Abstract
Background
Calcium-binding proteins (CBPs) are instrumental in the control of Ca2+ signaling. They are the fastest players within the Ca2+ toolkit responding within microseconds to [Ca2+] changes. The CBPs compete for Ca2+ which plays a direct role in modulating Ca2+ transients and the resulting biochemical message. The kinetic properties of the CBPs have to be known to have a good understanding of Ca2+ signaling.
Scope of review
Most techniques used to measure binding kinetics are too slow to accurately determine the fast kinetics of most CBP. Furthermore, many CBPs bind Ca2+ in a cooperative way, which should be incorporated in the kinetic modeling. Here we will review a new ultra-fast in vitro technique for measuring Ca2+ binding properties of CBPs following flash photolysis of caged Ca2+. Compartmental modeling is used to resolve the kinetics of fast cooperative Ca2+ binding to CBPs.
Major conclusions
Currently this technique has only been used to quantify the kinetics of three CBPs (calbindin, calretinin and calmodulin), but has already provided remarkable insights into the specific role that these kinetics in Ca2+ signaling.
General significance
The potential to gain novel insights into Ca2+ signaling by quantifying kinetics of other CBPs using this technique is very promising.
Keywords: Calcium binding kinetics, Calcium binding proteins, Flash photolysis, Calcium signaling, Cooperative binding
1 Introduction
1.1 The role of Calcium Binding Proteins in Ca2+ signaling
Calcium ions (Ca2+) are the signaling particles that function in the largest variety of biological signaling pathways. In all eukaryotic cells, Ca2+ signals play a crucial messenger role in the regulation of many processes including neurotransmission, muscle contraction, metabolism, cytoskeleton dynamics, gene transcription, cell cycle and cell death. Some of these Ca2+ signals are highly localized within a cell, while others are more global. Moreover, the regulation of Ca2+ practically covers the whole temporal spectrum over which biological processes are modulated, from (sub) milliseconds to years [1, 2]. Notably, some of the processes triggered by an increase in intracellular [Ca2+] in a given cell oppose one another. For example, neuronal growth cone outgrowth/exploration vs. growth cone retraction [3] and long-term potentiation (LTP) vs. long-term depression (LTD) [4, 5] are diametrically opposed processes. It is remarkable how a simple ion can regulate cellular functions in such a multitude of ways. How can changes in intracellular [Ca2+] modify cellular signaling over such a broad spectrum of processes with distinct temporal and spatial outcomes [6]? Since the serendipitous discovery of Ca2+ as an essential signaling ion in 1883 by Ringer [7] many strides have been made towards answering this question.
The structure of a Ca2+ signal can generally be described as follows: at resting conditions the intracellular [Ca2+] is kept low, around 100 nM. Upon an appropriate Ca2+ stimulus, so-called ON-mechanisms are activated [1, 2]. These ON-mechanisms (e.g., voltage- or ligand-gated ion channels in the plasma membrane or IP3 activated channels) let Ca2+ into the cytoplasm from the extracellular space or intracellular organelles (Ca2+ stores, i.e. sarco-endoplasmatic reticulum or mitochondria) causing a rapid increase in cytosolic [Ca2+]. If this “signal” is sufficiently large it will be “translated” into a biochemical message. When the [Ca2+] increases sufficiently, Ca2+ will bind to sensor Ca2+ binding proteins (CBPs), changing the physiological properties of these proteins. A hallmark for these sensor CBPs is a relatively large conformational change upon Ca2+ binding that is often accompanied by exposure to hydrophobic surfaces. Consequently, this allows interactions with specific ligands linked to subsequent biochemical regulation of downstream effectors [8]. Meanwhile, OFF-mechanisms work to lower the [Ca2+] in the cytoplasm to the resting concentrations [1, 2]. These OFF-mechanisms are the pumps and exchangers that transport the Ca2+ either back into the Ca2+ stores or to the extracellular space. Furthermore, there are buffering CBPs that rapidly bind free Ca2+, causing a seemingly immediate decrease in [Ca2+]. However, in later phases of the OFF-period, as the [Ca2+] decreases, these CBPs will release the bound Ca2+, causing a prolonged Ca2+ signal. Therefore, CBPs are not strictly OFF-mechanisms, but they are involved in shaping the amplitude and duration of the Ca2+ signal [1, 2, 9,10]. According to its specific function, each cell expresses a unique and specific set of Ca2+ signaling tools, i.e., ON and OFF components, to create the distinctive spatial and temporal Ca2+ signaling properties needed for the cells function [11]. Depending on their exact composition of their Ca2+ signaling toolkit, each Ca2+ signal system will produce Ca2+ transients varying from milliseconds to several hundreds of milliseconds. Various names are given to the Ca2+ transients, such as sparks, embers, quarks, puffs, blips or waves, depending on their exact spatial and temporal properties and the cell type in which they occur [2, 12]. Furthermore, the Ca2+ signals can be highly repetitive, forming Ca2+ oscillations [13]. In short, the spatiotemporal characteristics of short-lived and often highly localized changes in intracellular [Ca2+] result from a complex interplay between Ca2+ influx/extrusion systems, mobile/stationary CBPs, and intracellular sequestering mechanisms.
To understand the kinetics of cellular Ca2+ transients and their influence on the processes they regulate requires an in-depth knowledge of the Ca2+ sensitivities and binding properties of all the components involved. Upon an increase in [Ca2+], the CBPs are the first to respond as they immediately start binding Ca2+. Within each system, a Ca2+ signal will be interpreted and translated depending on the amplitude and temporal pattern of Ca2+ binding to the sensor CBPs. Both buffering and sensing CBPs are the fastest players within the Ca2+ toolkit and respond directly, working on a timescale of tens of microseconds to tens of milliseconds. The various CBPs are in an immediate competition to bind the freshly available Ca2+. On the other hand, the OFF components work on a somewhat slower timescale of tens of milliseconds to seconds and will not immediately reduce the [Ca2+] back to normal [14]. Hence, the competition for Ca2+ between the various CBPs within a system plays an essential and direct role in modulating the shape of Ca2+ transients and the outcome of the conveyed biochemical message [10]. Evidently, to have a good understanding of Ca2+ signaling, it is essential to know the properties of the CBPs that are involved in the studied process. A few key features of CBPs determine the spatiotemporal characteristics of Ca2+ signals and their transduction: the overall Ca2+ affinity of CBPs, their localization and concentration, their mobility inside cells, and their binding kinetics [15]. The latter of which are conceivably the most critical determinant of cellular Ca2+ signaling [16]. The lack of accurate data on the kinetic properties of CBPs gives rise to uncertainties in models studying intracellular Ca2+ signaling [10]. Two major obstacles make it challenging to accurately determine the kinetic properties of CBPs. First, the Ca2+ binding kinetics are very fast and, for accurate quantification, require the ability to measure changes in [Ca2+] (or any other parameter related to Ca2+ binding) with an accuracy of 10–100 μs. Conventional techniques used to measure binding kinetics to macromolecules, like stopped flow fluorimetry, have dead times >1ms [17], precluding accurate determination of the faster Ca2+ binding kinetics of CBPs. Secondly, many of the CBPs bind Ca2+ in a cooperative way, which is the ability to influence ligand binding at a site of a macromolecule by previous ligand binding to another site of the same macromolecule. There are four commonly used descriptions for cooperativity (for review see [18]): the Hill[19], the Adair-Klotz [20, 21], the Monod-Wyman-Changeux (MWC) [22], and the Koshland-Némethy-Filmer (KNF) [23] models. Yet all these models describe cooperativity only when the binding reactions are at equilibrium. Only under specific conditions [24–26] the MWC model was used to describe kinetics of cooperative binding. Such conditions do not hold for Ca2+ binding to CBPs. Furthermore, when using the MCW model with most CBPs the mathematical description becomes too complex for simple/practical interpretations [18, 27]. Over the last few years we have been working on overcoming these obstacles. We have developed an in vitro technique to measure the fast Ca2+-binding kinetics CBPs following flash photolysis of caged Ca2+ [28, 29]. In combination with compartmental kinetic modeling and a simple kinetic model for cooperative binding, we have begun to resolve the Ca2+ binding kinetics of some CBPs [28, 30, 31]. In this paper we will give a short overview some of the commonly used techniques that give insight into the Ca2+ binding kinetics of CBPs. We will then discuss our technique and describe various findings we have discovered while developing our technique that may be relevant to others using similar methods, such as measuring [Ca2+] with fluorescent dyes.
