Abstract
N363S and ER22/23EK polymorphisms observed within glucocorticoid receptor gene (NR3C1) may play an important role in the development of bronchial asthma. NR3C1 gene is associated with an altered sensitivity to GCs. The aim of the research project was to study the correlation between this NR3C1 gene polymorphisms and occurrence of asthma in the population of Polish asthmatics. Peripheral blood was obtained from 207 healthy volunteers and 221 asthma patients. Genotyping was carried out with PCR-RFLP method. In the groups of patients with uncontrolled moderate asthma and uncontrolled severe disease, the genotype distribution for the investigated polymorphisms was as follows: N363S-AA, AG, GG occurring with 0.881/0.073/0.046 frequency and ER22/23EK-GG, GA, AA occurring with 0.963/0.037/0.000 frequency. Chi-square analysis revealed a significantly different (P < 0.05) distribution between cases and controls for the N363S polymorphisms. The N363S polymorphism of NR3C1 gene is significantly associated with bronchial asthma, susceptibility to the development of moderate to severe form of uncontrolled bronchial asthma.
Keywords: Glucocorticoid receptor gene polymorphism, SNP (single nucleotide polymorphism), Inflammation, Resistance to steroids, NR3C1 gene
Introduction
Bronchial asthma is a hetereogeneous disease. The clinical presentation is a resultant of interactions between environmental and genetic factors [1]. Multifactor character of etiopathogenesis of bronchial asthma results in significant hereditary variability of phenotypic traits [2]. It should be emphasized that there is no single-gene model of asthma heredity, and the correlation of changes within a single gene with the clinical presentation is difficult to determine [3–5]. The role of genetic factors in etiopathogenesis of bronchial asthma is currently estimated to range from 36 to 94% [4, 6]. Studies of the human genome focus on ca. 100 loci, which may correlate with chronic inflammatory processes in the respiratory tract, atopy, bronchiolar hyperreactivity and production of inflammatory mediators [1]. Hereditary variability and structural changes within the single gene demonstrate relatively low risk associated with the pathogenesis of bronchial asthma (<5%) [6, 7]. Studies concerning the role of mutations and gene polymorphisms with respect to genotype-phenotype relations in bronchial asthma are conducted at present. Correlations between molecular changes in numerous gene loci and their consequences are analyzed at the proteomic level. It is noteworthy that the genetic spectrum involved in pathogenesis of the diseases includes a few chromosomes, and the loci correlating with the phenotype of asthma and atopy occupy about a half of the genome [1, 8]. The results of the search for gene or genes predisposing for the development of atopy or bronchial asthma obtained so far are not consistent, and research is still being conducted in this area (http://www.ginasthma.com) [9].
In view of contemporary research concerning the role of gene expression in bronchial asthma, the studies investigating the functional significance of glucocorticoid receptor gene (NR3C1 nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor)) in steroid-resistant or difficult-to-treat forms of the disease are particularly interesting [9–14]. Complete resistance to glucocorticoids is very rare—it has been estimated to affect less than 1:1000 bronchial asthma patients. Two types of resistance to glucocorticoids have been distinguished: type 1—resistance induced by cytokines and type 2—resistance associated with a mutation of polymorphisms of the NR3C1 gene [9, 10, 15–18]. It should be emphasized, however, that glucocorticoid resistance is observed in ca. 50% of patients with severe and difficult-to-treat asthma [9, 10, 15]. The area of genotype-phenotype correlations with respect to mutations and polymorphic variants of the NR3C1 gene extends far beyond the symptoms of bronchial asthma only [17, 19–24]. The NR3C1 gene is localized on chromosome 5q31-q32 and contains nine exons. As a result of alternative splicing of the NR3C1 gene, four isoforms of glucocorticoid receptor (GCRα, GCRβ, GCRδ, GCRγ) are formed, out of which only isoform α is active [25–29]. The glucocorticoid receptor consists of 777 amino acids and four domains can be distinguished in it: N-terminal AF1 (activation domain 1), DBD (DNA binding domain), NLS (hinge region), LBD (ligand-binding domain) i C-terminal AF2 (activation domain 2) [1, 9–11, 13, 15]. The activation domains determine correct biological mechanisms of receptor function as well as interactions with transcriptional factors and coactivator proteins [1, 9, 13, 15, 30].
It should be emphasized that the available reports, relatively scarce and sometimes inconsistent, concerning correlations between the particular polymorphic variants of the NR3C1 gene with resistance to glucocorticoids and development of severe/difficult-to-treat asthma, postulate further thorough research in this field.
The aim of the study
The aim of the presented study was to determine the correlations between polymorphism of N363S and ER22/23EK exon 2 of the NR3C1 gene and resistance to treatment, as well as with the severity profile and extent of control of the disease in asthma patients. Analysis of correlation between the prevalence of polymorphisms ER22/23EK and N363S of the NR3C1 gene, and increased sensitivity, or resistance to glucocorticoid therapy was undertaken in the study.
Materials and methods
The study was approved by the local ethics committee (Consent of Research Review Board at the Medical University of Lodz, Poland, No RNN/133/09/KE). At the commencement of the study, participants were invited to attend voluntarily. Before enrollment, written informed consent was obtained from every patient.
