Abstract
The objective of this research was to detect bovine GDF10 gene polymorphism and analyze its association with body measurement traits (BMT) of animals sampled from 6 different Chinese indigenous cattle populations. The populations included Xuelong (Xl), Luxi (Lx), Qinchuan (Qc), Jiaxian red (Jx), Xianang (Xn) and Nanyang (Ny). Blood samples were taken from a total of 417 female animals stratified into age categories of 12–36 months. Polymerase chain reaction–single strand conformation polymorphism (PCR–SSCP) was employed to find out GDF10 single polymorphism nucleotide (SNPs) and explore their possible association with BMT. Sequence analysis of GDF10 gene revealed 3 SNPs in total: 1 in exon1 (G142A) and 2 in exon3 (A11471G, and T12495C). G142A and T12495C SNPs are both synonymous mutation. They showed 2 genotypes namely respectively (GG, GA) and (PP and PB). A11471G SNP is a missense mutation leading to the change of Alanine to Threonine amino acid. It showed three genotypes namely AA, BB and AB. Analysis of association of polymorphism with body measurement traits at the three locus showed that there were significant effects on BMT in Qc, Jx and Ny cattle population. These results suggest that the GDF10 gene might have potential effects on body measurement traits in the above mentioned cattle populations and could be used for marker-assisted selection.
Keywords: Chinese indigenous cattle, GDF10, Body measurement traits, SNPs
Introduction
The principal use of beef cattle is for meat production. In China, industries of meat production have developed rapidly and recently showed an increasingly positive trend. Chinese yellow cattle are known to be good beef cattle because of peculiar qualities, such as strong trunk, crude feed tolerance, high stress resistance, good adaptability, fine beef flavor and so on. However, some drawbacks still stand, such as underdeveloped hind hip and slow growth. It’s then necessary to select important functional genes of beef cattle through marker-assisted selection in order to solve the problems of low efficiency in Chinese beef cattle breeding, increase economic benefits and promote the development of domestic cattle industry towards high quality and efficiency.
Bone morphogenetic proteins (BMPs) are members of the transforming growth factor-beta (TGF-β) superfamily [1], which can influence a variety of differentiation processes, including adipogenesis, myogenesis, chondrogenesis, hematopolesis and epithelial cell differentiation [2]. In this superfamily, BMPs were originally identified on the basis of their ability to induce ectopic bone formation when implanted within soft tissue in vivo [3].
Growth differentiation factor 10 (GDF10) is the regulator of cell growth and differentiation in both embryonic and adult tissues. It is one of the most important members of BMPs. [4]. Previous studies in murine allowed GDF10 mRNA detection in both neonatal and adult bone samples with higher levels being detected in calvaria than in long bone [5, 6]. These results suggest that GDF10 gene may play multiple roles in regulating cell differentiation events and skeletal morphogenesis. In order to determine the biological function of GDF10, [7] carried out a detailed analysis of the expression pattern of GDF10. They found that during embryogenesis GDF10 is expressed prominently in developing skeletal structures both in the craniofacial region and in the vertebral column. Additionally, [8] suggested that GDF10 may have novel uncharacterized roles in transducing teratogenic signal in the limb. Furthermore they localized GDF10 in situ hybridization (ISH) to area of programmed cell death in the limb.
BMT are economically important in cattle breeding and their variations have been claimed to be associated with different factors among which genetic factors are predominant. Based on the role of GDF10 in bone morphogenetic as determined in mice and human, GDF10 could be an attractive candidate gene for BMT in bovine genetic improvement. Therefore, the objective of this study is to detect SNP of GDF10 gene in different Chinese indigenous cattle population and explore their possible association with BMT.
Materials and methods
Experimental animals
A total of 417 female animals stratified into age categories of 12 months to 36 months which comprises:
Xl cattle (n = 50, Dalian) blood samples collected from Dalian province;
Lx cattle (n = 62, Shandong) blood samples collected from Luxi cattle breeding farm, Heze City Shandong Province;
-
Qc cattle (n = 148, Shaanxi) blood samples collected from Shaanxi province
Yuanzhong farm and five animal husbandry and technology companies;
Jx cattle (n = 71, Henan) blood samples collected from Pingdingshan city, Henan province;
Xn cattle (n = 38, Henan) blood samples collected from Zhumadian Biyang County, Henan Province;
Ny cattle (n = 48, Henan Province) blood samples collected from cattle conservation farm, Nanyang, Henan Province were randomly selected from breeding population and used to analyze the GDF10 allelic frequencies.
DNA isolation
Animals blood samples were obtained from the 417 animals and treated with 2% heparin then stored at −80°C. DNA samples were extracted from blood samples according to standard procedures [9].
