Abstract
Methylmercury (MeHg) is a ubiquitous toxicant that targets the developing fetal nervous system. MeHg interacts with the Notch signaling pathway, a highly-conserved intercellular signaling mechanism required for normal development. Notch signaling is conveyed by activation of the genes in the Enhancer of Split (E(spl)) locus in Drosophila. We have previously shown that acute high doses of MeHg upregulate several E(spl) genes in Drosophila neural-derived C6 cells. Furthermore, MeHg induction of E(spl) can occur independent of the Notch receptor itself. We now show that MeHg, unlike inorganic mercury (HgCl2), preferentially upregulates E(spl)mδ and E(spl)mγ in Drosophila C6 cells. This is distinct from Delta ligand-induced Notch signaling in which no induction of E(spl)mδ is seen. MeHg is also seen to specifically upregulate E(spl)mδ in Drosophila embryos where HgCl2 showed no such effect. Additionally, treatment of embryos with MeHg caused a consistent failure in axonal outgrowth of the intersegmental nerve (ISN). This ISN phenotype was partially replicated by genetic activation of the Notch pathway, but was not replicated by increasing expression of E(spl)mδ. These data suggest a role for Notch signaling and the E(spl)mδ target gene in MeHg toxicity, however, the site of action for E(spl)mδ in this system remains to be elucidated.
Keywords: methylmercury, Notch, Enhancer of split, Drosophila, intersegmental nerve, embryo
Introduction
Methylmercury (MeHg) is a ubiquitous environmental toxin that preferentially targets the developing nervous system. Because of its apparent specificity for neural tissue, signaling pathways in neural development may be important targets in MeHg toxicity. Several studies in both mammalian and invertebrate systems now support the hypothesis that the Notch pathway is a potential target for MeHg. Notch is a fundamental cell-cell signaling pathway that directs cell fate decisions during neurogenesis. Being first elucidated in Drosophila, it is now well understood that signals through Notch receptors cause activation of downstream effectors; those of the Enhancer of Split [E(spl)] gene locus in flies and the Hairy/Enhancer of Split (HES) genes in mammals (de-la-Concha et al., 1988; Jarriault et al., 1998; Preiss et al., 1988). The E(spl) locus in flies consists of 11 genes in a single 50kb locus. Seven of these E(spl) genes, E(spl)mδ, E(spl)mγ, E(spl)mβ, E(spl)m3, E(spl)m5, E(spl)m7, and E(spl)m8, are basic helix-loop-helix transcriptional repressors. While different E(spl) genes are known to be preferentially expressed in various developing tissues, manipulations in Drosophila demonstrate that all the E(spl) genes are capable of responding to Notch signals (Jennings et al., 1999; Nellesen et al., 1999; Wech et al., 1999; Wurmbach et al., 1999). Signals at the level of the Notch receptor are propagated by cleavage and activation by members of the ADAM family of metalloproteases. The potential for MeHg to stimulate ADAM activity initially led to the hypothesis that MeHg could ultimately induce Notch signals (Bland and Rand, 2006). This was supported by evidence that E(spl)mγ and E(spl)mβ show a dose-dependent increase in transcription with MeHg applied to Drosophila neural cells in culture (Bland and Rand, 2006). In subsequent studies we have shown that stimulation of E(spl) genes in Drosophila cells by MeHg can occur despite knockdown of Notch receptor expression (Rand et al., 2008). These observations suggest MeHg can act through a more direct mechanism, bypassing the receptor to stimulate transcription of Notch targets.
In this study, using MeHg exposures to Drosophila C6 neural derived cells in culture in addition to exposures of the whole animal at various developmental stages, we confirm a specific action of MeHg toward the E(spl)mδ gene. We also demonstrate that E(spl)mδ, in stark contrast to the other E(spl)s, is not responsive to Notch signals propagated by its cognate ligand, Delta, in the C6 neural cell line, allowing us to elucidate the MeHg specific action on this gene target. A specific effect of MeHg relative to inorganic mercury (HgCl2) in E(spl)mδ activation is observed in C6 cells and in embryos, however, mercury induction of E(spl)mδ was not seen at later developmental stages. MeHg treated embryos exhibit an overt defect in formation of the intersegmental nerve (ISN). Increasing Notch pathway activity in neurons by driving expression of the Notch intracellular domain (NICD) under the control of the pan-neural elav promoter causes a similar defect in ISN outgrowth, however driving expression of E(spl)mδ in neurons did not elicit an ISN phenotype.
Our findings indicate that MeHg specifically effects E(spl)mδ in vitro and in vivo during Drosophila embryogenesis. Our data shows specificity in gene activation by MeHg compared to other stressors, such as HgCl2, highlighting the potential for E(spl)mδ to mediate some MeHg-induced changes in developmental signaling in the embryo. However, neuron-specific expression of E(spl)mδ did not replicate a characteristic ISN MeHg phenotype, which could be partially replicated with neuron-specific Notch activation. These results point to novel non-canonical Notch pathway mechanisms that contribute to MeHg toxicity in the embryonic nervous system.
Methods
Cell Culture
Drosophila bg2-c6 cells (C6 cells), a neural cell line obtained from the Drosophila Genomics Resource Center (Ui et al., 1994) were cultured in Shields and Sang M3 medium with added bactopeptone and yeastolate (BPYE), supplemented with bovine serum, insulin, and penicillin/streptomycin at 25°C in a humidified incubator.
Cell culture mercury treatments
Stock solutions of methylmercury (MeHg chloride, Aldrich 442534) and mercury chloride (HgCl2, Sigma-Aldrich 215465) were prepared in dimethyl sulfoxide (DMSO) at 50mM. Mercury treatments were made at concentration ranging from 0-100μM. DMSO concentration was adjusted to be equivalent across control and mercury treatments and never exceeded 0.1% DMSO. Cells were plated at 80% confluence in standard medium and allowed to adhere and recover for one hour. They were then washed three times with M3 medium lacking serum or antibiotics (M3-) and the medium was replaced with M3-with added mercury or DMSO control. Cells were treated for three hours, after which the medium was removed and cells were harvested for either RNA extraction with Trizol reagent (Invitrogen) or viability assays were performed.