1.2 Ca2+ Buffering Capacity (κ) is a Description of Ca2+ binding Kinetics
One of the practical ways to quantify Ca2+ buffering in a cell is the buffering capacity (κ), which is the ratio of buffer-bound Ca2+ to free Ca2+ upon a change in total Ca2+[32, 33]:
where S is the endogenous buffer. For example, if κs=23, then out of every 24 ions entering a compartment, 23 will be bound by S (i.e, ~4% of Ca2+ entering remains unbound). This number gives important insights into several aspects of Ca2+ signaling. For instance, the size of the Ca2+ influx required to reach a certain free [Ca2+] can be determined using κ. As it is defined in the equation above κ does not reveal anything about the dynamics of Ca2+ buffering. However, the κ that is generally used in literature does. It is not trivial to determine the theoretical κ in a (sub) cellular compartment because it requires a small known change in [Ca2+]total and a measurement of the resulting Ca2+ signal. To evoke a change in intracellular [Ca2+], one can stimulate the cell to open Ca2+ permeable channels. In many structures, such as dendritic spines, it is impossible to use a technique (e.g., voltage clamping) to precisely determine the evoked Ca2+ influx (e.g., by measuring the Ca2+ current). Hence, it is impossible to exactly determine the amount of Ca2+ entering the structure. Estimates may be made based on the number of expected open channels and the driving force for Ca2+ over the whole time course of the Ca2+ influx. But unfortunately, often there are no exact data on the number of Ca2+ permeable channels open following stimulation, or the exact time-course of the membrane potential, hence the driving force for Ca2+. Another approach is to measure the Ca2+ signal following the Ca2+ influx by using Ca2+-indicators such as fura-2 or Oregon Green BAPTA (OGB). However, these dyes act as exogenous Ca2+ buffers, which will have a significant impact on the Ca2+ signal itself. Therefore, an approach has been developed that gives a quantification of the buffer capacity of endogenous buffers by extrapolating a series of indicator concentrations to zero [32, 34, 35]. It can be derived that changes in [Ca2+] at equilibrium:
If Ca2+ binding to all the buffers (endogenous S, and exogenous B) is fast enough so that the binding reactions are always close to equilibrium, then by approximation:
The assumption that the buffers are fast enough to always be in equilibrium (i.e., easily follow the [Ca2+] increase induced by the ON-systems) automatically implies that
where κB can be approximated as [32, 34, 35]:
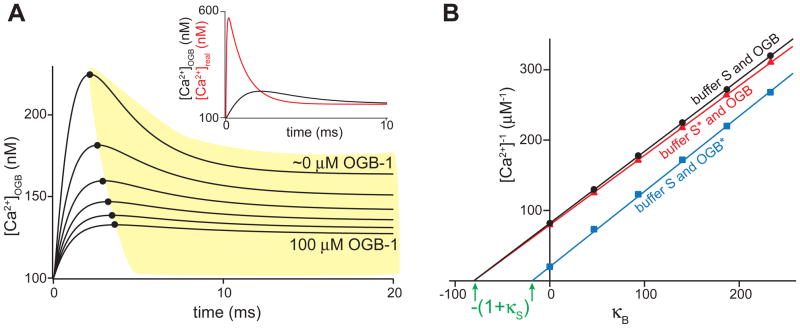
When 1/Δ[Ca2+]peak is plotted as a function of κB (which can be varied by using different concentrations of Ca2+ buffer/indicator) the function will intercept with the y-axis at 1/Δ[Ca2+]peak and with the x-axis at -(1+κS) (see figure 1A and B). The 1992 paper introducing this method has, thus far, been cited almost 500 times indicating that this method is widely used to determine κ. Generally, when buffering capacity is mentioned, it is the buffering capacity determined with this method. However, this method is based on the assumption that the system of which the capacity is determined buffers Ca2+ faster than the speed of the change in [Ca2+]total. Anything slower is grouped in with the extrusion mechanisms (OFF-mechanisms) [32]. In practice, this means that only the buffering that is faster than the measured [Ca2+] change is considered part of the buffer capacity. Hence, it is often also termed the fast buffer capacity. Since the [Ca2+] is measured with the exogenous buffer (i.e. the Ca2+-indicator), κ most often indicates the buffer capacity that is faster than the dye which was used to measure κ. This does not necessarily mean that the on–rate of that portion of the buffer is faster than that of the dye, as the absolute forward rate is determined by the product of the on-rate and the concentration of the buffer.
Figure 1.
Simulation of an experiment to measure Ca2+ buffering capacity. With a computer model developed in Berkeley Madonna, we simulated a compartment with 200 μM buffer S (Kd,S=1 μM, kon=1×108 M−1s−1) at [Ca2+]rest=100 nM. Buffer capacity is determined by measuring the change in [Ca2+] upon a 1 μM influx of Ca2+ (exponential step τ=0.5 ms) with OGB-1 (Kd,OGB=170 nM, kon=109). OGB-1 is often used to determine k. The experiment is performed with 100, 80, 60, 40, 20 and ~0 μM OGB-1. Zero OGB-1 is simulated by using 1 yM (10−24 M), and indicates the theoretical limit of what one could directly measure in the experiment with OGB-1. A) The [Ca2+]OGB traces, calculated from the measured OGB-1 signal, with the maxima ([Ca2+]OGB,peak) indicated by round markers. Inset shows the [Ca2+]OGB measured with ~0 μM OGB-1 curve in comparison to the actual [Ca2+] in red. OGB-1 cannot measure the actual [Ca2+] because it is limited by its kon. The measured decrease in [Ca2+]OGB, indicated with the yellow area, is caused by buffer S but is not considered part of κ when this method of determining κ is used. B) To determine κ, 1/[Ca2+]OGB,peak is plotted as a function of κB (black circles). Extrapolation onto the x-axis gives -(1+κS) (indicated with green arrows). The measured κ for the black line is 79. The experiments were also simulated with a 10-times faster, hypothetical OGB-1* (kon=1010 M−1s−1), which gives the blue squares, resulting in a κ of 18. This shows that the measurement of κ, with this method, is dependent on the speed of the used indicator. We also simulated a measurement with normal OGB-1 and with 96 μM buffer S* that has the same Kd as buffer S but is 3 times faster (kon=3×108 M−1s−1). These experiments give the red triangles and result in a κ of 79, just like the 200 μM buffer S. This shows that the measurement of κ with this method, is not a measure of the absolute buffer concentration/capacity, but only a measure of the fast buffer capacity.
It means that portion of the Ca2+ increase is buffered away before it can be detected by the dye and is therefore dependent on the on-rate of the dye and its concentration. A fast dye will give lower κ values than a slow dye (see figure 1B). Conversely, the value of κ, as it is generally used in the literature, is dependent on the concentration of the buffer, its Ca2+ affinity, and its speed (see figure 1B).
1.3 Measuring Ca2+ Binding Kinetics of CBPs in vitro
Thus far, we have established that κ is at least a quantification of the ‘fast’ buffer component. The buffer capacity is sometimes simplified as a portion of the Ca2+ signal that rapidly buffered before any message is conveyed (e.g. [36]). In that sense it would work like a simple attenuation factor. Maybe such simplifications are justified under certain conditions. However, it is unlikely that during periods of Δ[Ca2+] that are too fast for our detection methods, only buffering CBPs are active. For example, we have recently shown that the ubiquitous sensor CBP calmodulin (CaM) binds Ca2+ extremely fast and is likely to constitute a major part of the buffering capacities measured in cells. This confirms that at least some essential parts of Ca2+ signal transduction takes place during the initial fast phase of Δ[Ca2+], exactly during the period that is used to measure the amplitude of Ca2+ buffering and the quantification of κ.
The easiest and probably the most exact way to measure the binding kinetics of a CBP is to isolate the protein and study it in vitro under tightly controlled conditions. A great number of methods have been developed to study fast molecular interactions down to the picosecond range [17]. However, not every method is appropriate or practical to measure Ca2+ binding kinetics. Stopped flow and 43Ca2+-NMR are commonly used to accurately quantify Ca2+ binding kinetics (for description of the techniques see [17, 37]). For example, these techniques have been utilized multiple times to determine the Ca2+ binding kinetics of free CaM that was not bound to another protein or peptide. These measurements of CaM are relatively consistent, and most differences can be explained by differences in experimental temperature. In summary the finding are that CaM has two slow binding sites on its C-terminus (koff ~10 s−1 at room temperature) and two fast binding sites on its N-terminus (koff >1000 s−1 at room temperature) [38–45]. However, no distinctions could ever be detected between the two binding sites on either terminus using these techniques due to limited temporal resolution. The 43Ca2+-NMR is limited to measuring dissociation rates in the window of 10–105 s−1 [37]. In addition, the 43Ca2+-NMR works with the rare 43Ca isotope (natural occurrence <0.14%), making it somewhat costly. The stopped flow technique has a dead time of 1–2 ms at the initiation of the binding reactions that are to be quantified, limiting the maximally measurable dissociation rates at 103 s−1.
