Abstract
In recent years, biological web resources such as databases and tools have become more complex because of the enormous amounts of data generated in the field of life sciences. Traditional methods of distributing tutorials include publishing textbooks and posting web documents, but these static contents cannot adequately describe recent dynamic web services. Due to improvements in computer technology, it is now possible to create dynamic content such as video with minimal effort and low cost on most modern computers. The ease of creating and distributing video tutorials instead of static content improves accessibility for researchers, annotators and curators. This article focuses on online video repositories for educational and tutorial videos provided by resource developers and users. It also describes a project in Japan named TogoTV (http://togotv.dbcls.jp/en/) and discusses the production and distribution of high-quality tutorial videos, which would be useful to viewer, with examples. This article intends to stimulate and encourage researchers who develop and use databases and tools to distribute how-to videos as a tool to enhance product usability.
Keywords: screencast, vodcast, tutorial, YouTube, QuickTime, Flash
INTRODUCTION
Recent advances in life sciences technology have dramatically changed the research style from hypothesis-driven research (bottom-up style) to data-driven research (top-down style). Current ‘omics’ projects have produced vast amounts of data that have been stored in various online databases. Simultaneously, many types of web tools have been developed to analyze the stored data. Some of them are annually featured in the Nucleic Acid Research’s database issue and web server issue [1, 2]. Although the increase in available resources (databases and tools) has promoted life sciences research, this situation causes the following difficulties for researchers, especially ‘wet’ biologists: (i) What kinds of resources exist? (ii) Where are they? (iii) How can the resources be used and combined? and (iv) How does one interpret a result? To solve these issues, development of educational content as well as a system for navigation of web resources is required [3].
Traditional methods for distributing educational content include publishing textbooks and web documents. Although the contents of a textbook are sustainable, they quickly become obsolete because of frequent updates of web interfaces and improvement in web service functions. Web documents can more easily keep up with database and tool updates. However, it has become difficult to describe current web services in such static documents because of the evolution of web technology. Web 2.0, Flash and AJAX (Asymmetric JavaScript + XML) have led to the development of interactive and dynamic web services. This type of content would be better expressed in an animated environment rather than in a document. As a similar example, educational videos have been distributed using videotapes, CDs/DVDs and the Internet for >30 years in some clinical fields because it was excessively difficult to describe an actual procedure in writing [4–7]. However, their creation and distribution costs were high.
Rapid improvements in recent years in computer hardware, software and the Internet have reduced the publishing cost of multimedia content. A personal computer with a high-end CPU, extensive memory and large-capacity storage space enables users to produce and encode videos with relative ease. The latest releases of major OS packages include software for recording, editing and encoding videos, such as Windows Live Movie Maker (Microsoft Corporation, Redmond, WA, USA) and QuickTime (Apple Inc., Cupertino, CA, USA). The reasonable price of such software reduces the installation cost, and its user friendliness reduces the time required. In addition to the reduction in production cost, broadband networks have also reduced the distribution cost of multimedia content and have allowed experts to readily distribute video content in their field to anyone with an Internet connection. Many recent web browsers are by default equipped with video players, such as the Adobe Flash player (Adobe Systems Incorporated, San Jose, CA, USA) and QuickTime player (Apple Inc.); thus, one can easily view a video on web browser by simply clicking the play button.
In this article, we describe online video repositories for educational purposes, worldwide movements of distributing video tutorials created by major database and tool developers, and our recent activity in Japan. In addition, we propose distributing how-to videos to the developers and users of databases and tools to promote their usability and contribute to the scientific community.
ONLINE VIDEO REPOSITORIES
Several web services are already available for video distribution. YouTube is the most popular online video sharing service, and it contains many tutorial videos and lectures in many fields [8]. Similarly, there are repository services such as Dailymotion and Vimeo (for more examples, see the Wikipedia article entitled ‘List of video hosting services’) [9–11]. Most services are free to use, and any registered user can upload video. Live streaming services such as Ustream, Justin.tv and Stickam also exist [12–14]. As the term ‘live streaming’ suggests, these services provide live streaming services for lectures, workshops, seminars and meetings that are recorded and may be played back at a later time.
In the scientific field, the Journal of Visualized Experiments has been published since 2006 [15]. It is a peer-reviewed, PubMed-indexed journal devoted to the publication of biological research in a video format. SciVee offers a comprehensive set of rich media solutions to enhance the discovery and collaboration of knowledge [16]. It provides Video and Podcasts (standard videos and podcasts), PubCast (synchronized video abstracts of peer-reviewed articles), PaperCast (synchronized video abstracts of non-peer-reviewed articles), SlideCast (synchronized videos of slide presentations) and PosterCast (synchronized videos of posters or other conference presentations) in collaboration with scientists and researchers, as well as journals and publishers, societies, conference organizers, universities and research institutions. Dnatube is a community-based repository of scientific videos including educational materials, seminars and lectures [17]. This site has over 5000 videos and 30 000 community members. Individual videos can be found using keyword search, category tags and topics.