The study was conducted in a group of 221 patients with bronchial asthma. Asthma diagnosis was established according to GINA (The Global Initiative For Asthma) recommendations, based on clinical asthma symptoms and lung function test. The level of asthma severity and control was determined on the basis of GINA Report Guidelines. All the participants underwent subjective examinations (structuralized anamnesis including, besides the element of subjective examination, also the analysis of factors such as: gender, obesity, tobacco smoking, duration of bronchial asthma, allergy to house dust mites, animal fur, mould spores, cockroaches allergens, hypersensitivity to non-steroid anti-inflammatory drugs (NSAIDs), etc., in order to determine their role in the development of resistance to glucocorticoids, as well as to establish whether they are primary or secondary to genetic factors in character) and objective examinations. The results of pulmonary function tests and allergologic tests were obtained from the individual medical records of the patients. If there were no results of spirometry or allergologic tests available, such examinations were additionally performed during the recruitment visit. Subjects suffering from clinically significant exacerbations, using drugs which might induce resistance to glucocorticoids (such as rifampicin, phenobarbital, phenytoin, effedrine), subjects with signs of viral infections, either generalized, or affecting the respiratory tract, as well as subjects failing to comply with the doctor’s recommendations, were excluded from the patient group. The control arm included a group of 207 healthy adults, who met the following criteria: no history or symptoms of either bronchial asthma, or other pulmonary diseases, no history or symptoms of allergy, no history or symptoms of atopic dermatitis, no history or signs of hypersensitivity to aspirin, negative results of skin tests for 12 common allergens, no first-degree relatives with bronchial asthma or atopic disorders.
According to the standards developed by the Polish Society for Pulmonary Diseases, the best result of three spirometry maneuvers was selected for the analysis of obstructive disorders and disease severity. The correlation analysis took into consideration FEV1 (forced expiratory volume) expressed in liters, FEV1% (A/N%—percentage ratio of the measured to expected value) expressed as per cent of the expected value and FEV1% FVC index (FEV1 to FVC ratio—forced vital capacity) expressed as absolute numbers. Spirometry tests were conducted in the Outpatient Department according to ERS (European Respiratory Society)/ATS (American Thoracic Society) standards, and allergological tests according to the guidelines of EAACI (European Academy of Allergy and Clinical Immunology).
The level of asthma control was assessed with Asthma Control Test, which is clear and easy for patients. The Asthma Control Test (ACT™) consists of five questions. It was developed by Nathan et al. in cooperation with general practitioners and specialists in diagnostics and therapy of asthma. Bronchial asthma control level was calculated on the basis of the following patients’ results obtained in ACT: 00–19 points—no asthma control, 20–24 points—partially controlled asthma and 25 points—well-controlled asthma.
The study was carried out on 428 participants. The control group consisted of 138 women (66.7%) and 69 men (33.3%). The youngest participant was 18 years old and the oldest one 85 years old. The mean age in the control group was 46.06 years, the median 47.50, SD ± 16.05. The case group consisted of 140 women (63.3%) and 81 men (36.7%). The youngest participant was 19 years old and the oldest one 81 years old. The mean age in the case group was 48.94 years, the median 51.00, SD ± 15.78. Non-severe asthma was diagnosed in 147 patients (66.52%) whereas 74 (33.48%) had a severe form of the disease. Comprehensive comparison of the analyzed groups is presented in Tables 1 and 2. The whole group of patients participating in the study was stratified and subjected to statistical analysis.
Table 1.
Descriptive statistics of the analyzed parameters in controls and cases (general asthma population in the presented study)
| Parameter | Control group | Group of bronchial asthma patients |
|---|---|---|
| N | 207 | 221 |
| Gender | 138 women: 69 men | 140 women: 81 men |
| Gender | 66.7% women: 33.3% men | 63.3% women: 36.7% men |
| Mean age | 46.06 years | 48.94 years |
| SD for age | ±16.05 years | ±15.78 years |
| Min. age | 18 years | 19 years |
| Max. age | 85 years | 81 years |
| Kurtosis | −0.94 | −0.96 |
| Skewness | 0.10 | −0.19 |
| Median | 47.50 | 51.00 |
| FEV1 (L) mean ± SD | 2.95 ± 0.78 | 2.20 ± 0.84 |
| FEV1 (L) median | 2.85 | 2.17 |
| FEV1 (%) mean ± SD | 96.17 ± 12.69 | 73.13 ± 19.38 |
| FEV1 (%) median | 96 | 74 |
Table 2.
Descriptive statistics of the analyzed parameters in cases (general asthma population in the presented study)
| Parameter | Group of bronchial asthma patients (number) | Group of bronchial asthma patients (%) |
|---|---|---|
| Asthma | 221 | 100.00 |
| Non-severe asthma | 147 | 66.52 |
| Severe asthma | 74 | 33.48 |
| Chronic mild asthma | 34 | 15.38 |
| Chronic moderate asthma | 113 | 51.14 |
| Chronic severe steroid-dependent asthma | 66 | 29.86 |
| Chronic severe steroid-resistant asthma | 8 | 3.62 |
Venous blood samples were collected from the participants onto EDTAK3, DNA was obtained from peripheral blood leukocyte fraction. The genetic material was isolated using QIAamp DNA Blood Mini Kit (QIAGEN Inc.) according to the guidelines provided by the manufacturer. The investigated polymorphisms were analyzed using PCR-RFLP method.
