively, the above findings suggest that SENP2-Tg mice gradually developed hypertrophic and dysfunctional hearts with aging.
Figure 5. Hearts of aging SENP2-Tg mice were hypertrophic and fibrotic.
A–C. SENP2-Tg mice exhibited smaller cardiomyocyte size compared with WT mice at embryonic stages. Representative WGA-TRITC staining of E14.5 heart sections (A–B) and statistical analysis (C) are shown. Surface areas were measured in 10–20 randomly selected fields from each individual heart sample (n=5 per group for WT and SENP2-Tg mice) using Software ImageJ (National Institutes of Health). Bar, 500 μm. *, p<0.05. The average surface area calculated for WT cardiomyocytes was taken as 100%. Magnification, 40×. D–F. SENP2-Tg mice developed cardiac hypertrophy with aging. Sections of hearts from mice with over one year of age were WGA-TRITC stained and the surface areas were measured as described above (n=4 per group for WT and SENP2-Tg mice). Representative data are shown in D–E and statistical analysis is shown in F. Bar, 200 μm. **, p<0.001. Magnification, 20×. G–H. Aging SENP2-Tg mouse heart presented fibrosis (Masson's trichrome staining). Bar, 200 μm. Magnification, 20×.
3.5 Defect in cardiomyocyte proliferation during embryogenesis in the SENP2-Tg mice
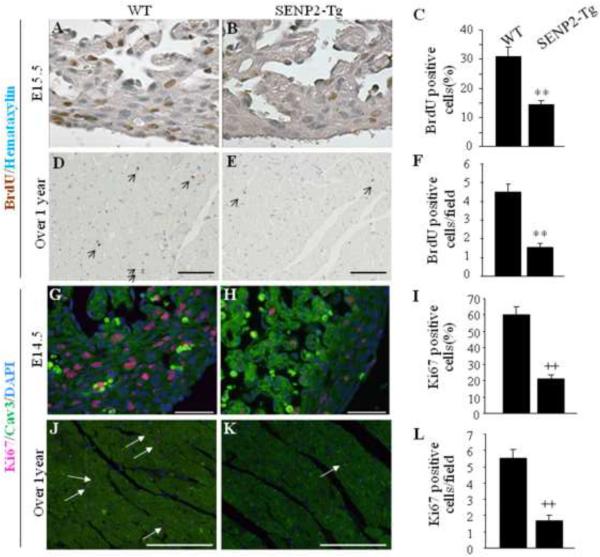
The SUMO pathway is involved in regulating cell division. To gain insight into the molecular basis of the cardiac phenotypes observed in SENP2-Tg mice, DNA synthesis of cardiomyocytes was first evaluated by BrdU staining performed on both early stage (E15.5) and aging heart (over one year of age) sections prepared from the age-matched WT and SENP2-Tg mice. As shown in Fig. 6A–B, the number of BrdU positive nuclei was substantially decreased in the SENP2-Tg embryonic heart compared with WT (Fig. 6C, WT vs Tg: ~30% vs. ~14%, p<0.001), indicative of impaired cardiomyocyte division during heart development. The decreased BrdU staining in the SENP2-Tg hearts persisted with aging (Fig. 6D–F, p<0.001). Next, staining for Ki67, an indicator of proliferating cells, was performed on the sections of both E14.5 embryonic hearts and aging hearts (over one year of age). As expected, the number of dividing cardiomyocytes in the SENP2-Tg mouse hearts at E14.5 was significantly lower than that of the WT (Fig. 6G–I, WT ~60% vs. SENP2-Tg ~20%, p<0.001). A similar phenomenon was also observed in the aging mice (Fig. 6J–L, p<0.001), although the number of dividing cardiomyocytes in the aging WT hearts was significantly decreased compared with that in the E14.5 WT hearts, as expected. Since the SUMO pathway is also involved in regulating apoptosis, we performed TUNEL staining on the same heart samples used for BrdU studies. No significant difference in the TUNEL staining between WT and SENP2-Tg hearts was observed at E14.5 (Supplemental Fig. 1A–B'). The results were further confirmed by cleaved caspase-3 staining (Supplemental Fig. 1C–D'). Taken together, these findings indicate that overexpression of SENP2 negatively affected cardiomyocyte division but did not impact cell survival.
Figure 6. Defect in cardiomyocyte proliferation in SENP2-Tg mice.
A–C: DNA synthesis of cardiomyocyte was decreased in SENP2-Tg mice at E15.5 embryonic stage. Representative data from BrdU staining on E15.5 heart sections (A–B) and statistical analysis (C) are shown. Statistics in C represents the scores of 100 cells per section. A–B, 40× magnification. n=3 per group. **, p<0.001. D–F: SENP2-Tg mouse hearts exhibited decreased BrdU positive cardiomyocytes at age of over one year. Representative BrdU staining of heart sections from aging WT and SENP2-Tg mice (D–E) and statistical analysis (F) are shown. Scale bar, 100 μm. Statistics in F represents the scores of 5 randomly selected fields per section. D–E, 20× magnification. n=3 for each group. **, p<0.001. G–I: SENP2-Tg mouse hearts exhibited decreased Ki67 positive cardiomyocytes at E14.5 embryonic stage. Representative data from Ki67 staining on E14.5 heart sections (G–H) and statistical analysis (I) are shown. Statistical analysis in panel I represents the average scores of 100 cells per section. G–H, 40× magnification. Scale bar, 50μm. ++, p<0.001. J–L: SENP2-Tg mouse hearts exhibited decreased Ki67 positive cardiomyocytes at age of over one year. Representative data from Ki67 staining on sections of aging WT and SENP2-Tg hearts (J–K) and statistical analysis (L) are shown. Statistical results in L represent the scores from at least four randomly selected fields per section. J–K, 20× magnification. n=3 per group. ++, p<0.001. Arrows in D, E, J and K indicate BrdU (panel D–E) or Ki67 (panel J–K) positive cells.
