“Say what you see”.
(The Geographical History of America or the relation of Human Nature to the Human Mind, Gertrude Stein, 1936)
Laxus oneistus’ identity tag
L. oneistus, named from the Greek oneistos meaning “most useful”, is a marine nematode that occurs in subtidal coarse sand from below the waterline. So far, it has only be collected from the sediment of the barrier reef surrounding the island of Carrie Bow Cay, in Belize (Ott et al. 1995). When extracted from the sediment, it tends to curl around its body or that of other conspecifics, due to its thigmotactic instinct, leaving only its anterior-most part protruding from such tight aggregations (Fig. 1a). 18S rRNA-gene based phylogeny placed L. oneistus into a group of closely related genera classified as the Stilbonematinae subfamily (Ott et al. 2004a, b). The group is monophyletic and forms a distinct clade within the Desmodoridae (Kampfer et al. 1998; Bayer et al. 2009). All the known stilbonematids establish ectosymbioses with thiotrophic Gammaproteobacteria (see below). A single layer of rod-shaped bacteria, in the case of L. oneistus, covers the cuticle of the whole nematode, except for an anterior region, which in males can be up to 1/10 of their body length (ca. 1 mm). For simplicity, we refer to the ectosymbiont-free region of the L. oneistus body as the anterior (A) region and to the ectosymbiont-associated region as the posterior (P) region. Notably, the cuticle thins at the bacterial-coat onset (Urbancik et al. 1996) (Fig. 2). Besides the bacterial coat, two additional morphological characters unify all stilbonematids: a weak pharynx, glandular rather than muscular (Hoschitz et al. 2001), and unique epidermal organs known as glandular sense organs (GSOs; Fig. 1c, d and g) (Bauer-Nebelsick et al. 1995). Hundreds of GSOs underlie the worm cuticle throughout the A-P axis. They are separated from the cuticle only by an epidermal layer, which is thin and does not possess a basal lamina. Two gland cells (A and B) and a sensory neuron are the basic components of the GSO (Bauer-Nebelsick et al. 1995). This bears a central, pear-shaped canal (Fig. 1c, e and g) where secretory products can accumulate. The canal crosses the epidermis and cuticle and terminates in the pore of a bristle-like structure called the seta (Fig. 1b e and g). Therefore, a continuum exists between the GSO canal and the nematode surface. The P GSOs are crucial to the L. oneistus ectosymbiosis because they express and secrete the Ca2+-dependent sugar-binding protein Mermaid. This C-Type Lectin Domain-containing protein (CTLD) is mannose-, galactose-, and N-acetylglucosamine-specific (Bulgheresi et al. 2006; Zhang et al. 2006; Nabatov et al. 2008). Moreover, its carbohydrate recognition domain (CRD) is structurally and functionally similar to the CRD of the human dendritic cell-specific immunoreceptor DC-SIGN (Bulgheresi et al. 2006; Zhang et al. 2006; Nabatov et al. 2008; Mittal et al. 2009). The surface of the A cuticle is both free of bacteria and Mermaid lectin. Adding recombinant Mermaid causes L. oneistus ectosymbiont to aggregate or to detach from the worm. This suggests an involvement of this CTLD in both symbiont-symbiont and host-symbiont attachment. Different recombinant Mermaid isoforms vary in their affinity for particular stilbonematid symbionts. In particular, DDA-type Mermaids—which possess D, D and A at position 105, 108 and 109, respectively—appear to be the most expressed isoforms in L. oneistus. DDA-type Mermaids show a higher affinity for the ectosymbiont of L. oneistus than for that coating the co-occuring stilbonematid Stilbonema majum. On the other hand, GDA-type Mermaids are expressed by S. majum but not by L. oneistus and agglutinate the S. majum ectosymbiont more efficiently than that of L. oneistus. These data suggest that the expression of different Mermaid isoforms may mediate the attachment of a specific ectosymbiont to a given stilbonematid species.
Fig. 1.
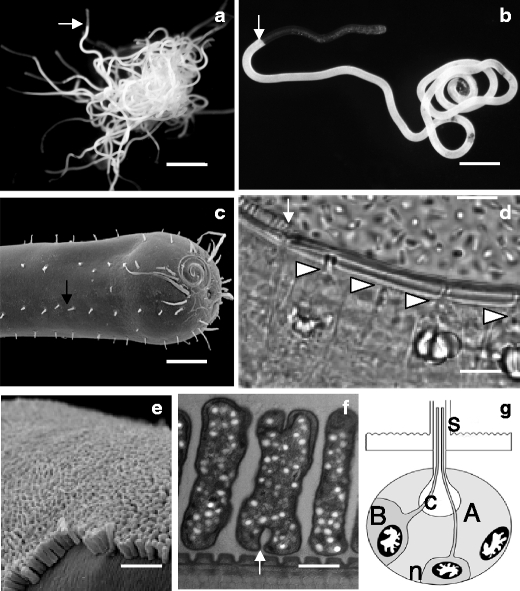
The stilbonematid nematode L. oneistus. a Light microscope image of an aggregation of approximately 50 living individuals; white arrow points to the beginning of the bacterial coat in one L. oneistus individual; scale bar is 2 mm. b Light microscope image of a single nematode; white arrow points to the beginning of the bacterial coat; scale bar is 150 μm. c Scanning electron microscope image of the anterior cuticle from which numerous setae protrude, black arrow points to one of them; scale bar is 20 μm. d Light microscope image of glandular sense organs (GSOs); the arrow points to the beginning of the bacterial coat, the arrowheads point to the canals of four GSOs; scale bar is 10 μm. e Scanning electron microscope image of the bacterial coat; scale bar is 3 μm. f Transmission electron microscope image of bacterial ectosymbionts; arrow points to one bacterium undergoing binary fission; scale bar is 0.5 μm. g Schematic representation of a GSO according to Bauer-Nebelsick et al. 1995 depicting its basic components: the A and B gland cells, the neuronal cell (n), the canal (C) and the seta (s). a and b are by Ulrich Dirks, c by Mario Schimak, d by Joerg A. Ott, e and f by Nikolaus Leisch
Fig. 2.
