Abstract
We have compared DNA methylation in normal colon mucosa between colon cancer patients and patients without cancer. We identified significant differences in methylation between the two groups at 114 – 874 genes. The majority of the differences are in pathways involved in the metabolism of carbohydrates, lipids and amino acids. We also compared transcript levels of genes in the insulin-signaling pathway. We found that the mucosa of cancer patients had significantly higher transcript levels of several hormones regulating glucose metabolism and significantly lower transcript levels of a glycolytic enzyme and a key regulator of glucose and lipid homeostasis. The se differences suggest that the normal colon mucosa of cancer patients metabolizes dietary components differently than the colon mucosa of controls. Because the differences identified are present in morphologically normal tissue, they may be diagnostic of colon cancer and/or prognostic of colon cancer susceptibility.
Keywords: DNA methylation, colon mucosa, metabolic differences, insulin signaling, epigenetic biomarkers
Introduction
One of the goals of human genome sequencing and whole genome association analyses is to identify genes involved in common human disease. Although there have been multiple successes (e.g., type 2 diabetes 1, 2, asthma 3, 4 and other common diseases5–7), there has been little translational impact of identifying “common disease genes” because the relative risk of developing disease for carriers of each risk allele is small (odds ratio averaging 1.18 for SNPs at 10 loci with very strong association in type 2 diabetes8, for example). Given the multifactorial nature of common disease, this fact is not surprising, but it begs the question of what additional information could be added to genetic risk data to increase the predictive power for any particular disease. In this vein, there is great interest in the potential for various measures of “epigenotype” to add predictive value to genetic risk data9, 10.
The value of epigenetic information in this venture is potentially three-fold. First, systemic epigenetic differences between individuals (i.e., those differences that result from stochastic, environmental or genetic factors that act very early in development) can help explain differences in gene expression between individuals of identical genotype at the affected locus. Second, systemic epigenetic differences that can be detected in an easily accessible tissue may serve as surrogate markers of gene activity in tissues that are inaccessible to analysis. Third, tissue-specific epigenetic differences between individuals may provide a mechanistic link between the genetic and environmental factors that contribute to disease risk.
Colon cancer accounts for more than 10% of all invasive cancer in the United States and more than 100,000 new cases are diagnosed annually11. It is the third most common type of cancer in both men and women and is the third leading cause of cancer-related death11. Only a small fraction (~5%) of all colon cancer is caused by highly penetrant inherited mutations 12 and only a minority of cases (up to 35%13) appear to be influenced by heritable factors that have not yet been identified. Moreover, there is compelling evidence linking environmental influences such as western diets and cigarette smoking with increased risk of colon cancer14–17. Colon cancer is the ideal disease for studying both epigenetic differences between individuals and the epigenetic changes caused by the environment. Furthermore, epigenetic alterations have been associated with both increased risk of disease 18–20 and tumor progression21, 22.
The genetic and epigenetic changes observed in colon tumors have been characterized in great detail by multiple laboratories23–29. These differences have largely been characterized in colon cancer patients, comparing colon tumors to adjacent normal colonic mucosa from the same patient. These studies lack true controls (i.e. patients without colon cancer) and do not reveal whether there is anything distinctive about the normal mucosa of colon cancer patients compared to the colonic mucosa of patients without cancer. We hypothesize that the normal colonic mucosa of cancer patients is, in fact, not “normal” but “epigenetically predisposed” to cancer because of the acquisition of multiple somatically heritable chromatin modifications, including differences in DNA methylation. The goal of our study was to identify DNA methylation differences that distinguish the “normal” colonic mucosa of cancer patients from colon mucosa of individuals who do not have cancer and to determine whether these differences reflect an environmental interaction associated with colon cancer. The design of our current study allows us to identify epigenetic changes that may represent “field defects” that are found in the normal mucosa of colon cancer patients that are unlikely to be a result of the aberrant cellular machinery of the tumor cells.
Methods
Tissue collection
The “normal” mucosa specimens from patients with colon cancer were collected from colon tissue removed in the operating room. Normal appearing colonic mucosa away from the tumor tissue was sharply removed and the samples were snap frozen prior to DNA and RNA isolation. Patients with a known history of FAP (familial adenomatous polyposis) or HNPCC (hereditary non-polyposis colon cancer) were excluded.
Normal colon mucosa control specimens were collected from patients undergoing screening colonoscopy. Each patient was interviewed prior to the procedure by one of the investigators (M.L.S. or B.P.S.). Patients who reported a personal or family history of colon cancer were excluded. Patients with a personal history of colon polyps or inflammatory bowel disease were also excluded. After providing informed consent, each patient underwent complete colonoscopy by a board-certified gastroenterologist. During that procedure, mucosal biopsies were obtained with a radial jaw large capacity biopsy forceps (Boston Scientific). Specimens were placed into RNALater RNA Stabilization Reagent (Ambion, USA), and stored at 4° C prior to DNA and RNA isolation.
DNA and RNA isolation
Tissue samples were rinsed with sterile saline and blotted dry prior to nucleic acid extraction. DNA was extracted using standard phenol-chloroform techniques. The isolated DNA was dissolved in 10mM TrisCl(pH 8.0). Samples were quantified by spectrophotometry and stored at −80°C until ready for use. RNA was isolated using TRIzol Reagent (Invitrogen Corporation, USA) according to the manufacturer’s instructions. The RNA samples were purified using the Clean All RNA/DNA Clean Up Kit (NorgenBiotekCorp., Ontario, Canada). The isolated RNA was dissolved in Milli-Q water, quantified by spectrophotometry, and stored at −80°C until ready for use.
