Abstract
A new tetracenomycin analogue, 8-demethyl-8-(4′-keto)-α-l-olivosyl-tetracenomycin C, was generated through combinatorial biosynthesis. Streptomyces lividans TK 24 (cos16F4) was used as a host for expression of a “sugar plasmid” (pKOL) directing the biosynthesis of NDP-4-keto-l-olivose. This strain harbors all of the genes necessary for production of 8-demethylt-etracenomycin C and the sugar flexible glycosyltransferase ElmGT. To the best of our knowledge, this report represents the first characterization of a tetracenomycin derivative decorated with a ketosugar moiety. Also, as far as we know, 4-keto-l-olivose has only been described as an intermediate of oleandomycin biosynthesis, but has not been described before as an appendage for a polyketide compound. Furthermore, this report gives further insight into the substrate flexibility of ElmGT to include an NDP-ketosugar, which is unusual and is rarely observed among glycosyltransferases from antibiotic biosynthetic pathways.
Keywords: Glycosyltransferase, ketosugar, elloramycin, combinatorial biosynthesis, Streptomyces, polyketide
Streptomycetes produce a variety of glycosylated polyketide compounds with antibiotic and antitumor properties. The appended sugar moiety/moieties contribute tremendously to the biological activity of parent compounds. The alteration of these appended sugar(s) can improve or diversify the biological activities of these compounds. As such, combinatorial biosynthesis is one promising strategy for altering deoxysugar moieties. This can be achieved through either gene deletion or heterologous expression of deoxysugar gene clusters from other pathways. Provided that the endogenous glycosyltransferase is flexible enough to accommodate the altered NDP-deoxysugar donor substrates, novel derivatives can be generated.1
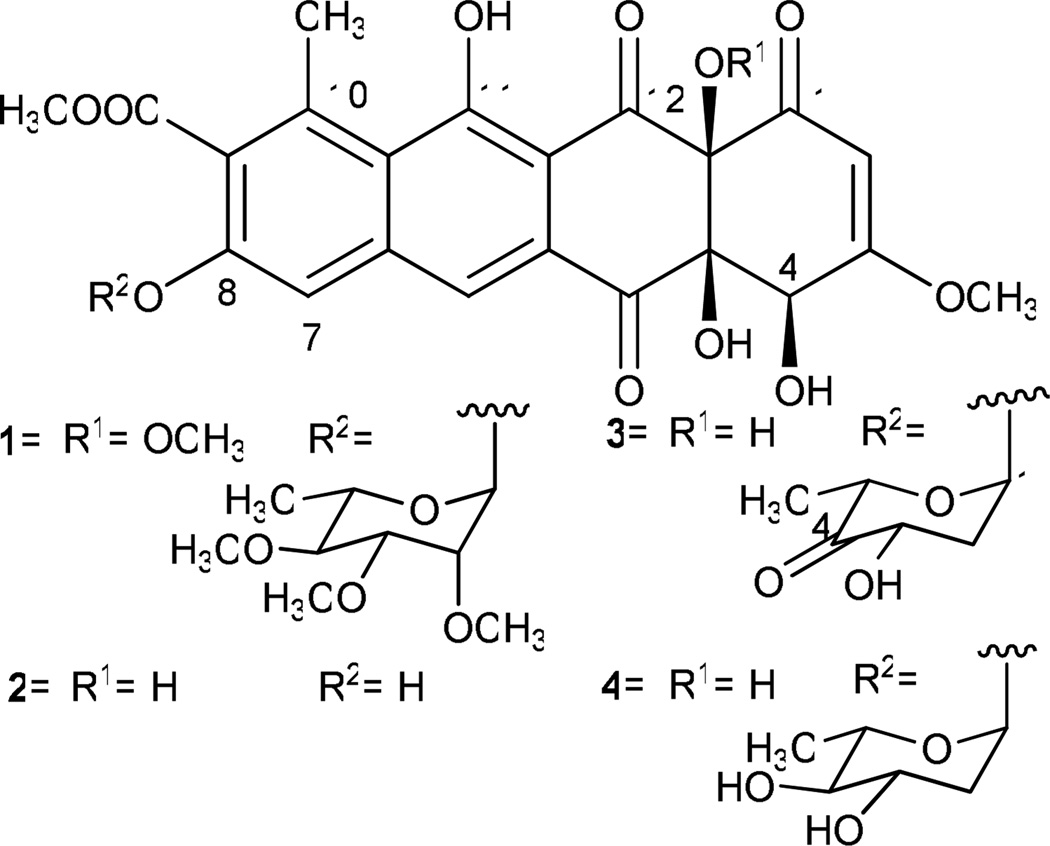
Elloramycin (1) and the structurally-related tetracenomycins are anthracycline-like polyketide compounds with mild antitumor activity.2,3 Elloramycin is produced by Streptomyces olivaceus Tü2353, and it features 12a-O-methyl moiety and 2′, 3′, 4′-tri-O-methyl-α-l-rhamnose O-glycosidically linked at 8-position (Figure 1). In previous studies, the entire gene cluster responsible for production of 8-demethyl-tetracenomycin C (2) (8-DMTC) was cloned into cosmid 16F4, and by expressing the cosmid in various Streptomyces sp., tetracenomycins with novel deoxysugar moieties were produced.4–6 It was then discovered that the