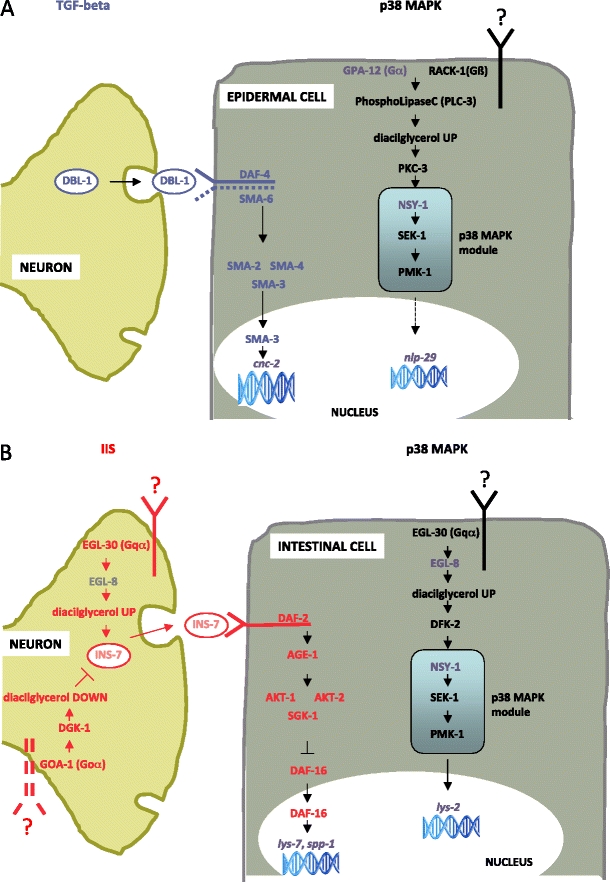
Schematic representation of signaling pathways regulating epithelial immune response in nematode epidermal (a) and intestinal (b) cells, adapted from Tan and Shapira 2011. The scheme is based on C. elegans immune response. GPA-12, NSY-1, cnc-2, lys-2, lys-7, nlp-29, and spp-1 are displayed in grey as transcripts corresponding to L. oneistus orthologs have not been identified yet. The p38 MAPK pathway (black receptors, letters, and arrows) regulates the expression of nlp-29, an antimicrobial peptide (AMP) encoding gene in the epithelial cell. A second AMP, encoded by cnc-2, is regulated by neuronally expressed dbl-1 that actives the epidermal cell TGF-β pathway (blue letters). The G-protein-coupled receptors that engage epithelial Goα signaling to modulate epidermal cell immune response is not known. b In the intestinal epithelial cell, the IIS pathway (red letters, receptors and arrows) is regulated primarily by the insulin-like ligand released by the neuron. IIS activity regulates DAF-16 subcellular localization. A separate G-protein signaling pathway (black receptors, letters and arrows) modulates the activity of the p38 MAPK module through phospholipases such as EGL-8, which determines the level of diacylglycerol, and protein kinase D (DFK-2). The G-protein-coupled receptors that engage epithelial Goα and neuronal Goα and Gqα signaling to modulate intestinal cell immune response are not known. See Appendix and Tables 1 and 2 for details
The reason for the P-specific secretion of the lectin Mermaid is unclear. Although no differences among L. oneistus A and P GSOs were reported (Bauer-Nebelsick et al. 1995), a recent, more detailed morphological analysis lead to the identification of three types of setae, with the longest type being restricted to the P region (A. Schmidt, unpublished). In another desmodorid nematode (Desmodora ovigera) the histochemical properties of the GSOs underlying long setae differs from those of the GSOs underlying short setae. Moreover, both in D. ovigera and in Laxus cosmopolitus histochmical differences between somatic and sexual GSOs have been found (I. Eichinger, unpublished). Taken together, it is possible that L. oneistus bears different GSO types and that these are differently distributed along the worm A-P axis. Consequently, the P-specific Mermaid secretion may reflect the A-P distribution of a specific GSO type. Thorough ultrastructural analysis is needed to clarify this. Moreover, the identification and localization analysis of additional GSO-specific transcripts and proteins will help reveal the function and physiology of these enigmatic organs.