Bisulfite conversion and methylation assay
The EZ DNA Methylation-Gold Kit™ (Zymo Research, USA) was used to convert unmethylated genomic DNA cytosine to uracil. Site-specific CpG methylation was analyzed in the converted DNA template (5μl at 50ng/μl) using the Infinium Assay (Illumina Inc., USA), the HumanMethylation27 BeadChip, and a BeadArray Reader according to the manufacturer’s instructions. The HumanMethylation27 BeadChip targets 27,578 CpG’s, the vast majority of which lie within the proximal promoter regions of transcription start sites of 14,475 consensus coding sequences in the NCBI database (Genome Build 36). Methylation data were analyzed on the GenomeStudio Data Analysis Software (Illumina Inc., USA) as well as SPSS version 16.0 (SPSS Inc., Chicago, IL).
Pyrosequencing validation of methylation assay
Primers were designed for the genes of interest using PyroMark Assay Design Software version 2.0 (Qiagen, USA). The PyroMark Gold Q96 Kit (Qiagen, USA) was used to test 500 ng of bisulfite converted DNA from samples and internal controls according to the manufacturers recommendations. Analysis was conducted using the PSQ 96 HS instrument and the PyroMark Q96 MD Software (Qiagen, USA).
Transcriptome profiling
RNA integrity was tested using the 2100 Bioanalyzer 600 Nano RNA Chip (Agilent Technologies, USA). The six RNA samples with the highest quality from cancer and non-cancer patients were pooled. All six of the RNA samples from non-cancer patients had RIN values in excess of 8. Two of the RNA samples from cancer patients had RIN values in excess of 8 but the remaining four samples had RIN values of 3.5, 4.0, 4.2 and 4.3. However, we tested whether the (Cthousekeeping gene−Ctgene of interest) was related to RIN value in the individual samples by comparing Ct values for CEBPA, SLC2A1 and GAPDH and demonstrating that neither Ct values nor relative rank of transcript level were related to RIN and therefore concluded that differences between groups could not be explained by RNA sample quality in these particular samples. In addition, we assayed transcript levels between cancer and control groups for nine of the genes with the largest difference in an independent sample of cancer and control individuals by Realtime RTPCR (see below). The Superscript III Reverse Transcriptase Protocol (Invitrogen Corporation, USA) was used to create cDNA. One microgram of cDNA from each group was added to a 96 well RT Profiler PCA Array System: Human Insulin Signaling Pathway array plate (SABiosciences, USA). Analysis was performed using the Excel based template provided by SABiosciences.
Quantitative real time RT-PCR validation of transcript levels
Gene-specific TaqMan probes (Applied Biosystems, USA) were used to quantify steady state mRNA levels of CEBPA, G6PC, IGFBP1, LEP, PTPRF, RETN, SERPINE1, SLC2A1 and INS in 21 additional samples of normal colon mucosa from cancer patients and 21 additional samples of normal colon mucosa from patients without cancer or polyps. GAPDH was used as the reference housekeeping gene. The cDNA and TaqMan mix were amplified under the following conditions: 50°C for 2 minutes, 95°C for 10 minutes, 45 cycles of 95°C for 15 seconds and 60°C for 60 seconds. A melting curve analysis of the PCR products was performed to verify their specificity and identity. Raw CT values were used to compare relative gene expression levels using the ΔΔCT method.
Results
DNA Methylation Profiling Identifies an Epigenetic Signature of Cancer in the Normal Colon Mucosa of Cancer Patients
Normal colon mucosa (see Methods) from 30 cancer patients and 18 controls was selected for quasi-genome-wide DNA methylation analysis. Approximately twice as many cancer patients as controls were selected to gain statistical power 30. All of the colon mucosa specimens used in our analysis were from the right side of the colon (proximal to the hepatic flexure, see Methods). There was no difference in the mean age of cancer patients and control patients (65.6±11.6 vs. 61.3±11.6, p=0.222) or in the distribution of sex between the two groups (Female: 55.6% of control vs. 56.7% of cancer, p=0.588).
DNA was extracted by procedures that are standard for human tissues (see Methods) and 500ng of each DNA sample was treated with sodium bisulfite and monitored for conversion using a commercially available assay (see Methods). Site-specific DNA methylation was assayed using “HumanMethylation27 BeadChip” arrays, which contain probes for 27,578 CpG sites in 14,495 genes (see Methods).
Signals significantly above background were detected for more than 27,561 CpGs in all 48 samples. We compared mean beta-values (“beta-value” is the fraction of a particular CpG site that is methylated, which may range from 0 to 1; raw beta values were background normalized to correct for any differences in signal intensity between arrays) at each CpG site between the 30 cancer patients and the 18 controls.
We used three different metrics to identify significant differences in site-specific mean methylation level between the normal colonic mucosa of cancer patients and the colon mucosa of controls: 1) a Bonferroni-corrected P-value of 1.8 × 10−6 (i.e., 0.05/27,578 to correct for the number of individual CpG sites being tested) identified 119 sites in 114 genes (Table 1); 2) a Benjamini-Hochberg false-discovery rate 31 of 0.05 identified 909 sites in 873 genes (all of the genes identified in the Bonferroni-corrected data were also identified in the Benjamini-Hochberg false discovery rate screen; the additional 759 genes are shown in Supplementary Table 1), and; 3) a requirement that any candidate gene must show a significant difference (P≤0.05) between cancer and control groups for at least three CpGs in each gene identified 299 sites in 65 genes (an average of 4.5 CpGs per gene, Table 2).
Table 1.