elloramycin glycosyltransferase, ElmGT, was responsible for appending these foreign NDP-deoxysugars to 8-DMTC. Recent studies by Salas and co-workers have involved cloning several “deoxysugar plasmids,” which combine deoxysugar genes from different pathways into a single operon for expression in Streptomyces species. By expression of these sugar plasmids in a Streptomyces host harboring cos16F4, it was discovered that ElmGT demonstrated remarkable flexibility towards accepting L- and D- neutral and branched sugar substrates in generating a library of tetracenomycin derivatives.7–12 Recently, mutation of active site residues of ElmGT has been shown to modulate transfer of specific deoxysugars.13 Here we report the isolation and characterization of a new tetracenomycin decorated with a 4′-keto-l-olivose moiety.
Figure 1.
Elloramycin and tetracenomycin analogues discussed in this communication. Elloramycin (1); 8-demethyl-tetracenomycin C (2); 8-demethyl-8-(4'-keto)-α-l-olivosyl-tetracenomycin C (3); 8-demethyl-8-α--olivosyl-tetracenomycin C (4).
In order to generate a plasmid for expression of a ketosugar, the 4-ketoreductase gene (oleU) was removed from plasmid pLN2 (Rodriguez, et al., 2002). NheI and SpeI were used to digest pLN2, thereby releasing the oleU fragment. DNA gel electrophoresis was carried out to remove the oleU gene and the resulting fragment (~14kb) was rescued and re-ligated to generate pKOL. In this construct, all of the sugar genes are under strong ermE* promotion. The plasmid map for this construct is depicted in Figure S1 (see Supplementary Information). This plasmid was introduced into the S. lividans (cos16F4) strain through protoplast transformation as described previously.14–20
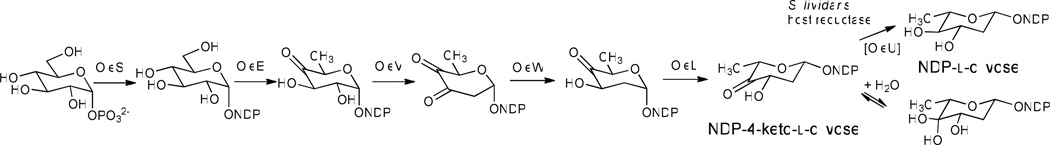
The activities of OleS (NDP-glucose-synthase), OleE (NDP-glucose-4, 6-dehydratase), OleV (2, 3-dehydratase), OleL (3, 5-epimerase), and OleW (3-ketoreductase) catalyze the conversion of NDP-d-glucose to NDP-4-keto-2, 6-dideoxy-d-glucose during the biosynthesis of NDP-l-olivose (Figure 2). The 4-ketoreduction step catalyzed by OleU represents the last step of the NDP-l-olivose biosynthetic pathway. Thus, the deletion of oleU from pLN2 in pKOL would lead to the accumulation of NDP-4-keto-l-olivose, which could be utilized by ElmGT as an alternative donor substrate when the natural substrate (TDP-l-rhamnose) is not available. To test this hypothesis, the pKOL. plasmid was expressed in the S. lividans (cos16F4) strain.