L. oneistus ectosymbiont’s identity tag
The L. oneistus ectosymbiont is a 0.6 × 2.1 μm rod aligned perpendicularly to the nematode surface and forms a bacterial layer that resembles a columnar epithelium (Fig. 1e and f). The frequency of dividing bacteria is almost double in the vicinity of bacteria-free areas (Polz et al. 1992). According to 16S-rRNA gene phylogeny, all the characterized stilbonematid ectosymbionts belong to the marine oligochaete and nematode thiotrophic symbiont (MONTS) cluster that includes all gammaproteobacterial sulfur-oxidizers stably associated with these invertebrates (Polz et al. 1994; Bayer et al. 2009; Bulgheresi et al. 2011; Heindl et al. 2011). The MONTS are a basal group of Gammaprotobacteria (Harald R. Gruber-Vodicka, unpublished data) that likely originated from marine free-living bacteria because their closest relatives are the free-living sulfur purple bacteria of the Chromatiaceae (Heindl et al. 2011). Evolutionary transitions from free-living to mutualistic lifestyles are indeed common among bacteria (Sachs et al. 2011). The 16S rRNA-genes of the ectosymbionts of three stilbonematid species PCR amplified from pelagic seawater samples are also part of the MONTS cluster (Heindl et al. 2011). However the L. oneistus 16S rRNA-gene has never been amplified from the marine environment (unpublished data). In addition to their 16S rRNA-gene based phylogenetic placement, uptake of 14C bicarbonate (Schiemer et al. 1990) and the presence of RuBisCo enzymatic activity indicate autotrophy [(Polz et al. 1992). Note that L. oneistus was referred to as Catanema sp. in publications predating Ott et al. (1995)]. Enzymatic activity of ATP sulfurylase and sulfite oxidase as well as the presence of elemental sulfur in symbiotic but not in aposymbiotic L. oneistus (Polz et al. 1992) and the cloning of the alpha subunit of the adenosine-5′-phosphosulfate (APS) reductase gene (aprA; (Bayer et al. 2009)) support sulfur-oxidation by the L. oneistus ectosymbiont. Therefore, this may profit from the nematode’s migration through the sulfide gradient that exists in the marine sediment (Ott et al. 1991). It has long been hypothesized that adult stilbonematids feed on their ectosymbionts. This is mainly because stable carbon isotope ratios of stilbonematids resemble those reported from free-living thiobacilli (Ott et al. 1991). Nevertheless, ectosymbiont ingestion has never been observed. Only the guts of Leptonemella sp. and S. majum were found to contain ectosymbiont-like bacteria displaying signs of lysis (Ott et al. 1991). However, apart from digestion, ectosymbiont-produced organic compounds could directly diffuse through the worm cuticle [see for example (Rutherford and Webster 1974)].
An inventory of L. oneistus putative immunity pathways based on the Caenorhabditis elegans-pathogen model
The presence of a specific bacterial phylotype on the cuticle of each described stilbonematid species is exceptional in the nematode phylum. From the approximately 4,000 described marine nematode species (Miljutina et al. 2010) less than 2%—comprising members of the closely related families Epsilonematidae, Draconematidae and Desmodoridae—show at least occasional microbial epigrowth (J.A. Ott, unpublished observation). I hypothesize that L. oneistus and its ectosymbiont are adapted to each other in one of two, non-mutually exclusive, ways: (1) the ectosymbiont down-regulates host immunity and produces antimicrobials to protect the immunodepressed host from the attack of deleterious microbes; (2) the ectosymbiont is subject to a similar immune reaction that a pathogen would trigger, but does not succumb to it. In this last scenario the ectosymbiont may even exploit components of the host defense to (a) establish or strengthen its physical contact to the host or (b) regulate its proliferation in a way that facilitates contact persistence.
The immune system of the model nematode Caenorhabditis elegans was reviewed recently (Alper et al. 2007; Schulenburg et al. 2008; Irazoqui et al. 2010) (Ewbank and Zugasti 2011; Tan and Shapira 2011) (See Appendix and Fig. 2). Although C. elegans is a model organism for the study of host-microbe interactions, research to date has focused exclusively on interactions with pathogenic microbes. C. elegans is able to change its behavior in order to avoid deleterious microorganisms (Pradel et al. 2007). If this does not exempt it from their contact, C. elegans will rely on its epithelial immunity to respond to pathogens because it does not have circulating immune cells. Different microbes trigger distinct, specific immune reactions so that C. elegans can discern between non-pathogenic versus pathogenic, and also between different classes of microbes (See Appendix). The specificity of this custom made immune response may arise at the recognition level or at the effector level, but may also be achieved through differential immune regulation (e.g. different microbes cause a different degree of activation of one or more signaling pathway(s) or a different integration among pathways) (Schulenburg et al. 2008). Remarkably, C. elegans epithelial immunity does not rely on Toll-like receptors, despite their undisputed role in mammalian immunity (Pujol et al. 2001). Moreover, many genes encoding Toll-NF-kB pathway components are lacking not only from C. elegans but also from all the available nematode genomes (Irazoqui et al. 2010).
Epithelial immunity elicited in the epidermal cells differs from that in the intestinal cells. Although the p38 MAPK pathway (Kim et al. 2002) plays a central role in both epithelial tissues, two additional signaling pathways are integrated into it and are responsible for its neuronal modulation: the TGF-β-signaling pathway, in epidermal immunity, and the insulin/IGF-1 signaling (IIS) pathway, in intestinal immunity (Garsin et al. 2003) (Fig. 2).
Epidermal immune defense
It is unclear if L. oneistus epidermal cells immunologically react to the presence of bacteria attached to the worm’s intact cuticle. Although an intact nematode cuticle may be “impermeable” to Microbe-Associated Molecular Patterns (MAMPs), there is a continuum between the GSO lumen and the nematode surface (see Section 1). Therefore, it is conceivable that the GSO gland cell(s) sense and respond to bacteria instead of, or in addition to, the epidermal cells. Moreover, it is also possible that secretion of immune effectors by each GSO may be locally modulated by its neuronal cell.