Some universities and organizations also administer a video repository server, especially for providing lecture videos that are part of OpenCourseWare (OCW). The Massachusetts Institute of Technology (MIT) hosts MIT OCW and MIT World, and the University of Tokyo provides UT OCW [18–20]. Academic Earth provides online courses of the world’s top scholars from Harvard University and Stanford University among other top academic institutions [21]. YouTube also has a special channel for education from colleges and universities named YouTube EDU, and another channel, Technology, Entertainment, Design (TED), delivers interesting lectures by respected individuals [22, 23]. A complete list of OCW websites is found at the OCW Consortium Website, and other useful services are listed in the Wikipedia article entitled ‘List of educational video websites’ [24, 25].
In addition to repository-type services, delivery-type services named vodcasts (video podcasts) are available via Really Simple Syndication (RSS) technology. If a user subscribes to a vodcast program in a vodcast player such as iTunes, the contents of the program are automatically updated when new content arrives. Since the vodcast programs can be transferred to portable devices such as the iPod, iPhone or iPad, the user can watch them anytime, anywhere. Although vodcast programs are mainly focused on news, entertainment and fashion, educational programs are also provided. Indeed, some institutes have already used the podcast/vodcast for education [26–28]. Apple collects and webcasts educational contents via the iTunes store called iTunes U [29].
VIDEO TUTORIALS PROVIDED BY RESOURCE DEVELOPERS AND USERS
As noted earlier, the publishing of tutorial videos by some providers has increased as the creation and distribution costs of videos have decreased. For example, National Center for Biotechnology Information provides tutorial videos of some services both on the YouTube channel and on their server such as dbGaP, the database of Genotypes and Phenotypes, that archives and distributes the results of studies that have investigated the interaction of genotype and phenotype and PubMed that is a database of citations and abstracts for biomedical literature from MEDLINE and additional life sciences journals [30–35]. Some projects in the European Bioinformatics Institute also distributed how-to videos for tools such as Ensembl that is genome databases for vertebrates and other eukaryotic species, QuickGO that is a fast web-based browser for Gene Ontology (GO) terms and annotations, and GOA, Gene Ontology Annotation, that provides high-quality GO annotations to proteins in the UniProt Knowledgebase and International Protein Index [36-41].
Not only service providers in national institutes but also individual service providers including relatively small communities distributed tutorial videos. Galaxy, a collaboration system for genomic research, is a highly functional and complex system, but the procedure is easily understandable because the developers provide tutorial videos on their website [42, 43]. Taverna, which is an open source and domain-independent workflow management system (a suite of tools used to design and execute scientific workflows and aid in silico experimentation), is also described in the tutorials in a video format [44, 45]. ATTED-II, which provides co-regulated gene relationships to estimate gene function, has YouTube channel for tutorials [46, 47]. There are many video tutorials provided by the database and tool developers.
In addition, educators and users of web resources who do not develop any databases or tools also contribute to the scientific community by providing tutorial videos. BITS, BioInformatics Tutorials Series, is a collaboration work of the MIT Engineering and Science Libraries and Harvard’s Countway Library [48, 49]. BIREC, Bioinformatics Information Resource and eLearning Center, also provides tutorial videos [50]. OpenHelix provides over 100 well-organized tutorial suites including videos on web-based bioinformatics and genomic resources [3, 51]. It also has many tutorial videos in ‘Tip of this week’ tagging articles in the blog section [52]. In addition to videos provided by organizations, a YouTube search by database or tool name will provide many tutorial videos produced by volunteers.
TogoTV: ONLINE TUTORIAL VIDEO DISTRIBUTION TRIAL IN JAPAN
To bridge the gap between service providers and users, we created and distributed tutorial videos of databases and web tools. We describe in this article, a methodology for making and distributing videos and elaborate on this methodology with examples. TogoTV (‘Togo’ means ‘integration’ in Japanese; pronunciation symbol is [toɯgoɯ]) that is one of the services in the Integrated Database Project in Japan (Figure 1) is a portal site of tutorial and lecture videos about bioinformatics resources [53–55]. Although the original TogoTV site is mostly written in Japanese [53], there is the English interface for international users [54]. The site contains our original videos and third-party videos from publicly available website such as YouTube. All contents provided by us are distributed under the Creative Commons Attribution 2.1 Japan license and also provided as vodcasts that can be viewed using a portable device and on YouTube. Although most of the contents are described in Japanese, there are 19 original programs in English, most of which explain a service developed in the Integrated Database Project such as TogoWS [56], which provides an integrated SOAP and REST APIs for interoperable bioinformatics Web services and OReFiL [57], which is an online resource finder for life science. We plan to expand our own English contents so as to enable our service to be used all over the world.
Figure 1:
Screenshot of the MEXT Integrated Database Project portal (http://lifesciencedb.jp/en/). Paper icons and TV icons zoomed in the call-out following service names are linked to PDF documents and tutorial videos, respectively. MEXT, Ministry of Education, Culture, Sports, Science and Technology of Japan.