Body measurements traits
The traits measured, as described previously [10], included BL, BH, HH, RL, HW, CD and CC. In order to minimize systematic error, the same person was assigned to measure one of the seven traits in all animals.
Primers design
Based upon the bovine GDF10 gene (GenBank accession No, NC_007329.3), 10 pairs of PCR primers have been designed by Primer Premier 5.0 software to amplify a DNA sequence from GDF10 gene exon1, 2 and 3.
Polymerase chain reaction condition
Polymerase chain reaction (PCR) amplifications were performed in 20 μl reaction mixture containing 50 ng DNA template, 10 pM of each primer, 0.20 mM dNTP, 2.5 mM MgCl2 and 0.5 U Taq DNA polymerase (TaKaRa, Dalian, China).
Single strand conformation polymorphism (SSCP) and sequencing
PCR products were analyzed by single-strand conformation polymorphisms (SSCP). Aliquots of 4 μl of above PCR products were mixed with 8 μl of the denaturing solution (95% formamide, 25 mM EDTA, 0.025% xylene-cyanole and 0.025% bromophenol blue), incubated at 98°C for 10 min and then chilled in ice. Denatured DNA was loaded in 8% to 16% PAGE gel according to the size of the pair of primer used. The gel was run at constant voltage of 121 V for a period of time depending on the concentration of the gel. The gel was stained with 0.1% silver nitrate and visualized with 2% NaOH solution (containing 0.1% formaldehyde) according to [11]. To confirm the results based on PCR–SSCP technique, the PCR products from the mix DNA template were sequenced in both directions. DNAMAN (version 6.0) software was used to analyze the sequences.
Statistical analyses
Genotypic frequencies, allelic frequencies, Hardy–Weinberg equilibriums, gene homozygosity (Ho), gene heterozygosity (He), effective allele numbers (Ne) and polymorphism information content (PIC) were statistically analyzed according to the previous approaches of [12, 13]. The association between SNP marker genotypes of GDF10 gene and records of body measurement traits (BL, WH, HH, RL, HW, CD, CC) were analyzed by the software SPSS (version 17.0) according to the following statistical linear model:
where Yijk is the observation for the body measurement traits, μ is the overall mean for each trait, Gj is the genotype effect, Ai is the fixed effect of age and Eijk is the random error.
Results
PCR–SSCP analysis of the GDF10 gene
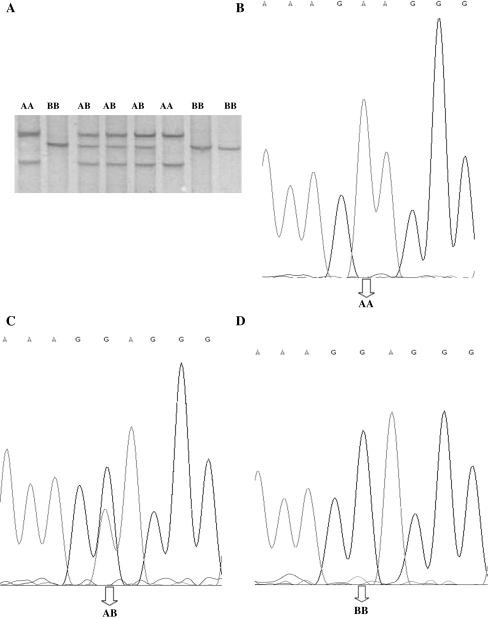
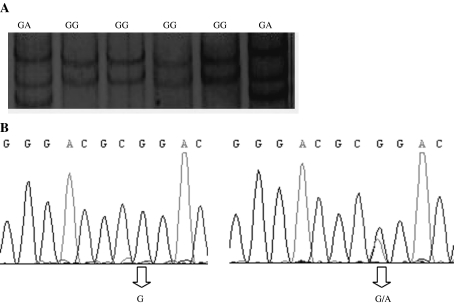
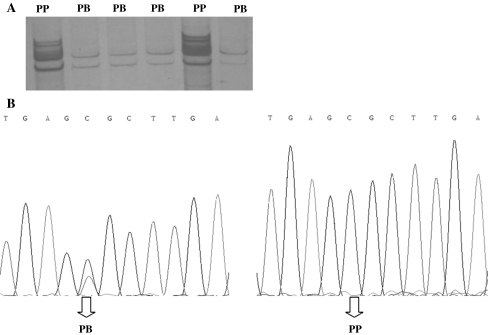
10 pairs of PCR primers have been designed to amplify a DNA sequence from GDF10 gene exon1, 2 and 3. Sequence analysis of GDF10 gene revealed 3 SNPs in total, 1 in exon1 (G142A) and 2 in exon3 (A11471G and T12495C). A11471G SNP showed 3 genotypes namely AA, AB and BB (Fig. 2a), whereas both G142A SNP and T12495C SNP showed two genotypes namely GG and GA (Fig. 1a) and PB and PP (Fig. 3a) respectively.