Adult L. oneistus express transcripts encoding for the p38 MAPK module components SEK-1 and PMK-1 and a TGF-β activated MAP3K protein (adult L. oneistus transcriptomic data are available at http://genepool.bio.ed.ac.uk/GP_Partigene/2008075_SilviaBulgheresi/; clusters encoding for orthologs of C. elegans immunity pathway components are listed in Table 1). Putative p38 MAPK module activators expressed in L. oneistus may include heteromeric G protein component β RACK-1 and protein kinase C PLC-3, as well as a Tribbles homolog 1 (C. elegans NIPI-3). As anticipated, the neuronally activated TGF-β pathway is integrated into the p38 MAPK pathway in the C. elegans epidermal immunity. L. oneistus expresses transcripts encoding for all the TGF-β pathway basic components: DBL-1, a TGF-β type I receptor, and Smad proteins 2, 3, 4 and 6. At the immune effector level, transcripts encoding for caenacin or immune-related neuropeptide-like proteins (NLPs) have not been found in the available transcriptomic data (See also Section 4 and Table 2).
Table 1.
C. elegans immunity pathway components (adapted from Schulenburg and Boehnisch, 2008), H. sapiens orthologs, and L. oneistus clusters encoding for putative orthologs. Isogroup abundance indicates the number of sequences making up each cluster (when more than one cluster is present, the number of sequences making up each cluster were added and the total number is displayed); isogroup abundance may be directly correlated to transcript abundance. 454 transcriptomic data generated from symbiotic L. oneistus adults is available at: http://genepool.bio.ed.ac.uk/GP_Partigene/2008075_SilviaBulgheresi/; 14,747 putative genes (11,368 contigs and 3,379 singletons) corresponding to 5,888,206 bases were identified
| C. elegans | H. sapiens | L. oneistus ortholog-encoding clusters | Isogroup abundance |
|---|---|---|---|
| TGF-beta pathway | |||
| Neuron | |||
| DBL-1 | TGF--β ligand | LOP04675 | 3 |
| Epithelial cell | |||
| SMA-6 | Type I TGF-β receptor | LOP07766 | 1 |
| SMA-2 | Smad protein | LOP03197 | 1 |
| SMA-3 | Smad protein | LOP03388 | 2 |
| SMA-4 | Smad protein | LOP06423 | 1 |
| Insulin/IGF-1 (IIS) pathway | |||
| Neuron | |||
| GOA-1 | G protein α subunit | LOP05008 | 2 |
| DGK-1 | Diacylglycerol kinase -β | LOP03176, LOP07534 | 2 |
| Epithelial cell | |||
| DAF-2 | Insulin/IGF-1 receptor | LOP06243 | 1 |
| AGE-1 | Phosphatidylinositol 3-kinase | LOP04446, LOP08838, LOP07119 | 3 |
| AKT-1/AKT-2 | Rac Ser/Thr protein kinase | LOP05995, LOP09110, LOP06152 | 5 |
| SGK-1 | Serum/glucocorticoid regulated kinase 1 | LOP05995, LOP09110 | 3 |
| DAF-16 | FOXO family transcription factor | LOP10192, LOP07765 | 5 |
| P38 MAPK pathway | |||
| Epidermal cell | |||
| RACK-1 | G protein β subunit | LOP01842 | 9 |
| PLC-3 | Phospholipase C γ | LOP05069 | 2 |
| PKC-3 | Protein kinase C iota type | LOP11897 | 2 |
| Intestinal cell | |||
| EGL-30b | G protein G(q) α subunit | LOP06585, LOP05008 | 4 |
| DKF-2 | Ser/Thr protein kinase D | LOP07419, LOP10773 | 4 |
| NSY-1 | ASK1 MAPKKK | LOP04342a, LOP09080a | 5 |
| SEK-1 | MKK3, MKK6 MAPKK | LOP05076, LOP05850, LOP07733, LOP02761 | 7 |
| PMK-1 | p38 MAPK | LOP01689, LOP03611, LOP07256 | 13 |
aNo NSY hortologue was identified; clusters encode for a ortholog of human TGF-beta activated kinase MAPKKK7
bIn C. elegans the Gqα signaling component EGL-30 is also expressed in the neuron and can modulate intestinal immunity
Table 2.
L. oneistus clusters encoding for putative orthologs of C. elegans immune effectors. See Table 1 legend for explanation of isogroup abundance and information about the used transcriptomic data
| C. elegans | L. oneistus ortholog-encoding clusters | Isogroup abundance |
|---|---|---|
| Caenopores (saposin-like proteins; SPPs) | ||
| SPP-10 | LOP01886,LOP01470,LOP01471 | 72 |
| Lysozymes | ||
| LYS-8 | LOP02195 | 18 |
| LYS-5 | LOP03710 | 6 |
| LYS-6 | LOP07511,LOP06316 | 5 |
| LYS-10 | LOP11094 | 3 |
| LYS-4 | LOP03904 | 2 |
| C type lectin domain-containing proteins (CTLD) | ||
| CLEC-50 | LOP07024,LOP02441,LOP00362,LOP00364,LOP00366,LOP00367,LOP00371,LOP00372,LOP00373,LOP00376,LOP00368,LOP00369,LOP00375,LOP00377,LOP02333,LOP00378,LOP00379,LOP00380,LOP00381,LOP01037,LOP01038 | 442 |
| CLEC-178 | LOP04079,LOP01041,LOP01042 | 78 |
| CLEC-48 | LOP05939,LOP02725 | 18 |
| CLEC-56 | LOP01721 | 4 |
| CLEC-11 | LOP07307 | 2 |
| CLEC-3 | LOP03907 | 2 |
| CLEC-150 | LOP07069 | 2 |
| CLEC-10 | LOP05031 | 1 |
| FIPR proteins | ||
| FIP-1-like | LOP04451 | 4 |
| Glycine-rich proteins | ||
| Glycine-rich protein | LOP00858,LOP00856 | 22 |
Intestinal immune defense
Apart from covering its surface, it is possible that the ectosymbiont may, at some L. oneistus developmental stage, proliferate and persist in its gut. As already mentioned above, the pharynx of the stilbonematids is weak and, in contrast to C. elegans, they do not possess an efficient grinder-like organ, which can efficiently triturate the bacteria. In L. oneistus, the gene encoding for protein kinase DFK-2 is transcribed and this could activate the p38 MAPK module in intestinal cells. Additionally, the IIS pathway might play a role in mediating microbial recognition in the gut. Although we did not identify transcripts encoding for neuronally secreted insulin-like ligands such as INS-7 (see Appendix), those encoding for orthologs of the insulin/IGF-1 receptor DAF-2, the Ser/Thr kinases AKT-1/AKT-2/SGK-1 and DAF-16 were all present. The stress protective transcription factor DAF-16 was long hypothesized to be the only capable to confer pathogen resistance to C. elegans (see Section 4 and Table 2 for putative L. oneistus immune effectors expressed in intestinal immune response).
