Abstract
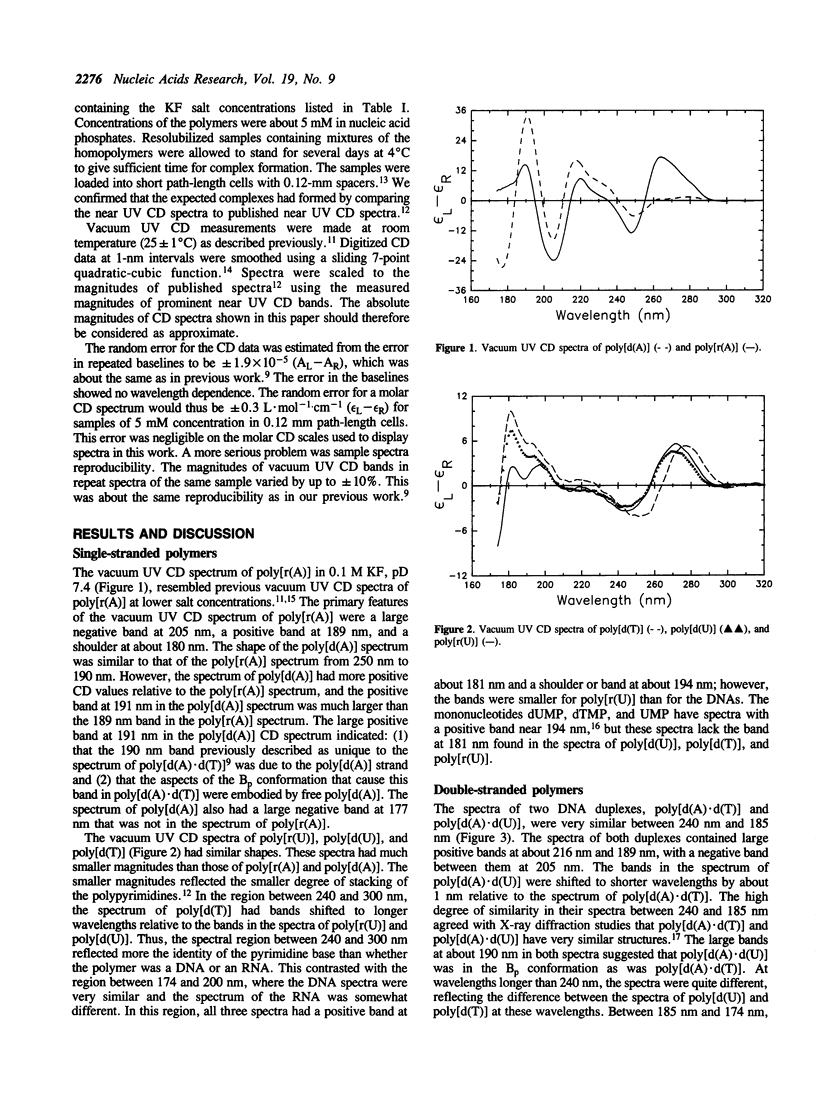
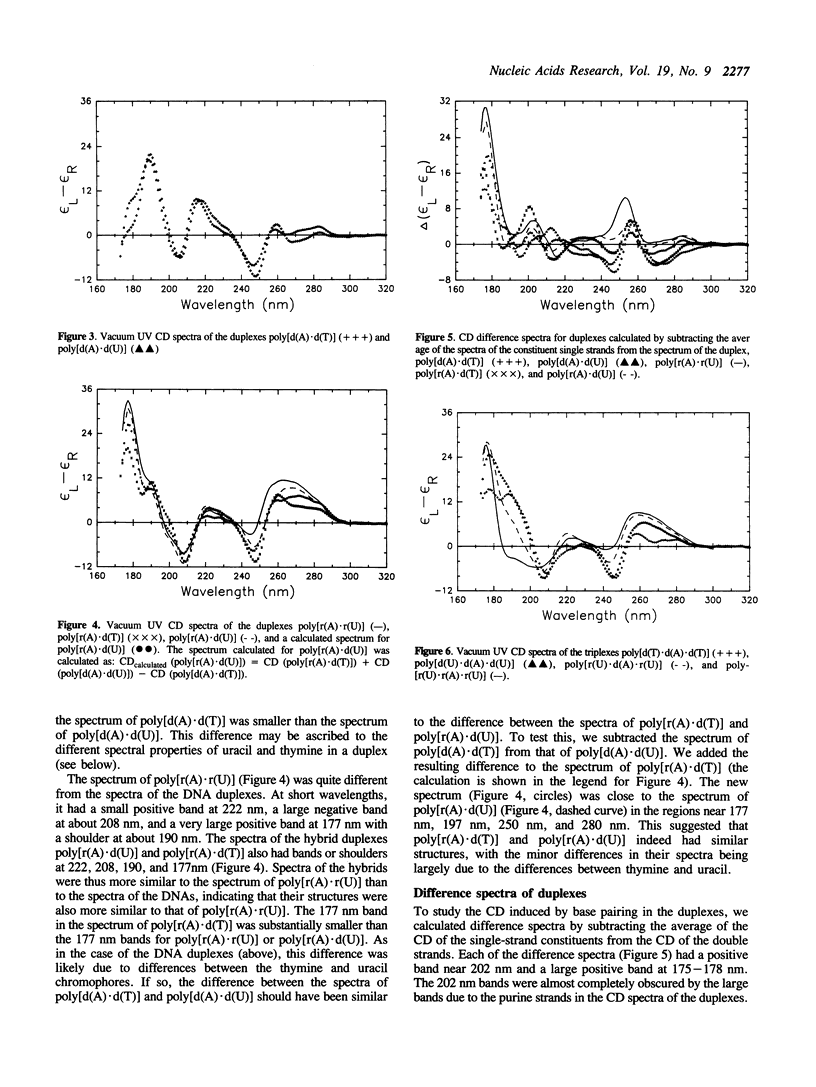
Vacuum UV circular dichroism (CD) spectra were measured down to 174 nm for five homopolymers, five duplexes, and four triplexes containing adenine, uracil, and thymine. Near 190 nm, the CD bands of poly[d(A)] and poly[r(A)] were larger than the CD bands of the polypyrimidines, poly[d(T)], poly[d(U)], and poly[r(U)]. Little change was observed in the 190 nm region upon formation of the duplexes (poly[d(A).d(T)], poly[d(A).d(U)], poly[r(A).d(T)], poly[r(A).d(U)], and poly[r(A).r(U)]) or upon formation of two of the triplexes (poly[d(T).d(A).d(T)] and poly[d(U).d(A).d(U)]). This showed that the purine strand had the same or a similar structure in these duplexes and triplexes as when free in solution. Both A.U and A.T base pairing induced positive bands at 177 and 202 nm. For three triplexes containing poly[d(A)], the formation of a triplex from a duplex and a free pyrimidine strand induced a negative band centered between 210 and 215 nm. The induction of a band between 210 and 215 nm indicated that these triplexes had aspects of the A conformation.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Antao V. P., Gray D. M., Ratliff R. L. CD of six different conformational rearrangements of poly[d(A-G).d(C-T)] induced by low pH. Nucleic Acids Res. 1988 Jan 25;16(2):719–738. doi: 10.1093/nar/16.2.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnott S., Bond P. J., Selsing E., Smith P. J. Models of triple-stranded polynucleotides with optimised stereochemistry. Nucleic Acids Res. 1976 Oct;3(10):2459–2470. doi: 10.1093/nar/3.10.2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnott S., Bond P. J. Structures for Poly(U)-poly(A)-poly(U)triple stranded polynucleotides. Nat New Biol. 1973 Jul 25;244(134):99–101. doi: 10.1038/newbio244099a0. [DOI] [PubMed] [Google Scholar]