Making videos
Currently, two types of video are provided: (i) tutorial videos of databases and tools (screencasts) and (ii) lecture videos of symposiums and workshops (live action). For screencast videos, a screen where the database or tools were operated was captured and edited using screencast software equipped with a caption-adding function, such as Camtasia Studio (TechSmith Corporation, Okemos, MI, USA) for Windows and DesktopToMovie (Pencil Software, Okinawa, Japan; only a Japanese-language version is available) for Mac [58–60]. Recently, Camtasia:Mac (TechSmith Corporation) has been released, and we recommend its use rather than DesktopToMovie. For live-action videos, a lecture was recorded using a digital video camera or voice recorder, and then the source media was edited or embedded with presentation slides using tools such as Final Cut Pro (Apple Inc.) or iMovie (Apple Inc.). It is also possible to output presentations in Keynote (Apple Inc.) to videos. After capturing and editing, the source media was encoded in QuickTime format (.mov) and MPEG-4 format (.m4v) for distribution via websites and vodcast, respectively. The video compression type was set to H.264, and the sound format was specified to AAC if an audio track was included. For encoding in the QuickTime format, the ‘Prepare for Internet Streaming’ option was set to ‘Fast Start’ rather than ‘Fast Start—Compressed Header’ because the compressed header file format is impossible to play on Flash players. Other useful software packages for screencasting are listed in the article of Wikipedia entitled ‘Comparison of screencasting software’ [61].
To create user-friendly and high-quality tutorials, we suggest the following points: plan the tutorial; do a run-through before recording; edit adequately; pause at essential points; make the duration as short as possible and keep effects to a minimum. To capture a video smoothly, it is important to create a plan and run through it before recording. Editing costs may increase considerably if these preparatory steps are skipped. Here, editing involves deleting unnecessary frames and loading animation frames, thus ultimately reducing video downloading time, user viewing time and also file size. At key operating points, it is necessary to pause the animation; viewers need time to understand and absorb the information. In TogoTV videos, we insert a pause of about 5–10 s, depending on the situation. We also recommend that the video duration be made as short as possible and that animation effects be suppressed to a minimum. Most TogoTV contents fit in a 5-min video, except for lectures. Excessive production not only increases the production cost but also conceals the essence of the video. In general, since a dynamic video tends to increase file size, suppression of excessive animation will reduce the file size. Most video repositories have upload limitations based on video length and file size.
The most important thing when creating a video is to create a high-quality video that would be useful to viewers. When one creates video easily without any consideration of the quality of the product, it would be a waste of viewer’s time and content creator’s time and would add to the already overwhelming ‘noise’ of available training materials. Because both creating and viewing video are time consuming, one needs to create a video carefully. In a case of TogoTV, we have adopted an internal review in order to ensure quality. From planning to drafting, reviewing and publishing takes about a week in our case.
Video distribution
We used a blog to distribute videos. This allows the video creation date to be clear to viewers since the web service interface changes often. In addition, it is possible to easily implement a comment-posting system and an update-notification system via RSS. We selected tDiary for the original TogoTV site [62]. It is a blog kit written in the Ruby programming language with the ability to easily add functions using plug-in programs such as update notification (makerss.rb) via RSS feeds. In addition to plug-in programs included with tDiary, we activated third-party plug-in programs, e.g. opening/closing caption texts that appear in the video (netabare.rb). We also developed plug-in programs to display the view-count ranking, query word trends by keyword search from Google and video recommendations (similar to the Amazon recommendation system). We chose WordPress for English interface of TogoTV [63]. It has smart interface compared with that of tDiary and can also be easily extended using plug-in programs.
We used JW Player (LongTail Video, New York, NY, USA), an open source software program for playing Flash video on web browsers [64]. The latest version of JW Player supports HTML5 video elements; thus, a user can view a video directly on an iPod, iPhone or iPad. The H264 Streaming Module for Apache was installed on the server for video streaming on the hypertext transfer protocol (HTTP) instead of setting up a real-time messaging protocol (RTMP) server [65]. This module provides the ability to start a video at any specified point.
A screenshot of the TogoTV home page is shown in Figure 2 [54]. There are links for video categories, most-watched videos and discrete tutorial videos sorted by creation date, similar to a blog. Every video has category tags to enable the searching of a video by categories, such as ‘Genome analysis’, ‘Visualization’ and ‘Sequence analysis’. By clicking a date or thumbnail image, an individual video screen page will open (Figure 3). In the video screen page, there are description of the video, summary text of the story, and all captions or slides in the video in addition to the video player. Clicking the thumbnail on the video player plays the video from the beginning, and clicking a summary text or the thumbnail of a slide plays the video from the specific time at which the caption or slide appears.
Figure 2:
Screenshot of the (A) international version of and (B) original version of TogoTV website. All contents are sorted by publishing date. There are list of category tags, daily/weekly/monthly ranking of playing video (only for the original site), publishing date, description, thumbnail, summary of the story and link to all captions in the video. When you click a date or thumbnail, a new page will open for playing the video (Figure 3A). When you click a sentence of summary, the video will start corresponding scene.
Figure 3:
Screenshot of TogoTV’s tutorial programs. Clicking the thumbnail in the center of the page will start the video. When the summary text of a video is clicked, the video will start the scene corresponding to the summary. There are links to recommended videos that are related playing video at the bottom of the page. (A) This video is entitled ‘How to use BodyParts3D/Anatomography 2010’. (B) This video is entitled ‘How to make probeID list for microarray using BioMart’.
We also distribute videos as vodcasts, so anyone can download and watch the videos on their own computer or handheld device, such as an iPod, iPhone or iPad via iTunes by subscribing to TogoTV vodcast. The vodcast distribution website was constructed using iWeb, which was a part of the iLife package (Apple Inc.). YouTube videos are also provided to maximize the reach of the videos, and it is possible to watch the videos even when the original TogoTV server is down for maintenance.