An inventory of putative L. oneistus immune effectors based on the C. elegans-pathogen model
In C. elegans the activation of immune pathways leads to the production and secretion of the following antimicrobial proteins and peptides (Ewbank and Zugasti 2011): caenopores (or SPPs), lysozymes, lectins, antibacterial factors (ABF) peptides, NLPs, caenacins, fungus-induced proteins (FIPs), FIP-related proteins (FIPRs) and glycine-rich secreted proteins (GRSPs). Of these, ABFs are constitutively expressed and the last five classes are epidermally expressed.
At least 33 C. elegans SPPs were identified (Roeder et al. 2010). They are pore-forming proteins structurally and functionally similar to vertebrate granulysin and NK-lysin and L. oneistus express one of them, the spp-10 ortholog.
Lysozymes are thought to be secreted in the C. elegans intestinal lumen where they may directly act on both Gram-negative and positive bacteria (Mallo et al. 2002; O’Rourke et al. 2006; Schulenburg et al. 2008). Of the 15 known in C. elegans, five putative orthologs are expressed by L. oneistus (lys-4,-5,-6,-8, and 10). C. elegans lys-8—which might be the most abundantly expressed in L. oneistus—is up-regulated in the intestine upon infection by the Gammaproteobacterium Serratia marcescens (Mallo et al. 2002).
C. elegans lectins might (a) bind microbial surface sugars and be involved in their recognition, (b) make microbes more susceptible to be phagocytosed by immune cells, (c) have antimicrobial activity, and/or (d) mask host epitopes targeted by microbial effectors (Schulenburg et al. 2008). The regulation of their expression is very complex, microbe-dependent and controlled by several signal transduction pathways (Mallo et al. 2002; O’Rourke et al. 2006; Alper et al. 2007). Among the lectins, 278 CTLD proteins were identified, with the most numerous class (class I) being characterized by a single CTLD (141 proteins, 99 of which are secreted (Schulenburg et al. 2008)). Other classes carry additional CTLDs, CUB domains, a Caenorhabditis-specific CW domain, von Willebrand factors (VWA), or a proline-rich repeat (PRR). Each CTLD protein class contains pathogen-induced genes and silencing of clec-17, clec-60, and clec-86 shows a direct role in immune defense (O’Rourke et al. 2006).
L. oneistus express orthologs of eight C. elegans CTLD proteins. In particular, a clec-50-like gene appears to be very abundantly expressed (442 isogroups grouped in 21 clusters). As observed for lys-8, C. elegans clec-50 is up-regulated in the intestine upon S. marcescens infection (Mallo et al. 2002). The C. elegans CTLD most similar to Mermaid is CLEC-178. Although the L. oneistus clec-178 ortholog appears to be abundantly expressed, mermaid transcripts are absent from the transcriptomic data and only present in a previously generated EST dataset (http://www.nematodes.org/NeglectedGenomes/NEMATODA/Laxus_oneistus/index.html). One possible explanation for this very surprising finding is that mermaid genes are not constantly and abundantly expressed in the adult L. oneistus. Given that the Mermaid proteins appear to be very stable (Bulgheresi et al., 2006) transcription of their corresponding genes in the P GSOs may be limited to particular developmental stages, those in which the ectosymbiosis must be established. This would also explain why the ectosymbiont is not able to recolonize bacteria-free areas artificially produced on adult nematodes (Hentschel et al. 1999).
Other potential immune effectors conserved in both C. elegans and L. oneistus are FIPRs and GRSPs. We did not identify L. oneistus transcripts encoding for orthologs of C. elegans antibacterial factors (ABFs), immune-related NLPs or caenacins. In C. elegans, the first are mostly effective against Gram+, whereas the expression of NLPs and caenacins is triggered by cuticle-infecting fungi, but not by bacteria.