3.6 SUMO-1 played a critical role in the development of cardiac structural defects in SENP2-Tg mice
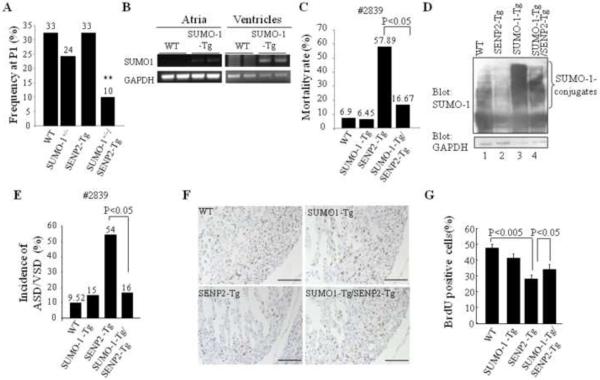
Since SENP2 deconjugated all SUMO isoforms and the balance of SUMO-1 conjugation and deconjugation was important for normal cardiac structural morphogenesis (36), we assessed if SUMO-1 played a critical role in the phenotypic development observed in SENP2-Tg mice. The compound SENP2-Tg/SUMO-1+/− mice, which were generated from crossbreeding between SENP2-Tg mice and SUMO-1+/− mice, were present at much lower frequency at P1 (~10%) than the expected Mendelian ratio (25%, p<0.01, Fig. 7A), suggesting that haploinsufficiency of SUMO-1 led to embryonic lethality in the presence of the increased level of SENP2. Next, we evaluated whether the SUMO-1 transgene would suffice to rescue the CHDs present in the SENP2-Tg mice. We generated double transgenic SUMO-1-Tg/SENP2-Tg (double Tg) mice by crossbreeding SENP2-Tg (line #2839) and SUMO-1-Tg mice, (the latter designed to express flag-tagged SUMO-1 transgene driven by the α-MHC promoter (36)). The SUMO-1 transgene was expressed in both the atrium and the ventricle at E16.5 (Fig. 7B), and the double Tg line displayed a much lower mortality rate (16.67%) compared with the single SENP2-Tg mice (57.89%, p<0.05, Fig. 7C); the improved survival correlated well with improved SUMO-1 conjugation (Fig. 7D, upper panel). Histological analysis (Fig. 7E) revealed that the penetrance of cardiac defects was reduced in the double Tg mice compared with the single SENP2-Tg mice analyzed between E14.5 and P2 (SENP2-Tg vs double Tg: 54.55% vs. 16.67%, P<0.05); this reduced CHD penetrance correlated well with improved survival rate in double Tg mice. Also, improved BrdU incorporation in the double Tg heart was observed in comparison with that in the single SENP2-Tg hearts (Fig. 7F–G). Collectively, these findings indicate a critical role for SUMO-1 in the initiation of CHDs caused by overexpression of SENP2. Interestingly, overexpressed SUMO-1 transgene failed to rescue growth retardation or to alleviate cardiac dysfunction observed in the surviving SENP2-Tg mice (Supplemental Fig. 2 and data not shown). Thus, growth retardation/cardiac dysfunction was caused via pathway(s) independent of SUMO-1 conjugation, at least in SENP2-Tg mice.
Figure 7. SUMO-1 played a critical role in the development of cardiac structural malformation in SENP2-Tg mice.
A. Overexpression of SENP2 in the presence of SUMO-1 haploinsufficiency led to embryonic lethality. The compound SENP2-Tg/SUMO-1+/− mice obtained from crossbreeding between SENP2-Tg mice (line #2839) and SUMO-1+/− mice were present at lower P1 frequency than the expected Mendelian rate (25%). The P1 frequency was analyzed from 7 litters comprising a total of 49 animals. The number shown above each bar indicates the P1 frequency of each corresponding group. **, P<0.01. Chi-square test was used for statistical analysis. B. The transcripts of transgene flag-tagged SUMO-1 were detected in both atria and ventricles of SUMO-1-Tg mouse hearts at E16.5 but not in the WT littermate hearts. GAPDH was used as a control. C. Mortality rate of double SUMO-1-Tg/SENP2-Tg mice was substantially decreased compared with that of single SENP2-Tg mice. Rescue data from line #2839 were compiled from 15 litters of 97 animals in total. Fisher's Exact Probability Test was used for statistical analysis. D. Improved modification of SUMO-1 in double SUMO-1-Tg/SENP2-Tg mouse heart. Western blots were performed on heart extracts from the WT, SENP2-Tg, SUMO-1-Tg and double SUMO-1-Tg/SENP2-Tg mice. Compare SUMO-1 conjugation (upper panel) in lane 2 and lane 4. GAPDH (lower panel) served as a control. E. Decreased penetrance of cardiac defects in the double SUMO-1-Tg/SENP2-Tg mice. The data were compiled from 76 available animals of 10 litters with ages ranging from E14.5 to P2. Fisher's Exact Probability Test was used for statistical analysis. The number shown above each bar indicates the incidence of cardiac defects of that corresponding group. F–G. Improved cardiomyocyte proliferation in the double SUMO-1-Tg/SENP2-Tg mouse hearts. BrdU staining was performed on sections of E14.5 embryonic hearts of the WT, SUMO-1-Tg, SENP2-Tg and double SUMO-1-Tg/SENP2-Tg mice, respectively (F) and statistical analysis was shown in G. n=4 for each group. Representative data were shown. Magnification, 20×. Bar, 200 μm.