Figure 2.
Hypothesized deoxysugar biosynthetic pathway of pKOL.. OleU, which was encoded in the parent plasmid pLN2 but not encoded in this construct, is indicated in brackets.
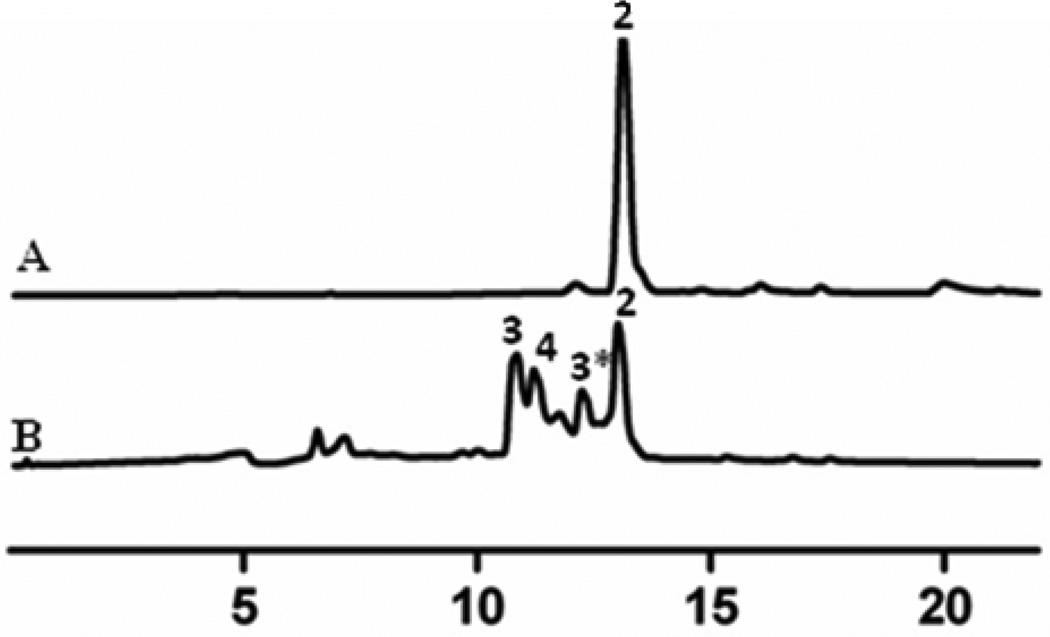
Interestingly, expression of pKOL in S. lividans (cos16F4) resulted in the accumulation of two major peaks with Rt (10.76 and 11.26 min., respectively), in addition to 8-DMTC, when ethyl acetate extracts were analyzed by HPLC/MS (Figure 3). Both peaks showed UV absorption typical for a tetracenomycin-type compound. Low resolution ESI/MS revealed a peak of 585 amu (−ve mode) pertaining to the molecular ion of the compound and another pseudomolecular ion of a hydrated species at 603 amu (−ve mode) indicating the addition of water to the molecule. Under aqueous conditions, ketosugars can easily interconvert from the keto form to a hydrate form. The second peak possessed an m/z value of 587 in (−ve) ESI mode. This mass data suggested the presence of a glycosylated tetracenomycin in which the sugar was fully reduced. Both peaks possessed a fragmentation ion corresponding to the 8-DMTC aglycone (m/z 457, M-H−).
Figure 3.
HPLC analyses of the metabolites: trace A, 8-demethyl-tetracenomycin C (2) (Rt. 12.98 min.) isolated from S. lividans (cos16F4); trace B, metabolites isolated from the S. lividans (cos16F4)/pKOL mutant, 8-demethyl-(4’-keto)-α-l-olivosyl-tetracenomycin C (3 (Rt. 10.76 min.) and (Rt. 12.26 min.) 3* denote hydrated and keto forms, respectively) and 8-demethyl-α-l-olivosyltetracenomycin C (4) (Rt. 11.26 min.)
