Abstract
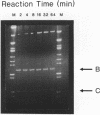
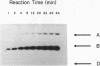
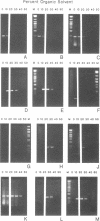
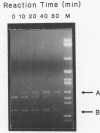
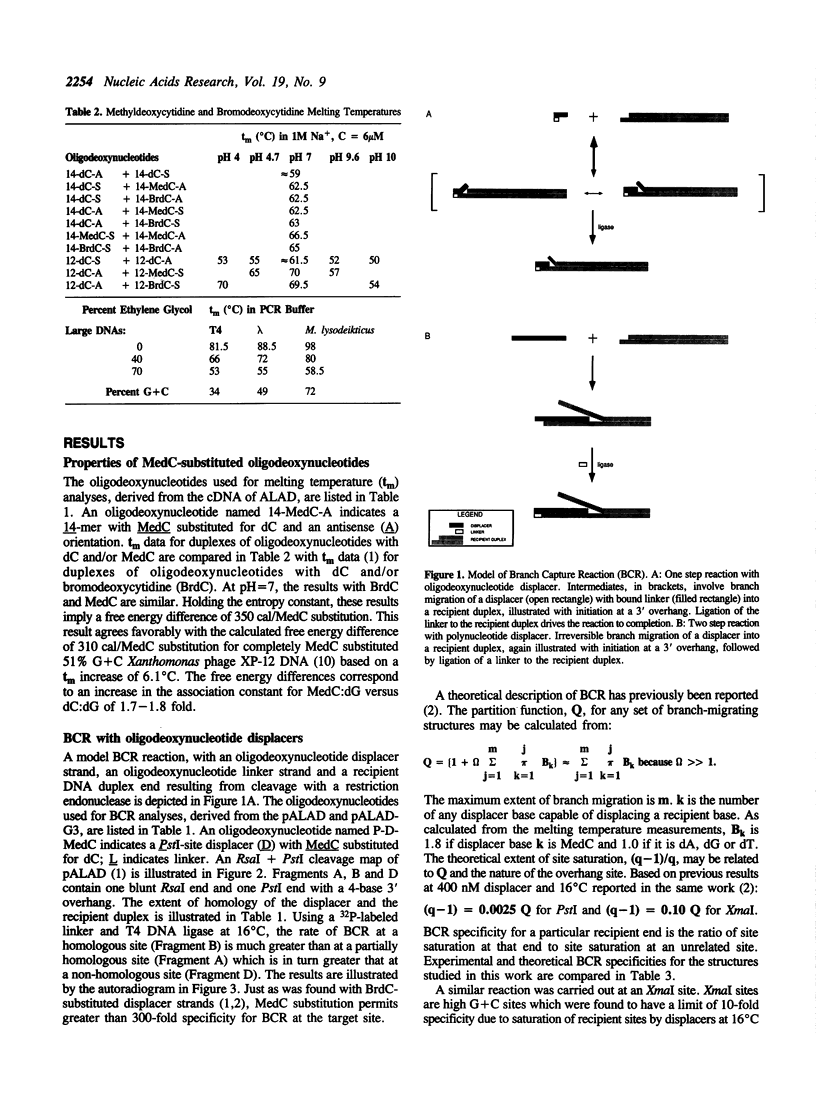
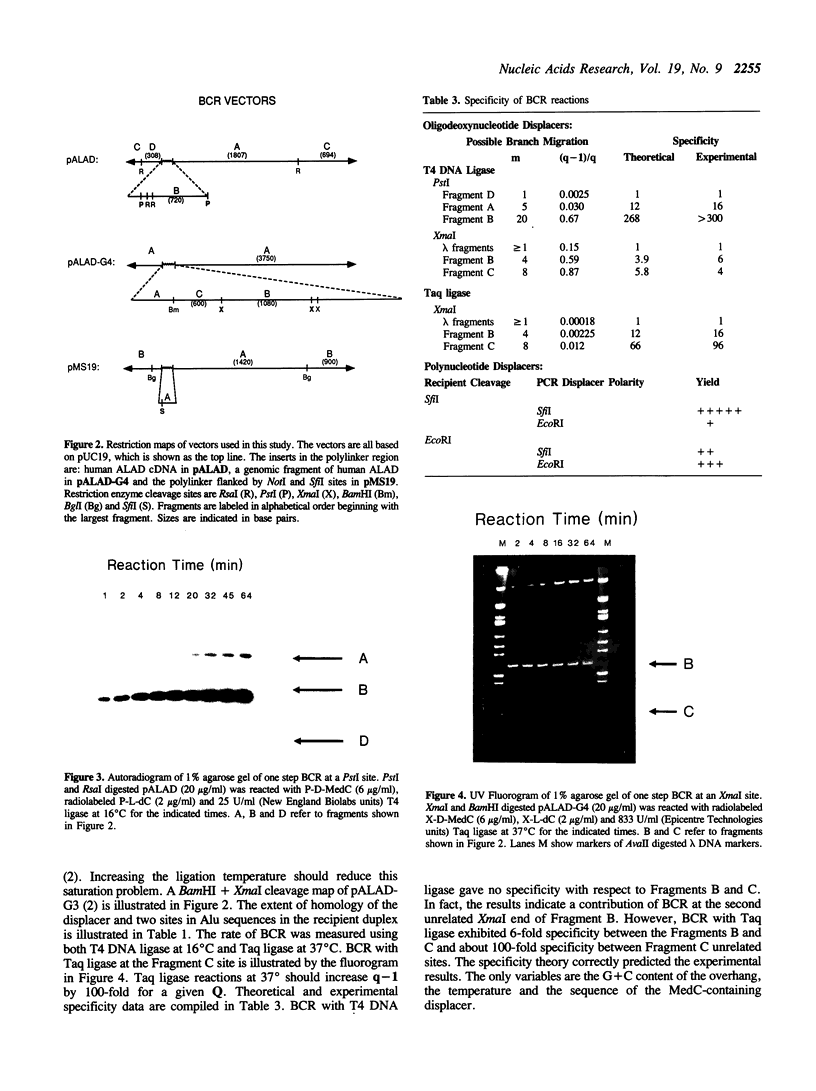
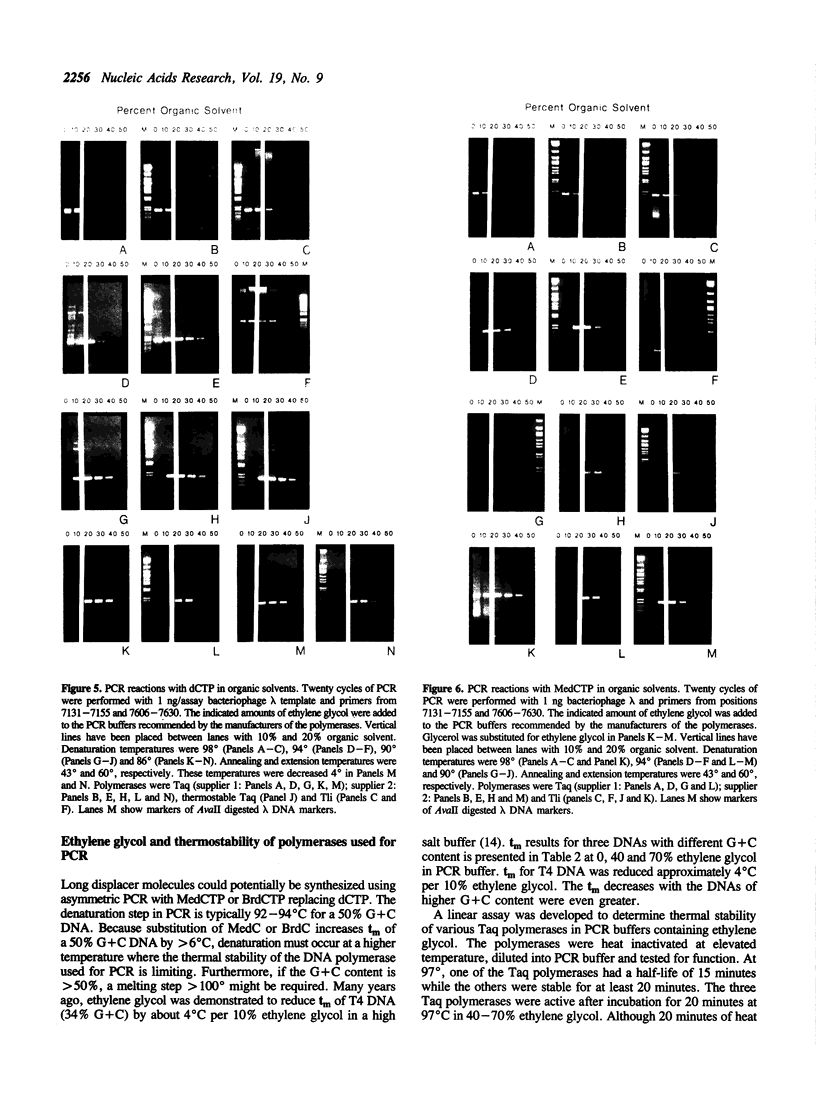
Branch capture reactions (BCR) contain three DNA species: (i) a recipient restriction fragment terminating in an overhang, (ii) a displacer strand containing two adjacent sequences, with one complementary to the overhang and to contiguous nucleotides within the recipient duplex and (iii) a linker which is complementary to the second displacer sequence. Branched complexes containing all three species may be captured by ligation of the linker to the recipient overhang. The use of 5-MedC in the displacer facilitates BCR. High temperature ligation with a thermostable enzyme increased specificity for ligation to the correct recipient in a complex mixture of restriction fragments. Displacer synthesis by PCR permitted separate reactions of formation of stable displacement complexes and of high-temperature ligation. Ethylene glycol-containing buffer permitted PCR with 5-MedCTP or high G + C products using thermostable polymerases. BCR may be used to modify the ends of one recipient DNA duplex in a population of duplex DNA fragments. Modification of the recipient could be used to facilitate detection, affinity chromatography or cloning. By using PCR to obtain a BCR displacer, the sequence non-homologous to the recipient duplex may be expanded to include the sequence of a selectable marker, thus facilitating chromosome walking.