Fig. 2.
a SSCP electrophoresis pattern of GDF10 gene exon3 (11471 bp). b–d The sequencing map of the novel SNP of bovine GDF10 exon3 (11471 bp locus)
Fig. 1.
a Electrophoresis patterns of PCR–SSCP exon 1 of the bovine GDF10. b Sequencing map of G142A mutation in GDF10 gene exon1
Fig. 3.
a SSCP electrophoresis pattern of GDF10 gene exon3 (12495 bp locus). b The sequencing map of the novel SNP of bovine GDF10 exon3 (12495 bp locus)
Genetic analysis of bovine GDF10 gene exon1 and χ2 test
GDF10 exon1 analysis showed that there was a Ğ > A synonymous mutation at 142-bp position (Fig. 1b). Genetic diversity of the locus was calculated (Tables 1, 2); mutant allele A was present in five populations out of 6, and it was less frequent than the wild allele G in the population involved. GG genotype frequency ranged from 0.5968 (Lx) to 1.0000 (Ny). In all populations except Ny, GA genotype existed and its frequency ranged from 0.1200 (Xl) to 0.4032 (Lx) (Table 1). The result in Table 2 shows that Jx and Lx population’s heterozygosity, polymorphism information content and effective number of allele’s values were higher than that of the other populations. This meant that Jx and Lx polymorphism and genetic variation were the highest; according to the classification of PIC (low polymorphism if PIC value <0.25, medium polymorphism if 0.25< PIC value <0.5, and high polymorphism if PIC value >0.5), Lx and Jx showed medium polymorphism levels whereas Xl, Qc and Xn showed low polymorphism levels. The χ2 test showed that genotypic distributions in all the populations involved agreed with Hardy–Weinberg equilibrium (P > 0.05).
Table 1.
Genotype and allele frequencies at 142 bp locus of GDF10 gene exon1
| Population | Genotypic frequencies | Total | Allelic frequencies | χ2(HW*) | ||
|---|---|---|---|---|---|---|
| GG | GA | G | A | |||
| Xl | 0.8800 (44) | 0.1200 (06) | 50 | 0.9400 | 0.0600 | 0.5747 |
| Lx | 0.5968 (37) | 0.4032 (25) | 62 | 0.7984 | 0.2016 | 2.7554 |
| Qc | 0.7905 (117) | 0.2095 (31) | 148 | 0.8953 | 0.1047 | 1.0598 |
| Jx | 0.6056 (43) | 0.3944 (28) | 71 | 0.8028 | 0.1972 | 3.0844 |
| Xn | 0.7105 (27) | 0.2895 (11) | 38 | 0.8553 | 0.1447 | 0.2400 |
| Ny | 1.0000 (48) | 0.0000 (00) | 48 | 1.0000 | 0.0000 | 0 |
Table 2.
Population genetic indexes at the GDF10 exon 1 (142 bp) region
| Population | Gene homozygosity | Gene heterozygosity | Effective allele numbers | Polymorphic information content |
|---|---|---|---|---|
| (Ho) | (He) | (Ne) | (PIC) | |
| Xl | 0.8872 | 0.1128 | 1.1271 | 0.1064 |
| Lx | 0.6781 | 0.3219 | 1.4748 | 0.2701 |
| Qc | 0.8125 | 0.1875 | 1.2308 | 0.1699 |
| Jx | 0.6834 | 0.3166 | 1.4633 | 0.2665 |
| Xn | 0.7524 | 0.2476 | 1.3290 | 0.2169 |
| Ny | 1.0000 | 0.0000 | 1.0000 | 0.0000 |
Association of polymorphism with body measurement traits at GDF10 exon 1 (142 bp) locus
Analysis of association of polymorphism with BMT at GDF10 exon 1 (142 bp) locus showed that there were significant effects on HW, CD, (P < 0.01) and CC (P < 0.05) in the Jx cattle population (Table 3). GG genotype had the higher mean value for all the traits involved and might be the favorable genotype.
Table 3.