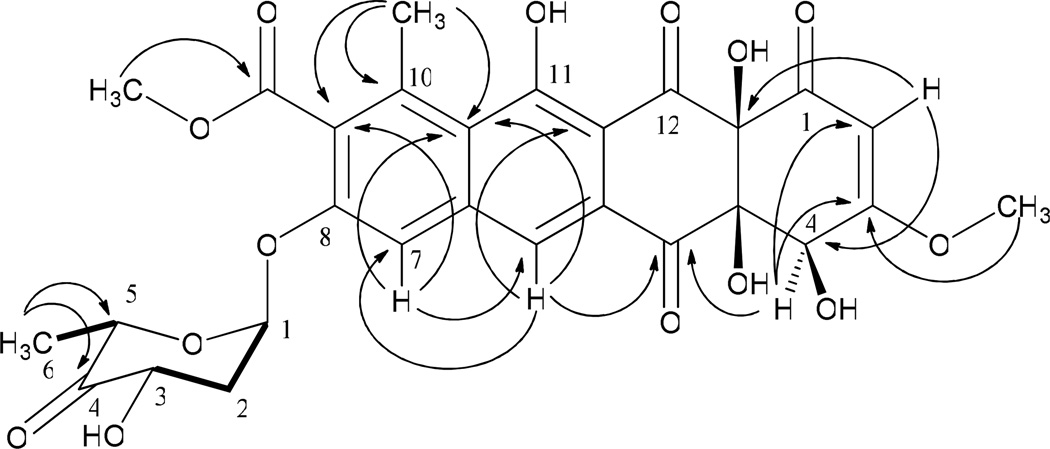
This strain was fermented in a large scale fermentation (7.2 L) for isolation and spectroscopic characterization of the metabolites. The structure of 8-demethyl-8-(4′-keto)-α-l-olivosyl-tetracenomycin C (3) was solved through NMR and mass spectral analyses. The (−) HR-ESI MS of 3 showed two peaks at 585.1267 amu and 603.1363 amu, which corresponded to the molecular formula of its keto form (C28H26O14, calcd. molecular weight 585.1317 [M-H−]) and its hydrate form (C28H28O15, calcd. molecular weight 603.1423 amu [M-H−]), respectively. 1H NMR data of 3 revealed 2 singlets for two aromatic protons (δ 7.94 and δ 7.66) and two methoxy signals corresponding to the 3-OCH3 (δ 3.81) and 9-OCH3 (δ 3.97) of 8-DMTC, respectively (Table 1). The anomeric proton of the sugar appeared as a broad singlet (δ 6.16), which suggested its α configuration. A pair of protons at δ 2.13 and δ 2.24 corresponded to the C-2′ methylene protons. A 4′-H signal was not observed, indicating the presence of keto or a hydrated-keto group. The splitting of the 3′-H (dd, J= 12.0, 6.5 Hz) at δ 4.68 indicated a large diaxial coupling with 2′-Ha and an axial-equatorial coupling with 2′-He. The 5′-H appeared as a quartet (J= 6.5 Hz) at δ 4.39, which indicated coupling with 6′-CH3. The 1H, 1H-COSY exhibited two spin systems for the sugar moiety, one stretching from 1′-H to 3′-H, and the other stretching from 5′-H to 6′-CH3, which strongly indicates the presence of a carbonyl at C-4′ (Figure 4). The 13C showed a carbonyl signal at δ 207.4, which indicates that 3 is present predominantly in the ketosugar form when measured in Methanol-d4. The 2D-HMBC demonstrated a correlation between the 6’-CH3 protons (δ 1.12) and the 4’-C carbonyl moiety (δ 207.4), which unambiguously assigned it to this position (Figure 4). These data suggested the structure of 3 as 8-demethyl-8-(4′-keto)-α-l-olivosyl-tetracenomycin C. Compound 4 was eluted at the identical retention time when co-injected with standard 8-demethyl-8-α-l-olivosyl-tetracenomycin. The identity of 4 as 8-demethyl-8-α-l-olivosyl-tetracenomycin was further confirmed through the comparison of 1H NMR data with published 1H NMR data.9
Table 1.