Concluding remarks
Based on the available L. oneistus transcriptomic data and by using the C. elegans-pathogen model system as a reference, a few preliminary conclusions can be drawn: 1) L. oneistus constitutively express the signaling pathway components necessary for these nematodes to react to the presence of its ectosymbiont or to other microbes. In particular, transcripts encoding for all the members of the TGF-β pathway, which is central in C. elegans epidermal immunity, are present. 2) More conservation seems to exist among signaling pathways working in C. elegans and L. oneistus than among the downstream effectors that they regulate. This is not surprising as antimicrobial compounds are notorious for being poorly conserved. 3) L. oneistus appear to constitutively express immune effector genes, which are up-regulated by C. elegans upon neuronal activation of the TGF-β pathway by the Gammaprotebacterium Serratia marcescens (such as clec-50 and lys-8); it is conceivable that L. oneistus clec-50 and lys-8 orthologs are expressed in response to the (gammaproteobacterial) ectosymbiont. 4) The L. oneistus GSOs underlie both P and A cuticle. Therefore, throughout the A-P axis, the presence/absence of bacteria could trigger differential secretion of immune effectors by A and P GSOs. This is possible, given that each GSO may secrete immune effectors in response to a local signal. The establishment of gene silencing in L. oneistus is necessary to prove these conclusions. At the same time, understanding the molecular basis of C. elegans interaction with non pathogenic microbes would help elucidate the molecular base of nematode-bacterium symbioses.
Acknowledgments
I would like to thank the National Science Foundation Nemasym Research Network for supporting my participation to its 3rd Meeting in Corvallis, Oregon, USA. This invited review paper was inspired by my oral presentation at that meeting. I am extremely grateful to Mark Blaxter for supporting the analysis of the L. oneistus transcriptome and to Stephen Bridgett for assembling the 454 transcriptome and making it available to the public by creating the Partigene web interface. Finally I would like to thank an anonymous reviewer for helping me improve the accuracy and readability of this manuscript. I am supported by the Austrian Science Foundation (FWF) grant # P22470.
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
Appendix. Epidermal and intestinal immunity in C. elegans
A few pathogenic bacteria and fungi are known to infect C. elegans by adhering to its cuticle (Zugasti and Ewbank 2009). D. coniospora attach to- and pierce the nematode cuticle. After penetrating the epidermis, hyphae then develop and grow inside the host, eventually killing it (Dijksterhuis et al. 1990). In the case of p38 MAPK-mediated epidermal immunity, the binding of an unknown ligand to an unknown receptor results in successive activation of heterotrimeric G protein, protein kinase (s) C (such as PKC-3) and the p38 MAPK module (NSY-1/SEK-1/PMK-1). Activation of the module results in the expression of antimicrobial peptides such as the neuropeptide-like peptide (NLP; see Section 4) nlp-29. Neuronally secreted DBL-1 may also trigger epidermal immunity. The identity of the DBL-1 secreting neurons is unknown. In this case, DBL-1 receptor-regulated Smad proteins (Sma-6, 2, -3, and -4) would activate (an) unknown transcription factor(s) in the epidermal cell and these factors, in turn, would switch on expression of the caenacin (see Section 4) cnc-2.
Most bacterial pathogens colonize the intestine of C. elegans and affect defense gene expression in its gut. In intestinal defense, the p38 MAPK cassette can be activated by the protein kinase DKF-2 and it regulates the expression of immunity genes such as the lysozyme encoding lys-2 (see Section 4). As it is the case for epidermal immunity, we do not know the ligand(s) and the G protein-coupled receptor(s) leading to DFK-2 activation. In addition to the p38 MAPK pathway, the IIS pathway functions in intestinal immunity. This is a very conserved regulator of metabolism, stress resistance and ageing, with increasingly recognized roles in metazoan immune homeostasis (Peng 2010) (Becker et al. 2010). The activation of the IIS pathway in the intestinal cell is controlled by INS-7, a ligand of the insulin/IGF-1 receptor DAF-2 (Kawli and Tan 2008). The levels of neuronally secreted INS-7 are in turn regulated by both Goα signaling, which includes EGL-30 and EGL-8 (see Table 1) and Gqα signaling, which includes GOA-1 and DGK-1 (see Table 1). If INS-7 is abundant, DAF-2 activation triggers a phosphorylation cascade involving the phosphatidylinositol 3-kinase protein AGE-1, and lipid and serine/threonine kinases (AKT-1/AKT-2/SGK-1). These phosphorylation events lead to the phosphorylation and cytoplasmic retention of the FoxO forkhead transcription factor homologue DAF-16. Cytoplasmic DAF-16 can not ignite intestinal cell immune defense. On the other hand, if INS-7 secretion and, consequently, DAF-2 function is reduced, DAF-16 is translocated into the nucleus and this triggers the expression of antimicrobial genes, such as the intestinal lysozyme LYS-7 and the saposin-like protein SPP-1 (lysozymes and SPPs are discussed in Section 4).