Exponential amplification of DNA segments for N363S polymorphism was carried out using a forward primer (5′-CCA GTA ATG TAA CAC TGC CCC-3′) and a reverse primer (5′-TTC GAC CAG GGG AAG TTC AGA-3′) according to standard PCR protocol. Starter binding to complementary DNA matrix sites was conducted at 56°C. Amplified DNA sequences of 357 bp length were obtained. The material was incubated with FastDigest ® Tsp509I (TasI) restriction enzyme (Fermentas International INC) at 65°C for 1 h. DNA fragments with 135, 73, 70, 60 and 19 bp, were obtained as a set of representative, typical (wild type) alleles, whereas segments of 135, 92 (73 + 19 bp), 70 and 60 bp, constituted the set of polymorphic alleles. RFLP products were separated by electrophoresis on 40% polyacrylamide gel 1:20, stained with ethidine bromide and observed in UV light. Representative, typical homozygotes, as well as heterozygotes were sequenced and used as internal control.
Exponential amplification of DNA segments for ER22/23EK polymorphism was carried out using a forward primer (5′-TGC ATT CGG AGT TAA CTA AAA AG-3′) and a reverse primer (5′-ATC CCA GGT CAT TTC CCA TC-3′) according to standard PCR protocol. Starter binding to complementary DNA matrix sites was conducted at 56°C. Amplified DNA sequences of 446 bp length were obtained. The material was incubated with Mnl I restriction enzyme (Fermentas International INC) at 37°C for 20 h. DNA fragments with 149 and 163 bp (and shorter fragments containing 50, 49 and 35 base pairs), were obtained as a set of representative, typical (wild type) alleles, whereas segments of 163 and 184 bp (and shorter fragments containing 50 and 49 base pairs), constituted the set of polymorphic alleles. RFLP products were separated by electrophoresis on 40% polyacrylamide gel 1:60, stained with ethidine bromide and observed in UV light. Representative, typical homozygotes, as well as heterozygotes were sequenced and used as internal control.
The obtained results were subjected to descriptive statistical analysis with calculation of arithmetic means and standard deviations. The significance of differences between mean values was determined by means of χ2, with P < 0.05 adopted as the significance level.
Results and findings
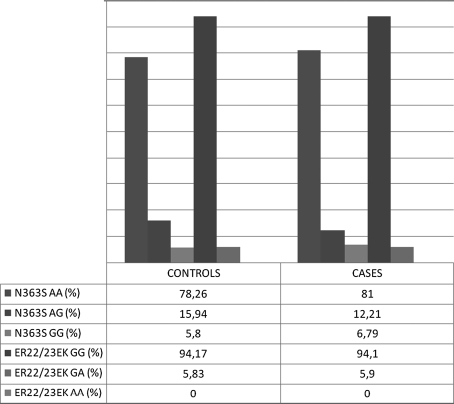
In healthy, non-atopic population, within the N363S gene, a polymorphism of N363S: AA, AG, GG occurring with 0.783/0.159/0.058 frequency, and ER22/23EK: GG, GA, AA occurring with 0.942/0.058/0.000, were identified. In cases (general asthma population analyzed in study) two polymorphisms were identified in exon 2 at 1,220 and 198 position in the NR3C1 gene: N363S (AA, AG, GG occurring with 0.810/0.122/0.068 frequency) and ER22/23EK (GG, GA, AA occurring with 0.941/0.059/0.000 frequency).
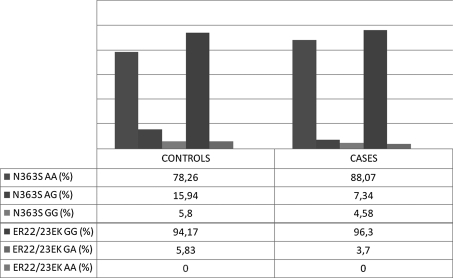
The group of patients with moderate and severe asthma stratified with respect to the extent of control of the disease according to ACT™ is described in Table 3. The distribution of genotypes for the studied polymorphisms among moderate uncontrolled and severe uncontrolled asthma patients was as follows: N363S-AA, AG, GG occurring with 0.881/0.073/0.046 frequency and ER22/23EK-GG, GA, AA occurring with 0.963/0.037/0.000 frequency.
Table 3.