3.7 Deconjugation of SUMO substrates in SENP2-Tg hearts
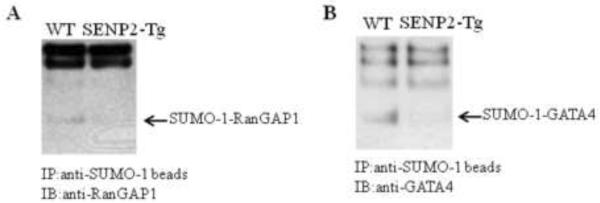
Given the phenotypes, i.e. defective cell cycle, cardiac structural defects, and globally decreased sumoylation observed in the SENP2-Tg hearts, we determined if SENP2 was able to demodify sumoylation of some particular SUMO substrates in vivo. Co-immunoprecipitation was carried out on heart lysates purified from age-matched WT and SENP2-Tg mice using agarose-conjugated SUMO-1 antibody for pulldown and desired specific antibodies for detection. As shown in Fig. 8A, the level of SUMO-1-conjugated RanGAP1, the first identified SUMO substrate and a critical factor in regulating cell cycle, was decreased in the SENP2-Tg heart compared with that in the WT heart. Also, GATA4, an important transcription factor for normal cardiac development and function, was also desumoylated in the SENP2-Tg mouse heart (Fig. 8B). These observations suggest that SENP2 indeed demodified a number of SUMO-conjugated substrates examined in vivo.
Figure 8. Desumoylation of SUMO substrates in SENP2-Tg hearts.
Co-immunoprecipitation (IP) was performed on the whole tissue extracts from age-matched WT and SENP2-Tg mouse hearts using agarose-conjugated anti-SUMO-1 antibody for pulldown; subsequently, immunoblot (IB) was performed using anti-RanGAP1 (A) or anti-GATA4 antibodies (B) for detection. Arrows indicate SUMO-1-conjugated RanGAP1 (A) and SUMO-1-conjugated GATA4 (B). Note the substantial decrease in the level of SUMO-1 conjugated RanGAP1 and GATA4 in SENP2-Tg heart compared with those in WT heart.
3.8 Functional interaction between the SUMO pathway component SENP2 and Nkx2.5
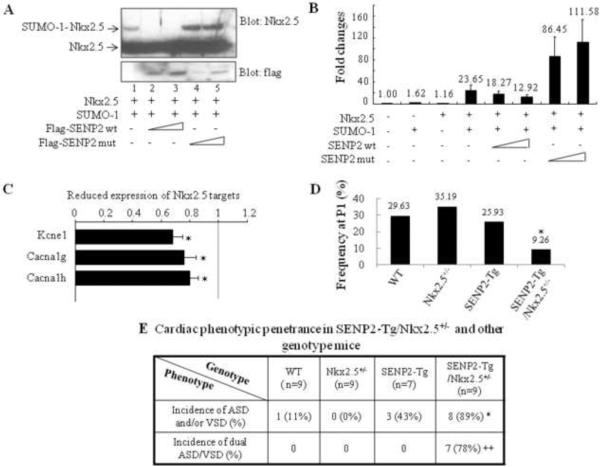
Mutations of Nkx2.5, the cardiac specific homeodomain-containing protein, have been causally linked to CHDs (49, 50), and sumoylation of Nkx2.5 greatly enhanced its transcriptional activity (35). Human Nkx2.5 mutants exhibited altered sumoylation (39). In addition, heterozygous Nkx2.5+/−:SUMO-1+/− mice showed more severe cardiac phenotypes (36). Thus, we further investigated whether functional interaction between Nkx2.5 and the SUMO pathway components occurred in vivo. First, we tested whether Nkx2.5 was a substrate for deconjugation by SENP2. As shown in Fig. 9A, co-transfection of SUMO-1 with Nkx2.5 produced a SUMO-1-conjugated Nkx2.5 species (a retarded migratory band, as expected). This effect was blocked by the delivery of SENP2 wt (two dosages), but was enhanced by the presence of the SENP2 catalytically inactive mutant (SENP mut). Expression of the SENP2 mut appeared to have a dominant negative effect, inhibiting the activity of endogenous SENP2. The activity of the ANF promoter, which is a target of Nkx2.5, was elevated by the combination of Nkx2.5 and SUMO-1, although delivery of either SUMO-1 or Nkx2.5 alone did not exert significant effects at the tested dosages (Fig. 9B). Neither SENP2 wt nor SENP2 mut alone significantly influenced ANF promoter activity (data not shown); however, expression of SENP2 wt resulted in a dose-dependent suppression of ANF promoter activation achieved by the combined expression of Nkx2.5 and SUMO-1. On the contrary, SENP2 mut enhanced promoter activation in the presence of both Nkx2.5 and SUMO-1. This observation demonstrates the inhibitory effect of SENP2 on Nkx2.5 transcriptional activity. Consistent with this, microarray analysis showed that a number of Nkx2.5 target genes (i.e., Kcne1, Cacna1g and h, (51)) were downregulated in the E16.5 SENP2-Tg hearts (Fig. 9C, p<0.05, n=3 for WT and SENP2-Tg, respectively).
Figure 9. Functional interaction between the SUMO pathway component, SENP2, and Nkx2.5.