Genes in which mean colon mucosa methylation levels differ between cancer patients and controls at the Bonferroni-corrected P≤1.8 × 10−6. Gene names in bold have largest mean differences between groups and were used to select individuals for analysis of insulin signaling pathway transcript levels.
| SEPT4 | CD55 | FBXO6 | IL1B | NIP | SLC16A3 |
| ACAD11 | CDK9 | FLJ14346 | INPP5D | NT5E | SLC43A3 |
| AHNAK | CGB5 | FLJ20186 | IRF5 | PARVB | SLCO1C1 |
| ALAS1 | CGB8 | FLJ30294 | ISG20L2 | PDCD1 | SPRR2D |
| ALOXE3 | CGI-69 | FLJ39822 | ITLN1 | PDPK1 | SULT1C2 |
| ALPP | CHCHD1 | FLJ43855 | JAG2 | PHLDB2 | SUSD3 |
| AP1S1 | CHFR | FSD1NL | KCNQ1 | PTD004 | TBC1D5 |
| APOA1 | CHID1 | FXYD7 | KIAA1913 | RAB11FIP5 | TCF8 |
| AQP7 | COL13A1 | GABRR2 | KRTHB6 | RAD23A | TCN2 |
| ARFGAP1 | CORO6 | GATA2 | KSP37 | RAMP1 | TEKT3 |
| ARHGAP11A | CUGBP2 | GCNT1 | LTC4S | RASSF5 | TFR2 |
| ASAHL | DDX49 | GGTLA1 | MAPK10 | RPS3A | TMEM55B |
| ATP9A | DLK1 | GK2 | MAPK15 | RPUSD1 | TNFRSF4 |
| BCDIN3 | DUSP5 | GNRH2 | MGC7036 | RUFY3 | TSSK6 |
| BRF1 | ELK4 | GP1BB | MGC9712 | S100A3 | UNC13D |
| C12orf24 | ENPEP | GSTP1 | MUC5B | SEMA3B | USHBP1 |
| C1QC | EPHA1 | HIST1H2AJ | NDUFS2 | SEMA6B | VAV1 |
| CCL8 | EXOC6 | HIST2H4 | NEK6 | SERPING1 | ZC3H11A |
| CCT6A | FAM105A | HS747E2A | NGFR | SGSH | ZNF248 |
Table 2.
Genes in which mean colon mucosa methylation levels differ between cancer patients and controls at three or more CpGs (P< 0.05 at each CpG). Gene names in bold have largest mean differences between groups and were used to select individuals for analysis of insulin signaling pathway transcript levels.
| ABCB4 | CCND2 | FEN1 | KLK10 | PEG10 | SLC22A18 |
| ALX4 | CDH13 | GALR1 | LOC129285 | POLR2G | SMPD3 |
| ATP10A | CDKN2A | GATA4 | LOX | PPP1R9A | SNRPN |
| BCDIN3 | CHFR | GNAS | MAGEL2 | PSMB6 | SYK |
| BCL2 | CTSZ | GNMT | MEST | PTPRO | THRB |
| BIK | DAPK1 | GPX3 | MGMT | PYCARD | TNFRSF10C |
| BRAF | DIRAS3 | GRB10 | MLH1 | RAB32 | UBE3A |
| C12orf24 | DLX5 | H19 | MSX1 | RB1 | VHL |
| CALCA | DNAJC18 | IGF2 | NNAT | RUNX3 | WT1 |
| CASP8 | EDNRB | INS | OBFC2B | SEMA3B | ZNF512 |
| CCND1 | ERBB2 | KCNQ1 | OSBPL5 | SERPINB5 |
We have used the latter ad hoc but “common sense” approach to identifying candidate genes that were differentially methylated in children conceived through assisted reproduction32, 33, as well as individuals with diabetic nephropathy34 and have shown that many of the methylation differences so identified are also correlated with differences in mean transcript level between groups10, 32. We note that adopting the criterion of three or more differentially methylated CpGs distinguished the 65 candidate genes (Table 2) and 299 CpG sites from only 1,588 CpGs on the array that could fulfill the criterion of three or more CpGs per candidate gene. It is noteworthy that nearly 20% of the 1,588 CpGs that could have been different between cancer patients and controls were found to be significantly different because these 1,588 CpGs are concentrated in genes selected on the basis of perceived importance in cancer or development. Furthermore, of the 114 genes that fulfill the Bonferroni-correction requirement in Table 1, only five are represented on the array by three or more CpGs (BCDIN3, C12orf24, CHFR, KCNQ1, SEMA3B). It is noteworthy that even though selection in Table 1 is for a single CpG to be different at the Bonferroni-corrected P-value, all five genes exhibit significant differences at three or more CpGs, suggesting that the methylation differences between groups observed at single CpG sites in Table 1 are robust over greater distances. In fact, inspection of data on all of the CpGs interrogated in the 65 genes in Table 2 show numerous cases in which multiple CpGs, spread over hundreds to thousands of base pairs are similarly and significantly differently methylated between cancer mucosa and controls (Supplementary Table 4).