Characteristics of the group of patients with moderate and severe asthma with respect to the extent of control of the disease according to ACT™
| Parameter | Moderate bronchial asthma | Severe bronchial asthma |
|---|---|---|
| N | 113 | 74 |
| ACT™ score 25 | 8 (7.08%) | 2 (2.70%) |
| ACT™ score 24–20 | 49 (43.36%) | 19 (25.68%) |
| ACT™ score ≤19 | 56 (49.56%) | 53 (71.62%) |
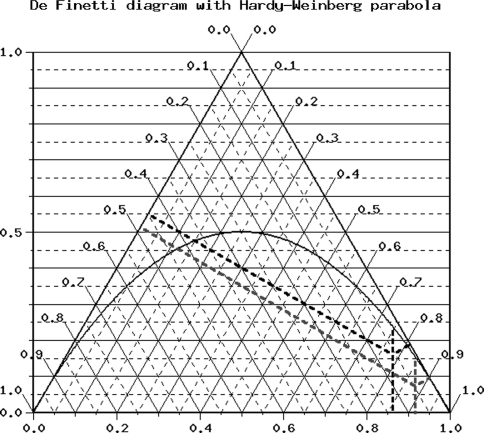
The study demonstrated numerous and strong correlations between the prevalence of moderate uncontrolled and severe uncontrolled asthma and the control group for polymorphism N363S of the NR3C1 gene. Such correlations were not observed for the second analyzed polymorphism-ER22/23EK. Distribution of N363S polymorphisms of NR3C1 gene was consistent with Hardy–Weinberg equilibrium. Distribution of ER22/23EK polymorphisms of NR3C1 gene was not consisted with Hardy–Weinberg equilibrium [31–33]. Table 4 and Fig. 1 illustrate de Finetti distributions of the investigated genotypes [31, 33–35]. Graph 1 presents the percentage frequencies of allelic variants of the investigated polymorphisms for the control group and the group of patients with moderate and severe asthma (ACT™ score ≤ 19). Graph 2 presents genotypes of the analyzed SNPs in the control population and the whole population of asthma patients.
Table 4.
Description of Hardy–Weinberg equilibrium for control and cases groups (moderate and severe asthma, ACT™ score ≤19)
| SNPs | Tests for deviation from Hardy–Weinberg equilibrium | |
|---|---|---|
| Controls | Cases | |
| N363S | nAA = 162 (153.92) | nAA = 96 (91.74) |
| nAG = 33 (49.15) | nAG = 8 (16.51) | |
| nGG = 12 (3.92) | nGG = 5 (0.74) | |
| f_a1 = 0.86 ± 0.020 | f_a1 = 0.92 ± 0.023 | |
| F = 0.32862 | F = 0.51556 | |
| P = 2.268e−06 (Pearson) | P = 7.343e−08 (Pearson) | |
| P = 0.000035 (Llr) | P = 0.000058 (Llr) | |
| P = 0.000035 (Exact) | P = 0.000084 (Exact) | |
| ER22/23EK | nGG = 194 (194.17) | nGG = 104 (104.04) |
| nGA = 12 (11.65) | nGA = 4 (3.93) | |
| nAA = 0 (0.17) | nAA = 0 (0.04) | |
| f_a1 = 0.97 ± 0.008 | f_a1 = 0.98 ± 0.009 | |
| F = −0.03000 | F = −0.01887 | |
| P = 0.666773 (Pearson) | P = 0.844547 (Pearson) | |
| P = 0.548476 (Llr) | P = 0.783524 (Llr) | |
| P = 1.000000 (Exact) | P = 1.000000 (Exact) | |
Tests for deviation from Hardy–Weinberg equilibrium were performed using free software provided by Institut für Humangenetik, Helmholtz Zentrum München, Deutsches Forschungszentrum für Gesundheit und Umwelt (Germany)
The tests for association are adapted from Sasieni (1997) [36]
f_a1 Frequency of allele 1 ± SD
F Inbreeding coefficient
P (Pearson) Pearson’s goodness-of-fit chi-square (degree of freedom = 1)
P (Llr) Log likelihood ratio chi-square (degree of freedom = 1)
P (Exact) Exact test
n number
Fig. 1.
Description of diagram: black line controls; blue line cases. De Finetti diagram with Hardy–Weinberg parabola was drawn using free software provided by Institut für Humangenetik, Helmholtz Zentrum München, Deutsches Forschungszentrum für Gesundheit und Umwelt (Germany). (Color figure online)
Graph 1.
Descriptive statistics of the analyzed allele of N363S and ER22/23EK polymorphisms of NR3C1 gene in controls and cases (moderate and severe asthma, ACT™ score ≤19)
Graph 2.
Descriptive statistics of the analyzed allele of N363S and ER22/23EK polymorphisms of NR3C1 gene in controls and cases (general asthma population in the presented study)
Significant statistical differences between the investigated genotypes were demonstrated (P < 0.05). Significantly more frequent occurrence of allele A in N363S polymorphism of the NR3C1 gene was observed in the population of patients with severe and moderate uncontrolled asthma in comparison with the control group (OR = 1.774, CI: 95% confidence interval, CI = 1.016–3.098, χ2 = 4.15, P = 0.041). In the investigated N363S polymorphism, allele G was associated with lower probability of development of the disease (OR = 0.564, CI: 95% confidence interval, CI = 0.323–0.984, χ2 = 4.15, P = 0.041). For genotype AG versus AA, a significant correlation was demonstrated between patients and the control group with respect to the investigated SNP (OR = 0.409, CI: 95% confidence interval, CI = 0.182–0.922, χ2 = 4.88, P = 0.027). Additionally, for the investigated N363S polymorphism a statistically significant correlation was demonstrated for the following allele variants: [AG + GG] versus [AA] (OR = 0.487, CI: 95% confidence interval, CI = 0.250–0.950, χ2 = 4.59, P = 0.032). The details of the analysis of genotypes for the investigated SNP N363S of the NR3C1 gene are presented in Table 5 [34–36].