When the TogoTV service began, YouTube restricted upload file size and duration time up to 1 GB and 10 min, respectively. Above all, the video quality was extremely low, for example, it was difficult to read captions. Thus, we decided to use our own distribution server. There are many features that have made possible by preparing our own server such as recommendations, the ability to play from specific time and updating a video without changing URL. In addition, search of YouTube video hits more noise because the YouTube contains various genres, not limited to educational videos. However, YouTube now allows high-definition quality videos, and limitations of file size and duration time increased up to 2 GB and 15 min, respectively [66]. In addition, the time limitation of upload videos can be derestricted if users have complied with the YouTube community guidelines and copyright rules [67]. For these reasons, when distributing tutorial videos, we recommend using YouTube rather than a private server. Indeed, YouTube contains many useful tutorial videos. When we searched YouTube ‘biomart’ as a query, 42 videos were found. In the search result, 22 videos were noise that was unconcerned to biology, and 13 videos including one lecture video were uploaded by us (account: togotv). The rest included five videos provided by one of the service provider (account: EnsemblHelpdesk) and two videos provided by BioMart users. YouTube provides video-embedding code, so anyone can embed a video on any website. Clicking the embed button below a video shows the embedding code as well as options and skins for embedding.
When we publish a new video, an announcement is distributed using RSS and Twitter [68, 69]. The Twitter notification is a reposting of the RSS feed using Twitterfeed [70].
Update of content
Since the databases and tools are constantly evolving, it is important to keep up with updating the corresponding tutorial videos. Speedy update of the content is one of advantages of TogoTV; it cannot do in a paper device. Update of the resources is constantly monitored using a news release, RSS and DBCLS Database Catalog, which is a service developed in the Integrated Database Project [71]. An entry in this catalog contains ‘Last Modified’ date information that is automatically collected by a crawling program. When a resource is updated significantly or changed its interface, we replace an old tutorial video with a recreated video as soon as possible. In May 2011, we have 310 tutorial videos in TogoTV (excluding lecture videos), and a total of 54 videos are updated ones.
Examples of tutorial videos in TogoTV
How to use BodyParts3D/anatomography
Anatomography is a 3D rendering tool for human anatomy and has been developed as part of the Integrated Database Project [72]. A user can generate anatomical images by selecting body parts stored in the BodyParts3D database and setting their opacities, colors and viewpoint [73]. The image is useful for communication between physicians and patients, and it can be generated as a heat map of the human body based on an organ name and a numeric value such as organ-specific gene expression data and cancer mortality. Figure 3A shows a screenshot of the BodyParts3D/anatomography tutorial video [74]. This video describes how to build a 3D image, how to manipulate viewpoint and size, how to set opacities and colors and how to output to an image file. Videos of other services provided by the Integrated Database Project are also available at the project page [55]. A TV icon after the service name (Figure 1) provides a link to a tutorial video.
How to use BioMart
We provide how-to videos of not only our own services but also useful tools all over the world. BioMart, a query-oriented data management system, is one of the most important tools in genome science [75]. Users can submit various queries to retrieve lists of interest from BioMart. A screenshot of the tutorial video entitled ‘How to make probeID list for microarray using BioMart’ is shown in Figure 3B [76]. In this video, the process for creating an ID conversion list for microarray analysis on the BioMart central portal website is introduced [77]. From all genes in the mouse genome, genes that have corresponding entries in the Affymetrix mouse430 2 GeneChip are considered for further analysis. Genes with the Affymetrix GeneChip ID mentioned above are associated with the Agilent ProbeID and RefSeq ID via the Ensembl Gene ID. The results are downloadable in the tab separated value format with GNU-zip (.gzip) compression.
Lectures
Lectures are also distributed from TogoTV. Currently, we broadcast eight lectures in English. One is a video of a lecture about this service (TogoTV) held at the 2007 Annual Conference of the Japanese Society for Bioinformatics (JSBi2007, Figure 4A) [78]. The second lecture is about Gendoo [79], a functional profiling tool for gene and disease features using the Mesh vocabulary, held at JSBi2008 [80]. The third is about copyright and data sharing in science entitled ‘Copyright in the Digital Age and Its Impact on Scientific Data Sharing’ by Professor Lawrence Lessig from Harvard Law School from the ‘Balancing Intellectual Property Protection and Data Sharing in Science’ symposium (Figure 4B) [81]. The others are about processing of large genomic data from ‘Workshop on Parallel and Distributed Processing of Large Genome Data’ [82].
Figure 4:
Screenshot of TogoTV’s lecture programs. There are the slides appeared in the presentation at the bottom of the page. When you click a slide, the video will start corresponding scene. (A) Video of voice-over on slides. (B) Live-action video.
In addition to hosting TogoTV, we also organize workshops, which is called AJACS (All Japan Annotators/Curators/System DB administrators) workshop, for Japanese users of biological databases and tools for educational purposes [83]. We have published a lecture video with an online handout for participants to review and for interested parties who could not attend. Currently, there are 63 lecture videos in Japanese from 13 workshops.