References
- Alper S, McBride SJ, Lackford B, Freedman JH, Schwartz DA. Specificity and complexity of the Caenorhabditis elegans innate immune response. Mol Cell Biol. 2007;27(15):5544–5553. doi: 10.1128/MCB.02070-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer-Nebelsick M, Blumer M, Urbancik W, Ott JA. The Glandular Sensory Organ of Desmodoridae (Nematoda)-Ultrastructure and Phylogenetic Implications. Invertebrate Biology. 1995;114(3):211–219. doi: 10.2307/3226876. [DOI] [Google Scholar]
- Bayer C, Heindl NR, Rinke C, Lücker S, Ott JA, Bulgheresi S. Molecular characterization of the symbionts associated with marine nematodes of the genus Robbea. Environmental Microbiology Reports. 2009;1(2):136–144. doi: 10.1111/j.1758-2229.2009.00019.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T, Loch G, Beyer M, Zinke I, Aschenbrenner AC, Carrera P, Inhester T, Schultze JL, Hoch M. FOXO-dependent regulation of innate immune homeostasis. Nature. 2010;463(7279):369–373. doi: 10.1038/nature08698. [DOI] [PubMed] [Google Scholar]
- Bulgheresi S, Schabussova I, Chen T, Mullin NP, Maizels RM, Ott JA. A new C-type lectin similar to the human immunoreceptor DC-SIGN mediates symbiont acquisition by a marine nematode. Appl Environ Microbiol. 2006;72(4):2950–2956. doi: 10.1128/AEM.72.4.2950-2956.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulgheresi S, Gruber-Vodicka HR, Heindl NR, Dirks U, Kostadinova M, Breiteneder H, Ott JA (2011) Sequence variability of the pattern recognition receptor Mermaid mediates specificity of marine nematode symbioses. ISME J.5(6):986-98 [DOI] [PMC free article] [PubMed]
- Dijksterhuis J, Veenhuis, M, Harder W (1990) Ultrastructural study of adhesion and initial stages of infection of the nematode by conidia of Drechmeria coniospora. Mycological Research 94:1–8
- Ewbank JJ, Zugasti O. C. elegans: model host and tool for antimicrobial drug discovery. Dis Model Mech. 2011;4(3):300–304. doi: 10.1242/dmm.006684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garsin DA, Villanueva JM, Begun J, Kim DH, Sifri CD, Calderwood SB, Ruvkun G, Ausubel FM. Long-lived C. elegans daf-2 mutants are resistant to bacterial pathogens. Science. 2003;300(5627):1921. doi: 10.1126/science.1080147. [DOI] [PubMed] [Google Scholar]
- Heindl NR, Gruber-Vodicka HR, Bayer C, Luecker S, Ott JA, Bulgheresi S (2011) First detection of thiotrophic symbiont phylotypes in the pelagic marine environment. FEMS Microbiol Ecol 77:223–227 [DOI] [PubMed]
- Hentschel U, Berger EC, Bright M, Felbeck H, Ott JA (1999) Metabolism of nitrogen and sulfur in ectosymbiotic bacteria of marine nematodes (Nematoda, Stilbonematinae). Mar Ecol Prog Ser 183:149–158
- Hoschitz M, Bright M, Ott JA. Ultrastructure and reconstruction of the pharynx of Leptonemella juliae (Nematoda, Adenophorea) Zoomorphology. 2001;121:95–107. doi: 10.1007/PL00008498. [DOI] [Google Scholar]
- Irazoqui JE, Urbach JM, Ausubel FM. Evolution of host innate defence: insights from Caenorhabditis elegans and primitive invertebrates. Nat Rev Immunol. 2010;10(1):47–58. doi: 10.1038/nri2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampfer S, Sturmbauer C, Ott JA. Phylogenetic analysis of rDNA sequences from adenophorean nematodes and implications for the adenophorea-secernentea controversy. Invertebrate Biology. 1998;117(1):29–36. doi: 10.2307/3226849. [DOI] [Google Scholar]
- Kawli T, Tan MW. Neuroendocrine signals modulate the innate immunity of Caenorhabditis elegans through insulin signaling. Nat Immunol. 2008;9(12):1415–1424. doi: 10.1038/ni.1672. [DOI] [PubMed] [Google Scholar]
- Kim DH, Feinbaum R, Alloing G, Emerson FE, Garsin DA, Inoue H, Tanaka-Hino M, Hisamoto N, Matsumoto K, Tan MW, Ausubel FM. A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science. 2002;297(5581):623–626. doi: 10.1126/science.1073759. [DOI] [PubMed] [Google Scholar]
- Mallo GV, Kurz CL, Couillault C, Pujol N, Granjeaud S, Kohara Y, Ewbank JJ. Inducible antibacterial defense system in C. elegans. Curr Biol. 2002;12(14):1209–1214. doi: 10.1016/S0960-9822(02)00928-4. [DOI] [PubMed] [Google Scholar]
- Miljutina MA, Miljutin DM, Mahatma R, Galeron J. Deep-sea nematode assemblages of the Clarion-Clipperton Nodule Province (Tropical North-Eastern PAcific) Mar Biodivers. 2010;40:1–15. doi: 10.1007/s12526-009-0029-0. [DOI] [Google Scholar]
- Mittal R, Bulgheresi S, Emami C, Prasadarao NV. Enterobacter sakazakii targets DC-SIGN to induce immunosuppressive responses in dendritic cells by modulating MAPKs. J Immunol. 2009;183(10):6588–6599. doi: 10.4049/jimmunol.0902029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabatov AA, de Jong MA, de Witte L, Bulgheresi S, Geijtenbeek TB. C-type lectin Mermaid inhibits dendritic cell mediated HIV-1 transmission to CD4+ T cells. Virology. 2008;378(2):323–328. doi: 10.1016/j.virol.2008.05.025. [DOI] [PubMed] [Google Scholar]