Table 5.
Description of tests for association (CI 95% confidence interval) for control and cases groups (ACT™ score ≤19)
| SNP | Tests for association (CI 95% confidence interval) | ||||
|---|---|---|---|---|---|
| Allele freq. difference | Heterozygous | Homozygous | Allele positivity | Armitage’s trend test | |
| N363S | Risk allele G | ||||
| [G] vs. [A] | [AG] vs. [AA] | [GG] vs. [AA] | [AG + GG] vs. [AA] | Common odds ratio | |
| Odds ratio = 0.564 | Odds ratio = 0.409 | Odds ratio = 0.703 | Odds ratio = 0.487 | Odds ratio = 0.721 | |
| CI = [0.323–0.984] | CI = [0.182–0.922] | CI = [0.240–2.057] | CI = [0.250–0.950] | ||
| χ2 = 4.15 | χ2 = 4.88 | χ2 = 0.42 | χ2 = 4.59 | χ2 = 3.01 | |
| P = 0.04170 | P = 0.02710 | P = 0.51829 | P = 0.03220 | P = 0.08297 | |
| Risk allele A | |||||
| [A] vs. [G] | [AG] vs. [GG] | [AA] vs. [AG] | [AA + AG] vs. [GG] | Common odds ratio | |
| Odds ratio = 1.774 | Odds ratio = 0.582 | Odds ratio = 1.422 | Odds ratio = 1.280 | Odds ratio = 1.366 | |
| CI = [1.016–3.098] | CI = [0.159–2.131] | CI = [0.486–4.160] | CI = [0.439–3.732] | ||
| χ2 = 4.15 | χ2 = 0.68 | χ2 = 0.42 | χ2 = 0.21 | χ2 = 3.01 | |
| P = 0.04170 | P = 0.41054 | P = 0.51829 | P = 0.65044 | P = 0.08297 | |
Tests for association were performed using free software provided by Institut für Humangenetik, Helmholtz Zentrum München, Deutsches Forschungszentrum für Gesundheit und Umwelt (Germany)
The tests for association are adapted from Sasieni (1997) [36]
f_a1 Frequency of allele 1 ± SD
F Inbreeding coefficient
P (Pearson) Pearson’s goodness-of-fit chi-square (degree of freedom = 1)
P (Llr) Log likelihood ratio chi-square (degree of freedom = 1)
P (Exact) Exact test
n number
The frequencies of genotypes in the group of patients with partially controlled and controlled asthma in comparison with the control group for N363S polymorphism of the NR3C1 gene were also analyzed demonstrating no statistically significant correlation (P > 0.05).
The analysis of ER22/23EK polymorphism of the NR3C1 gene revealed the absence of any mutants (genotype GG). Only wild homozygotes and heterozygotes were observed. No correlations between the variants of ER22/23EK polymorphism in the study group and the control were found (P > 0.05). No correlations were demonstrated taking into consideration the phenotype of asthma severity and extent of control of the disease in the analyzed populations.
Discussion
In view of the presented results, the key role is played by the analysis of factors influencing the functional significance of the described polymorphisms. Structural changes within the NR3C1 gene lead to the formation of various alleles. Single nucleotide polymorphisms (SNPs), involving substitution of a nucleotide by another one, commonly occur in the. N363S and ER22/23EK SNPs affect the coding domains of the NR3C1 gene, significantly changing the sense of the nucleotide sequence and regulatory regions of the gene, and modifying its expression. The occurrence of SNPs results in variability of the analyzed gene. The observed polymorphic changes in the length of restriction fragments N363S and ER22/23EK (RFLP, Restriction Fragment Length Polymorphism) are located in the coding and regulatory regions of the NR3C1 gene, causing modifications of function or synthesis of glucocorticoid receptor. This undoubtedly affects modification of phenotypic characteristics of bronchial asthma, which may be associated with partial or complete resistance to anti-inflammatory medications. The loss of control of asthma symptoms by means of glucocorticoids is associated with chronic inflammatory condition of the respiratory tract, more frequent exacerbations, persistent bronchial obstruction and marked deterioration of the patients’ quality of life.
The discussed mutations, or polymorphism, significantly affect gene transcription and RNA splicing process. Recent studies by van Rossum and Lamberts demonstrated a correlation of ER22/23EK polymorphism with resistance to glucocorticoids [37]. It is located in exon 2 and consist of two nucleotide transitions connected with each other in codons 22 and 23 [37]. Transition in codon 22 causes no amino acid substitution, whereas transition in codon 23 causes substitution of arginine with lysine, which may lead to a change in the tertiary structure of the receptor domain responsible for activation of transcription [38]. Transition in codon 23 may also cause a change in the secondary structure of glucocorticoid receptor mRNA, and, consequently, initiation of translation from 1 or 27 methionine [39, 40]. Moreover, the aforementioned substitution in codon 23 may affect the stability of glucocorticoid receptor mRNA [39, 40]. Association of that polymorphism with resistance to glucocorticoids, as well as with more favorable metabolic profile in healthy population has also been described [39, 40].