Cell viability assay
Cell viability was determined by dual staining with calcein and ethidium using the LIVE/DEAD Viability/Cytotoxicity Kit for mammals (Invitrogen) as per product instructions. Briefly, cells were treated with various concentrations of MeHg or HgCl2 for 3hr, then treated with 2μM calcein AM and ethidium homodimer and incubated for 30min. Cells were washed, then plated and counted for green and red cells. Breakdown of calcein AM by esterases causes green fluorescence and marks living cells, while disruption of cell membranes in death allows permeability to ethidium homodimer causing nuclear red fluorescence. The ratio of green to red cells, normalized to control cell treatments and treatments eliciting 100% cell death, were used to determine the measure of cell viability. (n>150 cells per single treatment across seven concentrations).
Fly stocks and crosses
Unless otherwise stated, fly stocks were maintained on standard cornmeal-yeast-molasses food at 25°C. Lines used include Canton S, elav-GAL4 (Bloomington Drosophila Stock Center, #458), UAS-Notchintra (gift from Cedric Wesley, University of Wisconsin Laboratory of Genetics), and UAS-E(spl)mδ (Bloomington Drosophila Stock Center, #26677). Standard crosses were performed between virgin female Gal4 driver lines and male UAS responder lines to generate F1 progeny to be tested.
Embryo mercury treatments
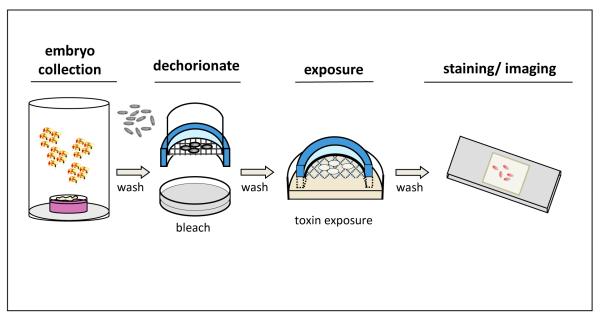
Cages of adult flies were allowed to lay eggs on a grape agar plate with yeast paste smeared on the center. (See Fig. 4) Unless stated otherwise the embryo collection occurred over a two hour laying period. Embryos were then aged on the grape plates at 25°C; standard aging time was two hours. Embryos were dechorionated using a standard protocol. Briefly, embryos were washed from the plate using tap water and a brush into a nytex basket, then rinsed to remove yeast; baskets were transferred to 50% bleach (~3.8% sodium hypochlorite) for three minutes, then rinsed in tap water to remove bleach. Embryos were transferred to separate nytex baskets for each treatment, which were in turn placed into petri dishes containing phosphate buffered saline (PBS) with added mercury or DMSO control. These dishes were covered to prevent evaporation and the embryos were allowed to incubate for 16-18 hours or various times where indicated. A schematic of this novel in vitro method to dose embryos with toxins can be found in Figure 4.
Figure 4. A schematic representation of in vitro dosing of Drosophila embryos with toxins.
Embryos collected from adults laying on grape-agar plates are dechorionated in dilute bleach. Embryos are then placed in baskets designed to optimize air exposure while being immersed in a solution containing the toxin of interest. After a period of developmental exposure, embryos can then be processed for RNA isolation or for fixation, staining and, imaging.
Immunostaining
Immunostaining was performed as previously described (Rand et al., 2009). Treated embryos were fixed in 500 μL of a 50:50 mix of 8% paraformaldehyde in 0.1 M PIPES, 2 mM EGTA, 1 mM MgSO4, pH 6.9 (PEM) with heptane by rocking for 25 min. Vitelline membranes were subsequently removed by discarding the lower PEM layer and adding 750 mL MeOH, then vortexing for 30 s. Settled embryos were collected, washed and stored at −20C in methanol until staining. For immunostaining, embryos were permeablized in phosphate buffered saline (PBS) with 1% BSA, 0.1% triton X-100 (PBT). Subsequent blocking, primary and secondary antibody incubations were done in PBT with 5% each of donkey and goat serum. Primary antibodies used were: mouse anti-elav (9F8A9), mouse anti-notch (9C6), and rat anti-FasII (1D4) (Obtained from the Developmental Studies Hybridoma Bank, Univ. of Iowa). Secondary antibodies used were: Alexa488-conjugated goat anti-rat and Alexa555-conjugated goat anti-mouse (Jackson ImmunoResearch). Embryos were visualized by fluorescence microscopy on a Leitz Orthoplan 2 microscope equipped with a Spot One digital camera and associated acquisition software (MVI, Avon, MA). Images were assembled in Adobe Photoshop, GIMP, and Microsoft PowerPoint.
RNA Extraction and quantitative PCR
After mercury treatments, embryos were transferred with a paintbrush into microcentrifuge tubes containing PBS supplemented with 0.1% Triton-X and 1% bovine serum albumin (PBT). The PBT was removed with a pipette and replaced with Trizol reagent (Invitrogen). Embryos were homogenized in Trizol and processed for RNA isolation. RNA samples were treated with Turbo DNAse (Ambion) and reversed transcribed using SSII reverse transcriptase (Invitrogen). RNA was subsequently removed with RNAse H (USB). cDNA samples were assayed for gene expression via qRT-PCR using SybrGreen dye (Bio-Rad). Data were normalized to the ribosomal protein RP49 housekeeping gene and analyzed using the comparative Ct method (Livak and Schmittgen, 2001). Primer sequences for E(spl)mδ, E(spl)mγ, E(spl)mβ, E(spl)m2, E(spl)m3, E(spl)m7, Notch, and the RP49 control were taken from (Rand et al., 2008). The sequences used for Hsp70Ab were Forward: TGAGAGTGATAAGAATGTTTCGAT and Reverse: AGTCTACAAAACATTAAATGACCAAGTT. For Hsp70Bc the sequences were Forward: ATCAGCAGGGAGCGGGAGCA and Reverse: TCACTTTTAAAAACTTAAGCCGAAA.