Least square means and standard errors of the body measurement traits obtained for the genotypes of the GDF10 gene polymorphism in Jx cattle population at locus G142A
| Genotype | Traits (cm, Means ± SE) | ||||||
|---|---|---|---|---|---|---|---|
| BL | BH | HH | RL | HW | CD | CC | |
| GG | 137.605 ± 3.568 | 125.843 ± 2.324 | 126.433 ± 2.699 | 43.910 ± 1.653 | 41.345 ± 1.537A | 64.900 ± 2.134A | 175.119 ± 2.948a |
| GA | 135.186 ± 3.463 | 124.090 ± 2.256 | 123.938 ± 2.619 | 42.271 ± 1.604 | 38.488 ± 1.492B | 60.443 ± 2.071B | 171.071 ± 2.861b |
a,bMeans with different superscripts were significantly different (P < 0.05)
A,BMeans with different superscripts were significantly different (P < 0.01)
Genetic analysis of GDF10 gene exon3 and effect of SNP on body measurement traits
Analysis of GDF10 gene exon3 showed that there were 2 mutations namely A11471B and P12495B. Genetic diversity of the loci was calculated.
Exon3 (11471 bp)
Genetic analysis
Analysis of GDF10 gene exon3 showed that there was A > B missense mutation leading to the change of Alanine to Threonine amino acid at 11471 bp position (Fig. 2b–d). Genetic diversity of the locus was calculated (Tables 4, 5). In the six populations involved, AB genotype frequency of Xl and Lx were the highest, Ny population had the lowest value; AA genotype frequency of Lx and Ny were the highest, while the lowest value was that of Xl. In all populations, except in Lx, the BB genotype existed (Table 4). Concerning the others, BB genotype frequency of Xn was the lowest; Qc population had the highest value.
Table 4.
Genotype and Allele frequencies at 11471 bp locus of GDF10 gene exon3
| Population | Genotypic frequencies | Total | Allelic frequencies | χ2 (HW*) | |||
|---|---|---|---|---|---|---|---|
| AA | AB | BB | A | B | |||
| Xl | 0.1800 (9) | 0.5000 (25) | 0.3200 (16) | 50 | 0.4300 | 0.5700 | 0.0200 |
| Lx | 0.5161 (32) | 0.4839 (30) | 0.0000 (0) | 62 | 0.7581 | 0.2419 | 4.9810 |
| Qc | 0.3446 (51) | 0.3311 (49) | 0.3243 (48) | 148 | 0.5101 | 0.4899 | 16.8647 |
| Jx | 0.3239 (23) | 0.3803 (27) | 0.2958 (21) | 71 | 0.5141 | 0.4859 | 4.0499 |
| Xn | 0.3684 (14) | 0.3421 (13) | 0.2895 (11) | 38 | 0.5395 | 0.4605 | 3.6872 |
| Ny | 0.5208 (25) | 0.1667 (8) | 0.3125 (15) | 48 | 0.6042 | 0.3958 | 20.3764 |
Table 5.
Population genetic indexes at the GDF10 exon3 (11471 bp) region
| Population | Gene homozygosity | Gene heterozygosity | Effective allele numbers | Polymorphic information content |
|---|---|---|---|---|
| (Ho) | (He) | (Ne) | (PIC) | |
| Xl | 0.5098 | 0.4902 | 1.9616 | 0.3701 |
| Lx | 0.6332 | 0.3668 | 1.5793 | 0.2995 |
| Qc | 0.5002 | 0.4998 | 1.9992 | 0.3749 |
| Jx | 0.5004 | 0.4996 | 1.9984 | 0.3748 |
| Xn | 0.5031 | 0.4969 | 1.9876 | 0.3734 |
| Ny | 0.5217 | 0.4783 | 1.9168 | 0.3639 |
Table 5 showed that among the six populations, gene homozygosity at this locus was classified in descending order: Lx > Ny > Xl > Xn > Jx > Qc.
The polymorphic information content values are in the following order: Qc > Jx > Xn > Xl > Ny > Lx. All population showed medium polymorphism. The χ2 test showed that, except Xl population, genotypic distributions in all the populations involved agreed with Hardy–Weinberg equilibrium (P > 0.05).
Association of polymorphism with body measurement traits at GDF10 exon3 (11471 bp) locus
Significant difference has been found between genotypes in Qc, Jx and Ny population.
In Qc population there was significant difference between BB and AA genotypes and also between BB and AB genotypes with BL, RL, HW, and CC. On the other hand BH was significantly different between AB and AA genotypes and also AB and BB genotype. However in Jx population significant difference appeared between BB and AB genotypes with BL, CD and CC. In the same way there was significant difference between BB and AB genotype and also between AA and AB genotypes with BH and HH.
In Ny population significant difference was found between AB and BB genotypes with BL, BH and HH.
In most of the cases AB genotype had the highest mean value and might be the beneficial genotype (Table 6).