One, or more of the discussed SNPs may occur in isolation or in combination, affecting significantly the NR3C1 gene transcription and RNA splicing process.
It is interesting that N363S and ER22/23EK SNPs consistently modify the N-terminal AF1 glucocorticoid receptor domain, thus affecting activation of the receptor function and the potential for interactions with transcriptional factors [1]. As a result of alternative splicing of mRNA, defective glucocorticoid receptor isoforms may be formed. Polymorphisms N363S and EK22/23EK of exon 2 of the NR3C1 gene are known to modify the protein structure within the AF1 domain, affecting the antigen properties of the receptor [1].
The in vivo studies conducted by Huizenga et al. demonstrated a correlation of N363S polymorphism with increased sensitivity to glucocorticoids [30]. In the Huizenga et al. performed study, it was observed that individuals carrying this N363S polymorphism (allele G) seemed healthy at clinical examination but had a higher sensitivity to exogenously administered glucocorticoids [32]. In the presented study, it was shown that appearance of allele A in the investigated N363S polymorphism correlated with statistical significance with increased risk of more severe phenotypic form of asthma and loss of the possibility to reduce its symptoms with anti-inflammatory drugs. Obviously, the severity of the disease and the possibility of its control are affected by multiple factors, both environmental and genetic. The analyzed population of patients (moderate and severe uncontrolled asthma) was selected from among all bronchial asthma patients. Significant statistical correlations were obtained only for the selected phenotypic subgroup of patients, and not for the whole studied population. No similar correlations were observed for the whole study group.
Thus, it can be concluded that N363S SNP may determine only the disease phenotypes from moderate to severe, but only uncontrolled forms (ACT™ score ≤19). Statistical analysis demonstrated significant correlations only for the patients with uncontrolled or insufficiently treated patients enrolled in the study, which may suggest decreased sensitivity of the glucocorticoid receptor. In fact, patients with asthma control score according to ACT™ ≤19 are regarded as treated not intensively enough. Such patients should be shifted to more intensive treatment, as they suffer from more severe asthma than indicated by the profile of the received medication. This phenomenon reflects underestimation of severe asthma. Incorrect qualification of patients to the particular categories may falsify the actual percentage distribution of bronchial asthma severity grades. The above may be important for interpretation of genetic investigations. The RFLP marker for N363S SNP seems therefore to have a clinical significance. It may be useful for identification of patients with severe asthma profile, requiring more intensive treatment from the very beginning.
Nevertheless, it should be stated that bronchial asthma is a disease determined by multiple factors and that asthmatics constitute a heterogeneous group of patients.
Conclusions
A correlation was demonstrated between N363S polymorphism of the NR3C1 gene and moderate/severe uncontrolled bronchial asthma phenotype. It was confirmed in a large group of subjects that N363S SNP may play an important role in the development of difficult-to-treat/treatment-resistant disease. No such correlation was demonstrated for the second investigated polymorphism.
As it is observed not only in bronchial asthma, but also in many other diseases (rheumatoid arthritis, leukemias, graft rejection and others), steroid resistance requires further thorough studies, addressing in particular the role of polymorphisms of the NR3C1 gene, which may become the target for novel therapies. Evaluation of its usefulness in this respect requires further analyses.
Acknowledgments
I am heartily thankful to Damian Tworek, MD, PhD, Oszajca Katarzyna PhD, Janiszewska Grażyna PhD, Wieteska Łukasz MA, Jarmakowska Katarzyna MD, Elgalal Anna MD, PhD, Antczak-Marczak Monika MD, PhD, Zielińska Ewa MD, PhD, Guzek Andrzej LIC, whose cooperation, guidance and support from the initial to the final level enabled me to develop an understanding of the subject. Lastly, I offer my regards and blessings to all of those who supported me in any respect during the completion of the project. The study was partially financed from the Grant No N N402 374638 of The Minister of Science and Higher Education of the Polish Republic received by Michał Panek, MD and Prof. Piotr Kuna. The study was partially financed from the grant received by Michał Panek, MD within the framework of a project supporting innovative doctoral studies, entitled: “Scholarships supporting innovative doctoral studies” based on funds provided by the EU European Social Fund and Polish state budget allocation for Integrated Operational Plan for Regional Development according to Lodz Region Innovation Strategy-RSI LORIS. Program title: ZPORR. Sector: 2.6 Regional Innovation Strategies and Transfer of Knowledge. Project title: “Scholarships supporting innovative doctoral studies”. Project number: Z/2.10/II/2.6/1/09. The project is financed in 75% from the European Social Fund and in 25% from the state budget.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
The contributions of the first two authors are equivalent.
Contributor Information
Michał Panek, Phone: +42-677-69-49, Phone: +42-677-69-39, Email: michalmp@poczta.onet.pl.
Tadeusz Pietras, Phone: +42-678-75-05, Phone: +42-677-66-99, FAX: +42-678-21-29, Email: cital200@wp.pl.
Paweł Górski, Phone: +42-678-75-05, Phone: +42-677-66-99, FAX: +42-678-21-29, Email: pawel.gorski@umed.lodz.pl.
Piotr Kuna, Phone: +42-677-69-49, Phone: +42-677-69-39, Email: pkuna@barlicki.internetdsl.pl.