Statistics and user feedback
In May 2011, we provided over 450 videos, most of which are in Japanese. TogoTV is accessed 20 000 times per month from 5000 unique IP addresses and videos are played 4000 times per month. According to the analysis of IP addresses, accesses from ‘.jp’ and ‘.com’ domain accounted for about 33% and 31%, respectively. The rest included unknown domain (28%), ‘.net’ (5%) and ‘.edu’ (1%) domains. Although contents of TogoTV are mainly Japanese, there are accesses from outside of Japan. The analysis of access was carried out using AWStats, and we excluded the bot programs from the analysis [84]. Videos accessed on YouTube measured through the YouTube API are played about 5000 times per month. Since TogoTV was started in July 2007, videos have been watched >250 000 times. The tutorial videos of basic tools and databases such as primer3, reactome, blast, clustalw, and Ensembl are the popular contents in TogoTV [36, 85–88].
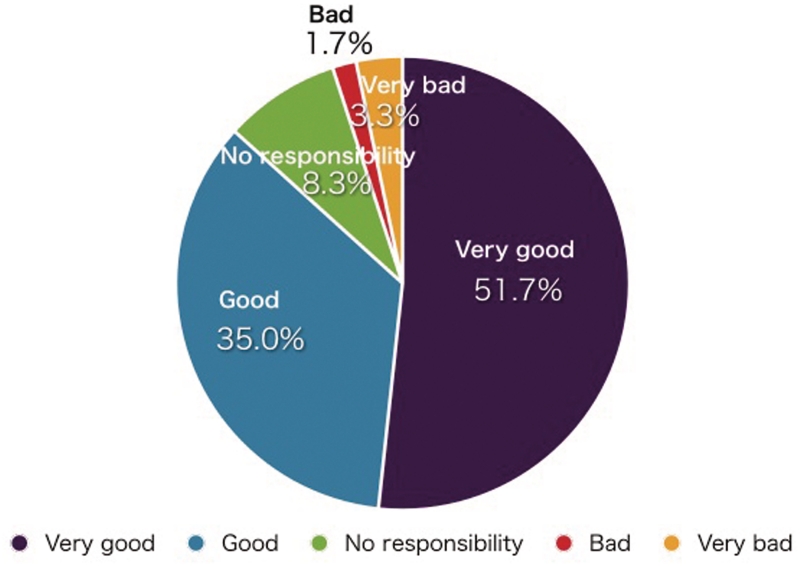
Reactions of most viewers are positive judging from the responses received during interviews at lectures and exhibitions. According to some instructors who used our videos for their bioinformatics lectures, their students’ understanding increased considerably. Figure 5 shows a part of the results of the Integrated Database Project user feedback investigation in FY2008–2009. More than 85% of reviewers reacted positively to our service.
Figure 5:
User feedback about TogoTV. External evaluation about the services provided by the Integrated Database Project was carried out via web from September to October 2009 [89]. There were 117 respondents including principal investigators, corporate researchers, postdocs and PhD students in total, and 60 peoples evaluated TogoTV. The question was ‘For this service, please give us feedback on how to proceed in the future’. Respondents chose from five options: ‘Because it seems pretty useful, I suggest it should be driven forward’ (Very good); ‘Because it seems useful in its own way, I suggest it may be driven forward’ (Good); ‘I do not have responsibility to assess it’ (No responsibility); ‘Because it seems unuseful, I suggest it need not be driven forward’ (Bad); and ‘Because it seems completely unuseful, it should be dropped’ (Very bad).
CONCLUSION
It is possible to provide tutorials in the video format because the continued development of computer hardware and software and the Internet. Various service developers have created and distributed tutorial videos via their own server or YouTube; our contribution was the launching of TogoTV, which provides over 450 videos and is one of the most active site collecting and maintaining tutorial videos. As pointed out by Williams et al. [3], tutorial videos are not always in depth enough to provide a full understanding of the resource to users. But it is effective for them to touch on use of web resources. Further development of computer and web infrastructure will accelerate this movement in the future.
It would be helpful if database and tool developers publish how-to videos as well as documents to encourage greater use. Even if one is just a user of web resources and not a service developer or provider, the creation and distribution of a tutorial video based on experience is useful for numerous researchers. The most important thing is to create a high-quality video that would be useful to viewers. To show summary text, captions and dialogues of a video as well as description is also useful in determining whether or not to watch the video. The creation of tutorial videos in a community is particularly useful for sharing and standardizing the annotation and curation process.
We believe that providing tutorial videos created by database and tool providers as well as users will promote research activities and help to distribute the knowledge of database and tool handling in research communities. Thus, we propose that everyone who produces and uses web resources create tutorial videos and share them.
Key Points.
Improvements of computer technology and the Internet enable creation and distribution of a tutorial video easily.
Some major database and tool developers provided tutorial videos via their website and/or YouTube.
We developed TogoTV, a website where tutorial videos of bioinformatics databases, tools and lectures are distributed, and this attempt acquired a good reputation.
Let us create and share tutorial videos all together.
FUNDING
This work was supported by the Integrated Database Project of Ministry of Education, Culture, Sports, Science and Technology in Japan [07046015, 10100921].