1H and 13C NMR of 8-Demethyl-8-(4’-keto)-α-L-olivosyl-tetracenomycin C (3) in comparison with the reported data of 8-Demethyl-8-α-l-olivosyl-tetracenomycin C (4),9 δ in ppm relative to TMS, (multiplicity, J/Hz).
| Position | 8-Demethyl-8-(4'-keto)-α-l-olivosyl- tetracenomycin C (3)a |
8-Demethyl-8-α-l-olivosyl-tetracenomycin C (4)b |
||
|---|---|---|---|---|
| δC (125 MHz) | δH (500 MHz) | δC (100 MHz) | δH (400 MHz) | |
| 1 | 193.0 | - | 190.8 | - |
| 2 | 100.8 | 5.62 (s) | 100.2 | 5.66 (s) |
| 3 | 176.1 | - | 175.6 | - |
| 3-OCH3 | 57.8 | 3.81 (s) | 57.5* | 3.87 (s) |
| 4 | 70.7 | 4.88 (d, 1.5) | 70.6 | 5.11 (d, 7.0) |
| 4-OH | - | - | - | 4.96 (d, 7.0) |
| 4a | 85.9 | - | 85.2 | - |
| 4a-OH | - | - | - | 5.18 (s) |
| 5 | 194.8 | - | 194.0 | - |
| 5a | 141.5 | - | 141.2 | - |
| 6 | 122.1 | 7.91 (s) | 121.6 | 8.10 (s) |
| 6a | 129.6 | - | 131.0** | - |
| 7 | 112.4 | 7.55 (s) | 112.0 | 7.80 (s) |
| 8 | 155.4 | - | 155.6 | - |
| 9 | 130.9 | - | 129.3 | - |
| 9-OCH3 | 53.4 | 3.97 (s) | 53.0* | 4.00 (s) |
| 9-CO | 169.5 | - | 168.0 | - |
| 10 | 139.5 | - | 138.6 | - |
| 10-CH3 | 21.3 | 2.79 (s) | 21.3 | 2.87 (s) |
| 10a | 122.6 | - | 122.0** | - |
| 11 | 167.8 | - | 167.8 | - |
| 11-OH | - | - | - | 14.02 (s) |
| 11a | 110.6 | - | 110.2 | - |
| 12 | 198.2 | - | 198.0 | - |
| 12a | 84.6 | - | 83.8 | - |
| 12a-OH | - | - | - | 5.81 (s) |
| 1′ | 96.4 | 6.16 (s) | 97.0 | 6.07 (d, 3.0) |
| 2′ | 37.3 | 2.24 (ddd, 15.0, 12.0, 3.0, He), | 38.2 | 2.32 (ddd, 16.0, 6.0, 2.0, He), |
| 2.13 (dd, 15.0, 6.5, Ha) | 1.88 (ddd, 14.0, 12.0, 3.0, Ha) | |||
| 3′ | 70.6 | 4.68 (dd, 12.0, 6.5) | 69.0 | 3.95 (dddd, 11.0, 6.0, 6.0, 6.0) |
| 3′-OH | - | - | - | 4.25 (s) |
| 4′ | 207.4 | - | 70.6 | 3.14 (dd, 9.0, 9.0) |
| 4′-OH | - | - | - | 4.38 (s) |
| 5′ | 72.5 | 4.39 (q, 6.5) | 78.4 | 3.67 (dq, 10.0, 6.0) |
| 6′ | 14.3 | 1.12 (keto, d, 6.5) | 18.3 | 1.22 (d, 6.0) |
| 1.16 (hydrate, d, 6.5) | ||||
Methanol-d4,
Acetone-d6,
* and ** signals were mutually wrongly assigned in cited publication, however, assignments were corrected here for better comparison
Figure 4.
1H-1H-COSY ( ), and selected HMBC (→) correlations of 8-demethyl-8-(4'-keto)-α-l-olivosyl-tetracenomycin C (3).
), and selected HMBC (→) correlations of 8-demethyl-8-(4'-keto)-α-l-olivosyl-tetracenomycin C (3).
To evaluate the microbiological activity of 3, 1 mg mL−1 methanolic solutions of 1 and 3 were prepared for disc diffusion assays against E. coli XL1-blue, Streptomyces prasinus, and Mucor meihei strains. Streptomyces prasinus NRRL B-2712 was chosen because it was previously shown to be the most susceptible gram positive organism to 1.2 3 demonstrated no detectable activity against E. coli or Mucor meihei, but demonstrated a diameter halo against Streptomyces prasinus of 12±2 mm as compared to the 25±2 mm of 1. This finding indicates that the 4’-ketosugar modification of 3 diminishes some of the antimicrobial effect observed with 1. This is consistent with similar modifications of the sugar moiety of 8-demethyl tetracenomycin C, as many derivatives have resulted in poorer antimicrobial activity.10 Very possibly, the O-methyl groups of the L-rhamnose moiety of 1 are essential for its cytotoxicity. The possible anticancer activity of 3 is currently being evaluated against several cancer cell lines in our lab for a future report.