Janusz Szemraj, Phone: +42-678-06-20, Phone: +42-678-24-65, Email: jszemraj@csk.am.lodz.pl.
References
- 1.Droszcz W. Astma. Warszawa: Wydawnictwo Lekarskie PZWL; 2007. [Google Scholar]
- 2.Duffy D, Martin N, Battistutta D, et al. Genetics of asthma and hay fever in Australian twins. Am Rev Respir Dis. 1990;142:1351–1358. doi: 10.1164/ajrccm/142.6_Pt_1.1351. [DOI] [PubMed] [Google Scholar]
- 3.Cooke R, Vander Veer A. Human sensitization. J Immunol. 1916;1:201–305. [Google Scholar]
- 4.National Asthma Education and Prevention Program Expert Panel Report 3: guidelines for the diagnosis and management of asthma full report (2007) National Heart, Lung, and Blood Institute. National Institute of Health, Bethesda
- 5.Ober C. Perspectives on the past decade of asthma genetics. J Allergy Clin Immunol. 2005;116:274–278. doi: 10.1016/j.jaci.2005.04.039. [DOI] [PubMed] [Google Scholar]
- 6.Anderson G. Endotyping asthma: new insights into key pathogenic mechanisms in a complex, heterogeneous disease. Lancet. 2008;372:1107–1119. doi: 10.1016/S0140-6736(08)61452-X. [DOI] [PubMed] [Google Scholar]
- 7.Kupczyk M. Epigenetics of bronchial asthma. Terapia. 2011;4:65–69. [Google Scholar]
- 8.Heinzmann A, Deichmann K. Genes for atopy and asthma. Curr Opin Allergy Clin Immunol. 2001;1:387–392. doi: 10.1097/01.all.0000011050.36626.ec. [DOI] [PubMed] [Google Scholar]
- 9.Barnes P. Glucocorticosteroids. W: Asthma: basic mechanisms and clinical management. San Diego: Academic Press; 1998. [Google Scholar]
- 10.Lane S, Lee T. Mechanisms of corticosteroid resistance in asthmatic patients. Int Arch Allergy Immunol. 1997;113:193–195. doi: 10.1159/000237544. [DOI] [PubMed] [Google Scholar]
- 11.Leung D, Bloom J. Update on glucocorticoid action and resistance. J Allergy Clin Immunol. 2003;111:3–22. doi: 10.1067/mai.2003.97. [DOI] [PubMed] [Google Scholar]
- 12.Bray P, Cotton R. Variations of the human glucocorticoid receptor gene (NR3C1): pathological and in vitro mutations and polymorphisms. Hum Mutat. 2003;21:557–568. doi: 10.1002/humu.10213. [DOI] [PubMed] [Google Scholar]
- 13.DeRijk R, Schaaf M, de Kloet E. Glucocorticoid receptor variants: clinical implications. J Steroid Biochem Mol Biol. 2002;81:103–122. doi: 10.1016/S0960-0760(02)00062-6. [DOI] [PubMed] [Google Scholar]
- 14.de Lange P, Koper J, Brinkmann A, et al. Natural variants of the beta-isoform of the human glucocorticoid receptor do not alter sensitivity to glucocorticoids. Mol Cell Endocrinol. 1999;153:163–168. doi: 10.1016/S0303-7207(99)00072-6. [DOI] [PubMed] [Google Scholar]
- 15.Panek M, Pietras T, Kupryś-Lipińska I. The analysis of the factors influencing the development of glucocorticoid resistance in the etiopathogenesis of severe bronchial asthma. Postępy Biochem. 2010;56:373–382. [PubMed] [Google Scholar]
- 16.de Lange P, Koper J, Huizinga N, et al. Differential hormone-dependent transcriptional activation and repression by naturally accuring human glucocorticoid receptor variants. Mol Endocrinol. 1997;11:1156–1164. doi: 10.1210/me.11.8.1156. [DOI] [PubMed] [Google Scholar]
- 17.Ruiz M, Lind U, Gafvels M, et al. Characterization of two novel mutations in the glucocorticoid receptor gene in patients with primary cortisol resistance. Clin Endo. 2001;55:363–371. doi: 10.1046/j.1365-2265.2001.01323.x. [DOI] [PubMed] [Google Scholar]
- 18.Huizenga A, de Lange P, Koper J, et al. Five patients with biochemical and/or clinical generalized glucocorticoid resistance without alterations in the glucocorticoid receptor gene. J Clin Endo Met. 2000;85:2076–2081. doi: 10.1210/jc.85.5.2076. [DOI] [PubMed] [Google Scholar]
- 19.Dobson M, Redfern C, Unwin N, et al. The N363S polymorphism of the glucocorticoid receptor: potential contribution to central obesity in men and lack of association with other risk factors for coronary heart disease and diabetes mellitus. J Clin Endo Met. 2001;86:2270–2274. doi: 10.1210/jc.86.5.2270. [DOI] [PubMed] [Google Scholar]
- 20.Huizenga N, Koper J, de Lange P, et al. A polymorphism in the glucocorticoid receptor gene may be associated with an increased sensitivity to glucocorticoids. J Clin Endo Met. 1998;83:144–151. doi: 10.1210/jc.83.1.144. [DOI] [PubMed] [Google Scholar]