Table 6.
Least square means and standard errors of the body measurement traits obtained for the genotypes of the GDF10 gene polymorphism in Qc, Jx and Ny cattle population in exon3 locus (11471 bp)
| P | G | Traits (cm, Means ± SE) | ||||||
|---|---|---|---|---|---|---|---|---|
| BL | BH | HH | RL | HW | CD | CC | ||
| QC | AA | 129.362 ± 2.137a | 117.586 ± 1.428b | 122.822 ± 1.582 | 42.983 ± 0.855a | 41.345 ± 1.038a | 63.724 ± 1.042 | 165.983 ± 2.957a |
| AB | 130.019 ± 2.215a | 119.056 ± 1.480a | 123.333 ± 1.640 | 42.648 ± 0.886a | 41.519 ± 1.076a | 63.537 ± 1.080 | 165.148 ± 3.064a | |
| BB | 122.609 ± 2.399b | 116.652 ± 1.603b | 123.435 ± 1.777 | 40.087 ± 0.960b | 38.048 ± 1.165b | 63.087 ± 1.170 | 155.870 ± 3.320b | |
| JX | AA | 129.407 ± 1.786AB | 118.926 ± 1.356a | 124.852 ± 1.428a | 42.074 ± 0.769 | 41.907 ± 0.906 | 64.963 ± 1.137ab | 167.593 ± 1.836ab |
| AB | 133.091 ± 1.978A | 118.364 ± 1.502a | 124.045 ± 1.582a | 40.378 ± 0.852 | 42.682 ± 1.004 | 68.182 ± 1.260a | 168.182 ± 2.034a | |
| BB | 124.813 ± 2.319B | 114.063 ± 1.761b | 119.750 ± 1.855b | 40.813 ± 1.000 | 41.188 ± 1.177 | 63.688 ± 1.478b | 161.750 ± 2.385b | |
| NY | AA | 129.379 ± 1.746ab | 118.483 ± 1.398ab | 124.828 ± 1.398ab | 41.690 ± 0.752 | 41.879 ± 0.881 | 65.034 ± 1.138 | 167.517 ± 1.819 |
| AB | 132.762 ± 2.051a | 118.286 ± 1.562a | 123.190 ± 1.643a | 42.095 ± 0.884 | 42.095 ± 1.035 | 67.048 ± 1.337 | 166.667 ± 2.138 | |
| BB | 125.267 ± 2.427b | 114.667 ± 1.848b | 120.600 ± 1.944b | 42.067 ± 1.046 | 42.067 ± 1.225 | 65.267 ± 1.582 | 163.667 ± 2.529 | |
P population, G genotype
a,bMeans with different superscripts were significantly different (P < 0.05)
A,BMeans with different superscripts were significantly different (P < 0.01)
Exon3 (12495 bp)
Genetic analysis
Analysis of GDF10 gene exon3 showed that there was P > B synonymous mutation at 12495 bp position (Fig. 3b). Among the 6 populations, PB genotype frequency of Xl was the highest while that of Qc was the lowest (Table 7). Gene homozygosity at this locus classified in descending order is: Qc > Ny > Jx > Xn > Lx > Xl. The polymorphic information content values are in the following order: Xl > Lx > Xn > Ny > Jx > Qx. Xl, Lx, Xn, Ny and Jx showed medium polymorphism while Qc showed low polymorphism (Table 8). The χ2 test showed that genotypic distributions in all the populations involved agreed with Hardy–Weinberg equilibrium (P > 0.05).
Table 7.
Genotype and allele frequencies at 12495 bp locus of GDF10 gene exon3
| Population | Genotypic frequencies | Total | Allelic frequencies | χ2 (HW*) | ||
|---|---|---|---|---|---|---|
| PP | PB | P | B | |||
| Xl | 0.3200 (16) | 0.6800 (34) | 50 | 0.6600 | 0.3400 | 13.2691 |
| Lx | 0.3871 (24) | 0.6129 (38) | 62 | 0.6935 | 0.3065 | 12.1049 |
| Qc | 0.7973 (118) | 0.2027 (30) | 148 | 0.8986 | 0.1014 | 0.9328 |
| Jx | 0.4930 (35) | 0.5070 (36) | 71 | 0.7465 | 0.2535 | 8.1894 |
| Xn | 0.4211 (16) | 0.5789 (22) | 38 | 0.7105 | 0.2895 | 6.3073 |
| Ny | 0.5625 (27) | 0.4375 (21) | 48 | 0.7813 | 0.2188 | 2.5374 |
Table 8.