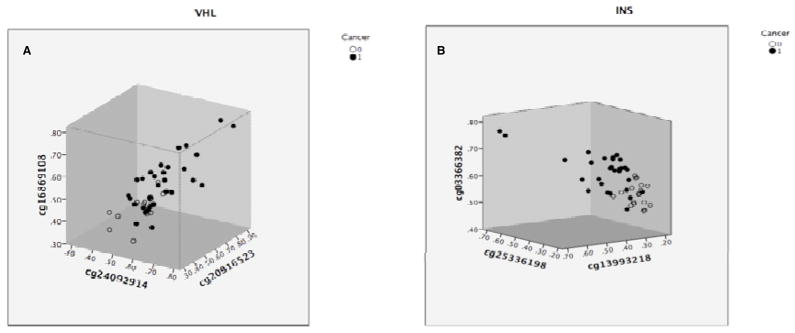
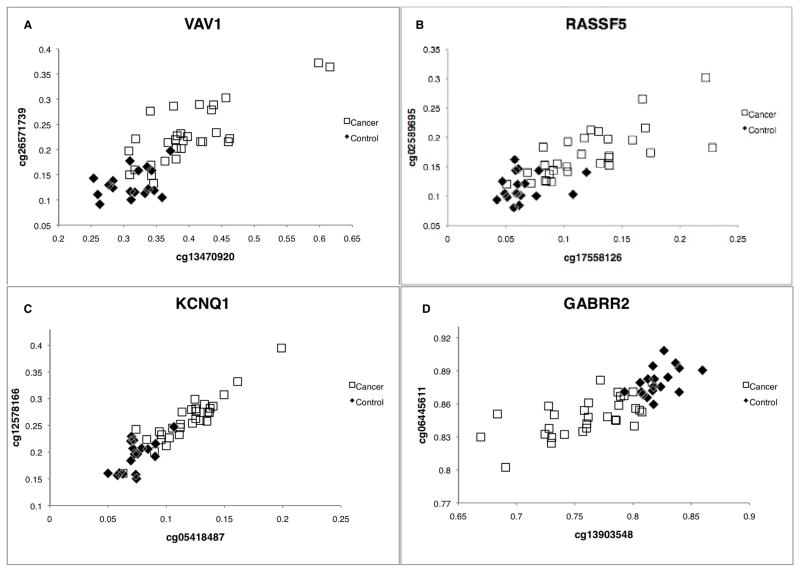
As a measure of the magnitude of the difference in methylation levels between the mucosa of cancer patients and the mucosa of controls and the discriminatory power of the approach, beta-values for individual patients are graphed at three CpGs for two of the most interesting genes in Table 2 (from the stand point of being cancer related and environment related), the tumor suppressor gene VHL and the gene encoding insulin (Figure 1). Similarly, individual beta-values at four of the genes which each have two CpGs that are significantly different in the Bonferroni-corrected gene list (Table 1), the oncogene VAV1, the oncogene RASSF5, the imprinted potassium channel gene KCNQ1 and the imprinted GABA receptor GABRR2, are shown in Figure 2. It should be noted that the strong correlation between methylation levels at different, but nearby (between 20 bp and 752 bp apart) CpGs that were also assayed on the array for five of the six genes shown in Figures 1 and 2 (the RASSF5 CpGs are 50 kb apart) suggests that the differences observed are representative of the actual level of methylation over the region (independently of external validation) and that the inter-individual differences observed are genuine. However, beta values for CpGs in four of the candidate genes (SLC16A3, VAV1 from Table 1 and INS, ZNF512 from Table 2) were validated independently by bisulfite pyrosequencing (see Methods and Supplementary Figure 1. Supplementary Figure 2 shows beta-values for individual patients at two CpGs in bisulfite pyrosequencing validated candidates SLC16A3 and ZNF512.).
Figure 1.
Methylation levels at three CpGs within the VHL gene (A) and the INS gene (B) in normal colon mucosa from patients with colon cancer (solid circles) and matched controls (open circles). CpGs were selected on the basis that mean methylation levels differed significantly between groups at P<0.05.
Figure 2.
Methylation levels at two CpGs that differ significantly between groups at P< 1.8 × 10−6 within the VAV1 gene (A) the RASSF5 gene (B) and the imprinted genes KCNQ1 (C) and GABRR2 (D). Methylation levels plotted for normal colon mucosa from patients with colon cancer (open squares) and matched controls (filled diamonds).
Functions of Genes that Are Differentially Methylated in Cancer Mucosa vs. Control Mucosa
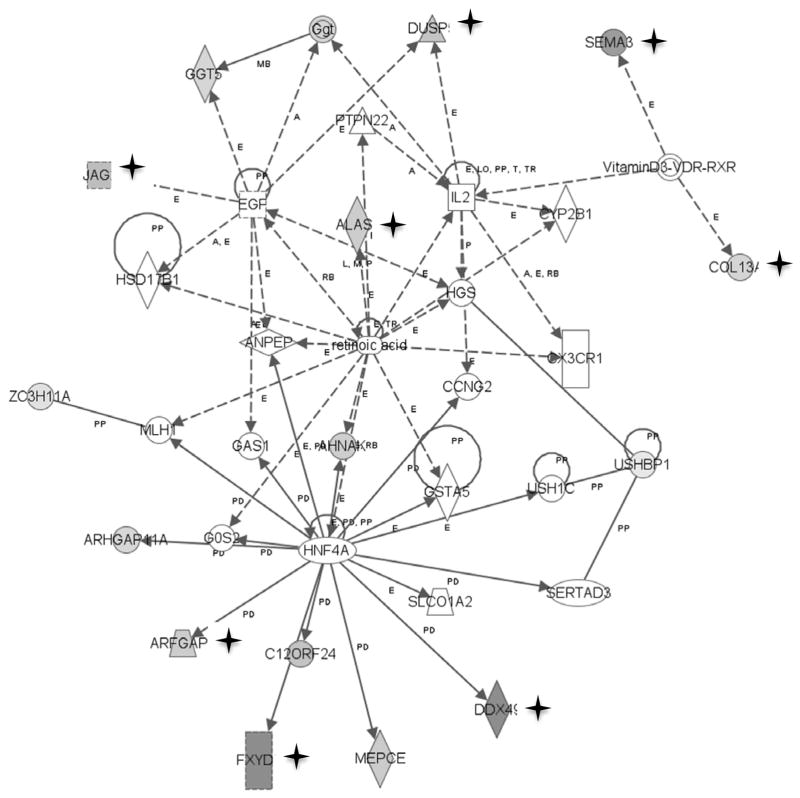
We performed Ingenuity Pathway Analysis (Ingenuity Systems, Inc., see Methods) with the candidate genes identified using each of the three metrics (Bonferroni correction, Benjamini-Hochberg false discovery rate, three or more CpGs different in the same candidate gene) to identify potential functional pathway differences between the normal colon mucosa of cancer patients and controls. Forty-nine of the 114 genes identified using the Bonferroni corrected P-value are found in pathways involved in carbohydrate and lipid metabolism and small molecule biochemistry (Supplementary Table 2). One of the top three networks (Figure 3) is involved in both lipid metabolism and cell growth and proliferation. Interestingly, this network also has a link to vitamin D metabolism and high levels of vitamin D are suspected to be preventive of colon cancer 35. As expected if the selection criteria are robust, the top genes identified using the Benjamini-Hochberg false discovery rate selection yield similar pathways (we used only the top 114 genes, by P-value, rather than all 873 genes, to determine whether the two metrics yielded comparable results; Supplementary Table 3). Of the top four networks obtained using each of the Bonferroni and Benjamini-Hochberg selected gene sets (Supplementary Tables 2 and 3) each of the eight networks has between 13 and 22 of the input gene list present and a network score of greater than 20 (probability that the molecules are unrelated by function <10−20) the top function of three networks is carbohydrate metabolism, the top function of two networks is lipid metabolism, three networks are involved in small molecule biochemistry, one network in amino acid metabolism and one of the eight networks (Figure 3) also has cell growth and proliferation as a top function.