- 21.Karl M, Lamberts S, Koper J, et al. Cushing′s disease preceded by generalized glucocorticoid resistance: clinical consequences of a novel, dominant-negative glucocorticoid receptor mutation. Proc Assoc Am Phys. 1996;108:296–307. [PubMed] [Google Scholar]
- 22.Muegge K, Willimans T, Kant W, et al. Interleukin-1 costimulatory activity on the interleukin-2 promoter via AP-1. Science. 1989;246:249–251. doi: 10.1126/science.2799385. [DOI] [PubMed] [Google Scholar]
- 23.Karl M, Lamberts S, Detera-Wadleigh S, et al. Familial glucocorticoid resistance caused by a splice site deletion in the human glucocorticoid receptor gene. J Clin Endo Met. 1993;76:683–689. doi: 10.1210/jc.76.3.683. [DOI] [PubMed] [Google Scholar]
- 24.Lamberts S, Koper J, Biemond P, et al. Cortisol receptor resistance: the variability of its clinical presentation and response to treatment. J Clin Endo Met. 1992;74:313–321. doi: 10.1210/jc.74.2.313. [DOI] [PubMed] [Google Scholar]
- 25.Murray R, Granner D, Mayes P, et al. Biochemia harpera. Wydawnictwo Lekarskie PZWL. 1998;38:493. [Google Scholar]
- 26.Encío I, Detera-Wadleigh S. The genomic structure of the human glucocorticoid receptor. J Biol Chem. 1991;266:7182–7188. [PubMed] [Google Scholar]
- 27.Murray R, Granner D, Mayes P, et al. Biochemia harpera . PZWL. 1998;39:524–525. [Google Scholar]
- 28.Rivers C, Levy A, Hancock J, et al. Insertion of an amino acid in the DNA-binding domain of the glucocorticoid receptor as a result of alternative splicing. J Clin Endocrinol Metab. 1999;84:4283–4286. doi: 10.1210/jc.84.11.4283. [DOI] [PubMed] [Google Scholar]
- 29.Adcock I, Lane S. Corticosteroid-insensitive asthma: molecular mechanisms. J Endocrinol. 2003;178:347–355. doi: 10.1677/joe.0.1780347. [DOI] [PubMed] [Google Scholar]
- 30.Huizenga N, Koper J, de Lange P, et al. A polymorphism in the glucocorticoid receptor gene may be associated with and increased sensitivity to glucocorticoids in vivo. J Clin Endocrinol Metab. 1998;83:144–151. doi: 10.1210/jc.83.1.144. [DOI] [PubMed] [Google Scholar]
- 31.Cannings C, Edwards A. Natural selection and the de Finetti diagram. Ann Hum Genet. 1968;31:421–428. doi: 10.1111/j.1469-1809.1968.tb00575.x. [DOI] [PubMed] [Google Scholar]
- 32.Elston R, Forthofer R. Testing for Hardy–Weinberg equilibrium in small samples. Biometrics. 1977;33:536–542. doi: 10.2307/2529370. [DOI] [Google Scholar]
- 33.Mendell N, Simon G. A general expression for the variance–covariance matrix of estimates of gene frequency: the effects of departures from Hardy–Weinberg equilibrium. Ann Hum Genet. 1984;48:283–286. doi: 10.1111/j.1469-1809.1984.tb01026.x. [DOI] [PubMed] [Google Scholar]
- 34.Smith C. A note on testing the Hardy–Weinberg law. Ann Hum Genet. 1970;33:377–383. doi: 10.1111/j.1469-1809.1970.tb01664.x. [DOI] [Google Scholar]
- 35.Weir B. Statitical data analysis II: methods for discrete population genetic data. Massachusetts: Sinauer Associates; 1996. [Google Scholar]
- 36.Sasieni P. From genotypes to genes: doubling the sample size. Biometrics. 1997;53:1253–1261. doi: 10.2307/2533494. [DOI] [PubMed] [Google Scholar]
- 37.van Rossum E, Lamberts S. Polymorphisms in the glucocorticoid receptor gene and their associations with metabolic parameters and body composition. Recent Prog Horm Res. 2004;59:333–357. doi: 10.1210/rp.59.1.333. [DOI] [PubMed] [Google Scholar]
- 38.de Lange P, Koper J, Huienga N, et al. Differential hormone-dependent transcriptional activation and repression by naturally occurring human glucocorticoid receptor variants. Mol Endocrinol. 1997;11:156–1164. doi: 10.1210/mend.11.8.9949. [DOI] [PubMed] [Google Scholar]
- 39.van Rossum E, Voorhoeve P, Te Velde S, et al. The ER22/23EK polymorphism in the glucocorticoid receptor gene is associated with a benefi cial body composition and muscle strength in young adults. J Clin Endocrinol Metab. 2004;89:4004–4009. doi: 10.1210/jc.2003-031422. [DOI] [PubMed] [Google Scholar]
- 40.Bulas M, Trelińska J, Stolarska M, et al. Association of steroid receptor gene (NR3C1) polymorphism with clinical course of lymphoproliferative disorders in children—preliminary results. Onkol Pol. 2008;2:77–81. [Google Scholar]