Larval and adult mercury treatments
Embryos collected from an overnight laying in a population cage were transferred to bottles of standard cornmeal-molasses food supplemented with 15 μM MeHg or DMSO (control). Embryos were allowed to develop until they reached the late third instar wandering stage (approximately 4 days). At this stage they were harvested and disrupted for RNA isolation using an RNeasy mini kit (Qiagen).
Adult flies were cultured for three days on food containing concentrations of MeHg up to 100μM. The flies were then frozen using liquid nitrogen and broken apart using a vortexer. Heads were collected using a sieve and subsequently homogenized in Trizol reagent. RNA transcript levels were assayed by qRT-PCR as described above.
Results
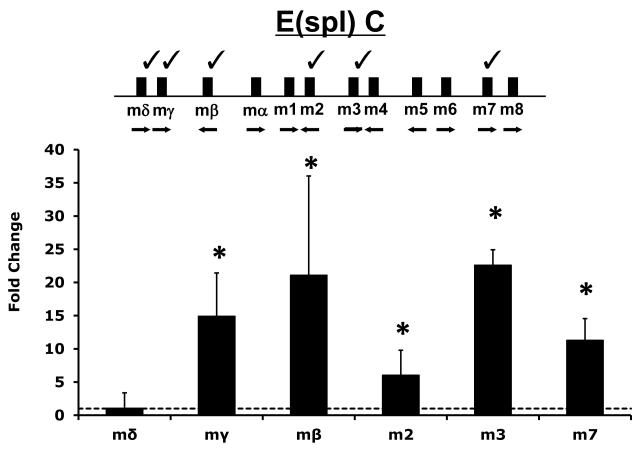
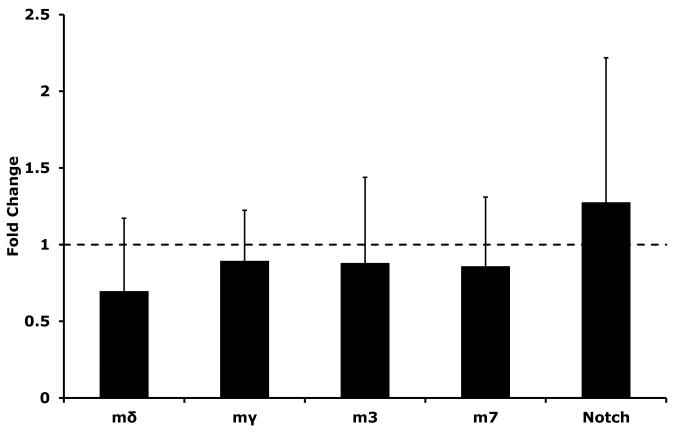
We have previously demonstrated an ability of MeHg to induce E(spl) gene expression in cultured Drosophila C6 cells (Rand et al., 2008). In this previous study EDTA was used as a proxy to invoke Notch signaling in C6 cells for comparative effects on E(spl) expression. Since notable differences between EDTA-induced and MeHg-induced E(spl) activation was observed, we wished to further validate these effects with respect to Notch signaling. To examine Notch signaling explicitly we co-cultured the C6 cells on fixed preparations of cells expressing Delta, the endogenous ligand of the Notch receptor, in a previously established assay in our laboratory (Delwig and Rand, 2008). We then analyzed E(spl) expression exclusively in the Notch expressing cells via qRT-PCR (Fig. 1). This treatment caused the greatest induction of E(spl)mβ and E(spl)m3 (>20-fold), and a nearly 15-fold induction of E(spl)mγ. In contrast, E(spl)mδ was not upregulated by Delta induced Notch signaling. Overall, Delta induced Notch signaling under these conditions gives a similar pattern of E(spl) induction as was seen with EDTA earlier (Rand et al., 2008) indicating this profile is representative of Notch signaling in this cell line.
Figure 1. E(spl) gene expression in Delta ligand induced Notch signaling.
Organization of the Enhancer of split (E(spl)C) locus is diagramed and select genes analyzed in this study are indicated with a “check mark”. Delta induced E(spl) expression was determined with a previously described co-culture assay (Delwig and Rand, 2008). Briefly, S2 cells stably expressing the Notch ligand Delta (Dl-S2 cells) or control S2 parent cells lacking the ligand were cultured as a monolayer then stabilized through brief fixation. C6 cells, which express Notch endogenously, but not the Delta ligand, were then co-cultured with either the fixed Dl-S2 cells or control S2 cells. E(spl) gene expression was measured via qRT-PCR. Data represent the fold change in E(spl) expression in C6 cells due to Delta exposure, Error bars indicate standard deviation. (n=6 (m2, m3, m7, mδ) to 12 (mβ and mγ) independent experimental determinations, * = p<0.01 by Student’s t-test).
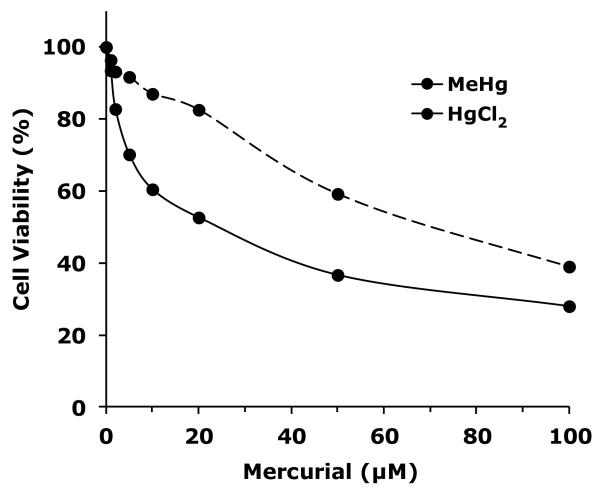
We next sought to compare MeHg effects with inorganic mercury to determine if the E(spl) activation profile of MeHg is unique or shares properties with other mercurials. We first determined levels of toxicity of MeHg and HgCl2 toward C6 cells. Cell viability subsequent to MeHg and HgCl2 exposures was determined using dual calcein/ethidium staining (Fig. 2). A dose dependent decrease in cell viability was observed with MeHg, which proved more potent than HgCl2. MeHg exhibited an approximate 20% cell death (80% viability) at 4μM and an approximate 50% reduction in viability at 20μM. HgCl2 proved weaker showing an approximate 20% reduced viability at 20μM and 50% reduced viability with 100μM HgCl2. Similar results were obtained using alternative determinations with Trypan Blue reagent (Data not shown). These doses were implemented in subsequent assays of acute exposure effects on E(spl) activation.