Figure 3.
Ingenuity Pathway Analysis of genes that are differentially methylated (P<1.8 × 10−6) between normal mucosa of cancer patients and controls. Genes in shaded symbols denoted by a star are significantly more methylated in cancer patients; genes in shaded symbols without a star are significantly less methylated in cancer patients. The top functions of this network are lipid metabolism, small molecule biochemistry, cellular growth and proliferation.
Overlap with Genes Previously Identified as Differentially Methylated in Colon Cancer
Relatively few of the colon mucosa differences identified in cancer patients in our study are among the large number of genes that have been shown to be differentially methylated between colon tumors and matched colon mucosa of cancer patients by others23–26. In other words, the major methylation differences that we have identified between the mucosa of the cancer patients and the mucosa of the controls are not concentrated solely in the set of genes that become altered during tumor development. Only two (Table 3, column 1) of the 77 cancer-specifically methylated genes described by Widschwendter et al.23 are among the 114 genes identified (Table 1) as significantly different using the Bonferroni corrected P-value of 1.8 × 10−6. Only nine (Table 3, column 2) of Widschwendter et al.’s 77 genes are present among the 874 genes identified using a Benjamini-Hochberg false discovery rate of 0.05. Although the CpGs on the Illumina array used in our experiment are concentrated mainly in promoter regions and do not interrogate many of the “CpG Island Shores” described by Irizarry et al.26, only nine (Table 3, column 3) of the 114 genes in Table 1 are among the more than 2,700 identified as significantly differently methylated in the study of Irrizary et al.26. As expected for a candidate gene list that is enriched in cancer-associated genes (Table 2), nearly one-quarter (16 of 66) of the genes at which “normal” mucosa from colon cancer patients differs significantly at three or more CpGs are among those described by Irizarry et al.26 (Table 3, column 4).
Table 3.
Genes that differ in methylation between normal mucosa of cancer patients and normal mucosa of controls and also differ between normal mucosa of cancer patients and colon tumors.
| Bonferroni vs. Widschwendter et al. (2007) | Benjamini-Hochberg vs. Widschwendter et al. (2007) | Bonferroni vs. Irizarry et al. (2009) | Genes with three CpGs different vs. Irizarry et al. (2009) |
|---|---|---|---|
| CHFR | BCL2 | ALOXE3 | ALX4 |
| GSTP1 | CHFR | CUGBP2 | BCL2 |
| ESR1 | DLK1 | CALCA | |
| GATA4 | DUSP54 | DLX5 | |
| GSTP1 | GATA2 | EDNRB | |
| IGF2 | RASSF5 | GALR1 | |
| MSHR | SEMA6B | GATA4 | |
| RUNX3 | SUSD3 | GNAS | |
| SFRP4 | ZNF248 | IGF2 | |
| MEST | |||
| MGMT | |||
| MSX1 | |||
| PTPRO | |||
| RB1 | |||
| THRB | |||
| WT1 |
Transcript Levels of Insulin Signaling Pathway Genes Are Altered in Normal Colon Mucosa from Patients with Cancer
If the DNA methylation differences we observe in genes involved in carbohydrate, lipid and amino acid metabolism are indicative of altered metabolic function in the normal colon mucosa of cancer patients, we might expect to observe differences in the expression of key components of important metabolic pathways. We used a commercially available PCR array (see Methods) to compare transcript levels of 89 genes in the insulin signaling pathway in the normal mucosa of six cancer patients and six matched controls, pooled (Methods). The cancer patients were selected on the basis of the greatest DNA methylation differences, compared with controls, in a selection of 10 genes from the Bonferroni candidates (Table 1) and “three CpG” candidates (Table 2). The insulin signaling pathway was selected for analysis because it is central to the metabolism of carbohydrates and Ingenuity Pathway Analysis suggested that methylation of genes in the insulin signaling pathway is altered in the normal mucosa of cancer patients (Supplementary Tables 1 and 2).