A. SENP2 wt, but not the catalytic mutant, deconjugated SUMO-1-Nkx2.5 conjugates. Sumoylation assays were performed on HeLa cell extracts containing overexpressed proteins, as indicated. Upper panel: blotted with anti-Nkx2.5 antibody to detect both free and conjugated Nkx2.5 (arrows); lower panel: blotted with anti-flag antibody to detect SENP2 wt and mutant. B. SENP2 wt, but not the catalytic mutant, suppressed Nkx2.5 activity in a dose-dependent manner. Transactivation assays were performed on HeLa cells transfected with the ANF-Luc construct together with single or combined expression of vectors, as indicated. Relative activation level (fold change) was calculated based on the luciferase activity of the control (empty vector alone), which was taken as 1. The dosage for each of those expression vectors were used as follows: Nkx2.5: 0.5 μg; SUMO-1: 0.75 μg; SENP2 wt: 50 and 100 ng; SENP2 mutant: 50 and 100 ng. The number shown above each bar indicates the fold change of the luciferase activity of that particular group. C. Down-regulation of a number of Nkx2.5 target genes in SENP2-Tg hearts was shown in comparison with those of WT hearts. The microarray assays of RNA samples from E16.5 SENP2-Tg and WT hearts were performed as described in the Materials and Methods. n=3 per group, with each sample carried out in duplicate. D–E. SENP2-Tg/Nkx2.5+/− mice exhibited a lower P1 frequency (D) and a higher incidence of cardiac defects, including the dual ASD/VSD phenotype (E), compared with those of single Nkx2.5+/− and SENP2-Tg mice. Mice were obtained from crossbreeding between SENP2-Tg mice of line #2839 and Nkx2.5+/− mice. The data were then compiled from 13 litters comprising a total of 75 animals (D) and from 7 litters comprising a total of 34 embryos with developmental stages ranging from E16.5 to E17.5 (E). The total animal number (n) of each genotype group analyzed in E was shown. Chi-square test was used for statistical comparison. *, p<0.05; ++, p<0.005.
To further assess the functional interaction between SENP2 and Nkx2.5 in vivo, the SENP2-Tg mouse line #2839 was crossed with the Nkx2.5+/− mouse line. As shown in Fig. 9D, the compound SENP2-Tg/Nkx2.5+/− mice exhibited lower P1 frequency (9.26%) than the expected 25% of Mendelian rate (p<0.05). Also, the penetrance of cardiac structural defects in the SENP2-Tg/Nkx2.5+/− mouse embryos analyzed between E15.5 and E17.5 was significantly higher than the single SENP2-Tg mice (Fig. 9E, 89% vs. 43%, p<0.05). In addition, the incidence of the dual ASD/VSD phenotype—a more severe cardiac defect—in the SENP2-Tg/Nkx2.5+/− mouse embryos was significantly greater than that of the single SENP2-Tg mice (Fig. 9E, p<0.01). These results, combined with the SENP2 wt and mut transfection data, indicate a functional interaction between Nkx2.5 and the SUMO conjugation pathway component SENP2 in vivo.
4 Discussion
SUMO conjugation is a highly conserved process that contributes to the maintenance of normal cell function. Our previous work demonstrated the importance of SUMO-1 conjugation to normal heart development using a SUMO-1 knockout mouse model (36). In the present study, we further corroborated the essentiality of this posttranslational modification to heart development and function using an additional approach by establishing a murine model in which desumoylation was substantiated specifically in heart.
Balanced sumoylation is required for normal cardiac structural morphogenesis
Sumoylation and desumoylation is a reversible and dynamic process, interruption of which is implicated in abnormal organogenesis, i.e., development of cleft lip/palate (52, 53). Our recent work revealed a critical contribution of SUMO-1 to early cardiac morphogenesis (36), which was further corroborated by the findings provided in the present study demonstrating that i) an embryonic lethal phenotype resulted from SENP2 overexpression in SUMO-1 haploinsufficient mice and ii) the SUMO-1 transgene successfully reduced the incidence of cardiac defect phenotypes in the SENP2-Tg mice; this phenomenon was accompanied by improved SUMO-1 conjugation and cardiomyocyte proliferation. The defect in the cardiac myocyte cycle was associated with cardiac structural defects observed in the SENP2-Tg mice. Consistent with the observation of decreased cell division in the SENP2-Tg hearts, SUMO-1 conjugation to RanGAP1, a factor critical for cell cycle progression (54), was also suppressed. It will be of great interest to explore whether SUMO conjugation to other targets (e.g. RanBP2 and TopoII alpha) that are also important cell cycle regulators (55, 56) is negatively influenced in the sumoylation-deficient hearts.
Our findings revealed an important role for SUMO-1 in the formation of cardiac structures, but the contribution of SUMO-2/3 to heart development, particularly to the septation process, is still unclear. We noted that SUMO-1 expression did not completely rescue the cardiac phenotype in line #2837 mice (Supplemental Fig. 3), which (prior to rescue) exhibited more severe phenotypes and higher mortality rate. This incomplete rescue may suggest that the conjugation of SUMO-2/-3 and/or SENP2's non-catalytic activity was involved in the initiation of the CHDs observed in the SENP2-Tg mice. Alternatively, the difference in the rescue effects rendered by the SUMO-1 transgene in these two lines may only reflect differences in the expression level and/or spatial localization of the SENP2 transgene. Additionally, it is also possible that introduction of extra SUMO-1 resulted in random conjugation to targets with a failure of extra SUMO-1 in binding the appropriate substrate(s) required for normal heart development.