Figure 2. Cell viability of Drosophila neural-derived cells after mercurial treatment.
Drosophila C6 cells were treated for three hours with various doses of MeHg or HgCl2. Viability was determined with Calcein AM hydrolysis and ethidium homodimer permeability (see methods).
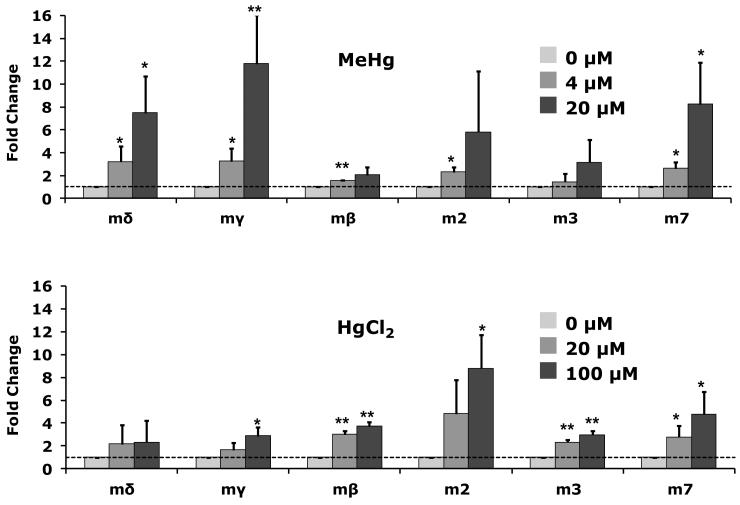
With MeHg treatment of C6 cells E(spl)mδ, E(spl)mγ, and E(spl)m7 showed the greatest fold-induction (7 to 12-fold, Fig. 3) of six representative E(spl) genes spanning the E(spl) locus. E(spl)mβ and E(spl)m3 showed less substantial increases (less than 4-fold) with MeHg treatment, while E(spl)m2 approached a 6-fold induction. In contrast, cells treated with HgCl2 showed less than 3-fold response in E(spl)mδ and E(spl)mγ, and less than 5-fold induction in E(spl)m7 (Fig. 3). E(spl)m2 responded to HgCl2 treatment with a nearly 9-fold change with 100μM MeHg (Fig. 3). These data show a differential response of individual genes in the E(spl) locus with MeHg versus HgCl2 exposure.
Figure 3. E(spl) gene induction by mercurial treatment.
Drosophila C6 cells were treated for three hours with the indicated concentrations of MeHg or HgCl2. E(spl) gene expression determined by qRT-PCR is expressed in fold change over control treatments. (n = 3 independent experiments; * = p<0.05, ** = p<0.01 by Student’s t-test).
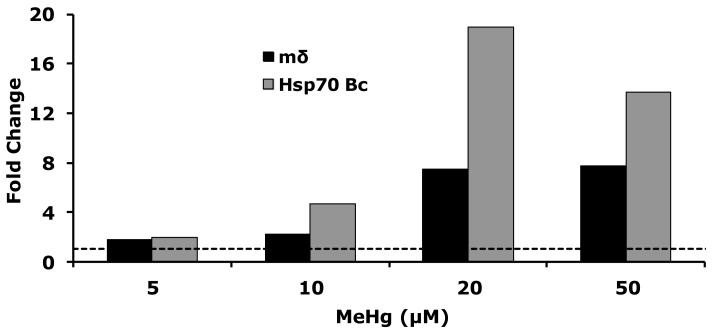
The apparent unique effect of MeHg on E(spl) gene expression in vitro prompted us to investigate similar effects in vivo. We have previously established an ability to dose Drosophila embryos with MeHg cultured in vitro (Rand et al., 2009). This methodology is summarized in Figure 4. The method takes advantage of the unique property that fly embryos denuded of their outer chorion layers of the eggshell are permeable to MeHg and are also able to continue development suspended in a defined culture media (see methods and Figure 4). Using this technique we evaluated the dose-response of embryos to MeHg by assaying gene expression using qRT-PCR and monitoring the response of a ubiquitous stress response gene, Hsp70. The Hsp70 Bc gene showed a robust increase in expression with increasing levels of MeHg, confirming the entry of the MeHg in embryonic tissues (Fig. 5). In parallel we probed E(spl)mδ gene expression which was seen to increase across all concentrations of MeHg. A more than seven-fold increase in E(spl)mδ was seen at 20μM MeHg, which appeared to be sustained at 50μM MeHg. From these data we chose to treat embryos with 50μM MeHg in subsequent analyses to ensure we were above a threshold in effect.
Figure 5. Dose response of E(spl) mδ in Drosophila embryos after MeHg treatment.