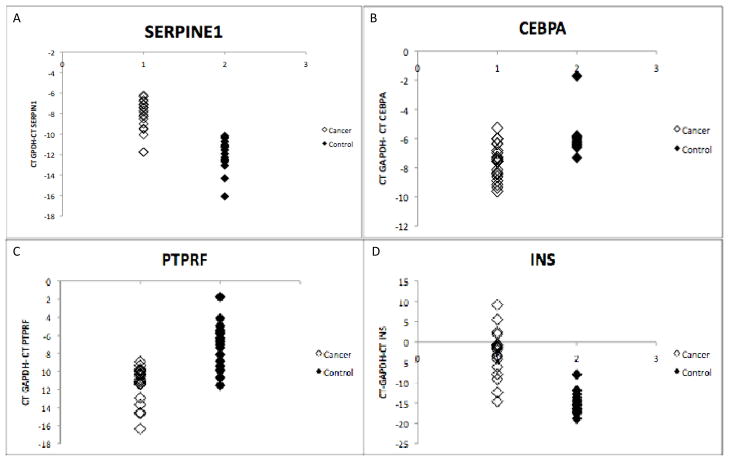
Of the 89 genes in the insulin-signaling pathway assayed for transcript level, the twenty genes showing the greatest difference between cancer and control (higher or lower) are shown in Table 4. Among the genes showing the greatest increase in transcript level in cancer mucosa are the hormones LEP and INS. Among the genes showing the greatest decrease in transcript level in cancer patients are a transcription factor (CEBPA) that is intimately involved in glucose homeostasis, a protein tyrosine phosphatase receptor (PTPRF) involved in metabolic regulation and an enzyme in the gluconeogenesis pathway (G6PC). Independent validation of the pooled-sample array result was attempted for nine of the genes in Table 4 (LEP, SERPINE1, CEBPA, SLC2A, G6PC, IGFBP1, INS, RETN and PTPRF) using additional individuals from cancer and control groups (none of the individuals in the validation were analyzed on the array, see Methods) by Realtime RTPCR. Six (LEP, G6PC, SERPIN1, CEBPA, PTPRF and INS) of the nine candidate genes tested confirmed significant differences between groups of cancer and control patients (Table 4) and individual transcript levels for four of these (two in which transcript levels are higher in cancer mucosa and two in which transcript levels are lower) are illustrated in Figure 4. The three genes for which significance was not reached also exhibited differences in the same direction as the pooled samples examined on the array (Table 4).
Table 4.
Top 20 genes whose transcript level was highest or lowest in normal mucosa of colon cancer patients compared with normal colon mucosa of patients without cancer, selected from the 89 genes profiled on the PCR array. Independent validation of nine candidates by Realtime RTPCR was performed for additional individuals from each group who were not analyzed on the PCR array and significant differences were confirmed for six genes. Significant P-values are shown in bold.
| Higher Expression | Lower Expression | ||||
|---|---|---|---|---|---|
| Symbol | Fold Change on Array | Fold Change in Validation, P-value | Symbol | Fold Change on Array | Fold Change in Validation, P-value |
| LEP | 242 | ~ cancer specific, 0.03 | PTPRF | 0.4 | 0.05, <10−5 |
| PRKCG | 26.3 | PPP1CA | 0.3 | ||
| IRS4 | 24.3 | PDPK1 | 0.3 | ||
| IGFBP1 | 23.7 | 2.4, 0.29 | PCK2 | 0.3 | |
| GCK | 16.0 | GSK3A | 0.3 | ||
| INS | 15.5 | >1,000, <10−6 | G6PC | 0.2 | 0.2, 0.01 |
| PRL | 13.0 | ACOX1 | 0.2 | ||
| TG | 12.2 | CEBPA | 0.2 | 0.35, <10−3 | |
| SERPINE1 | 8.8 | 12.3, <10−6 | HK2 | 0.2 | |
| RETN | 7.6 | 1.6, 0.34 | SLC2A1 | 0.1 | 0.77, 0.40 |
Figure 4.
Realtime RTPCR analysis of transcript level in normal colon mucosa of individual cancer and control patients for SERPINE1 (A; n=19 cancer, 19 control; P<10−6), CEBPA (B; n=21 cancer, 20 control; P<10−3), PTPRF (C; n=19 cancer, 20 control; P<10−5) and INS (D; n=18 cancer, 18 control; P<10−6).
These results suggest that the gene-specific DNA methylation differences we observe between the normal mucosa of cancer patients and the normal mucosa of controls result in differences in the ability of the two sources of normal mucosa to metabolize dietary components.
Discussion
Our findings indicate that there are major differences in DNA methylation between the normal mucosa of cancer patients and the normal mucosa of controls. The major targets of these differences are genes involved in metabolism of carbohydrates, lipids, amino acids and other small molecules. Our limited analysis of transcript levels in the insulin-signaling pathway corroborate that such differences result in quantitative differences in gene expression in important metabolic pathways. The methylation differences observed between the normal mucosa of cancer patients and the normal mucosa of controls are distinct from the differences found when the normal colon mucosa of cancer patients is compared with the colon tumors of the same patients. These differences suggest that the normal colon mucosa of cancer patients metabolizes dietary components differently than the colon mucosa of controls.
It is tempting to use the individual gene methylation differences observed to predict how they might affect transcription of each gene in each individual. Of the twenty genes profiled on the PCR array, only two exhibit significant between-group differences in methylation levels by the criteria considered in this study. Three CpGs in INS are significantly more methylated in cancer mucosa than control mucosa (Figure 1 and Table 2). Two of the CpGs are located within 250 bp 5′ to the transcription start site (but are not in a CpG island) and the third is in the first exon within 75 bp of the start site, however, INS is expressed at higher levels in cancer mucosa than control mucosa (Table 4 and Figure 4). On the other hand, a CpG in an island 5′ to the PDPK1 transcription start site is significantly more methylated in cancer mucosa than control mucosa (Table 1) and PDPK1 is expressed at lower levels in cancer mucosa (Table 4). Of the six genes for which we validated significant differences in transcript level (Table 4), two (LEP and SERPINE1) have two CpGs in CpG islands adjacent to the start site that differ in the expected direction and one (G6PC) is interrogated by only a single CpG (that is not in an island) but this CpG also differs in the expected direction. One CpG in PTPRF is within 500 bp of the transcription start, but not in an island, and is more methylated in cancer mucosa than controls and PTPRF is expressed at lower levels in cancer mucosa (Table 4 and Figure 4). Both of the CpGs interrogated in CEBPA are in a CpG island but one is within the single exon and the other is 3′ to the coding sequence. Both of these CpGs are less methylated in cancer mucosa than control mucosa but CEBPA is expressed at lower levels in cancer (Table 4 and Figure 4). We have, however, observed a positive correlation between DNA methylation and transcript level at this gene in a previous study32. Overall, we note that while approximately 50% of human genes exhibit an inverse correlation between transcription and DNA methylation36, inter-individual methylation differences, in cis, account for only a small fraction of inter-individual variance in transcript level (approximately 10–15%10, which is same fraction accounted for by genetic variation, in cis (reviewed in37) and cases of a positive correlation between methylation and transcript level also exist (e.g.32).