- O’Rourke D, Baban D, Demidova M, Mott R, Hodgkin J. Genomic clusters, putative pathogen recognition molecules, and antimicrobial genes are induced by infection of C. elegans with M. nematophilum. Genome Res. 2006;16(8):1005–1016. doi: 10.1101/gr.50823006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott JA, Novak R, Schiemer F, Hentschel U, Nebelsick M, Polz M. Tackling the sulfide gradient: a novel strategy involving marine nematodes and chemoautotrophic ectosymbionts. P.S.Z.N. I: Marine Ecology. 1991;12(3):261–279. doi: 10.1111/j.1439-0485.1991.tb00258.x. [DOI] [Google Scholar]
- Ott JA, Bauer-Nebelsick M, Novotny V. The genus Laxus Cobb, 1894 (Stilbonematinae: Nematoda): description of the two species with ectosymbiotic chemoautotrophic bacteria. Proceedings of the Biological Society of Washington. 1995;108:508–527. [Google Scholar]
- Ott JA, Bright M, Bulgheresi S. Marine microbial thiotrophic ectosymbioses. Oceanography and marine biology—An Annual Review. 2004;42:95–118. doi: 10.1201/9780203507810.ch4. [DOI] [Google Scholar]
- Ott JA, Bright M, Bulgheresi S. Symbioses between marine nematodes and sulfur-oxidizing chemoautotrophic bacteria. Symbiosis. 2004;36(2):103–126. [Google Scholar]
- Peng SL. Forkhead transcription factors in chronic inflammation. Int J Biochem Cell Biol. 2010;42(4):482–485. doi: 10.1016/j.biocel.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polz MF, Felbeck H, Novak R, Nebelsick M, Ott JA. Chemoautotrophic, sulfur-oxidizing symbiotic bacteria on marine nematodes: morphological and biochemical characterization. Microbial Ecology (Historical Archive) 1992;24(3):313–329. doi: 10.1007/BF00167789. [DOI] [PubMed] [Google Scholar]
- Polz MF, Distel DL, Zarda B, Amann R, Felbeck H, Ott JA, Cavanaugh CM. Phylogenetic analysis of a highly specific association between ectosymbiotic, sulfur-oxidizing bacteria and a marine nematode. Appl Environ Microbiol. 1994;60(12):4461–4467. doi: 10.1128/aem.60.12.4461-4467.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradel E, Zhang Y, Pujol N, Matsuyama T, Bargmann CI, Ewbank JJ. Detection and avoidance of a natural product from the pathogenic bacterium Serratia marcescens by Caenorhabditis elegans. Proc Natl Acad Sci USA. 2007;104(7):2295–2300. doi: 10.1073/pnas.0610281104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol N, Link EM, Liu LX, Kurz CL, Alloing G, Tan MW, Ray KP, Solari R, Johnson CD, Ewbank JJ. A reverse genetic analysis of components of the Toll signaling pathway in Caenorhabditis elegans. Curr Biol. 2001;11(11):809–821. doi: 10.1016/S0960-9822(01)00241-X. [DOI] [PubMed] [Google Scholar]
- Roeder T, Stanisak M, Gelhaus C, Bruchhaus I, Grotzinger J, Leippe M. Caenopores are antimicrobial peptides in the nematode Caenorhabditis elegans instrumental in nutrition and immunity. Dev Comp Immunol. 2010;34(2):203–209. doi: 10.1016/j.dci.2009.09.010. [DOI] [PubMed] [Google Scholar]
- Rutherford TA, Webster JM. Transcuticular uptake of glucose by the entomophilic nematode, Mermis nigrescens. J Parasitol. 1974;60(5):804–808. doi: 10.2307/3278905. [DOI] [PubMed] [Google Scholar]
- Sachs JL, Skophammer RG, Regus JU. Evolutionary transitions in bacterial symbiosis. Proc Natl Acad Sci USA. 2011;108(Suppl 2):10800–10807. doi: 10.1073/pnas.1100304108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiemer F, Novak R, Ott JA. Metabolic studies on thiobiotic free-living nematodes and their symbiotic microorganisms. Marine Biology (Berlin) 1990;106(1):129–137. doi: 10.1007/BF02114683. [DOI] [Google Scholar]
- Schulenburg H, Boehnisch C. Diversification and adaptive sequence evolution of Caenorhabditis lysozymes (Nematoda: Rhabditidae) BMC Evol Biol. 2008;8:114. doi: 10.1186/1471-2148-8-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulenburg H, Hoeppner MP, Weiner J, 3rd, Bornberg-Bauer E. Specificity of the innate immune system and diversity of C-type lectin domain (CTLD) proteins in the nematode Caenorhabditis elegans. Immunobiology. 2008;213(3–4):237–250. doi: 10.1016/j.imbio.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Tan MW, Shapira M. Genetic and molecular analysis of nematode-microbe interactions. Cell Microbiol. 2011;13(4):497–507. doi: 10.1111/j.1462-5822.2011.01570.x. [DOI] [PubMed] [Google Scholar]
- Urbancik W, Bauer-Nebelsick M, Ott JA. The ultrastructure of the cuticle of Stilbonematinae (Nematoda, Desmodoridae).I. The somatic cuticle. Zoomorphology. 1996;116(2):51–64. doi: 10.1007/BF02526870. [DOI] [Google Scholar]
- Zhang P, Snyder S, Feng P, Azadi P, Zhang S, Bulgheresi S, Sanderson KE, He J, Klena J, Chen T. Role of N-acetylglucosamine within core lipopolysaccharide of several species of gram-negative bacteria in targeting the DC-SIGN (CD209) J Immunol. 2006;177(6):4002–4011. doi: 10.4049/jimmunol.177.6.4002. [DOI] [PubMed] [Google Scholar]
- Zugasti O, Ewbank JJ. Neuroimmune regulation of antimicrobial peptide expression by a noncanonical TGF-beta signaling pathway in Caenorhabditis elegans epidermis. Nat Immunol. 2009;10(3):249–256. doi: 10.1038/ni.1700. [DOI] [PubMed] [Google Scholar]