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beattie K. L., Wiegand R. C., Radding C. M. Uptake of homologous single-stranded fragments by superhelical DNA. II. Characterization of the reaction. J Mol Biol. 1977 Nov;116(4):783–803. doi: 10.1016/0022-2836(77)90271-6. [DOI] [PubMed] [Google Scholar]
- DasGupta C., Radding C. M. Polar branch migration promoted by recA protein: effect of mismatched base pairs. Proc Natl Acad Sci U S A. 1982 Feb;79(3):762–766. doi: 10.1073/pnas.79.3.762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M., Ehrlich K., Mayo J. A. Unusual properties of the DNA from Xanthomonas phage XP-12 in which 5-methylcytosine completely replaces cytosine. Biochim Biophys Acta. 1975 Jun 16;395(2):109–119. doi: 10.1016/0005-2787(75)90149-5. [DOI] [PubMed] [Google Scholar]
- Green C., Tibbetts C. Reassociation rate limited displacement of DNA strands by branch migration. Nucleic Acids Res. 1981 Apr 24;9(8):1905–1918. doi: 10.1093/nar/9.8.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyllensten U. B., Erlich H. A. Generation of single-stranded DNA by the polymerase chain reaction and its application to direct sequencing of the HLA-DQA locus. Proc Natl Acad Sci U S A. 1988 Oct;85(20):7652–7656. doi: 10.1073/pnas.85.20.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloman W. K., Wiegand R., Hoessli C., Radding C. M. Uptake of homologous single-stranded fragments by superhelical DNA: a possible mechanism for initiation of genetic recombination. Proc Natl Acad Sci U S A. 1975 Jun;72(6):2394–2398. doi: 10.1073/pnas.72.6.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honigberg S. M., Rao B. J., Radding C. M. Ability of RecA protein to promote a search for rare sequences in duplex DNA. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9586–9590. doi: 10.1073/pnas.83.24.9586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandpal R. P., Ward D. C., Weissman S. M. Selective enrichment of a large size genomic DNA fragment by affinity capture: an approach for genome mapping. Nucleic Acids Res. 1990 Apr 11;18(7):1789–1795. doi: 10.1093/nar/18.7.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. S., Davis R. W., Davidson N. A physical study by electron microscopy of the terminally reptitious, circularly permuted DNA from the coliphage particles of Escherichia coli 15. J Mol Biol. 1970 Feb 28;48(1):1–22. doi: 10.1016/0022-2836(70)90215-9. [DOI] [PubMed] [Google Scholar]
- Quartin R. S., Plewinska M., Wetmur J. G. Branch migration mediated DNA labeling and cloning. Biochemistry. 1989 Oct 31;28(22):8676–8682. doi: 10.1021/bi00448a002. [DOI] [PubMed] [Google Scholar]
- Quartin R. S., Wetmur J. G. Effect of ionic strength on the hybridization of oligodeoxynucleotides with reduced charge due to methylphosphonate linkages to unmodified oligodeoxynucleotides containing the complementary sequence. Biochemistry. 1989 Feb 7;28(3):1040–1047. doi: 10.1021/bi00429a018. [DOI] [PubMed] [Google Scholar]
- Radding C. M., Beattie K. L., Holloman W. K., Wiegand R. C. Uptake of homologous single-stranded fragments by superhelical DNA. IV. Branch migration. J Mol Biol. 1977 Nov;116(4):825–839. doi: 10.1016/0022-2836(77)90273-x. [DOI] [PubMed] [Google Scholar]