One of the other techniques used to directly measure Ca2+ binding kinetics is the T-jump method. This method is capable of quantifying kinetics much faster than that allowed by stopped flow or 43Ca2+-NMR, and was used to resolved the kinetics of EGTA, BAPTA, and several Ca2+-indicators (all have a kon of 108–109 M−1s−1) [46]. So far, the T-jump method has only been used to resolve the kinetics of molecules that bind a single Ca2+ion. A probable reason for this is that the mathematics needed to resolve multiple bindings is too complex to resolve, especially if cooperativity is involved.
1.4 Measuring Ca2+ Binding Kinetics of CBPs in vitro
Another way to determine the kinetics of a CBP is to compare it to Ca2+ buffers with known properties. For instance, in cells of animals in which a certain buffering CBP has been genetically knocked out, other buffers can be introduced to examine whether they can rescue the knockout phenotype. For example, it has been shown in calretinin (CR) knockout animals that, in cells that normally contain CR, the wildtype firing behavior could be rescued by introducing intracellular BATPA [47]. Similarly, the altered paired pulse depression in the synaptic transmission from a presynaptic terminal that normally contained the CBP parvalbumin (PV) was rescued when EGTA was introduced in cells from PV knockout animals [48]. From this it can be concluded that the buffering speed of CR is comparable to that of BAPTA, which is fast (kon=108–109 M−1s−1)[46], while the buffering speed of PV is comparable to that of EGTA, which is slow (kon=3×106–107 M−1s−1) [28, 46]. This replacement method is used very often and thought to give great direct insights into the functioning of a CBP under the physiological conditions of the intracellular milieu. However, a quantitative comparison with the kinetics of the artificial buffers is not possible because the absolute amount of free binding sites should be the same for a one-to-one comparison. Since, BAPTA (Kd=160 nM) and EGTA (Kd=70 nM) have a much higher affinity than most CBPs and the exact resting [Ca2+] is not always known, it is impossible to know if the concentrations of the EGTA/BAPTA used are really comparable to the normal situation with the physiological CBP in place. A study with artificial buffers that have affinities comparable to those of the studied CBPs would be more conclusive (e.g., see [49]).
Furthermore, a replacement study may give information for the CBP under the specific studied condition but does not resolve the general property of the CBP. For instance, overexpression of the slow buffer PV in oocytes induces elementary Ca2+ release events of Ca2+ puffs via Ca2+-induced Ca2+ release at low concentrations of IP3 [50]. This puff activity is comparable to the condition when EGTA is injected, but it is not seen in the presence of a fast buffer like BAPTA when these signals are more spatially uniform, or “globalized” [51]. The replacement experiment described above [46] and other replacement experiments performed in frog saccular hair cells [52] show that CR behaves like BAPTA. Therefore, one would expect that the introduction of CR in oocytes would lead to the “globalized” Ca2+ signaling observed in the presence of BAPTA. However, besides these “globalized” signals, CR can under certain conditions also produce puffs typical of the slow buffers that are never seen with BAPTA [53]. We measured with our new technique the exact Ca2+ binding kinetics of CR [30]. From these results we concluded that CR, due to its cooperative properties, it can function in a slow mode, comparable to EGTA and in a fast mode comparable to BAPTA. This probably explains CRs multiple calcium buffering behavior in oocytes [54].
All the techniques described above, and some more that we unfortunately don’t have space to discuss here, have given, and still give, invaluable new insights into the mechanisms of Ca2+ signaling. To further the quest of resolving the details of Ca2+ signaling we attempted to develop a technique to measure the Ca2+ binding kinetics of CBPs with a sufficiently high temporal resolution to give a clear insight into the “competition” for Ca2+ by the CBPs.
2 Measuring Ca2+ Kinetics With Flash-Photolysis
2.1 Uncaging Ca2+from DM-nitrophen
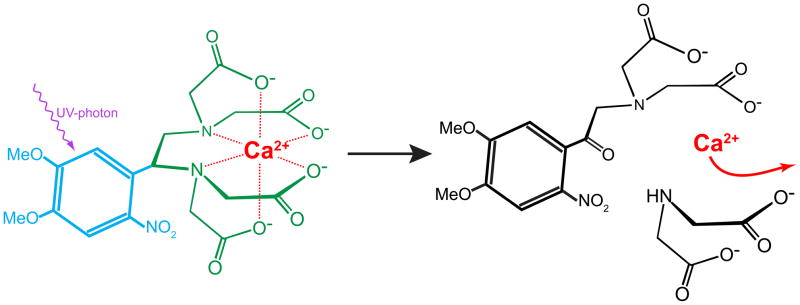
One of the fastest methods to measure reaction kinetics is flash-photolysis combined with spectroscopy [17]. The analysis of reaction kinetics using flash-photolysis is based on the activation of the reaction with a very short and intense light flash and observation of the initiated reaction. This means that at least one of the molecules within the system has to be photosensitive, and able to start the reaction by some form of photo conversion (e.g., radical formation or breaking up of the molecule forming new compounds). In 1988 Graham Ellis-Davies and Jack Kaplan developed DM-nitrophen (DMn, 1-(4,5-dimethoxy-2-nitrophenyl)-1,2-diaminoethane-N,N,N′,N′-tetraacetic acid) [55] which is a photo sensitive Ca2+ chealator. This so-called caged compound (or caged-Ca2+) made flash-photolysis a viable option for studying Ca2+ binding kinetics of CBPs. DMn (Kd ~5 nM) is an EDTA group, which strongly binds Ca2+, fused with a 2-nitophenyl group making the molecule light sensitive (see figure 2). When DMn captures a UV photon it breaks a covalent bond in the EDTA backbone, rapidly producing two photoproducts (PP) that have a ~6×105 times lower affinity for Ca2+ (~3 mM) than the unbroken DMn (see figure 2). Under the right conditions, this photo transition causes virtually any Ca2+ that was bound to the irradiated DMn molecule to be ‘released’ from a ‘cage’. The fragmentation of DMn into it PPs was reported to be very fast (photolysis of 1.1×104 – 8×104s−1 [56]). Therefore ‘ungaging’ of DMn should be able to step-like changes in [Ca2+] within tens of microseconds.
Figure 2.
Schematic view of DM-nitrophen. DMn is an EDTA molecule (green) with a photosensitive 2-nitophenyl group (blue) attached to it. When the 2-nitophenyl group absorbs a UV photon, the EDTA group breaks into two parts which have a much lower affinity for Ca2+, effectively causing the unbinding of Ca2+. Red dotted lines indicate coordination of the negative oxygen atoms with the chelated Ca2+.
2.2 Studying Ca2+ binding dynamics with DM-nitrophen
DMn (and molecules like it, see below) opened the possibility of studying the dynamics of fast Ca2+ binding, provided that the initiation of the studied reaction throughout the measurement space is instantaneous compared to the studied reaction kinetics. To achieve this requirement, Escobar et al., in the laboratory of Julio Vergara, used a pulsed laser that generated a 50 ns UV flash of sufficient power. In 1995 and 1997 they were the first ones that used flash photolysis of DMn to study Ca2+ binding kinetics of several Ca2+-indicators [57, 58]. We started collaboration with Julio Vergara to measure the kinetics of CBPs with this technique, and in 1999 we published our first success measuring the fast Ca2+ binding kinetics of calbindin D-28k (CB) [28]. CB is a buffering CBP with little or no cooperativity [31, 59] commonly found in neurons of all vertebrates and in the intestines, kidneys and pancreas of all mammals[60]. Since then we have been steadily improving this technique, making it more accurate, and including cooperative binding in the quantification of kinetics.