ElmGT represents one of the most flexible glycosyltransferases with respect to its ability to accommodate a large number of sugar donor substrates as compared to glycosyltransferases in other secondary metabolite biosynthetic pathways. ElmGT reportedly utilizes various NDP-d-sugars: d-olivose, d-mycarose, d-diolivose, d-amicetose, d-boivinose, d-digitoxose, and d-glucose. ElmGT also accepts a number of NDP-l-sugars: l-rhamnose, l-rhodinose, l-digitoxose, l-olivose, l-amicetose, l-mycarose and 4-deacetyl-l-chromose B. However, ElmGT has not been previously shown to accommodate NDP-ketosugars. In this context, production of 3 through the expression of pKOL in S. lividans (cos16F4) was interesting, especially, as earlier efforts to glycosylate 8-DMTC with NDP-4-keto-l-rhamnose or NDP-4-keto-l-mycarose were unsuccessful.9,10
The observed production of 4 along with 3 by S. lividans (cos16F4)/pKOL was unanticipated. However, we assume that a pathway-independent ketoreductase of S. lividans TK 24 might be responsible for the partial conversion of NDP-4-keto-l-olivose to NDP-l-olivose, and the latter is utilized by ElmGT as an alternate substrate to yield 4. Pathway independent reductions have been reported previously in the literature. Earlier in the pikromycin biosynthetic pathway, in which a d-quinovosyl macrolide was accumulated instead of the anticipated 4-keto-6-deoxy-d-glucosyl analogue when desI was inactivated in Streptomyces venezuelae.21 To search for possible sugar ketoreductases in Streptomyces lividans TK 24 that could be responsible for reduction of NDP-4-keto-l-olivose, thus possibly explaining the presence of 4, the amino acid sequence for OleU was compared to encoded proteins in the Streptomyces lividans genome using the protein BLAST public database. One such candidate was indicated in the search, SSPG_00655 (30% sequence identity/42% sequence similarity). This candidate has a domain that shows similarity to RfbD, which is a 4-ketohexulose reductase responsible for equatorial 4-ketoreduction of NDP-4’-keto-l-rhamnose in the NDP-l-rhamnose pathway (Figure S2, Supporting Information). This enzyme may be responsible for the 4-ketoreduction witnessed in 4. Despite the ability of SSPG_00655 or some other promiscuous reductase to reduce NDP-4-keto-l-olivose to NDP-l-olivose in S. lividans (Figure 2), enough of the ketosugar substrate was available to be accepted by ElmGT to flux towards 3. It is equally interesting that none of the elm pathway sugar O-methyltransferases (ElmMI, ElmMII, and ElmMIII) recognized the 4-keto-l-sugar moiety, even though these enzymes acted on an appended 8-O-l-olivose previously.
Despite the many glycosyltransferases discovered and explored, those which can attach ketosugars to their acceptor co-substrates are relatively scarce. MtmGIV (and possibly MtmGIII) from the mithramycin pathway has/have been shown to handle NDP-4-keto-d-olivose and NDP-4-keto-d-mycarose to generate novel premithramycin and mithramycin analogues in Streptomyces argillaceus strains in which mtmC (3-C-methyltransferase) and mtmTIII (4-ketoreductase) genes were disrupted.22 Like 3, the ketomithramycin derivative demonstrated weaker biological activity as compared to the parent mithramycin compound, possibly due to the ketosugar substitution.22 EryBV from the erythromycin pathway has been shown to accommodate NDP-4-keto-l-mycarose.23 Similarly, BgtfA from the balhimycin pathway is capable of utilizing NDP-4-keto-l-vancosamine as a natural sugar donor substrate.24 The unprecedented flexibility of ElmGT towards NDP-4′-keto-l-olivose reported in this communication is particularly encouraging for further studies towards understanding the structural role of ElmGT in binding deoxysugar substrates.