Acknowledgements
The authors would like to thank Yoko Ohmura, Yoko Yamaguchi, Kouichi Takewaka, Atsuko Chiba, Takuya Hashimoto, Takao Yokoyama, Yuki Okuda, Tatsuro Ohta, Haruko Hirukawa, Shoichiro Ohishi, Tomohiro Ono, Hayato Sakata, Tamayo Suizu, Hideyuki Takeda, Tetsuya Negoro, Maori Ito, Natsuki Kubo and Fumio Takahashi for creating videos, Yusuke Kumagae for the plug-in development and Takeru Nakazato for supervising web design. All contributors are presented on the TogoTV just like ‘end-title roll’ at http://togotv.dbcls.jp/movie/endroll_for_togotv.mov. They also thank Dr Ayumi Koso and Dr Mari Minowa for internationalization of the contents and summarization of user feedback.
Biographies
Shin Kawano is a Project Researcher at Database Center for Life Science (DBCLS). He produces TogoTV contents, maintains its web site and organizes workshops. His research focuses on bioinformatics approach to proteomics and glycomics.
Hiromasa Ono is a Project Senior Technical Support Specialist at DBCLS. He produces TogoTV contents and dictates research assistants. His research focuses on cellular dedifferentiation and acquisition of multipotency.
Toshihisa Takagi is a Professor at the University of Tokyo and the National Institute of Genetics. He is the head of the Integrated Database Project. His research interests include bioinformatics, ontology, text mining and database architecture.
Hidemasa Bono is a Project Associate Professor at DBCLS. He designed TogoTV project. He also develops gene expression database and analyzing tools. He is a specialist of transcriptome analysis.
References
- 1.Galperin MY, Cochrane GR. The 2011 Nucleic Acids Research Database Issue and the online Molecular Biology Database Collection. Nucleic Acids Res. 2011;39:D1–6. doi: 10.1093/nar/gkq1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brazas MD, Yamada JT, Ouellette BF. Providing web servers and training in Bioinformatics: 2010 update on the Bioinformatics Links Directory. Nucleic Acids Res. 2010;38:W3–6. doi: 10.1093/nar/gkq553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Williams JM, Mangan ME, Perreault-Micale C, et al. OpenHelix: bioinformatics education outside of a different box. Brief Bioinform. 2010;11:598–609. doi: 10.1093/bib/bbq026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cassie JM, Collins GF, Daggett CJ. The use of videotapes to improve clinical teaching. J Med Educ. 1977;52:353–4. doi: 10.1097/00001888-197704000-00015. [DOI] [PubMed] [Google Scholar]
- 5.Ruiz JG, Mintzer MJ, Issenberg SB. Learning objects in medical education. Med Teach. 2006;28:599–605. doi: 10.1080/01421590601039893. [DOI] [PubMed] [Google Scholar]
- 6.Vigeant D, Lefebvre H, Reidy M. The use of video as a pedagogic tool for the training of perioperative nurses: a literature review. Can Oper Room Nurs J. 2008;26:8–9. 14–5, 17–20. [PubMed] [Google Scholar]
- 7.Repanos C, Anderson D, Earnshaw J, et al. Manipulation of nasal fractures with local anaesthetic: a ‘how to do it’ with online video tutorial. Emerg Med Australas. 2010;22:236–9. doi: 10.1111/j.1742-6723.2010.01289.x. [DOI] [PubMed] [Google Scholar]
- 8. YouTube – Broadcast Yourself. http://www.youtube.com/ (24 May 2011, date last accessed)
- 9. Dailymotion – Online Videos, Music, and Movies. Watch a Video Today! http://www.dailymotion.com/ (24 May 2011, date last accessed)
- 10. Vimeo, Video Sharing For You. http://www.vimeo.com/ (24 May 2011, date last accessed)
- 11. List of video hosting services – Wikipedia the free encyclopedia. http://en.wikipedia.org/wiki/List_of_video_hosting_services (24 May 2011, date last accessed)
- 12. Ustream.tv: You’re On. http://www.ustream.tv/ (24 May 2011, date last accessed)
- 13. Justin.tv – Streaming live video broadcasts for everyone. http://www.justin.tv/ (24 May 2011, date last accessed)
- 14. Stickam – The Live Community, Live Streaming, Video Chat. http://www.stickam.com/ (24 May 2011, date last accessed)
- 15. JoVE: Journal of Visualized Experiments – a Video Journal for Biological and Medical Research. http://www.jove.com/ (24 May 2011, date last accessed)
- 16. SciVee | Making Science Visible. http://www.scivee.tv/ (24 May 2011, date last accessed)
- 17. DnaTube.com – Scientific Video Site. http://www.dnatube.com/ (24 May 2011, date last accessed)
- 18. Free Online Course Materials | MIT OpenCourseWare. http://ocw.mit.edu/ (24 May 2011, date last accessed)
- 19. MIT World | Distributed Intelligence. http://mitworld.mit.edu/ (24 May 2011, date last accessed)
- 20. UT OpenCourseWare. http://ocw.u-tokyo.ac.jp/english (24 May 2011, date last accessed)
- 21. Academic Earth | Online Courses | Academic Video Lectures. http://academicearth.org/ (24 May 2011, date last accessed)
- 22. YouTube – Education – YouTube EDU. http://www.youtube.com/edu (24 May 2011, date last accessed)
- 23. TED: Ideas worth spreading. http://www.ted.com/ (24 May 2011, date last accessed)