Population genetic indexes at the GDF10 exon3 (12495 bp)
| Population | Gene homozygosity | Gene heterozygosity | Effective allele numbers | Polymorphic information content |
|---|---|---|---|---|
| (Ho) | (He) | (Ne) | (PIC) | |
| Xl | 0.5512 | 0.4488 | 1.8142 | 0.3481 |
| Lx | 0.5749 | 0.4251 | 1.7394 | 0.3347 |
| Qc | 0.8178 | 0.1822 | 1.2227 | 0.1656 |
| Jx | 0.6215 | 0.3785 | 1.6090 | 0.3069 |
| Xn | 0.5886 | 0.4114 | 1.6988 | 0.3267 |
| Ny | 0.6582 | 0.3418 | 1.5193 | 0.2834 |
Association of polymorphism with body measurement traits at GDF10 exon3 (12495 bp) locus
Analysis of association of polymorphism with BMT at GDF10 exon3 (12495 bp) locus showed that there were significant effects on BL, BH and HH, (P < 0.05) in Qc cattle population (Table 9). PB had the higher mean value for all the traits involved and might be the favorable genotype.
Table 9.
Least square means and standard errors of the body measurement traits obtained for the genotypes of the GDF10 gene polymorphism in QC cattle population at exon3 (12495 bp) locus
| Genotype | Traits (cm, Means ± SE) | ||||||
|---|---|---|---|---|---|---|---|
| BL | WH | HH | RL | HW | CD | CC | |
| PP | 123.860 ± 1.102b | 143.160 ± 2.107b | 128.270 ± 0.983b | 40.378 ± 1.604 | 41.188 ± 1.170 | 65.022 ± 1.123 | 167.17 ± 1.819 |
| PB | 128.030 ± 1.285a | 147.236 ± 2.256a | 135.263 ± 1.793a | 42.271 ± 0.432 | 41.022 ± 1.118 | 65.367 ± 1.582 | 166.071 ± 2.861 |
a,bMeans with different superscripts were significantly different (P < 0.05)
Discussion
Identifying the QTL, which is responsible for the manifestation of economically important traits, will facilitate Chinese indigenous cattle breeding programs. Molecular genetic information is integral to bring about genetic improvement of animal species in order to have significant positive developments. Candidate gene approach is a very vital and crucial method to investigate associations of gene polymorphisms with economically important traits in farm animals [14]. Many previous studies have examined growth [15], skeletal muscle [16], physiological [17, 18], reproductive [19–21], meat quality [22] traits using the candidate gene approach in cattle in order to explain their associations.
Transforming growth factor (TGF) super family is a group of multifunctional proteins that are regulators of cell growth and differentiation in both embryonic and adult tissues [5, 23].Growth differentiation factor 10 (GDF10), also known as BMP-3b and closely related to bone morphogenetic protein 3 (BMP3), is one of the important members of this super family protein. It is able to induce endochondral bone formation [1] and possibly plays multiple roles in regulating cell differentiation events, including those involved in skeletal morphogenesis [5]. GDF10 has been discovered in rat and human femur tissue [23, 24]. Moreover, GDF10 has been localized in situ hybridization (ISH) to area of programmed cell death in the limb [8]. All of the above findings provided evidence that GDF10 may influence BMT.
In this study the possible relationship between GDF10 polymorphism and the seven body measurement traits, BL, BH, HH, RL, HW, CD and CC, was evaluated using blood samples from 417 cattle belonging to six different cattle populations. Sequence analysis of GDF10 gene revealed 3 SNPs in total, 1 in exon1 (G142A) and 2 in exon3 (A11471G and T12495C). A11471G SNP showed 3 genotypes (AA, AB and BB) whereas both G142A SNP and T12495C showed two genotypes namely (GG, AG) and (PB, PP) respectively. Association between SNPs and BMT has been analyzed. It seems that G142A SNP is associated with HW, CD and CC only in Jx cattle population. A11471G SNP is associated with BL, BH, RL, HW and CC in Qc cattle population, with BL, BH, HH, CD and CC in Jx cattle population and with BL and HH in Ny cattle population. T12495C SNP is associated with BL, WH and HH only in Qc cattle population. The association of BMT with T12495C SNPs in Qc cattle population and that of G142A SNP in Jx cattle population only may be correlated to other direct or indirect factors specific to the respective cattle population. At 142 bp locus, GG genotype appeared to be the beneficial genotype; at 11471 and 12495 bp respectively AB and PB appeared to be beneficial genotype. G142A and T12495C SNPs are all synonymous mutations whereas A11471G SNP appears to be a missense mutation inducing the replacement of an Alanine amino-acid by a Threonine amino-acid at the current locus. This change of amino acid may have an impact in terms of function of the protein produced by GDF10 gene. The absence of GA genotype in Ny cattle population at 142 bp locus and of BB genotype at 11471 bp locus in Lx cattle population can be explained in two ways: either GA and BB genotypes does not exist in the respective population or the size of sample used was small; Our results provide evidence that the GDF10 gene probably has potential effects on body measurement traits in the above mentioned cattle populations.