Balanced sumoylation is required for the maintenance of normal cardiac function
While overexpression of SUMO-1 successfully improved cardiac structural formation in SENP2-Tg mice, it failed to rescue growth retardation/cardiac dysfunction. This observation suggested that SENP2 has effects which are independent of repressed SUMO-1 conjugation. At both the embryonic stage and older ages (i.e., ~ one year), the SENP2-Tg mice showed similar defect in the cell cycle – reduction in the number of proliferating cardiomyocytes; however, the manifestations of cardiomyocyte phenotypes were different: smaller cardiomyocyte size was evident at the early stage while cardiomyocyte enlargement was associated with senescence and impaired heart function. In the aging SENP2-Tg mice, cardiomyopathy appeared to be manifested, and cardiac fibrosis was increased. The mechanisms underlying the size transformation of cardiomyocytes with aging is not clear and must be complex; however, it is likely that it involves the hypertrophic response to the long term “stimulation” exerted by the increased level of SENP2. Regulation of cardiomyocyte growth by the SUMO conjugation pathway must also be complex, as different phenotypes were exhibited despite a similar proliferation defect. Aging must trigger divergent mechanisms regulating cardiac growth in which distinct factors in the changing physiological milieu are expressed to influence SUMO conjugation. Since the aging double SENP2-Tg/SUMO-1-Tg mice exhibited cardiac hypertrophy and decreased cardiac function, as did the single SENP2-Tg mice (data not shown), we do not believe that the SUMO-1 conjugation is instrumental in the initiation of cardiac dysfunction associated with aging, at least in the murine model of SENP2 overexpression. Whether cardiomyopathy in the SENP2-Tg mice is attributable to decreased SUMO-2/3 conjugation remains unknown. Increased SUMO-1 conjugation in the heart efficiently rescued CHDs but not cardiac muscle disorders of the SENP2-Tg mice; thus, SUMO-1 conjugation is probably more involved in early cardiac structural morphogenesis, whereas the SUMO-2/3 conjugation is more important to the maintenance of normal cardiac function. However, this hypothesis requires further investigation.
The growth retardation of SENP2-Tg mice probably resulted from impaired cardiomyocyte maturation and/or cardiac dysfunction. To the best of our knowledge, global growth restriction originating from cardiac-specific expression of a transgene of interest was not common. In the α-MHC-Hoxb-7 Tg mice (57), skeletal malformation was observed, although the Hoxb-7 transgene was not detected in that region. Whether there are any other developmental defects in the SENP2-Tg mice, in addition to CHDs, is an open question and would be interesting to explore in the future.
Nkx2.5 is involved in the sumoylation deficiency-associated CHDs
Functioning as a desumoylation enzyme, SENP2 targeted a number of substrates, including GATA4. Therefore, the SENP2-elicited CHDs should be a net and/or a total effect of desumoylation of those SUMO substrates that are critical for cardiac morphogenesis, rather than a consequence of desumoylation of a single substrate. However, some SUMO substrate(s) may play more important roles than others in the development of sumoylation deficiency-associated phenotypes. Nkx2.5, a cardiac-specific homeodomain-containing transcription factor and a SUMO target (35), played a critical role in the development of CHDs in humans and in mice (49, 58). Our recent work demonstrated that a number of CHD-linked Nkx2.5 mutants exhibited altered sumoylation status (39). Thus, we hypothesized that Nkx2.5 could contribute to the development of CHDs associated with deficient SUMO conjugation pathway. To further understand the mechanisms underlying the evolution of CHDs that were triggered by overexpression of SENP2, we investigated the functional interaction between Nkx2.5 and SENP2 in vivo. We observed that i) SENP2 inhibited Nkx2.5 transcriptional activity by promoting deconjugation; ii) a number of Nkx2.5 targets were downregulated in the hearts of SENP2-Tg mice; iii) the compound SENP2-Tg/Nkx2.5+/− mice were born at a much lower frequency at P1 than either single SENP2-Tg or Nkx2.5+/− mice; iiii) the compound SENP2-Tg/Nkx2.5+/− mice exhibited a higher phenotypic penetrance and more severe cardiac phenotypes than either single SENP2-Tg or Nkx2.5+/− mice. Therefore, we concluded that overexpression of SENP2 initiated the development of CHDs at least partially by suppressing the transcriptional activity of Nkx2.5. Unfortunately, like for a number of studies in which sumoylation of endogenous substrates has not been possible to detect due to technical limitations (59, 60), we were unable to convincingly show the level of SUMO-conjugated Nkx2.5 in the hearts of WT or SENP2-Tg mice.
In conclusion, our findings support the notion that defects in the SUMO pathway, exemplified by the described SENP2-Tg mice, may contribute significantly to both CHDs and cardiac muscle disorders. Enhanced desumoylation activity in the heart promoted by SENP2 overexpression mainly influenced cardiomyocyte division. The functional interaction between the SUMO substrate, Nkx2.5, and the sumoylation pathway component, SENP2, presented a mechanism that at least partially accounted for the development of sumoylation deficiency-linked CHDs. Increased SUMO-1 activity effectively reduced the penetrance of cardiac structural malformation, but did not ameliorate cardiac dysfunction existent in SENP2-Tg mice; thus, SUMO-1 may play different roles in the early cardiac structural development than it does in late cardiac function. A future direction stemming from these studies will be to investigate the role of SUMO-1 conjugation in modulation of heart function under pathophysiological conditions such as ischemia, and to examine the respective contribution of SUMO-1 and SUMO-2/3 conjugation and the individual isopeptidases to cardiac development, function and disease.
Supplementary Material
Highlights.
-
>
We studied the effect of de-sumoylation on heart development in a transgenic model.
-
>
Increased desumoylation by overexpressed SENP2 in heart caused cardiac defects and dysfunction.
-
>
SENP2 overexpression also caused defect in cardiomyocyte proliferation.
-
>
SUMO-1 overexpression rescued cardiac structural phenotypes in SENP2-Tg mice.
-
>
Balanced SUMO conjugation is essential for normal heart development and function.