- 24. OCW Consortium – OpenCourseWare Websites. http://www.ocwconsortium.org/courses/ocwsites (24 May 2011, date last accessed)
- 25. List of educational video websites – Wikipedia, the free encyclopedia. http://en.wikipedia.org/wiki/List_of_educational_video_websites (24 May 2011, date last accessed)
- 26.Boulos MN, Maramba I, Wheeler S. Wikis, blogs and podcasts: a new generation of Web-based tools for virtual collaborative clinical practice and education. BMC Med Educ. 2006;6:41. doi: 10.1186/1472-6920-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jham BC, Duraes GV, Strassler HE, et al. Joining the podcast revolution. J Dent Educ. 2008;72:278–81. [PubMed] [Google Scholar]
- 28.Thapa MM, Richardson ML. Dissemination of radiological information using enhanced podcasts. Acad Radiol. 2010;17:387–91. doi: 10.1016/j.acra.2009.10.010. [DOI] [PubMed] [Google Scholar]
- 29. Apple – iTunes U – Learn anything, anywhere, anytime. http://www.apple.com/education/itunes-u/ (24 May 2011, date last accessed)
- 30.Cooper PS, Lipshultz D, Matten WT, et al. Education resources of the National Center for Biotechnology Information. Brief Bioinform. 2010;11:563–9. doi: 10.1093/bib/bbq022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tutorials. http://www.ncbi.nlm.nih.gov/education/tutorials/ (24 May 2011, date last accessed)
- 32. YouTube – NCBINLM’s Channel. http://www.youtube.com/ncbinlm (24 May 2011, date last accessed)
- 33.Zhang H, Morrison MA, Dewan A, et al. The NEI/NCBI dbGAP database: genotypes and haplotypes that may specifically predispose to risk of neovascular age-related macular degeneration. BMC Med Genet. 2008;9:51. doi: 10.1186/1471-2350-9-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. dbGaP Tutorial. http://www.ncbi.nlm.nih.gov/projects/gap/tutorial/dbGaP_demo_1.htm (24 May 2011, date last accessed)
- 35. PubMed Online Training. http://www.nlm.nih.gov/bsd/disted/pubmed.html (24 May 2011, date last accessed)
- 36.Flicek P, Amode MR, Barrell D, et al. Ensembl 2011. Nucleic Acids Res. 2011;39:D800–6. doi: 10.1093/nar/gkq1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. YouTube – EnsemblHelpdesk’s Channel. http://www.youtube.com/EnsemblHelpdesk (24 May 2011, date last accessed)
- 38. Huntley RP, Binns D, Dimmer E, et al. QuickGO: a user tutorial for the web-based Gene Ontology browser. Database 2009; doi:10.1093/database/bap010 (29 September 2009, date last accessed)
- 39. Video tutorials on the use of QuickGO. http://www.ebi.ac.uk/QuickGO/tutorial.html (24 May 2011, date last accessed)
- 40.Barrell D, Dimmer E, Huntley RP, et al. The GOA database in 2009-an integrated Gene Ontology Annotation resource. Nucleic Acids Res. 2009;37:D396–403. doi: 10.1093/nar/gkn803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gene Ontology Annotation (UniProtKB-GOA) | Tutorial | EBI. http://www.ebi.ac.uk/GOA/annotationexample.html (24 May 2011, date last accessed)
- 42.Goecks J, Nekrutenko A, Taylor J, et al. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Galaxy | Published Page | Screencasts. http://main.g2.bx.psu.edu/screencast (24 May 2011, date last accessed)
- 44.Hull D, Wolstencroft K, Stevens R, et al. Taverna: a tool for building and running workflows of services. Nucleic Acids Res. 2006;34:W729–32. doi: 10.1093/nar/gkl320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Videos | Taverna. http://prototype.taverna.org.uk/documentation/taverna-2-x/videos/ (24 May 2011, date last accessed)
- 46.Obayashi T, Nishida K, Kasahara K, et al. ATTED-II updates: condition-specific gene coexpression to extend coexpression analyses and applications to a broad range of flowering plants. Plant Cell Physiol. 2011;52:213–9. doi: 10.1093/pcp/pcq203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. YouTube – ATTEDable’s Channel. http://www.youtube.com/user/ATTEDable (24 May 2011, date last accessed)
- 48. BioInformatics Tutorial Series (BITS) – Research Guides at MIT Libraries. http://libguides.mit.edu/bits (24 May 2011, date last accessed)
- 49. Video Tutorials | Countway Library of Medicine. https://www.countway.harvard.edu/menuNavigation/libraryServices/classes/videoTutorials.html (24 May 2011, date last accessed)
- 50. Tutorials | BIREC. http://birec.org/tutorials (24 May 2011, date last accessed)
- 51. OpenHelix: List of available tutorials. http://www.openhelix.com/cgi/tutorials.cgi (24 May 2011, date last accessed)
- 52. Tip Of The Week | The OpenHelix Blog. http://blog.openhelix.eu/?category_name=tip-of-the-week (24 May 2011, date last accessed)
- 53. TogoTV (in Japanese). http://togotv.dbcls.jp/ (24 May 2011, date last accessed)
- 54. TogoTV | distributing tutorial videos of bioinformatics resources. http://togotv.dbcls.jp/en/ (24 May 2011, date last accessed)
- 55. Integrated Database Project – MEXT-LSDB. http://lifesciencedb.jp/en/ (24 May 2011, date last accessed)