Drosophila embryos (2-4 hours after egg laying) were dechorionated and soaked in buffer containing indicated concentration of MeHg for 16 hours. Gene expression for E(spl)mδ and the stress-response gene Hsp70 Bc determined by qRT-PCR is expressed in fold change over 0 μM MeHg treatments. (n>300 pooled embryos per each treatment)
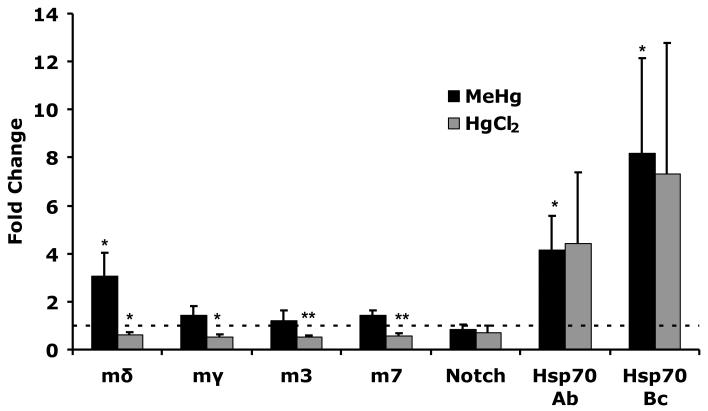
To assess the level of specificity with which MeHg acts on embryonic tissues we again compared E(spl) expression response to MeHg versus HgCl2 treatment (Fig. 6). After MeHg, treatment (50μM) E(spl)mδ consistently showed greater than three-fold upregulation in embryos across several trials. In contrast, none of the other E(spl) genes assayed showed a response to MeHg in embryos treated in vitro. In addition, Notch showed no change in expression in response to MeHg, indicating that increases in E(spl)mδ could not stem from increased receptor expression. An induction of two Hsp70 genes Hsp70Ab and Hsp70Bc was observed for MeHg, again confirming entry of the toxicant into the embryos. Treatment of embryos with 1mM HgCl2 did not cause any increase in E(spl) gene expression. In contrast, a modest decrease was seen in levels across all the E(spl) genes and Notch after HgCl2 treatment. Upregulation of Hsp70 genes after HgCl2 treatment indicated that the dose of HgCl2 used showed a similar degree of entry and overall toxic insult to that of MeHg. These data indicate that MeHg acts selectively on E(spl)mδ transcription in Drosophila embryos.
Figure 6. E(spl) gene induction in Drosophila embryos after mercurial treatment.
Drosophila embryos 2-4 hours after egg laying were soaked overnight in buffer containing 50μM MeHg or 1mM HgCl2. Gene expression determined by qRT-PCR is expressed in fold change over control treatments. Values are the mean (and SEM) of three independent treatments of embryo batches (n = 3 independent experiments; * = p<0.05, ** = p<0.01 by Student’s t-test).
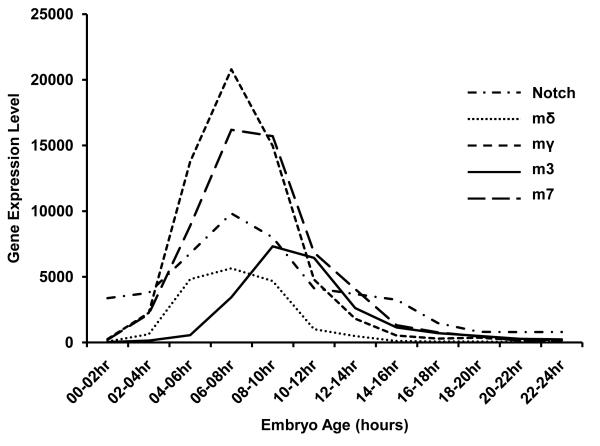
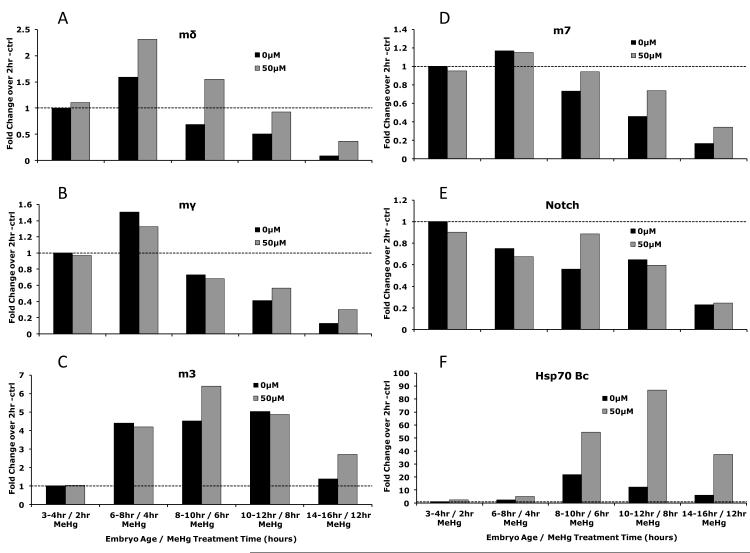
E(spl) gene expression is known to change over the course of embryogenesis (Tweedie et al., 2009). Recent data from gene expression arrays performed within the large scale ModEncode project (Tweedie et al., 2009) and publically available on Flybase (Graveley et al.) permit a comprehensive analysis of developmental expression of the E(spl) gene during normal embryogenesis. Transcript levels of E(spl)mδ, E(spl)mγ, and E(spl)m7 show a similar profile as Notch, which shows a peak of expression at 6-8 hours of development after egg laying (Fig. 7). In contrast, E(spl)m3 shows peak expression discernibly later, peaking at 8-10 hours AEL. The bell-shaped expression of the E(spl) genes prompted us to test whether the effect of MeHg on increasing E(spl)mδ in embryos was simply due to a developmental delay and shift in peak of gene expression versus an ectopic induction of gene expression. To achieve this we incubated batches of developmentally staged embryos with or without MeHg for various treatment intervals, and compared E(spl) expression via qRT-PCR. In untreated embryos the overall profile of E(spl) expression showed the characteristic increase followed by a decrease over the course of embryogenesis. For E(spl)mδ, E(spl)mγ, and E(spl)m7 a peak of expression between 6-8 hours after egg laying (AEL) was observed (Fig. 8). For E(spl)m3 this peak was seen between 8-10 hours AEL. Notch expression showed a gradual decline over the course of embryogenesis. With MeHg treatment, E(spl)mδ showed higher expression at each time point after 6hr AEL compared to untreated embryos, with more than 2-fold higher expression at the 8-10hr interval. Peak expression remained at the 6-8 hour AEL interval with MeHg, indicating the developmental delay was not substantial at this time point. Higher expression due to MeHg was not consistently observed for E(spl)mγ and E(spl)m3, while E(spl)m7 did show modest increases at time points after 8hrs AEL with MeHg treatment. Notch showed no consistent difference in expression due to MeHg treatment over these developmental periods (Fig. 8). Hsp70 Bc, like E(spl)mδ, showed increased expression due to MeHg treatment at every time point, confirming the access of MeHg to embryonic tissues at all stages.