Acknowledgements
The authors would like to thank Drs. Robert J. Schwartz and James T. Willerson for their support of the work. The authors are grateful to Mrs. Ling Qian for technical assistance. This work was supported in part by a grant from Texas Higher Education Coordinating Board (Grant No. 000089-0004-2007 to J.W.), a Beginning Grant-in-Aid from the American Heart Association (Grant No. 09BGIA2050016 to J.W.), and a grant from the National Institutes of Health (Grant No. 5R01HL102314-02 to J.C.). J.W. is the recipient of the start-up package (P30 grant) from the National Institutes of Health as a Newly Independent Investigator (NII).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DISCLOSURES None.
References
- 1.Bruneau BG, Nemer G, Schmitt JP, Charron F, Robitaille L, Caron S, et al. A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell. 2001;106(6):709–21. doi: 10.1016/s0092-8674(01)00493-7. [DOI] [PubMed] [Google Scholar]
- 2.Niessen K, Karsan A. Notch signaling in cardiac development. Circ Res. 2008;102(10):1169–81. doi: 10.1161/CIRCRESAHA.108.174318. [DOI] [PubMed] [Google Scholar]
- 3.Nie X, Deng CX, Wang Q, Jiao K. Disruption of Smad4 in neural crest cells leads to mid-gestation death with pharyngeal arch, craniofacial and cardiac defects. Dev Biol. 2008;316(2):417–30. doi: 10.1016/j.ydbio.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanaka M, Chen Z, Bartunkova S, Yamasaki N, Izumo S. The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development. Development. 1999;126(6):1269–80. doi: 10.1242/dev.126.6.1269. [DOI] [PubMed] [Google Scholar]
- 5.Garg V, Kathiriya IS, Barnes R, Schluterman MK, King IN, Butler CA, et al. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature. 2003;424(6947):443–7. doi: 10.1038/nature01827. [DOI] [PubMed] [Google Scholar]
- 6.Baldini A. Dissecting contiguous gene defects: TBX1. Curr Opin Genet Dev. 2005;15(3):279–84. doi: 10.1016/j.gde.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 7.Bentham J, Bhattacharya S. Genetic mechanisms controlling cardiovascular development. Ann N Y Acad Sci. 2008;1123:10–9. doi: 10.1196/annals.1420.003. [DOI] [PubMed] [Google Scholar]
- 8.Wang J. Cardiac function and disease: emerging role of small ubiquitin-related modifier. Wiley Interdiscip Rev Syst Biol Med. 2011;3(4):446–57. doi: 10.1002/wsbm.130. PMCID: 3110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J, Schwartz RJ. Sumoylation and regulation of cardiac gene expression. Circ Res. 2010;107(1):19–29. doi: 10.1161/CIRCRESAHA.110.220491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dasso M. Emerging roles of the SUMO pathway in mitosis. Cell Div. 2008;3:5. doi: 10.1186/1747-1028-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson ES, Blobel G. Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J Cell Biol. 1999;147(5):981–94. doi: 10.1083/jcb.147.5.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poulin G, Dong Y, Fraser AG, Hopper NA, Ahringer J. Chromatin regulation and sumoylation in the inhibition of Ras-induced vulval development in Caenorhabditis elegans. Embo J. 2005;24(14):2613–23. doi: 10.1038/sj.emboj.7600726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson ES. Protein modification by sumo. Annu Rev Biochem. 2004;73:355–82. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 14.Rosas-Acosta G, Russell WK, Deyrieux A, Russell DH, Wilson VG. A universal strategy for proteomic studies of SUMO and other ubiquitin-like modifiers. Mol Cell Proteomics. 2005;4(1):56–72. doi: 10.1074/mcp.M400149-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mukhopadhyay D, Dasso M. Modification in reverse: the SUMO proteases. Trends Biochem Sci. 2007;32(6):286–95. doi: 10.1016/j.tibs.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 16.Gong L, Millas S, Maul GG, Yeh ET. Differential regulation of sentrinized proteins by a novel sentrin-specific protease. J Biol Chem. 2000;275(5):3355–9. doi: 10.1074/jbc.275.5.3355. [DOI] [PubMed] [Google Scholar]
- 17.Hang J, Dasso M. Association of the human SUMO-1 protease SENP2 with the nuclear pore. J Biol Chem. 2002;277(22):19961–6. doi: 10.1074/jbc.M201799200. [DOI] [PubMed] [Google Scholar]
- 18.Mikolajczyk J, Drag M, Bekes M, Cao JT, Ronai Z, Salvesen GS. Small ubiquitin-related modifier (SUMO)-specific proteases: profiling the specificities and activities of human SENPs. J Biol Chem. 2007;282(36):26217–24. doi: 10.1074/jbc.M702444200. [DOI] [PubMed] [Google Scholar]
- 19.Yeh ET. SUMOylation and de-SUMOylation: Wrestling with life's processes. J Biol Chem. 2008 doi: 10.1074/jbc.R800050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng J, Bawa T, Lee P, Gong L, Yeh ET. Role of desumoylation in the development of prostate cancer. Neoplasia. 2006;8(8):667–76. doi: 10.1593/neo.06445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gong L, Yeh ET. Characterization of a family of nucleolar SUMO-specific proteases with preference for SUMO-2 or SUMO-3. J Biol Chem. 2006;281(23):15869–77. doi: 10.1074/jbc.M511658200. [DOI] [PubMed] [Google Scholar]