At this point, we cannot distinguish whether the epigenetic differences we observe are the result of pre-existing differences between control individuals and individuals who later go on to develop cancer, or are changes programmed by the tumor at distant sites in morphologically normal colon mucosa. In this regard, we note that at least some of the epigenetic differences we observe in the normal colon mucosa of cancer patients may be associated with “field cancerization” epigenetic events. For example, promoter methylation of the O6-methylguanine-DNA methyltransferase (MGMT) has been observed in a significant fraction of normal mucosa samples from cancer patients38 and we also observed MGMT methylation differences at multiple CpG sites in our experiment (Table 2). However, at least some epigenetic differences between cancer patients and controls are known to be pre-existing, such as “loss of imprinting” at IGF2/H19 18–20. Moreover, it does not, a priori, seem obvious why the predominant epigenetic pathways reprogrammed by colon tumors should be involved in metabolism of lipids and carbohydrates. On the other hand, there are epidemiological data that suggest a strong link between high fat diets and subsequent development of colon cancer 14–17, arguing that epigenetic reprogramming of lipid and carbohydrate pathways should occur before the development of cancer. If these metabolic differences do pre-exist and predispose individuals toward further genetic and epigenetic changes that may lead to cancer, the identification of the pathways involved could allow for novel dietary or pharmaceutical interventions in those patients at highest risk.
Although none of the eight examples of candidate gene methylation differences shown in Figures 1 and 2 and Supplementary Figure 2 completely discriminates the mucosa of cancer patients from the mucosa of controls, it is apparent that a collection of such markers (the number would depend on the degree of overlap of cancer and control distributions for each marker) would have very high diagnostic power, in aggregate, to distinguish colon mucosa of cancer patients from colon mucosa of controls and would fulfill the discriminatory demands of a clinical setting39. Furthermore, if a significant fraction of the differences we observe are systemic, similar to the constitutional “loss of imprinting” found at IGF2/H1918–20, then these markers may indicate a high probability that an individual will develop cancer and that this prediction could be made by assaying biomarker methylation levels in tissues such as peripheral blood or saliva. Even if all of the methylation differences observed are colon mucosa-specific and these differences accumulate over the lifetime of the individual, they can serve as sentinel markers at which differences may occur prior to the appearance of colon polyps. The potential addition of an objective biochemical measure of cancer risk to an invasive screening test that relies entirely on the un-aided eyes of the endoscopist to detect colon polyps would be an important diagnostic advance.
Supplementary Material
Acknowledgments
We thank Dr. John M. Daly and Dr. Daniel T. Dempsey for their ongoing support of our work, Dr. Benjamin Krevsky and the Department of Gastroenterology at Temple University Hospital, and Dr. Elin Sigurdson and Dr. Andrew Godwin from Fox Chase Cancer Center for their assistance with colon biopsies and tissue acquisition.
Grant Support
This work was funded in part by an NIH training grant (T32 CA 103652-05) and the Fels Institute for Cancer Research and Molecular Biology.
Footnotes
There are no potential conflicts of interest.
References
- 1.Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 2.Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet. 2006;38:320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 3.Denham S, Koppelman GH, Blakey J, Wjst M, Ferreira MA, Hall IP, et al. Meta-analysis of genome-wide linkage studies of asthma and related traits. Respir Res. 2008;9:38. doi: 10.1186/1465-9921-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dixon AL, Liang L, Moffatt MF, Chen W, Heath S, Wong KC, et al. A genome-wide association study of global gene expression. Nat Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 5.Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Libioulle C, Louis E, Hansoul S, Sandor C, Farnir F, Franchimont D, et al. Novel Crohn disease locus identified by genome-wide association maps to a gene desert on 5p13.1 and modulates expression of PTGER4. PLoS Genet. 2007;3:e58. doi: 10.1371/journal.pgen.0030058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang F, Sarginson J, Crombie C, Walker N, St Clair D, Shaw D. Genetic association between schizophrenia and the DISC1 gene in the Scottish population. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:155–159. doi: 10.1002/ajmg.b.30274. [DOI] [PubMed] [Google Scholar]
- 8.Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bjornsson HT, Sigurdsson MI, Fallin MD, Irizarry RA, Aspelund T, Cui H, et al. Intra-individual change over time in DNA methylation with familial clustering. JAMA. 2008;299:2877–2883. doi: 10.1001/jama.299.24.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turan N, Katari S, Coutifaris C, Sapienza C. Explaining inter-individual variability in phenotype: is epigenetics up to the challenge? Epigenetics. 2010;5:16–19. doi: 10.4161/epi.5.1.10557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.American Cancer Society. Colorectal Cancer Facts & Figures 2011–2013. Atlanta: American Cancer Society; 2011. [Google Scholar]
- 12.Jasperson KW, Tuohy TM, Neklason DW, Burt RW. Hereditary and familial colon cancer. Gastroenterology. 2010;138:2044–2058. doi: 10.1053/j.gastro.2010.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, et al. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 14.Willett WC. Diet and cancer. Oncologist. 2000;5:393–404. doi: 10.1634/theoncologist.5-5-393. [DOI] [PubMed] [Google Scholar]