In the TGF-β superfamily, growth differentiation factor 5 and BMP4 were found to be associated with body measurement traits [15, 25, 26]. Beside, BMP4 and GDF9 were found to be associated with litter size in goat [20, 27]. Based on the results obtained from the research it can be inferred that mutation in the coding regions of GDF10 gene has an effect on growth traits in Chinese indigenous cattle. Therefore, I recommend that further research will be necessary to use the SNPs for marker-assisted selection (MAS) in a larger population size. It is also necessary to investigate what is the impact of A12377G SNP on GDF10 protein function.
Acknowledgments
We would like to thank all the investigators, research assistants and laboratory technicians who have contributed to this study. We extend special thanks to Professor Zan, Dr Wang, Song Fubiao, Gaoli and Wang Hongcheng for their technical assistance. This work was funded by the National “863” Program of China (2006AA10Z1A1, 2008AA101010, 2010AA10Z101), Program for Changjiang Scholars and Innovative Research Team (IRT0940) and the “13115” Scientific and Technological Innovation Program of Shaanxi Province (2007ZDCY-01).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Wozney JM, Rosen V, Celeste AJ, Mitsock LM, Whitters MJ, Kriz RW, et al. Tissue growth and repair program, Genetics Institute, Inc., Cambridge, MA 02140. Science. 1988;242(4885):1528–1534. doi: 10.1126/science.3201241. [DOI] [PubMed] [Google Scholar]
- 2.Massegue J. The TGF-beta family of growth and differentiation factor. Cell. 1987;49:437–438. doi: 10.1016/0092-8674(87)90443-0. [DOI] [PubMed] [Google Scholar]
- 3.Urist MR. Bone: formation by autoinduction. Science. 1965;150:893–899. doi: 10.1126/science.150.3698.893. [DOI] [PubMed] [Google Scholar]
- 4.Hino J, Kangawa K, Matsuo H, Nohno T. Bone morphogenetic protein-3 family members and their biological functions. Front Biosci. 2004;9:1520–1529. doi: 10.2741/1355. [DOI] [PubMed] [Google Scholar]
- 5.Cunningham NS, Jenkins NA, Gilbert DJ, Copeland NG, Reddi, Lee SJ. Growth/differentiation factor-10: a new member of the transforming growth factor-beta super family related to bone morphogenetic protein-3. Growth Factors Res. 1995;12(2):99–109. doi: 10.3109/08977199509028956. [DOI] [PubMed] [Google Scholar]
- 6.Takao M, Hino J, Takeshita N, Konno Y, Nishizawa T, Matsuo H, et al. Identification of rat bone morphogenetic protein-3b (BMP-3b), a new member of BMP-3. Biochem Biophys Res Commun. 1996;219:656. doi: 10.1006/bbrc.1996.0289. [DOI] [PubMed] [Google Scholar]
- 7.Zhao R, Lawler AM, Lee SJ. Characterization of GDF-10 expression patterns and null mice. Dev Biol. 1999;212(1):68–79. doi: 10.1006/dbio.1999.9326. [DOI] [PubMed] [Google Scholar]
- 8.Eugene G, Barbara FH. Retinoic acid receptor gamma-induced misregulation of chondrogenesis in the murine limb bud in vitro. Toxicol Sci. 2008;106(1):223–232. doi: 10.1093/toxsci/kfn169. [DOI] [PubMed] [Google Scholar]
- 9.Sambrook J, Russell DW. Molecular cloning. A laboratory manual. 3. Beijing: Sci. Press; 2002. [Google Scholar]
- 10.Gilbert RP, Bailey DR, Shannon NH. Linear body measurements of cattle before and after 20 years of selection for postweaning gain when fed two different diets. J Anim Sci. 1993;71:1712–1720. doi: 10.2527/1993.7171712x. [DOI] [PubMed] [Google Scholar]
- 11.Zhang CL, Wang Y, Chen H, Lan XY. Enhance the efficiency of single-strand conformation polymorphism analysis by short polyacrylamide gel and modified silver staining. Anal Biochem. 2007;365:286–287. doi: 10.1016/j.ab.2007.03.023. [DOI] [PubMed] [Google Scholar]