- 22.Di Bacco A, Ouyang J, Lee HY, Catic A, Ploegh H, Gill G. The SUMO-specific protease SENP5 is required for cell division. Mol Cell Biol. 2006;26(12):4489–98. doi: 10.1128/MCB.02301-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mukhopadhyay D, Ayaydin F, Kolli N, Tan SH, Anan T, Kametaka A, et al. SUSP1 antagonizes formation of highly SUMO2/3-conjugated species. J Cell Biol. 2006;174(7):939–49. doi: 10.1083/jcb.200510103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen LN, Geoffroy MC, Jaffray EG, Hay RT. Characterization of SENP7, a SUMO-2/3-specific isopeptidase. Biochem J. 2009;421(2):223–30. doi: 10.1042/BJ20090246. [DOI] [PubMed] [Google Scholar]
- 25.Cheng J, Kang X, Zhang S, Yeh ET. SUMO-specific protease 1 is essential for stabilization of HIF1alpha during hypoxia. Cell. 2007;131(3):584–95. doi: 10.1016/j.cell.2007.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang X, Qi Y, Zuo Y, Wang Q, Zou Y, Schwartz RJ, et al. SUMO-specific protease 2 is essential for suppression of polycomb group protein-mediated gene silencing during embryonic development. Mol Cell. 2010;38(2):191–201. doi: 10.1016/j.molcel.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chiu SY, Asai N, Costantini F, Hsu W. SUMO-specific protease 2 is essential for modulating p53-Mdm2 in development of trophoblast stem cell niches and lineages. PLoS Biol. 2008;6(12):e310. doi: 10.1371/journal.pbio.0060310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dorval V, Fraser PE. SUMO on the road to neurodegeneration. Biochim Biophys Acta. 2007;1773(6):694–706. doi: 10.1016/j.bbamcr.2007.03.017. [DOI] [PubMed] [Google Scholar]
- 29.Aribi M. Candidate genes implicated in type 1 diabetes susceptibility. Curr Diabetes Rev. 2008;4(2):110–21. doi: 10.2174/157339908784220723. [DOI] [PubMed] [Google Scholar]
- 30.Wu F, Mo YY. Ubiquitin-like protein modifications in prostate and breast cancer. Front Biosci. 2007;12:700–11. doi: 10.2741/2094. [DOI] [PubMed] [Google Scholar]
- 31.Pauws E, Stanier P. FGF signalling and SUMO modification: new players in the aetiology of cleft lip and/or palate. Trends Genet. 2007;23(12):631–40. doi: 10.1016/j.tig.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 32.Zhang YQ, Sarge KD. Sumoylation regulates lamin A function and is lost in lamin A mutants associated with familial cardiomyopathies. J Cell Biol. 2008;182(1):35–9. doi: 10.1083/jcb.200712124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J, Feng XH, Schwartz RJ. SUMO-1 modification activated GATA4-dependent cardiogenic gene activity. J Biol Chem. 2004;279(47):49091–8. doi: 10.1074/jbc.M407494200. [DOI] [PubMed] [Google Scholar]
- 34.Wang J, Li A, Wang Z, Feng X, Olson EN, Schwartz RJ. Myocardin sumoylation transactivates cardiogenic genes in pluripotent 10T1/2 fibroblasts. Mol Cell Biol. 2007;27(2):622–32. doi: 10.1128/MCB.01160-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J, Zhang H, Iyer D, Feng XH, Schwartz RJ. Regulation of cardiac specific Nkx2.5 gene activity by sumo modificaiton. J Biol Chem. 2008 doi: 10.1074/jbc.M709748200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang J, Chen L, Wen S, Zhu H, Yu W, Moskowitz IP, et al. Defective sumoylation pathway directs congenital heart disease. Birth Defects Res A Clin Mol Teratol. 2011;91(6):468–76. doi: 10.1002/bdra.20816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen CY, Croissant J, Majesky M, Topouzis S, McQuinn T, Frankovsky MJ, et al. Activation of the cardiac alpha-actin promoter depends upon serum response factor, Tinman homologue, Nkx-2.5, and intact serum response elements. Dev Genet. 1996;19(2):119–30. doi: 10.1002/(SICI)1520-6408(1996)19:2<119::AID-DVG3>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 38.Wang J, Zhang H, Iyer D, Feng XH, Schwartz RJ. Regulation of cardiac specific nkx2.5 gene activity by small ubiquitin-like modifier. J Biol Chem. 2008;283(34):23235–43. doi: 10.1074/jbc.M709748200. PMCID: 2516993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim EY, Chen L, Ma Y, Yu W, Chang J, Moskowitz IP, et al. Expression of sumoylation deficient nkx2.5 mutant in nkx2.5 haploinsufficient mice leads to congenital heart defects. PLoS One. 2011;6(6):e20803. doi: 10.1371/journal.pone.0020803. PMCID: 3108998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moses KA, DeMayo F, Braun RM, Reecy JL, Schwartz RJ. Embryonic expression of an Nkx2-5/Cre gene using ROSA26 reporter mice. Genesis. 2001;31(4):176–80. doi: 10.1002/gene.10022. [DOI] [PubMed] [Google Scholar]
- 41.Evdokimov E, Sharma P, Lockett SJ, Lualdi M, Kuehn MR. Loss of SUMO1 in mice affects RanGAP1 localization and formation of PML nuclear bodies, but is not lethal as it can be compensated by SUMO2 or SUMO3. J Cell Sci. 2008;121(Pt 24):4106–13. doi: 10.1242/jcs.038570. [DOI] [PubMed] [Google Scholar]