- 15.Khan N, Afaq F, Mukhtar H. Lifestyle as risk factor for cancer: Evidence from human studies. Cancer Lett. 2010;293:133–143. doi: 10.1016/j.canlet.2009.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang PS, Chen TY, Giovannucci E. Cigarette smoking and colorectal cancer incidence and mortality: systematic review and meta-analysis. Int J Cancer. 2009;124:2406–2415. doi: 10.1002/ijc.24191. [DOI] [PubMed] [Google Scholar]
- 17.Limsui D, Vierkant RA, Tillmans LS, Wang AH, Weisenberger DJ, Laird PW, et al. Cigarette smoking and colorectal cancer risk by molecularly defined subtypes. J Natl Cancer Inst. 2010;102:1012–1022. doi: 10.1093/jnci/djq201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cui H, Cruz-Correa M, Giardiello FM, Hutcheon DF, Kafonek DR, Brandenburg S, et al. Loss of IGF2 imprinting: a potential marker of colorectal cancer risk. Science. 2003;299:1753–1755. doi: 10.1126/science.1080902. [DOI] [PubMed] [Google Scholar]
- 19.Cui H, Horon IL, Ohlsson R, Hamilton SR, Feinberg AP. Loss of imprinting in normal tissue of colorectal cancer patients with microsatellite instability. Nat Med. 1998;4:1276–1280. doi: 10.1038/3260. [DOI] [PubMed] [Google Scholar]
- 20.Nakagawa H, Chadwick RB, Peltomaki P, Plass C, Nakamura Y, de La Chapelle A. Loss of imprinting of the insulin-like growth factor II gene occurs by biallelic methylation in a core region of H19-associated CTCF-binding sites in colorectal cancer. Proc Natl Acad Sci U S A. 2001;98:591–596. doi: 10.1073/pnas.011528698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hiraoka S, Kato J, Horii J, Saito S, Harada K, Fujita H, et al. Methylation status of normal background mucosa is correlated with occurrence and development of neoplasia in the distal colon. Hum Pathol. 2010;41:38–47. doi: 10.1016/j.humpath.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 22.Kim MS, Louwagie J, Carvalho B, Terhaar Sive Droste JS, Park HL, Chae YK, et al. Promoter DNA methylation of oncostatin m receptor-beta as a novel diagnostic and therapeutic marker in colon cancer. PLoS One. 2009;4:e6555. doi: 10.1371/journal.pone.0006555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, et al. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 24.Hinoue T, Weisenberger DJ, Pan F, Campan M, Kim M, Young J, et al. Analysis of the association between CIMP and BRAF in colorectal cancer by DNA methylation profiling. PLoS One. 2009;4:e8357. doi: 10.1371/journal.pone.0008357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 26.Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang W, Press OA, Haiman CA, Yang DY, Gordon MA, Fazzone W, et al. Association of methylenetetrahydrofolate reductase gene polymorphisms and sex-specific survival in patients with metastatic colon cancer. J Clin Oncol. 2007;25:3726–3731. doi: 10.1200/JCO.2007.11.4710. [DOI] [PubMed] [Google Scholar]
- 28.Estécio MR, Gallegos J, Vallot C, Castoro RJ, Chung W, Maegawa S, et al. Genome architecture marked by retrotransposons modulates predisposition to DNA methylation in cancer. Genome Res. 2010;20:1369–1382. doi: 10.1101/gr.107318.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones S, Chen WD, Parmigiani G, Diehl F, Beerenwinkel N, Antal T, et al. Comparative lesion sequencing provides insights into tumor evolution. Proc Natl Acad Sci U S A. 2008;105:4283–4288. doi: 10.1073/pnas.0712345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andrieu N, Dondon MG, Goldstein AM. Increased power to detect gene-environment interaction using siblings controls. Ann Epidemiol. 2005;15:705–711. doi: 10.1016/j.annepidem.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 31.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J Royal Stat Soc. 1995;57:289–300. [Google Scholar]
- 32.Katari S, Turan N, Bibikova M, Erinle O, Chalian R, Foster M, et al. DNA methylation and gene expression differences in children conceived in vitro or in vivo. Hum Mol Genet. 2009;18:3769–3778. doi: 10.1093/hmg/ddp319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turan N, Katari S, Gerson LF, Chalian R, Foster MW, Gaughan JP, et al. Inter- and intra-individual variation in allele-specific DNA methylation and gene expression in children conceived using assisted reproductive technology. PLoS Genet. 2010;6:e1001033. doi: 10.1371/journal.pgen.1001033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sapienza C, Lee J, Powell J, Erinle O, Yafai F, Reichert J, et al. DNA methylation profiling identifies epigenetic differences between diabetes patients with ESRD and diabetes patients without nephropathy. Epigenetics. 2011;6:20–28. doi: 10.4161/epi.6.1.13362. [DOI] [PubMed] [Google Scholar]
- 35.Peterlik M, Grant WB, Cross HS. Calcium, vitamin D and cancer. Anticancer Res. 2009;29:3687–3698. [PubMed] [Google Scholar]
- 36.Rakyan VK, Down TA, Thorne NP, Flicek P, Kulesha E, Gräf S, et al. An integrated resource for genome-wide identification and analysis of human tissue-specific differentially methylated regions (tDMRs) Genome Res. 2008;18:1518–1529. doi: 10.1101/gr.077479.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheung VG, Spielman RS. Genetics of human gene expression: mapping DNA variants that influence gene expression. Nat Rev Genet. 2009;10:595–604. doi: 10.1038/nrg2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen L, Kondo Y, Rosner GL, Xiao L, Hernandez NS, Vilaythong J, et al. MGMT promoter methylation and field defect in sporadic colon cancer. J Natl Cancer Inst. 2005;97:1330–1338. doi: 10.1093/jnci/dji275. [DOI] [PubMed] [Google Scholar]
- 39.Moonesinghe R, Liu T, Khoury MJ. Evaluation of the discriminative accuracy of genomic profiling in the prediction of common complex diseases. Eur J Hum Genet. 2010;18:485–489. doi: 10.1038/ejhg.2009.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.