2.3 The basic setup
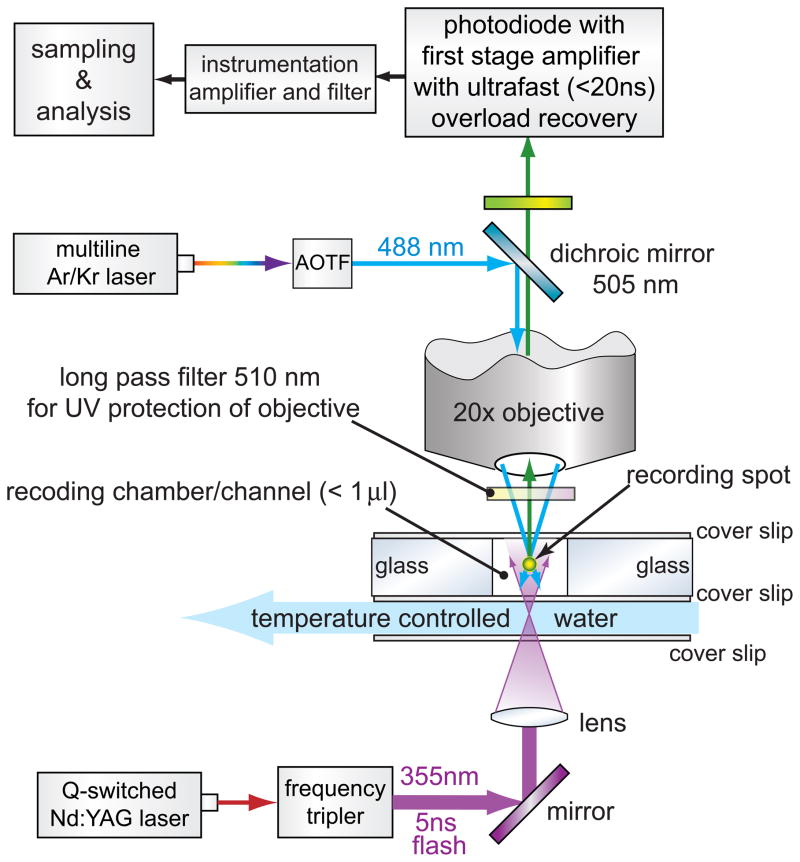
Since the first setup in 1995, we have made many changes; however, the basic principle stayed the same [28–31, 57, 58]. Figure 3 is a schematic layout of the current setup. It always consists of a small reaction chamber (currently 0.5–1 μL) in which epifluorescence is measured using an objective (currently 20×). The chamber is as small as possible so that the least amount of purified protein is used. There is a second optical path to deliver a short UV flash via a pulsed laser. In figure 3 the UV pathway is a direct one, but we have also successfully used flash delivery via an optic fiber. The laser should have enough power to initiate instantaneously (compared to the reaction kinetics, i.e., <1 μs) the uncaging of a sufficient amount of caged-Ca2+ molecules (DMn or other), so that change in [Ca2+] and the subsequent buffering is detectable with a fluorescent Ca2+-indicator. We currently use a frequency-tripled Nd:YAG laser (Surelite, Continuum, Santa Clara, CA) that can deliver enough energy in a single 5 ns UV flash (355 nm) to uncage at least 8% of the DMn. For most experiments, it should be sufficient to uncage up to ~2% per flash. It is important to have the ability to vary the output power of this UV laser (e.g., by using diffusion filters or by manipulating the timing of the pockels cell in the laser) to be able to repeat experiments with varying uncaging amplitudes. With 5mM DMn that is for 99% occupied with Ca2+ (at ~1μM [Ca2+]rest), uncaging can be detected with OGB-5N already at ~0.05% (~2 μM release). However, for complete experiments a range of uncaging levels needs to be measured (see below). Furthermore, we have added a system that heats the reaction chamber to the desired experimental temperature.
Figure 3.
Schematic overview of the latest uncaging setup we use. In a small (<1 μL), temperature controlled (35°C) recording chamber Ca2+ is uncaged from DM-nitrophen (DMn) with a 5 ns UV flash. Epifluorescence is used to measure the fluorescence of the Ca2+-indicator OGB-5N in a small recording spot in the middle of the chamber.
For an experiment a solution is made that contains, a Ca2+-cage (e.g., DMn), Ca2+, a Ca2+-indicator (e.g., OGB-5N) and a CBP of interest. Furthermore, the solution contains KCl and a pH buffer to set the solution to a physiologicalionic strength and pH mimicking the intracellular milieu. The [Ca2+]rest should be accurately titrated to a desired concentration so that enough Ca2+-cage is occupied with Ca2+ and the majority of the CBP’s binding sites is unoccupied. At the start of an experiment a sample of this solution is placed in the recording chamber. Then, while measuring the fluorescence of the Ca2+-indicator via the epifluorescence pathway, a UV flash is delivered starting the uncaging process. The resulting increase in [Ca2+] by release from the cage and ensuing decrease in [Ca2+] by Ca2+ binding to the CBP is reported by the fluorescence of the Ca2+-indicator (see figure 4). It is important in this process that the uncaging is homogeneous within the voxel where the [Ca2+] is recorded and a substantial volume around it, so that the recorded Ca2+ changes are only from the reactions occurring in that voxel. If uncaging is uneven, [Ca2+] gradients will occur and diffusion kinetics will contaminate the recording. To ensure that this will not happen, the recording spot should be relatively small compared to a large volume of UV illumination. To check if diffusion kinetics contaminate the recording, the recording spot should be moved to different locations within the uncaging area. The [Ca2+] transients should only change when the recording area is moved towards the outer edge of the uncaging area, were Ca2+ gradients are expected.
Figure 4.
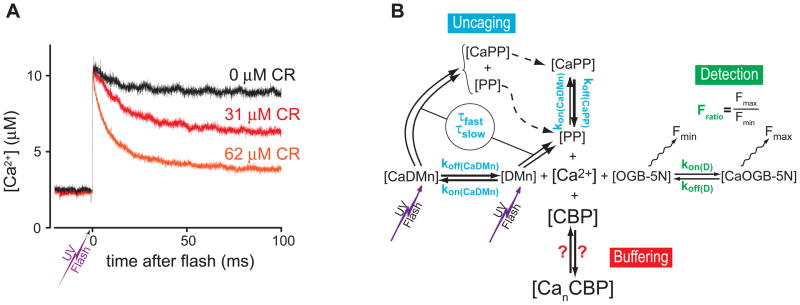
Ca2+ uncaging in vitro. A) Examples of changes in free [Ca2+] after photolysis of DMn in the absence of protein (upper trace) and in the presence of 31 μM and 62 μM CR (middle red and lower orange traces, respectively). The UV flash energies used to uncage DMn were of similar magnitude, resulting in an equivalent amount of uncaged-Ca2+. B) Scheme of all equilibrium reactions occurring in the measurement chamber after photolysis of caged-Ca2+ (DMn). The rate constants for DMn, its photoproducts (PP) and uncaging time constants (τf and τs) of DMn are independently determined (blue parameters) for each experiment. The rate constants and the Fratio for each batch of OGB-5N is also measured in an independent experiment (green parameters). The reaction parameters to be determined for describing the Ca2+ binding to a CBP are indicated by red question marks. The model/scheme for the buffering reaction is dependent on the used CBP, and is represented here by the simple equilibrium between [CBP] and [CanCBP], but can become quite complex, see text and [30, 31]. Figures adapted from figure 1 in reference [30]
2.4 Electronics
A simple and cost-effective solution to measure the fluorescence of the Ca2+-indicator is to use a photodiode in the focal plane of the emission pathway of the epifluorescence. It is important to use a photodiode that has a low capacity and a very fast rise time so that it can follow fast changes in fluorescence. Photodiodes with a smaller surface area will have a lower capacity. In addition a small enough surface area can work as a quasi pinhole and minimize the measuring voxel in the Z-direction. For our measurements we use the PIN-HR008 (0.8 pF, 0.04 mm2, UDT Sensors, Hawthorne, CA).
Another significant problem is that the high-energy UV flashes are so powerful that it will excite most, if not all, Ca2+-indicators that have their peak excitation wavelength in the visual spectrum (UV dyes cannot be used because then dye excitation could inadvertently photolyse Ca2+-cage). In our experience, this flash of UV-induced fluorescence is so bright that it will saturate the amplifier circuit that is strong enough to detect the ‘normal’ Ca2+ signals. For most amplifiers it takes considerable time (tens of milliseconds) to recover from such an overload. Therefore, special operational amplifiers should be used that can rapidly recover from such an overload, or have some overload protection. We generally use OPA699 (Burr-Brown, Texas Instruments), which has a very fast recovery time, resulting in a overall recovery/dead time of our system of 50–60 μs. For lower noise, but somewhat slower systems OPA637 and OPA111 (Burr-Brown, Texas Instruments) should also be sufficiently fast. For more information on photodiode amplifier circuits Burr-Brown has a very informative bulletin: focus.ti.com/lit/an/sboa035/sboa035.pdf.