Figure 7. Relative gene expression levels of select E(spl) genes and Notch during embryonic development.
Relative gene expression levels determined through the Mod Encode project (Graveley et al.) were adapted from FlyBase (Tweedie et al., 2009) and expressed graphically to allow comparison of peak expression timing. E(spl)mδ, E(spl)mγ, and E(spl)m7 coincide with Notch peak expression at 6-8 hours after egg laying. In contrast, E(spl)m3 expression occurs later peaking around 8-10 hours after egg laying.
Figure 8. Response of E(spl) genes in Drosophila embryos after MeHg treatment over the course of development.
Drosophila embryos were dechorionated and soaked in buffer containing 50μM MeHg for indicated lengths of time. Gene expression determined by qRT-PCR is expressed in fold change over control treatments for A) E(spl)mδ, B) E(spl)mγ, C) E(spl)m3, D) E(spl)m7, E) Notch, and F) HSP70 Bc. (Each data point is derived from >300 pooled embryos from a treatment sampled at the indicated developmental time points).
Observing that E(spl)mδ is upregulated in embryos after MeHg treatment we investigated whether or not the MeHg effect was penetrant at later developmental stages. First instar (L1) larvae were cultured on food containing 15μM MeHg and harvested for qRT-PCR after reaching the wandering third instar (L3) stage. Previous analyses have demonstrated that treatment of larvae with 15μM MeHg shows similar toxicity to 50μM treatments of embryos (Mahapatra et al., 2010; Rand et al., 2009). We then examined global transcript levels of E(spl)s and Notch from whole larval extracts using qRT-PCR (Fig. 9). No change in expression due to MeHg in E(spl)mδ or any E(spl) in the larvae was observed. These data suggest that Drosophila tissues at later developmental stages than the embryo are refractory to MeHg induced expression of E(spl).
Figure 9. E(spl) gene induction in Drosophila larvae after MeHg treatment.
Drosophila embryos were placed on standard cornmeal-molasses food containing 15μM MeHg or DMSO vehicle control. Development was allowed to progress to the late third instar larvae stage. Larvae were then homogenized, RNA extracted and gene expression determined by qRT-PCR, expressed in fold change over control treatments. (Error bars indicate standard deviation of three experimental replicates. No significant changes were found using Student’s t-test).
To further test whether the effect of MeHg on E(spl)mδ expression is specific to the embryo we tested E(spl) expression responses to MeHg in adult flies. Adult Drosophila were cultured for three days on food containing various concentrations of MeHg up to 100μM. RNA transcript levels in extracts prepared from isolated heads were assayed by qRT-PCR. We opted to examine this tissue since Notch activity and E(spl) expression is a strong determinant in neural tissues and the fly head is rich in brain tissue. We determined that no consistent changes in E(spl)mδ, or any of the E(spl) genes examined, were seen with MeHg treatments (data not shown). Altogether, the data from larval and adult assays indicate that the global effect of MeHg on E(spl)mδ expression is specific to embryos.
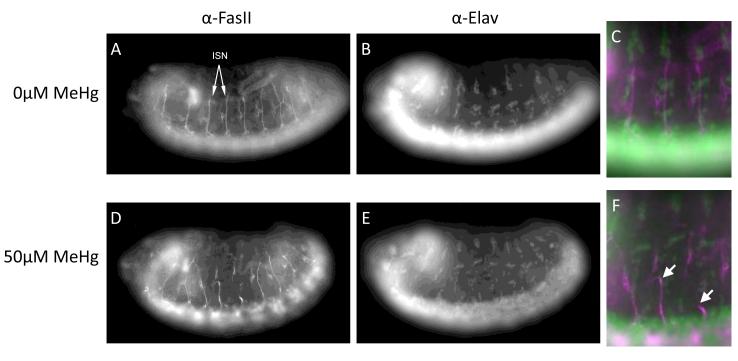
We next turned to examining the embryonic nervous system for phenotypes that characteristically reflected MeHg insult to development. Our previous studies have identified several features in the embryo CNS and PNS that reflect compromised development with MeHg exposure (Rand et al., 2009). Notably, neurite outgrowth of CNS and PNS axons visualized in the lateral field of the late stage embryo has been seen to be compromised (Rand et al., 2009). Using an antibody specific to the ISN, a bundle of four axons of central motor neurons, we observed that these axons frequently failed to develop properly in MeHg treated embryos (Fig. 10). This phenotype of MeHg exposure presents as stunted or misguided ISNs in treated embryos (Fig. 10 D-F) and is easily scored by the growth of axons relative to the position of elav-positive PNS neurons in the lateral field (Fig. 10 F). Interestingly, previous studies have identified a role for Notch signaling in the appropriate projection of the ISN neuron (Giniger et al., 1993).
Figure 10. MeHg treatment causes axonal disruption in embryos.
A-C) Control embryos at developmental stage 14 were co-stained to show the intersegmental nerve (ISN) and all neuronal nuclei using antibodies for FasII (A) and elav (B), respectively. D-F) Embryos were treated with MeHg (50μM) and stained for FasII (D) and elav (E); disruption of ISN outgrowth is indicated by arrowheads.