- 56.Katayama T, Nakao M, Takagi T. TogoWS: integrated SOAP and REST APIs for interoperable bioinformatics Web services. Nucleic Acids Res. 2010;38:W706–11. doi: 10.1093/nar/gkq386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamamoto Y, Takagi T. OReFiL: an online resource finder for life sciences. BMC Bioinformatics. 2007;8:287. doi: 10.1186/1471-2105-8-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schumacher G, Thurkettle MA. Camtasia studio: a teaching tool for academicians and clinicians. Comput Inform Nurs. 2007;25:130–4. doi: 10.1097/01.NCN.0000270038.29788.f1. [DOI] [PubMed] [Google Scholar]
- 59. TechSmith | Camtasia Screen Recorder Software, Home. http://www.techsmith.com/camtasia/ (24 May 2011, date last accessed)
- 60. DesktopToMovie (in Japanese). http://pencilsoftware.com/dtm.html (24 May 2011, date last accessed)
- 61. Comparison of screencasting software – Wikipedia, the free encyclopedia. http://en.wikipedia.org/wiki/Comparison_of_screencasting_software (24 May 2011, date last accessed)
- 62. tDiary | Download tDiary software for free at SourceForge.net. http://sourceforge.net/projects/tdiary/ (24 May 2011, date last accessed)
- 63. WordPress > Blog Tool and Publishing Platform. http://wordpress.org/ (24 May 2011, date last accessed)
- 64. LongTail Video | Home of the JW Player. http://www.longtailvideo.com/ (24 May 2011, date last accessed)
- 65. H264–Trac. http://h264.code-shop.com/trac/wiki (24 May 2011, date last accessed)
- 66. YouTube Blog: Upload limit increases to 15 minutes for all users. http://youtube-global.blogspot.com/2010/07/upload-limit-increases-to-15-minutes.html (24 May 2011, date last accessed)
- 67. YouTube Blog: Up, Up and Away – Long videos for more users. http://youtube-global.blogspot.com/2010/12/up-up-and-away-long-videos-for-more.html (24 May 2011, date last accessed)
- 68. TogoTV RSS feed. http://togotv.dbcls.jp/en/feed/ (24 May 2011, date last accessed)
- 69. TogoTV on Twitter. http://twitter.com/#!/togotv (24 May 2011, date last accessed)
- 70. Twitterfeed.com : feed your blog to twitter. http://twitterfeed.com/ (24 May 2011, date last accessed)
- 71. Integrated Database Project-MEXT-LSDB. http://lifesciencedb.jp/lsdb.cgi?gg=dbcatalog&lng=en (24 May 2011, date last accessed)
- 72. BodyParts3D. http://lifesciencedb.jp/bp3d/?lng=en (24 May 2011, date last accessed)
- 73.Mitsuhashi N, Fujieda K, Tamura T, et al. BodyParts3D: 3D structure database for anatomical concepts. Nucleic Acids Res. 2009;37:D782–5. doi: 10.1093/nar/gkn613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. TogoTV – How to use BodyParts3D/Anatomography 2010. http://togotv.dbcls.jp/20101117.html (24 May 2011, date last accessed)
- 75.Haider S, Ballester B, Smedley D, et al. BioMart Central Portal-unified access to biological data. Nucleic Acids Res. 2009;37:W23–7. doi: 10.1093/nar/gkp265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. TogoTV – How to make probeID list for microarray using Biomart. http://togotv.dbcls.jp/20090304.html (24 May 2011, date last accessed)
- 77. BioMart – MartView. http://www.biomart.org/biomart/martview/ (24 May 2011, date last accessed)
- 78. TogoTV – togoTV – JSBi2007. http://togotv.dbcls.jp/20071219.html (24 May 2011, date last accessed)
- 79.Nakazato T, Bono H, Matsuda H, et al. Gendoo: functional profiling of gene and disease features using MeSH vocabulary. Nucleic Acids Res. 2009;37:W166–9. doi: 10.1093/nar/gkp483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. TogoTV – Functional profiling of OMIM data using MeSH vocabulary – JSBi2008. http://togotv.dbcls.jp/20081219.html (24 May 2011, date last accessed)
- 81. TogoTV – Copyright in the Digital Age and Its Impact on Scientific Data Sharing. http://togotv.dbcls.jp/20100512.html (24 May 2011, date last accessed)
- 82. Workshop on Parallel and Distributed Processing of Large Genome Data. http://mlab.cb.k.u-tokyo.ac.jp/en/events/lgd/ (24 May 2011, date last accessed)
- 83. AJACS – MotDB (in Japanese). http://motdb.dbcls.jp/?AJACS (24 May 2011, date last accessed)
- 84. AWStats - Free advanced log file analyzer for web, ftp or mail statistics (GNU GPL). http://awstats.sourceforge.net/ (24 May 2011, date last accessed)
- 85.Untergasser A, Nijveen H, Rao X, et al. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007;35:W71–4. doi: 10.1093/nar/gkm306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Croft D, O'Kelly G, Wu G, et al. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 2011;39:D691–7. doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Altschul SF, Madden TL, Schäffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chenna R, Sugawara H, Koike T, et al. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Result – LSDB Evaluation Questionnaire (in Japanese). http://lifesciencedb.jp/feedback/hyouka20/?result#i760c229 (24 May 2011, date last accessed)