- 12.Nei M, Roychoudhury AK. Sampling variances of heterozygosity and genetic distance. Genetics. 1974;76:379–390. doi: 10.1093/genetics/76.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nei M, Li WH. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rothschild MF, Soller M. Candidate gene analysis to detect genes controlling traits of economic importance in domestic livestock. Probe Newsl Agric Genomic. 1997;8:13–20. [Google Scholar]
- 15.Fang X, Xu H, Zhang C, Chen H, Hu X, Gao X, Gu C, Yue W. Polymorphism in BMP4 gene and its association with growth traits in goats. Mol Biol Rep. 2009;36(6):1339–1344. doi: 10.1007/s11033-008-9317-1. [DOI] [PubMed] [Google Scholar]
- 16.Schulz RA, Yutzey KE. Calcineurin signaling and NFATactivation in cardiovascular and skeletal muscle development. Dev Biol. 2004;266:1–16. doi: 10.1016/j.ydbio.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 17.Emma JG, Stuart ED, Jagadish P, Someshwar Z (2010) Functional gene analysis suggests different acetogen populations in the bovine rumen and tammar walla by forestomach. Appl Environ Microbiol 76(23):7785–7795 [DOI] [PMC free article] [PubMed]
- 18.Li M, Chen Q, Sun G, Shi X, Zhao Q, Zhang C, Zhou J, Qin N. Characterization and expression of bone morphogenetic protein 4 gene in postnatal pigs. Mol Biol Rep. 2010;37(5):2369–2377. doi: 10.1007/s11033-009-9743-8. [DOI] [PubMed] [Google Scholar]
- 19.Chu M, Jia L, Zhang Y, Jin M, Chen H, Fang L, Di R, Cao G, Feng T, Tang Q, Ma Y, Li K. Polymorphisms of coding region of BMPR-IB gene and their relationship with litter size in sheep. Mol Biol Rep. 2011;38(6):4071–4076. doi: 10.1007/s11033-010-0526-z. [DOI] [PubMed] [Google Scholar]
- 20.Chu MX, Lu L, Feng T, Di R, Cao GL, Wang PQ, Fang L, Ma YH, Li K (2010) Polymorphism of bone morphogenetic protein 4 gene and its relationship with litter size of Jining Grey goats. Mol Biol Rep 37(8):3921–3929 [DOI] [PubMed]
- 21.Chu MX, Zhao XH, Zhang YJ, Jin M, Wang JY, Di R, Cao GL, Feng T, Fang L, Ma YH, Li K. Polymorphisms of BMPR-IB gene and their relationship with litter size in goats. Mol Biol Rep. 2010;37(8):4033–4039. doi: 10.1007/s11033-010-0062-x. [DOI] [PubMed] [Google Scholar]
- 22.Shin SC, Chung ER. Association of SNP marker in the leptin gene with carcass and meat quality traits in Korean cattle. Asian-Australas. J Anim Sci. 2007;20:1–6. [Google Scholar]
- 23.Hino J, Matsuo H, Kangawa K. Bone morphogenetic protein-3b (BMP-3b) gene expression is correlated with differentiation in rat calvarial osteoblasts. Biochem Biophys Res Commun. 1999;256(2):419–424. doi: 10.1006/bbrc.1999.0341. [DOI] [PubMed] [Google Scholar]
- 24.Hino J, Takao M, Takeshita N, Konno Y, Nishizawa T, Matsuo H, et al. cDNA cloning and genomic structure of human bone morphogenetic protein-3B (BMP-3b) Biochem Biophys Res Commun. 1996;223(2):304–310. doi: 10.1006/bbrc.1996.0889. [DOI] [PubMed] [Google Scholar]
- 25.Liu YF, Zan LS, Li K, Zhao SP, Ya PX, Qing L, et al. A novel polymorphism of GDF5 gene and its association with body measurement traits in Bos taurus and Bos indicus breeds. Mol Biol Rep. 2010;37:429–434. doi: 10.1007/s11033-009-9604-5. [DOI] [PubMed] [Google Scholar]
- 26.Zhong X, Zan LS, Wang HB, Liu YF. Polymorphic CA microsatellites in the third exon of the bovine BMP4 gene. Genet Mol Res. 2010;9(2):868–874. doi: 10.4238/vol9-2gmr732. [DOI] [PubMed] [Google Scholar]
- 27.Feng T, Geng CX, Lang XZ, Chu MX, Cao GL, Di R, Fang L, Chen HQ, Liu XL, Li N (2010) Polymorphisms of caprine GDF9 gene and their association with litter size in Jining Grey goats. Mol Biol Rep [Epub ahead of print] [DOI] [PubMed]