2.5 Reaction scheme
Figure 4A shows an example of uncaging data of 3 experiments under identical conditions (for [DMn]total, [Ca2+]rest, uncaging energy, etc.) 2 different concentrations of CBP present and with no CBP present. The most straightforward approach to determine the kinetics of a system is to fit the [Ca2+] decay with a set of exponential functions. However, the system underlying the kinetics is a bit more complex. Instead of just the binding of Ca2+ to the CBP, there are also equilibrium reactions with the Ca2+ indicator, the DMn that is not uncaged, and with the PPs (see figure 4B). For example, even when no CBP is present, a clear decay in the [Ca2+] is detected (Figure 1A, black trace), which is caused by some rebinding of uncaged Ca2+ to free DMn. Furthermore, upon flash delivery the uncaging process is initiated, but the uncaging process itself is not instantaneous compared to the Ca2+ binding by CBPs. Hence, to determine the kinetic parameters of the studied CBP from the fluorescence recordings we use a mathematical model built in an ordinary differential equation solver (Berkeley Madonna 8.0) that incorporates all of the reactions in the reaction chamber (figure 4B). Initially we used as kinetic parametes for DMn and the Ca2+ indicator values that were reported earlier [28]. For instance, for DMn a photolysis timeconstant of 20 μs was used. However, in a later study we found that DMn uncages with 2 time constants as depicted in figure 3B. Two thirds of the irradiated DMn indeed uncages with a τ of ~15 μs, but one third uncages much slower with a τ ~3 ms [29]. Furthermore, we found that variations in the properties Ca2+-indicators also occur (see below in 2.7 Choosing the right Ca2+-indicator). To assure correct modeling of the reactions in the reaction chamber, we determine the properties of the Ca2+ indicator and the DMn in separate experiments for each batch that we use.
2.6 Choosing the right Ca2+-cage
The fact that DMn uncages with two time constants, one of which is at least slow enough to interfere with the [Ca2+] decay, raises the question if DMn is the ideal Ca2+-cage to use for such experiments. Of the most commonly used and commercially available caged-Ca2+ compounds DMn seems to be the best choice to release caged-Ca2+ when rapid and large increases in [Ca2+] are desired. First of all, DMn has the highest affinity for Ca2+ (Kd=5 nM, the next best, NP-EGTA, has a Kd of 80 nM,), and its PPs have the lowest affinity for Ca2+ after photolysis [61, 62]. Consequently, about 95–99% of DMn is occupied with Ca2+ while the [Ca2+]rest is at 0.1–1 μM, and most of the bound Ca2+ will be released from the PPs. Much higher [Ca2+]rest than 1 μM would be useless, since most binding sites on CBPs will be occupied with Ca2+ at that concentration, making them unavailable to bind Ca2+ upon uncaging. Furthermore, DMn has a relatively high quantum efficiency of 0.18 [55, 61]. Moreover, choosing other Ca2+ cages might not solve the problem of two uncaging time constants at all. For example, the Ca2+-cage NP-EGTA also uncages with multiple and slower kinetics [29] [29]. Concerning Ca2+-cages in general, a few more facts need pointing out. First of all, we find that DMn is indeed very light sensitive, and special precautions need to be taken to prevent unwanted uncaging. All our experimentation rooms have special lighting that is >500nm (by using filters or LED lighting), and only in these rooms is DMn taken out of light-tight containers. Secondly, we found that the amount of DMn delivered by companies is generally inaccurate. We always make a stock solution directly in the vial delivered by the company and measure the accurate concentration via spectroscopy (εDMn=4330 M−1cm−1[55]) and find concentrations up to +/− 20% of the expected value. Most often it is lower than what would be expected from the amount indicated on the container. The accurate concentration of the total DMn is essential for a correct simulation of the experiment. Therefore, the [DMn] of every stock solution made should be verified individually.
2.7 Choosing the right Ca2+-indicator
To follow the [Ca2+] during the experiment, a Ca2+-indicator is used. This indicator has to be fast enough to follow the rapid [Ca2+] changes caused by Ca2+ binding to CBPs. Typically, the ion indicators have fast association constants that are always in the same range (kon 108–109 M−1s−1[46, 57]). The indicators that can follow a fast drop in [Ca2+] are indicators with a faster dissociation rate, i.e., the indicators with lower affinities. We generally use OGB-5N (Kd ~35 μM), but indicators like Rhod-FF and Fluo-4FF might also work. We have to make an extremely important note of caution here: over the past eight years, we have determined the properties of each individual factory batch of OGB-5N we used, and found that their properties vary considerably (see Table 1). Although the properties within a batch are very stable, we found that between batches the Kd varied from 34 to 46 μM and the Fratio (Fmax/Fmin) varied from from 10 to 40. Concurrently, we also found variations in the kinetics of the different batches of dye. We have no explanation for the variability between the batches other than the fact that specific contaminations might occur in different batches, as stated by the supplier (personal communication with Molecular Probes, Portland, OR). We found similar variations between the different batches of other Ca2+-indicators, like OGB-2 and Fluo-4. It is critical to have the correct properties of the dyes when quantifying data using such ion-indicators. Hence, we advise anyone using fluorescent probes for quantification purposes to measure the exact properties of each batch used. More details on how we measured those properties can be found in the text and supplements of these papers [29–31, 57].
Table 1.
Properties of different batches of OGB-5N
| Date | Batch | Kd (μM) | Fratio |
|---|---|---|---|
| Jan-03 | 34b1-2 | 39 | 11 |
| Aug-04 | 15c1-2 | 37 | 34 |
| Jan-08 | 29020W | 34 | 31 |
| Aug-08 | 39391A | 34 | 30 |
| Mar-09 | 531987 | 42 | 40 |
| Mar-11 | 25663W | 46 | 38 |
2.8 Modeling CBPs
For each solution of DMn and batch of OGB-5N we determine the properties in individual experiments. With these properties known, we can start fitting data from uncaging experiments that had a CBP present (question marks figure 4B). In the simplest case, the binding of Ca2+ to the CBP is non-cooperative and can be added to the system as a simple equilibrium for each binding site (B):
where B1 through Bn represent individual binding sites with different properties. To minimize the number of parameters and the degrees of freedom of the fit, different binding sites on a CBP are often considered similar. For instance CB, which has 4 non cooperative binding sites, can be simplified by assuming only one or two different types of binding sites [28, 31].
The modeling of CBPs gets a little more complicated if cooperative binding needs to be included. In order for cooperativity to occur, the properties of the binding sites are not static but depend on the binding state of the individual binding sites of the CBP. Thus far, details about what exactly happens with the CBP molecules and their binding sites during cooperativite binding remain unclear. Therefore, it is impossible to have an exact mathematical model for quantifying the kinetics of cooperative binding. The most commonly used mathematical descriptions of chemical equilibria, the Hill[19] and the Adair-Klotz[20, 21] equations, describe equilibria on a macroscopic level. Only the more complex MWC[22] and KNF[23] models are appropriate for describing binding kinetics of individual binding sites. The MWC and KNF models use two conformational states in which each binding sites can occur. The state of each site determines whether it has a a low affinity (called the tense or T-state) or a high affinity (called the relaxed or R-state) [18]. To model kinetics using either the MWC or KNF model we would need to incorporate the association and dissociation rate constants for the R and T states, as well as the transition rate constants (k+, k−) between these states. For instance for the MWC the reaction scheme for two binding sites with cooperativity looks like this:
Assuming that k+,1, k+,2, k+,3 and k−,1, k−,2, k−,3 easily relate to one and other they can be reduced to one k+ and one k− [30]. This means that for two identical sites that have cooperative binding the number of parameters to fit would at least be 6 (kon,T, koff,T, kon, R, Koff, R, k+, and k−). Similarly, KNF models that incorporate kinetics would at least need 6 kinetic constants to describe cooperative binding between two cooperative identical sites. To decrease the number of parameters to fit, and to make the model more intuitively comprehensible, we developed a different model for cooperative binding that allows for the description of kinetics. In our model each cooperative binding site can also exist in either the T or R state. However, we assume a direct allosteric influence between bindings in a cooperative set. The binding of Ca2+ to one site always leads to a transition T→R in the other site, and an unbinding of Ca2+ from one site always leads to a transition R→T in the other site. These transitions are immediate and occur simultaneously with the binding of the Ca2+. In this case, Ca2+ binding by two identical cooperative binding sites is described by:
So, with our model, only 4 transition rate constants are needed to describe the kinetics of two identical cooperative binding sites. For the fitting procedure it is important to minimize the number of variables to fit, so that degrees of freedom of the system are minimal, which increases the reliability of the fit. Furthermore, our model describes a simple two-step binding system that can easily translate to the macroscopic Adair-Klotz equation where the macroscopic binding constants K1 and K2 are:
We have used this model to fit data measured from the CBP calretinin (CR) which has four cooperative binding sites [63]. When comparing the fits with a MWC model describing CR, we found no differences and concluded that our new model was just as sufficient to describe the kinetics of CR[30].