- Rigas B., Welcher A. A., Ward D. C., Weissman S. M. Rapid plasmid library screening using RecA-coated biotinylated probes. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9591–9595. doi: 10.1073/pnas.83.24.9591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Stürzl M., Roth W. K. "Run-off" synthesis and application of defined single-stranded DNA hybridization probes. Anal Biochem. 1990 Feb 15;185(1):164–169. doi: 10.1016/0003-2697(90)90274-d. [DOI] [PubMed] [Google Scholar]
- Takahashi M., Yamaguchi E., Uchida T. Thermophilic DNA ligase. Purification and properties of the enzyme from Thermus thermophilus HB8. J Biol Chem. 1984 Aug 25;259(16):10041–10047. [PubMed] [Google Scholar]
- Thomas M., White R. L., Davis R. W. Hybridization of RNA to double-stranded DNA: formation of R-loops. Proc Natl Acad Sci U S A. 1976 Jul;73(7):2294–2298. doi: 10.1073/pnas.73.7.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock P. H., Wetmur J. G. Branch capture reactions: effect of recipient structure. Nucleic Acids Res. 1990 Jul 25;18(14):4207–4213. doi: 10.1093/nar/18.14.4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetmur J. G., Bishop D. F., Cantelmo C., Desnick R. J. Human delta-aminolevulinate dehydratase: nucleotide sequence of a full-length cDNA clone. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7703–7707. doi: 10.1073/pnas.83.20.7703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetmur J. G., Davidson N. Kinetics of renaturation of DNA. J Mol Biol. 1968 Feb 14;31(3):349–370. doi: 10.1016/0022-2836(68)90414-2. [DOI] [PubMed] [Google Scholar]
- Wetmur J. G. Hybridization and renaturation kinetics of nucleic acids. Annu Rev Biophys Bioeng. 1976;5:337–361. doi: 10.1146/annurev.bb.05.060176.002005. [DOI] [PubMed] [Google Scholar]