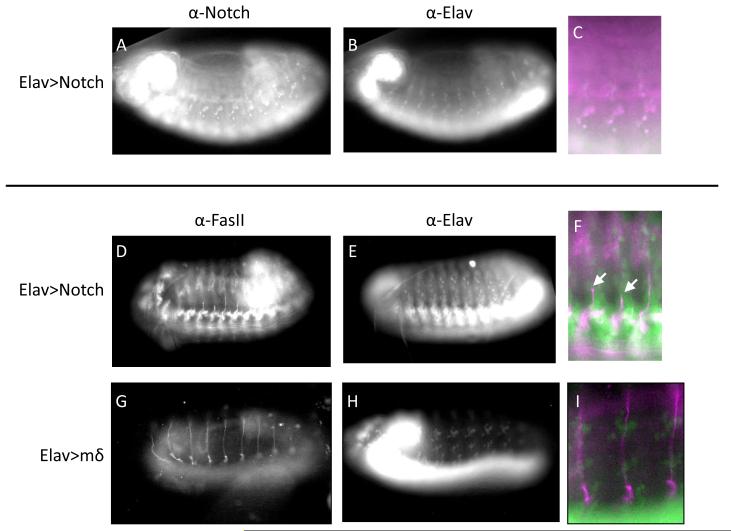
We therefore set out to test whether genetic manipulations of Notch in general, and E(spl)mδ in particular, could mirror effects of MeHg on ISN development. We first examined the effect of ubiquitous activation of Notch in post mitotic neurons by driving expression of NICD under control of the pan-neural elav promoter using the GAL4/UAS system (see methods). Ectopic expression of NICD in neurons could be detected with an antibody to the NICD (Fig. 11 A-C) and by qRT-PCR. In general, the potent activity of Notch was evident by an overall failure of a substantial number of embryos to develop to late stages. Of those that were able to develop to late stage a common feature was seen in a stunted outgrowth of the ISN (Fig. 11 D-F) akin to that seen with MeHg treatment. As E(spl)mδ was the most consistent E(spl) responder to MeHg we next determined if E(spl)mδ overexpression by the elav promoter could elicit an analogous MeHg-like ISN phenotype. Despite unambiguous overexpression of E(spl)mδ in elav-GAL4>UAS-E(Spl)mδ embryos (as determined by qRT-PCR, as no E(spl)mδ antibodies are available) no apparent phenotype in ISN development was observed (Fig. 11 G-I).
Figure 11. Notch pathway activation in neurons disrupts nerve outgrowth in embryos that is not induced with E(spl)mδ overexpression.
A-C) Embryos driving expression of the Notch intracellular domain (NICD), the active component of the Notch receptor, under control of elav show notch immunoreactivity in neurons. D-F) elav> NICD embryos stained for FasII (D) and elav (E) demonstrate stunted outgrowth of the ISN (arrow head). G-I) Embryos with targeted expression of the E(spl)mδ gene in neurons stained for FasII (G) and elav (H) show normal ISN outgrowth morphology.
Discussion
In this study we have shown that MeHg causes an upregulation of E(spl)mδ in Drosophila neural cells and, importantly, in embryos treated in vitro. This effect is specific to MeHg and does not occur with inorganic mercury. This E(spl)mδ specific response is starkly different from E(spl) activation profiles resulting from Notch receptor activation by the Delta ligand in the C6 neural cell line. MeHg elicits a characteristic failure in ISN axon outgrowth that, to some extent, can be induced with neural overexpression of Notch activity. However, targeted E(spl)mδ expression in neurons has no effect on ISN development. A role for E(spl)mδ in mediating MeHg effects via non-neuronal cells remains to be investigated. Altogether, these data highlight a novel and specific action for MeHg to target induced expression of a neurogenic signaling gene in conjunction with elicited neural developmental phenotypes. E(spl) genes, are basic helix-loop-helix (bHLH) transcriptional repressors.
These bHLH genes are the main effectors of Notch signaling in the nervous system, acting to repress transcription of proneural genes and generally preventing a default neuronal differentiation program from occurring. While various E(spl) genes have different expression patterns there is enough redundancy in expression and function to make discerning phenotypes difficult if only one E(spl) gene is perturbed. Nonetheless, E(spl)mδ siRNA injection in Drosophila embryos results a neurogenic phenotype, albeit with low frequency, consistent with a perturbation of Notch signaling (Nagel et al., 2004). The low penetrance of phenotypic effects of E(spl)mδ perturbation predict that its contribution to MeHg effects in the embryo will be subtle. It is thus not surprising that genetic manipulation of E(spl)mδ exclusively in neurons is not sufficient to replicate the MeHg-induced ISN phenotype. That genetic manipulation of the Notch pathway can replicate this phenotype suggests that there may be a central role for the Notch pathway in MeHg toxicity, potentially through the additive effects on E(spl)mδ and other E(spl) genes. Further study of MeHg in this system may not only elucidate the mechanisms of MeHg toxicity but also reveal unique mechanisms for the differential expression of E(spl) genes.
Our findings suggest that induction of E(spl)mδ by MeHg in vivo is restricted to the embryonic stage. This observation is consistent with the notion that cellular defense mechanisms (e.g. glutathione pathway) are not fully developed in early embryogenesis, as compared with the differentiated larval and adult tissues. This outcome grossly mimics the prefgerential toxicity of MeHg for the fetus versus adult in higher organisms. Alternatively, differences may stem from our method of administration of the MeHg, which differ between our embryonic and larval/adult treatments: larvae and adult flies consume MeHg with the food, and may be able to mitigate toxicity via the gut. In contrast, embryos are soaked in MeHg medium allowing for more direct contact with embryonic cells. Yet, at the doses used, larvae show similar lethality as embryos (Mahapatra et al., 2010). Assuming that embryo-specific induction of E(spl)mδ is not related to the route of MeHg administration it may help elucidate the mechanism for the specific neurodevelopmental toxicity in mice and humans.
In summary, we demonstrate that MeHg causes increased expression of E(spl)mδ in vivo in Drosophila embryos. We also demonstrated that while E(spl)mδ transcript upregulation correlates with MeHg neuronal phenotypes in embryos, E(spl)mδ overexpression restricted to neurons is not sufficient to replicate this characteristic axon outgrowth phenotype, pointing to a non-neuronal activity of E(spl)mδ in this toxicity model. These findings set the stage for investigating novel mechanisms of MeHg toxicity via non-canonical pathways of Notch signaling.
Highlights.
Methylmercury causes upregulation of the Notch target gene E(spl)mδ in Drosophila.