2.9 Fitting the data
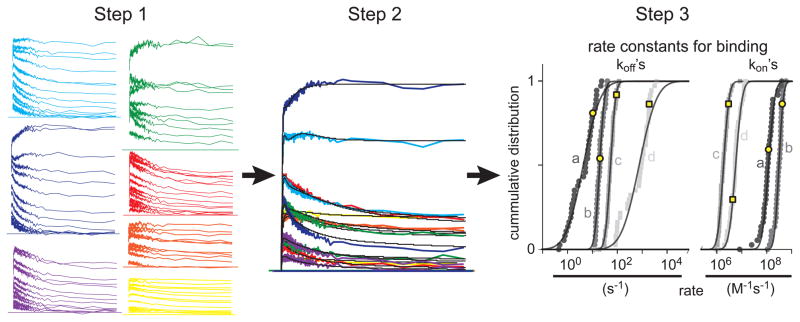
Though we have simplified the model with which the kinetics of cooperative binding can be quantified, most models that describe CBPs will still be fairly complex and will still have many variables to fit. For instance, CR has four cooperative sites and one independent site. In such a case, it is wise to explore how to simplify the model for the CBP. For instance, based on earlier findings on CR it is not unreasonable to assume that its cooperative binding sites function as two similar pairs of binding sites [30]. This compounds the number of parameters to fit for the cooperative sites of CR to 4, making a total of 6 when the independent site is also included. Although such simplifications are not always possible (e.g., for CaM [31]), it is a good first step to make the fitted parameters more reliable. Still 6 and sometimes more (e.g., 8 for CaM [31]) parameters result in too many degrees of freedom to accurately determine any of those parameters by fitting single curves as show in figure 4A. Therefore, we developed a procedure that significantly constrains the fit. In short, we obtain data under a variety of starting conditions for [Ca2+]rest, [DMn]total, [OGB-5N]total and [CBP]total. Furthermore, under each of these different conditions, we record many traces all initiated by different uncaging energies, i.e., different amounts of Ca2+ uncaging (figure 5, Step 1). This gives 100–200 different traces from which we compose random sets of 10–15 traces and fit them simultaneously (Figure 5, Step 2). This results in a parameter set (the fitted rate constants) that is constrained because the fit result has to describe all the different traces simultaneously with one single parameter set describing the CBP (Figure 5, Step 3, yellow markers). This procedure is then repeated 30–40 times with another random set of traces, every time resulting in a new value for each fitted parameter, creating a pool of 30–40 fit results for each parameter. Then, a distribution and average is determined for each parameter from this pool of fit results (Figure 5, Step 3). Combined with this procedure we use several other mechanisms to ensure that we probe whole physiologically feasible parameter domain and that we diminish the probability of assigning fit solutions to local error minima is. For more details please see [30, 31]
Figure 5.
Procedure that constrains the fits with a CBP model that has many degrees of freedom. In Step 1 many uncaging measurements are made with different uncaging energies at different starting conditions by varying [Ca2+]rest, [DMn]total, [OGB-5N]total and [CBP]total. These different conditions are represented as the colors in the traces under Step1. For instance for the yellow traces [Ca2+]rest, [DMn]total, [OGB-5N]total and [CBP]total are 1.9 μM, 5.6 mM, 50 μM and 123 μM, respectively while for the dark blue traces they are 0.4 μM, 3.6 mM, 100 μM and 47 μM, respectively. Of all the measured traces, a random group of traces is taken in Step 2 and fitted simultaneously. Each trace starts with it own set of starting conditions but they all have the same parameters that are fitted. In this case, the CBP is modeled with 2 types of binding sites that can occur in 2 states (R and T) making a total of 4 kon’s and 4 koff’s (a, b, c, d). The results of the binding sites and states (R or T) for this particular fit are plotted in the figure under Step 3 with yellow markers. Step 2 is repeated several times to create an average and a distribution for each fitted parameter as seen in Step 3.
3 The future of flash photolysis in biomolecular kinetics
3.1 Measurements beyond Ca2+ binding kinetics
There are few techniques that can replicate the speed of photolysis in which UV light is used to break a chemical bond that converts a biochemically inert molecule into one that is active, or simply releases a “caged” compound. In this special issue of Biochimica et Biophysica Acta on biochemical, biophysical and genetic approaches to intracellular Ca2+ signaling, we gave an overview on how we use photolysis to measure the fast Ca2+ binding properties of CBPs. However, we believe that the methods we applied here should also be applicable to measure reaction kinetics of a variety of biochemical processes. Ever since the first reported synthesis of a caged compound (ATP, [64]) the use of caged substances has been continuously developed, leading to the successful experimental use of more than 60 caged molecules of biological importance [65, 66], many of which are commercially available (e.g., Calbiochem, Invitrogen, Tocris, Probior, Enzo Life Sciences). Initially, only ions and small molecules could be caged such as nucleotides (and analogues), gasses (e.g., O2, CO and NO), simple neurotransmitters (glutamate, GABA and 5-HT), and simple intracellular messengers (e.g., DAG and IP3), as it is easier to shield their active domain. However, as new methods have been devised, it is now possible to specifically mask the active domain of much larger molecules, including proteins and oligonucleotides, with groups that can be removed with photolysis [65, 66]. The field of biochemical quantification/detection using fluorescent probes has also been developing steadily, resulting in many specific probes for ions (Invitrogen, Molecular Probes) and smaller molecules such as ATP [67], O2 [68] and NO [69]. In addition, spectroscopical techniques such as fluorescence correlation spectroscopy and fluorescence polarization/anisotropy provide quantitative tools to measure the formation of macromolecular complexes by probing their size. We hope that our work will be an incentive for expanding this type of quantification to other molecular interactions where fast kinetics play a decisive role.
3.2 Future of CBP kinetics
Up until now we used our technique to quantify the Ca2+ binding kinetics of CB, CR and CaM, which gave immediately some remarkable insights. We found that CR is comparable to EGTA around a low [Ca2+]rest (~100 nM), but shifts to a faster acting mode at higher [Ca2+]rest (~1 μM) and acts more like BAPTA. This leads to the remarkable property that the overall buffering speed of CR seemingly does not change over a wide rage physiological of [Ca2+], even as less CR binding sites are available at higher [Ca2+][30, 54]. For CaM, we found that the pair of binding sites on the N-terminus are much faster than ever estimated (or expected). The binding sites on the N-terminus have a ~10× lower affinity than the pair of binding sites on the C-terminus or the binding sites on CB. Nevertheless, upon Ca2+ influx, the N-terminal binding sites will initially win the competition for the Ca2+ because the N-terminus is the fastest. It is later during the Ca2+-signal transduction that Ca2+ leaves the N-terminus and trickles down to the higher affinity, and slower binding sites [31]. Furthermore, two very different types of cooperativity (on the N-terminus primarily caused by increasing kon in the T→R transition and on the C terminus primarily caused by decreasing koff in the T→R transition) provide a ‘two-step’ mechanism, one within the bulk solution and one within the nanodomain[70], to fully activate CaM. Although we have just started to quantify the kinetic properties of a few CBPs with a higher accuracy then ever before, the results, thus far, suggest that quantifying the kinetics of more CBPs will give a whole new perspective to Ca2+ signaling. There are many more CBPs that could and should be studied this way. Furthermore, the (regulating) effects of molecular interactions between other proteins and CBPs (e.g., CaM and Neurogranin or IQ domains[42, 43]), or the effects of molecular crowding on CBPs could be studies using this technique. This technique could potentially give many new insights into the mechanisms of Ca2+ signaling. With the three CBPs we quantified thus far, we have barely scratched the.
Highlights.
Kinetics of Ca2+ binding proteins (CBPs) are too fast to gauge with normal methods
The fast kinetics of CBPs can be determined using flash photolysis
Cooperativity should be included when quantifying binding kinetics of CBPs
The binding kinetics of CBPs play a key role in calcium signaling
Acknowledgments
The authors like to thank Ryan Jones and Kathleen Faas for proof reading the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Guido C. Faas, Email: gfaas@ucla.edu.
Istvan Mody, Email: mody@ucla.edu.