- 42.Wang J, Li A, Wang Z, Feng X, Olson EN, Schwartz RJ. Myocardin Sumoylation Transactivates Cardiogenic Genes in Pluripotent 10T1/2 Fibroblasts. Mol Cell Biol. 2007;27(2):622–32. doi: 10.1128/MCB.01160-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Monzen K, Shiojima I, Hiroi Y, Kudoh S, Oka T, Takimoto E, et al. Bone morphogenetic proteins induce cardiomyocyte differentiation through the mitogen-activated protein kinase kinase kinase TAK1 and cardiac transcription factors Csx/Nkx-2.5 and GATA-4. Mol Cell Biol. 1999;19(10):7096–105. doi: 10.1128/mcb.19.10.7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fujikura J, Yamato E, Yonemura S, Hosoda K, Masui S, Nakao K, et al. Differentiation of embryonic stem cells is induced by GATA factors. Genes Dev. 2002;16(7):784–9. doi: 10.1101/gad.968802. PMCID: 186328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Manabe I, Owens GK. Recruitment of serum response factor and hyperacetylation of histones at smooth muscle-specific regulatory regions during differentiation of a novel P19-derived in vitro smooth muscle differentiation system. Circ Res. 2001;88(11):1127–34. doi: 10.1161/hh1101.091339. [DOI] [PubMed] [Google Scholar]
- 46.Schoenfeld JR, Vasser M, Jhurani P, Ng P, Hunter JJ, Ross J, Jr., et al. Distinct molecular phenotypes in murine cardiac muscle development, growth, and hypertrophy. J Mol Cell Cardiol. 1998;30(11):2269–80. doi: 10.1006/jmcc.1998.0787. [DOI] [PubMed] [Google Scholar]
- 47.Makino S, Fukuda K, Miyoshi S, Konishi F, Kodama H, Pan J, et al. Cardiomyocytes can be generated from marrow stromal cells in vitro. J Clin Invest. 1999;103(5):697–705. doi: 10.1172/JCI5298. PMCID: 408125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang H, Saitoh H, Matunis MJ. Enzymes of the SUMO modification pathway localize to filaments of the nuclear pore complex. Mol Cell Biol. 2002;22(18):6498–508. doi: 10.1128/MCB.22.18.6498-6508.2002. PMCID: 135644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kasahara H, Lee B, Schott JJ, Benson DW, Seidman JG, Seidman CE, et al. Loss of function and inhibitory effects of human CSX/NKX2.5 homeoprotein mutations associated with congenital heart disease. J Clin Invest. 2000;106(2):299–308. doi: 10.1172/JCI9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tanaka M, Berul CI, Ishii M, Jay PY, Wakimoto H, Douglas P, et al. A mouse model of congenital heart disease: cardiac arrhythmias and atrial septal defect caused by haploinsufficiency of the cardiac transcription factor Csx/Nkx2.5. Cold Spring Harb Symp Quant Biol. 2002;67:317–25. doi: 10.1101/sqb.2002.67.317. [DOI] [PubMed] [Google Scholar]
- 51.Terada R, Warren S, Lu JT, Chien KR, Wessels A, Kasahara H. Ablation of Nkx2-5 at mid-embryonic stage results in premature lethality and cardiac malformation. Cardiovasc Res. 2011;91(2):289–99. doi: 10.1093/cvr/cvr037. PMCID: 3125071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alkuraya FS, Saadi I, Lund JJ, Turbe-Doan A, Morton CC, Maas RL. SUMO1 haploinsufficiency leads to cleft lip and palate. Science. 2006;313(5794):1751. doi: 10.1126/science.1128406. [DOI] [PubMed] [Google Scholar]
- 53.Song T, Li G, Jing G, Jiao X, Shi J, Zhang B, et al. SUMO1 polymorphisms are associated with non-syndromic cleft lip with or without cleft palate. Biochem Biophys Res Commun. 2008;377(4):1265–8. doi: 10.1016/j.bbrc.2008.10.138. [DOI] [PubMed] [Google Scholar]
- 54.Joseph J, Tan SH, Karpova TS, McNally JG, Dasso M. SUMO-1 targets RanGAP1 to kinetochores and mitotic spindles. J Cell Biol. 2002;156(4):595–602. doi: 10.1083/jcb.200110109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dawlaty MM, Malureanu L, Jeganathan KB, Kao E, Sustmann C, Tahk S, et al. Resolution of sister centromeres requires RanBP2-mediated SUMOylation of topoisomerase IIalpha. Cell. 2008;133(1):103–15. doi: 10.1016/j.cell.2008.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Azuma Y, Arnaoutov A, Anan T, Dasso M. PIASy mediates SUMO-2 conjugation of Topoisomerase-II on mitotic chromosomes. Embo J. 2005;24(12):2172–82. doi: 10.1038/sj.emboj.7600700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Argao EA, Kern MJ, Branford WW, Scott WJ, Jr., Potter SS. Malformations of the heart, kidney, palate, and skeleton in alpha-MHC-Hoxb-7 transgenic mice. Mech Dev. 1995;52(2–3):291–303. doi: 10.1016/0925-4773(95)98114-p. [DOI] [PubMed] [Google Scholar]
- 58.Moskowitz IP, Kim JB, Moore ML, Wolf CM, Peterson MA, Shendure J, et al. A molecular pathway including Id2, Tbx5, and Nkx2-5 required for cardiac conduction system development. Cell. 2007;129(7):1365–76. doi: 10.1016/j.cell.2007.04.036. [DOI] [PubMed] [Google Scholar]
- 59.Thakar K, Niedenthal R, Okaz E, Franken S, Jakobs A, Gupta S, et al. SUMOylation of the hepatoma-derived growth factor negatively influences its binding to chromatin. Febs J. 2008;275(7):1411–26. doi: 10.1111/j.1742-4658.2008.06303.x. [DOI] [PubMed] [Google Scholar]
- 60.Melchior F, Schergaut M, Pichler A. SUMO: ligases, isopeptidases and nuclear pores. Trends Biochem Sci. 2003;28(11):612–8. doi: 10.1016/j.tibs.2003.09.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.