Supplementary Material
Acknowledgements
This work was supported by the National Institutes of Health (grant CA 091901 to JR) and a predoctoral fellowship from the University of Kentucky graduate school to EN.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Notes
- 1.Thibodeaux CJ, Melancon CE, Liu HW. Angew. Chem.-Int. Ed. 2008;47:9814. doi: 10.1002/anie.200801204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drautz H, Reuschenbach P, Zahner H, Rohr J, Zeeck A. J. Antibiot. 1985;38:1291. doi: 10.7164/antibiotics.38.1291. [DOI] [PubMed] [Google Scholar]
- 3.Rohr J, Zeeck A. J. Antibiot. 1990;43:1169. doi: 10.7164/antibiotics.43.1169. [DOI] [PubMed] [Google Scholar]
- 4.Decker H, Rohr J, Motamedi H, Zahner H, Hutchinson CR. Gene. 1995;166:121. doi: 10.1016/0378-1119(95)00573-7. [DOI] [PubMed] [Google Scholar]
- 5.Wohlert SE, Blanco G, Lombõ F, Fernández E, Braña AF, Reich S, Udvarnoki G, Méndez C, Decker H, Frevert J, Salas JA, Rohr J. J. Am. Chem. Soc. 1998;120:10596. [Google Scholar]
- 6.Blanco G, Patallo EP, Braña AF, Trefzer A, Bechthold A, Rohr J, Méndez C, Salas JA. Chem Biol. 2001;8:253. doi: 10.1016/s1074-5521(01)00010-2. [DOI] [PubMed] [Google Scholar]
- 7.Rodriguez L, Aguirrezabalaga I, Allende N, Braña AF, Méndez C, Salas JA. Chem. Biol. 2002;9:721. doi: 10.1016/s1074-5521(02)00154-0. [DOI] [PubMed] [Google Scholar]
- 8.Fischer C, Rodriguez L, Patallo EP, Lipata F, Braña AF, Méndez C, Salas JA, Rohr J. J. Nat. Prod. 2002;65:1685. doi: 10.1021/np020112z. [DOI] [PubMed] [Google Scholar]
- 9.Rodriguez L, Oelkers C, Aguirrezabalaga I, Braña AF, Rohr J, Méndez C, Salas JA. J. Mol. Microbiol. Biotechnol. 2000;2:271. [PubMed] [Google Scholar]
- 10.Lombõ F, Gibson M, Greenwell L, Braña AF, Rohr J, Salas JA, Méndez C. Chem. Biol. 2004;11:1709. doi: 10.1016/j.chembiol.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 11.Perez M, Lombõ F, Zhu L, Gibson M, Braña AF, Rohr J, Salas JA, Méndez C. Chem. Commun. 2005;12:1604. doi: 10.1039/b417815g. [DOI] [PubMed] [Google Scholar]
- 12.Perez M, Lombõ F, Baig I, Braña AF, Rohr J, Salas JA, Méndez C. Appl. Environ. Microbiol. 2006;72:6644. doi: 10.1128/AEM.01266-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramos A, Olano C, Braña AF, Méndez C, Salas JA. J. Bacteriol. 2009;191:2871. doi: 10.1128/JB.01747-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kharel MK, Nybo SE, Shepherd MD, Rohr J. ChemBioChem. 2010;11:523. doi: 10.1002/cbic.200900673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics: John Innes Foundation. 2000 [Google Scholar]
- 16.Kharel MK, Pahari P, Lian H, Rohr J. Org. Lett. 2010;12:2814. doi: 10.1021/ol1009009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lombõ F, Gibson M, Greenwell L, Braña AF, Rohr J, Salas JA, Méndez C. Chem. Biol. 2004;11:1709. doi: 10.1016/j.chembiol.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 18.Bacterial strains and cloning techniques. Escherichia coli XL1 blue (Invitrogen) was used as a host for routine cloning experiments and was grown on LB agar or in LB broth at 37°C. The Plasmid MiniPrep GeneJET™ kit was used to purify plasmid DNA (Fermentas). Restriction enzymes were purchased from New England Biolabs and restriction digests were performed as per the manufacturers’ protocols. Streptomyces lividans TK24 and derivative strains were grown on M2 agar or in YEME liquid medium at 28 °C for 3 days for preparation of protoplasts following the previously reported protocol.14 When antibiotic selection of recombinant strains was required, 25 µg/mL apramycin, 25 µg/mL thiostrepton, or 100 µg/mL of ampicillin were used. Protoplast transformation was carried out to introduce the cosmid cos16F4 into Streptomyces lividans TK 24 following the standard protocol 15. An apramycin resistant colony was selected for the production of 8-demethyl-tetracenomycin and was used as host for the expression of a newly generated deoxysugar biosynthetic construct.