References
- 1.Berridge MJ, Bootman MD, Roderick HL. Calcium signalling: dynamics, homeostasis and remodelling. Nat Rev Mol Cell Biol. 2003;4:517–529. doi: 10.1038/nrm1155. [DOI] [PubMed] [Google Scholar]
- 2.Berridge MJ, Lipp P, Bootman MD. The versatility and universality of calcium signalling. Nat Rev Mol Cell Biol. 2000;1:11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- 3.Bolsover SR. Calcium signalling in growth cone migration. Cell Calcium. 2005;37:395–402. doi: 10.1016/j.ceca.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 4.Xia Z, Storm DR. The role of calmodulin as a signal integrator for synaptic plasticity. Nat Rev Neurosci. 2005;6:267–276. doi: 10.1038/nrn1647. [DOI] [PubMed] [Google Scholar]
- 5.Malenka RC, Bear MF. LTP and LTD: an embarrassment of riches. Neuron. 2004;44:5–21. doi: 10.1016/j.neuron.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 6.Burgoyne RD. Neuronal calcium sensor proteins: generating diversity in neuronal Ca2+ signalling. Nature reviews Neuroscience. 2007;8:182–193. doi: 10.1038/nrn2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ringer S. A further Contribution regarding the influence of the different Constituents of the Blood on the Contraction of the Heart. J Physiol. 1883;4:29–42. 23. doi: 10.1113/jphysiol.1883.sp000120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ikura M. Calcium binding and conformational response in EF-hand proteins. Trends Biochem Sci. 1996;21:14–17. [PubMed] [Google Scholar]
- 9.Markram H, Roth A, Helmchen F. Competitive calcium binding: implications for dendritic calcium signaling. J Comput Neurosci. 1998;5:331–348. doi: 10.1023/a:1008891229546. [DOI] [PubMed] [Google Scholar]
- 10.Neher E. Calcium buffers in flash-light. Biophys J. 2000;79:2783–2784. doi: 10.1016/S0006-3495(00)76517-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berridge MJ. Cardiac calcium signalling. Biochemical Society transactions. 2003;31:930–933. doi: 10.1042/bst0310930. [DOI] [PubMed] [Google Scholar]
- 12.Niggli E, Shirokova N. A guide to sparkology: the taxonomy of elementary cellular Ca2+ signaling events. Cell Calcium. 2007;42:379–387. doi: 10.1016/j.ceca.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 13.Iino M. Regulation of cell functions by Ca2+ oscillation. Advances in experimental medicine and biology. 2007;592:305–312. doi: 10.1007/978-4-431-38453-3_26. [DOI] [PubMed] [Google Scholar]
- 14.Miller RJ. The control of neuronal Ca2+ homeostasis. Prog Neurobiol. 1991;37:255–285. doi: 10.1016/0301-0082(91)90028-y. [DOI] [PubMed] [Google Scholar]
- 15.Schwaller B. The continuing disappearance of “pure” Ca2+ buffers. Cell Mol Life Sci. 2009;66:275–300. doi: 10.1007/s00018-008-8564-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams RJ. Calcium-binding proteins in normal and transformed cells. Cell Calcium. 1994;16:339–346. doi: 10.1016/0143-4160(94)90026-4. [DOI] [PubMed] [Google Scholar]
- 17.Bisswanger H. Enzyme kinetics : principles and methods. WILEY-VCH; Weinheim (Federal Republic of Germany): 2002. [Google Scholar]
- 18.Ricard J, Cornish-Bowden A. Co-operative and allosteric enzymes: 20 years on. Eur J Biochem. 1987;166:255–272. doi: 10.1111/j.1432-1033.1987.tb13510.x. [DOI] [PubMed] [Google Scholar]
- 19.Hill AV. The possible effects of the aggregation of the molecules of heamoglobin on its dissociation curves. J Physiol-London. 1910;40:iv–vii. [Google Scholar]
- 20.Adair GS. The hemoglobin system. VI. The oxygen dissociation curve of hemoglobin. Journal of Biological Chemistry. 1925;63:529–545. [Google Scholar]
- 21.Klotz IM, Hunston DL. Protein interactions with small molecules. Relationships between stoichiometric binding constants, site binding constants, and empirical binding parameters. J Biol Chem. 1975;250:3001–3009. [PubMed] [Google Scholar]
- 22.Monod J, Wyman J, Changeux JP. ON THE NATURE OF ALLOSTERIC TRANSITIONS: A PLAUSIBLE MODEL. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 23.Koshland DE, Jr, Nemethy G, Filmer D. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry. 1966;5:365–385. doi: 10.1021/bi00865a047. [DOI] [PubMed] [Google Scholar]
- 24.Kirschner K, Gallego E, Schuster I, Goodall D. Co-operative binding of nicotinamide-adenine dinucleotide to yeast glyceraldehyde-3-phosphate dehydrogenase. I. Equilibrium and temperature-jump studies at pH 8-5 and 40 degrees C. J Mol Biol. 1971;58:29–50. doi: 10.1016/0022-2836(71)90230-0. [DOI] [PubMed] [Google Scholar]
- 25.Hoggett JG, Kellett GL. Kinetics of the cooperative binding of glucose to dimeric yeast hexokinase P-I. Biochem J. 1995;305(Pt2):405–410. doi: 10.1042/bj3050405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woodruff NR, Neet KE. Beta nerve growth factor binding to PC 12 cells. Association kinetics and cooperative interactions. Biochemistry. 1986;25:7956–7966. doi: 10.1021/bi00372a026. [DOI] [PubMed] [Google Scholar]
- 27.Eigen M. Fast Reactions and Primary Processes in Chemical Kinetics. 5th Nobel Symposium, Interscience; New York. 1967. p. 333. [Google Scholar]
- 28.Nagerl UV, Novo D, Mody I, Vergara JL. Binding kinetics of calbindin-D(28k) determined by flash photolysis of caged Ca(2+) Biophys J. 2000;79:3009–3018. doi: 10.1016/S0006-3495(00)76537-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Faas GC, Karacs K, Vergara JL, Mody I. Kinetic properties of DM-nitrophen binding to calcium and magnesium. Biophys J. 2005;88:4421–4433. doi: 10.1529/biophysj.104.057745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Faas GC, Schwaller B, Vergara JL, Mody I. Resolving the fast kinetics of cooperative binding: Ca2+ buffering by calretinin. PLoS Biol. 2007;5:e311. doi: 10.1371/journal.pbio.0050311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Faas GC, Raghavachari S, Lisman JE, Mody I. Calmodulin as a direct detector of Ca2+ signals. Nature Neuroscience. 2011;14:301–304. doi: 10.1038/nn.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neher E, Augustine GJ. Calcium gradients and buffers in bovine chromaffin cells. J Physiol. 1992;450:273–301. doi: 10.1113/jphysiol.1992.sp019127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neher E. Usefulness and limitations of linear approximations to the understanding of Ca++ signals. Cell Calcium. 1998;24:345–357. doi: 10.1016/s0143-4160(98)90058-6. [DOI] [PubMed] [Google Scholar]
- 34.Lee SH, Rosenmund C, Schwaller B, Neher E. Differences in Ca2+ buffering properties between excitatory and inhibitory hippocampal neurons from the rat. J Physiol. 2000;525(Pt 2):405–418. doi: 10.1111/j.1469-7793.2000.t01-3-00405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Helmchen F, Imoto K, Sakmann B. Ca2+ buffering and action potential-evoked Ca2+ signaling in dendrites of pyramidal neurons. Biophys J. 1996;70:1069–1081. doi: 10.1016/S0006-3495(96)79653-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Friel DD, Chiel HJ. Calcium dynamics: analyzing the Ca2+ regulatory network in intact cells. Trends in Neurosciences. 2008;31:8–19. doi: 10.1016/j.tins.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 37.Drakenberg T. Calcium-43 NMR of calcium-binding proteins. Methods in molecular biology. 2002;173:217–230. doi: 10.1385/1-59259-184-1:217. [DOI] [PubMed] [Google Scholar]
- 38.Brown SE, Martin SR, Bayley PM. Kinetic control of the dissociation pathway of calmodulin-peptide complexes. The Journal of biological chemistry. 1997;272:3389–3397. doi: 10.1074/jbc.272.6.3389. [DOI] [PubMed] [Google Scholar]
- 39.Andersson T, Drakenberg T, Forsen S, Thulin E. Characterization of the Ca2+ binding sites of calmodulin from bovine testis using 43Ca and 113Cd NMR. European journal of biochemistry / FEBS. 1982;126:501–505. doi: 10.1111/j.1432-1033.1982.tb06808.x. [DOI] [PubMed] [Google Scholar]