Mercury chloride does not cause upregulation of E(spl)mδ.
Treating embryos with mercury in vitro replicated the E(spl)mδ expression changes.
Methylmercury treated embryos fail at axon outgrowth in the intersegmental nerve.
Expression of Notch but not E(spl)mδ in nerves mimics the axon outgrowth phenotype.
Acknowledgements
The Elav-9F8A9 antibody developed by G.M. Rubin, the C17.9c6 antibody developed by S. Artavanis-Tsakonas, and the 1D4 antibody developed by C. Goodman were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biology, Iowa City, IA 52242.
We are grateful to Julie Dao, Cecon Mahapatra, Amanda Burton, Ben Moody and other members of the Rand lab for technical help and discussion of data.
This work was supported by NIEHS R01-ES015550 awarded to M.D.R. and the UVM Environmental Pathology Training Grant NIEHS T32-ES07122.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bland C, Rand MD. Methylmercury induces activation of Notch signaling. Neurotoxicology. 2006;27:982–991. doi: 10.1016/j.neuro.2006.04.005. [DOI] [PubMed] [Google Scholar]
- de-la-Concha A, Dietrich U, Weigel D, Campos-Ortega JA. Functional interactions of neurogenic genes of Drosophila melanogaster. Genetics. 1988;118:499–508. doi: 10.1093/genetics/118.3.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delwig A, Rand MD. Kuz and TACE can activate Notch independent of ligand. Cell Mol Life Sci. 2008;65:2232–2243. doi: 10.1007/s00018-008-8127-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giniger E, Jan LY, Jan YN. Specifying the path of the intersegmental nerve of the Drosophila embryo: a role for Delta and Notch. Development. 1993;117:431–440. doi: 10.1242/dev.117.2.431. [DOI] [PubMed] [Google Scholar]
- Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, Brown JB, Cherbas L, Davis CA, Dobin A, Li R, Lin W, Malone JH, Mattiuzzo NR, Miller D, Sturgill D, Tuch BB, Zaleski C, Zhang D, Blanchette M, Dudoit S, Eads B, Green RE, Hammonds A, Jiang L, Kapranov P, Langton L, Perrimon N, Sandler JE, Wan KH, Willingham A, Zhang Y, Zou Y, Andrews J, Bickel PJ, Brenner SE, Brent MR, Cherbas P, Gingeras TR, Hoskins RA, Kaufman TC, Oliver B, Celniker SE. The developmental transcriptome of Drosophila melanogaster. Nature. 471:473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarriault S, Le Bail O, Hirsinger E, Pourquie O, Logeat F, Strong CF, Brou C, Seidah NG, Isra A., l Delta-1 activation of notch-1 signaling results in HES-1 transactivation. Mol Cell Biol. 1998;18:7423–7431. doi: 10.1128/mcb.18.12.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings BH, Tyler DM, Bray SJ. Target specificities of Drosophila enhancer of split basic helix-loop-helix proteins. Mol Cell Biol. 1999;19:4600–4610. doi: 10.1128/mcb.19.7.4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real- time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Maeder ML, Polansky BJ, Robson BE, Eastman DA. Phylogenetic footprinting analysis in the upstream regulatory regions of the Drosophila enhancer of split genes. Genetics. 2007;177:1377–1394. doi: 10.1534/genetics.107.070425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahapatra CT, Bond J, Rand DM, Rand MD. Identification of methylmercury tolerance gene candidates in Drosophila. Toxicol Sci. 2010;116:225–238. doi: 10.1093/toxsci/kfq097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel AC, Maier D, Krauss S, Mezger M, Preiss A. Neurogenic phenotypes induced by RNA interference with bHLH genes of the Enhancer of split complex of Drosophila melanogaster. Genesis. 2004;39:105–114. doi: 10.1002/gene.20033. [DOI] [PubMed] [Google Scholar]
- Nellesen DT, Lai EC, Posakony JW. Discrete enhancer elements mediate selective responsiveness of enhancer of split complex genes to common transcriptional activators. Dev Biol. 1999;213:33–53. doi: 10.1006/dbio.1999.9324. [DOI] [PubMed] [Google Scholar]
- Preiss A, Hartley DA, Artavanis-Tsakonas S. The molecular genetics of Enhancer of split, a gene required for embryonic neural development in Drosophila. Embo J. 1988;7:3917–3927. doi: 10.1002/j.1460-2075.1988.tb03278.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand MD, Bland CE, Bond J. Methylmercury activates enhancer-of-split and bearded complex genes independent of the notch receptor. Toxicol Sci. 2008;104:163–176. doi: 10.1093/toxsci/kfn060. [DOI] [PubMed] [Google Scholar]
- Rand MD, Dao JC, Clason TA. Methylmercury disruption of embryonic neural development in Drosophila. Neurotoxicology. 2009;30:794–802. doi: 10.1016/j.neuro.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tweedie S, Ashburner M, Falls K, Leyland P, McQuilton P, Marygold S, Millburn G, Osumi-Sutherland D, Schroeder A, Seal R, Zhang H. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 2009;37:D555–559. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ui K, Nishihara S, Sakuma M, Togashi S, Ueda R, Miyata Y, Miyake T. Newly established cell lines from Drosophila larval CNS express neural specific characteristics. In Vitro Cell Dev Biol Anim. 1994;30A:209–216. doi: 10.1007/BF02632042. [DOI] [PubMed] [Google Scholar]
- Wech I, Bray S, Delidakis C, Preiss A. Distinct expression patterns of different enhancer of split bHLH genes during embryogenesis of Drosophila melanogaster. Dev Genes Evol. 1999;209:370–375. doi: 10.1007/s004270050266. [DOI] [PubMed] [Google Scholar]
- Wurmbach E, Wech I, Preiss A. The Enhancer of split complex of Drosophila melanogaster harbors three classes of Notch responsive genes. Mech Dev. 1999;80:171–180. doi: 10.1016/s0925-4773(98)00212-3. [DOI] [PubMed] [Google Scholar]