- 19.Growth conditions and isolation of 3 and 4. One of the resulting transformants of recombinant strain S. lividans (cos16F4)/pKOL was grown on M2 agar medium at 28 °C supplemented with thiostrepton and apramycin for 5 days. For small scale production of tetracenomycins, an agar chunk of bacterial spores was inoculated in 250 mL Erlenmeyer flasks containing 100 mL of Soytone-Glucose (SG) medium supplemented with antibiotics for 4–5 days.16 25 mL of the culture was extracted with ethyl acetate and the organic layer was dried in vacuo. Extracts were then redissolved in methanol and were subjected to HPLC/MS analyses on a MicroMass ZQ 2000 (Waters) instrument equipped with HPLC (Waters alliance 2695 model) and photodiode array detector (Waters 2996). A Sun-Fire semi-prepC18 column (19×250 mm, 5 µm) and Symmetry C18 (4.6×250 mm, 5 µm) analytical column were used for semi-preparative and analytical scale separations, respectively. A gradient of acetonitrile and 0.1% formic acid in water was used to separate compounds as reported previously.17 Comparison of extract of Streptomyces lividans TK 24 (cos16F4)/(pKOL) strain to methanolic extracts of control strain Streptomyces lividans TK 24 (cos16F4)/ pEM4 resulted in the identification of new metabolites visualized by HPLC-MS (Figure 3). For larger scale production of glycosylated tetracenomycins, 7.2 L of SG medium and allocated into 800 mL portions in 9 baffled 2 L Erlenmeyer flasks. The production media was supplemented with antibiotics and inoculated with freshly prepared spores from M2 agar plates of the recombinant strains. Fermentation was carried out for 5 days at 28°C in an orbital shaker. The culture was harvested, mixed with celite (50 g/L fermentation broth), and then filtered. The mycelial cake was discarded, and the culture broth was passed through Amberlite® XAD16 resin (column dimensions 80 cm × 4 cm). The column was washed with 100% distilled water, and crude tetracenomycins were obtained by elution with 2 L of methanol. The solvent was dried in vacuo. This extract was separated via silica gel column chromatography using 200 mL gradients of dichloromethane and methanol from 0–100% of methanol, increasing in 5% each step. Fractions containing tetracenomycins were pooled, dried in vacuo, and further purified using semi-preparative HPLC as described previously17. Using this protocol, 14.4 mg and 12.2 mg of compounds 3 and 4 were isolated respectively.
- 20.Spectroscopic methods. (−) High resolution ESI spectra of pure compounds were recorded at the University of Wisconsin Biotechnology Center. NMR spectra were recorded on a Varian VNMR 500 MHz spectrometer using deuterated methanol as solvent.
- 21.Borisova SA, Zhao L, Sherman DH, Liu HW. Org. Lett. 1999;1:133. doi: 10.1021/ol9906007. [DOI] [PubMed] [Google Scholar]
- 22.Remsing LL, Garcia-Bernardo J, Gonzalez A, Künzel E, Rix U, Braña AF, Bearden DW, Méndez C, Salas JA, Rohr J. J. Am. Chem. Soc. 2002;124:1606. doi: 10.1021/ja0105156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salah-Bey K, Doumith M, Michel JM, Haydock S, Cortes J, Leadlay PF, Raynal MC. Mol. Gen. Genet. 1998;257:542. doi: 10.1007/s004380050680. [DOI] [PubMed] [Google Scholar]
- 24.Pelzer S, Sussmuth R, Heckmann D, Recktenwald J, Huber P, Jung G, Wohlleben W. Antimicr. Agents Chemother. 1999;43:1565. doi: 10.1128/aac.43.7.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.