- 40.Martin SR, Andersson Teleman A, Bayley PM, Drakenberg T, Forsen S. Kinetics of calcium dissociation from calmodulin and its tryptic fragments. A stopped-flow fluorescence study using Quin 2 reveals a two-domain structure. Eur J Biochem. 1985;151:543–550. doi: 10.1111/j.1432-1033.1985.tb09137.x. [DOI] [PubMed] [Google Scholar]
- 41.Teleman A, Drakenberg T, Forsen S. Kinetics of Ca2+ binding to calmodulin and its tryptic fragments studied by 43Ca-NMR. Biochim Biophys Acta. 1986;873:204–213. doi: 10.1016/0167-4838(86)90047-6. [DOI] [PubMed] [Google Scholar]
- 42.Putkey JA, Kleerekoper Q, Gaertner TR, Waxham MN. A new role for IQ motif proteins in regulating calmodulin function. J Biol Chem. 2003;278:49667–49670. doi: 10.1074/jbc.C300372200. [DOI] [PubMed] [Google Scholar]
- 43.Gaertner TR, Putkey JA, Waxham MN. RC3/Neurogranin and Ca2+/calmodulin-dependent protein kinase II produce opposing effects on the affinity of calmodulin for calcium. J Biol Chem. 2004;279:39374–39382. doi: 10.1074/jbc.M405352200. [DOI] [PubMed] [Google Scholar]
- 44.Ikura M, Hiraoki T, Hikichi K, Mikuni T, Yazawa M, Yagi K. Nuclear magnetic resonance studies on calmodulin: calcium-induced conformational change. Biochemistry. 1983;22:2573–2579. doi: 10.1021/bi00279a039. [DOI] [PubMed] [Google Scholar]
- 45.Bayley P, Ahlstrom P, Martin SR, Forsen S. The kinetics of calcium binding to calmodulin: Quin 2 and ANS stopped-flow fluorescence studies. Biochem Biophys Res Commun. 1984;120:185–191. doi: 10.1016/0006-291x(84)91431-1. [DOI] [PubMed] [Google Scholar]
- 46.Naraghi M. T-jump study of calcium binding kinetics of calcium chelators. Cell Calcium. 1997;22:255–268. doi: 10.1016/s0143-4160(97)90064-6. [DOI] [PubMed] [Google Scholar]
- 47.Gall D, Roussel C, Susa I, D’Angelo E, Rossi P, Bearzatto B, Galas MC, Blum D, Schurmans S, Schiffmann SN. Altered neuronal excitability in cerebellar granule cells of mice lacking calretinin. J Neurosci. 2003;23:9320–9327. doi: 10.1523/JNEUROSCI.23-28-09320.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Caillard O, Moreno H, Schwaller B, Llano I, Celio MR, Marty A. Role of the calcium-binding protein parvalbumin in short-term synaptic plasticity. Proc Natl Acad Sci U S A. 2000;97:13372–13377. doi: 10.1073/pnas.230362997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adler EM, Augustine GJ, Duffy SN, Charlton MP. Alien intracellular calcium chelators attenuate neurotransmitter release at the squid giant synapse. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1991;11:1496–1507. doi: 10.1523/JNEUROSCI.11-06-01496.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.John LM, Mosquera-Caro M, Camacho P, Lechleiter JD. Control of IP(3)-mediated Ca2+ puffs in Xenopus laevis oocytes by the Ca2+-binding protein parvalbumin. The Journal of physiology. 2001;535:3–16. doi: 10.1111/j.1469-7793.2001.t01-2-00003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dargan SL, Parker I. Buffer kinetics shape the spatiotemporal patterns of IP3-evoked Ca2+ signals. The Journal of physiology. 2003;553:775–788. doi: 10.1113/jphysiol.2003.054247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Edmonds B, Reyes R, Schwaller B, Roberts WM. Calretinin modifies presynaptic calcium signaling in frog saccular hair cells. Nat Neurosci. 2000;3:786–790. doi: 10.1038/77687. [DOI] [PubMed] [Google Scholar]
- 53.Dargan SL, Schwaller B, Parker I. Spatiotemporal patterning of IP3-mediated Ca2+ signals in Xenopus oocytes by Ca2+-binding proteins. J Physiol. 2004;556:447–461. doi: 10.1113/jphysiol.2003.059204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schwaller B. The continuing disappearance of “pure” Ca2+ buffers. Cellular and Molecular Life Sciences. 2009;66:275–300. doi: 10.1007/s00018-008-8564-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kaplan JH, Ellis-Davies GC. Photolabile chelators for the rapid photorelease of divalent cations. Proc Natl Acad Sci USA. 1988;85:6571–6575. doi: 10.1073/pnas.85.17.6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ellis-Davies GC, Kaplan JH, Barsotti RJ. Laser photolysis of caged calcium: rates of calcium release by nitrophenyl-EGTA and DM-nitrophen. Biophys J. 1996;70:1006–1016. doi: 10.1016/S0006-3495(96)79644-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Escobar AL, Velez P, Kim AM, Cifuentes F, Fill M, Vergara JL. Kinetic properties of DM-nitrophen and calcium indicators: rapid transient response to flash photolysis. Pflugers Arch. 1997;434:615–631. doi: 10.1007/s004240050444. [DOI] [PubMed] [Google Scholar]
- 58.Escobar AL, Cifuentes F, Vergara JL. Detection of Ca(2+)-transients elicited by flash photolysis of DM-nitrophen with a fast calcium indicator. FEBS Lett. 1995;364:335–338. doi: 10.1016/0014-5793(95)00425-9. [DOI] [PubMed] [Google Scholar]
- 59.Berggard T, Miron S, Onnerfjord P, Thulin E, Akerfeldt KS, Enghild JJ, Akke M, Linse S. Calbindin D28k exhibits properties characteristic of a Ca2+ sensor. J Biol Chem. 2002;277:16662–16672. doi: 10.1074/jbc.M200415200. [DOI] [PubMed] [Google Scholar]
- 60.Celio MR. Guidebook to the Calcium Binding Proteins. Oxford University Press; 1996. [Google Scholar]
- 61.Ellis-Davies GC. Caged compounds: photorelease technology for control of cellular chemistry and physiology. Nature methods. 2007;4:619–628. doi: 10.1038/nmeth1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ellis-Davies GC. Development and application of caged calcium. Methods Enzymol. 2003;360:226–238. doi: 10.1016/s0076-6879(03)60112-6. [DOI] [PubMed] [Google Scholar]
- 63.Schwaller B, Durussel I, Jermann D, Herrmann B, Cox JA. Comparison of the Ca2+-binding properties of human recombinant calretinin-22k and calretinin. J Biol Chem. 1997;272:29663–29671. doi: 10.1074/jbc.272.47.29663. [DOI] [PubMed] [Google Scholar]
- 64.Kaplan JH, Forbush B, 3rd, Hoffman JF. Rapid photolytic release of adenosine 5′-triphosphate from a protected analogue: utilization by the Na:K pump of human red blood cell ghosts. Biochemistry. 1978;17:1929–1935. doi: 10.1021/bi00603a020. [DOI] [PubMed] [Google Scholar]
- 65.Shigeri Y, Tatsu Y, Yumoto N. Synthesis and application of caged peptides and proteins. Pharmacol Ther. 2001;91:85–92. doi: 10.1016/s0163-7258(01)00148-6. [DOI] [PubMed] [Google Scholar]
- 66.Kao JP. Caged molecules: principles and practical considerations. Curr Protoc Neurosci. 2006;Chapter 6(Unit 6):20. doi: 10.1002/0471142301.ns0620s37. [DOI] [PubMed] [Google Scholar]
- 67.Li C, Numata M, Takeuchi M, Shinkai S. A sensitive colorimetric and fluorescent probe based on a polythiophene derivative for the detection of ATP. Angew Chem Int Ed Engl. 2005;44:6371–6374. doi: 10.1002/anie.200501823. [DOI] [PubMed] [Google Scholar]
- 68.Ribou AC, Vigo J, Salmon JM. Lifetime of fluorescent pyrene butyric acid probe in single living cells for measurement of oxygen fluctuation. Photochem Photobiol. 2004;80:274–280. doi: 10.1562/2004-03-11-RA-109. [DOI] [PubMed] [Google Scholar]
- 69.Ouyang J, Hong H, Shen C, Zhao Y, Ouyang C, Dong L, Zhu J, Guo Z, Zeng K, Chen J, Zhang C, Zhang J. A novel fluorescent probe for the detection of nitric oxide in vitro and in vivo. Free Radic Biol Med. 2008;45:1426–1436. doi: 10.1016/j.freeradbiomed.2008.08.016. [DOI] [PubMed] [Google Scholar]
- 70.Lee SJ, Escobedo-Lozoya Y, Szatmari EM, Yasuda R. Activation of CaMKII in single dendritic spines during long-term potentiation. Nature. 2009;458:299–304. doi: 10.1038/nature07842. [DOI] [PMC free article] [PubMed] [Google Scholar]