- Arnott S., Hukins D. W., Dover S. D., Fuller W., Hodgson A. R. Structures of synthetic polynucleotides in the A-RNA and A'-RNA conformations: x-ray diffraction analyses of the molecular conformations of polyadenylic acid--polyuridylic acid and polyinosinic acid--polycytidylic acid. J Mol Biol. 1973 Dec 5;81(2):107–122. doi: 10.1016/0022-2836(73)90183-6. [DOI] [PubMed] [Google Scholar]
- Arnott S., Selsing E. Structures for the polynucleotide complexes poly(dA) with poly (dT) and poly(dT) with poly(dA) with poly (dT). J Mol Biol. 1974 Sep 15;88(2):509–521. doi: 10.1016/0022-2836(74)90498-7. [DOI] [PubMed] [Google Scholar]
- Aymami J., Coll M., Frederick C. A., Wang A. H., Rich A. The propeller DNA conformation of poly(dA).poly(dT). Nucleic Acids Res. 1989 Apr 25;17(8):3229–3245. doi: 10.1093/nar/17.8.3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benevides J. M., Thomas G. J., Jr A solution structure for poly(rA).poly(dT) with different furanose pucker and backbone geometry in rA and dT strands and intrastrand hydrogen bonding of adenine 8CH. Biochemistry. 1988 May 17;27(10):3868–3873. doi: 10.1021/bi00410a051. [DOI] [PubMed] [Google Scholar]
- Causley G. C., Johnson W. C., Jr Polynucleotide conformation from flow dichroism studies. Biopolymers. 1982 Sep;21(9):1763–1780. doi: 10.1002/bip.360210907. [DOI] [PubMed] [Google Scholar]
- Chandrasekaran R., Radha A., Park H. S., Arnott S. Structure of the beta-form of poly d(A).poly d(U). J Biomol Struct Dyn. 1989 Jun;6(6):1203–1215. doi: 10.1080/07391102.1989.10506545. [DOI] [PubMed] [Google Scholar]
- Coll M., Frederick C. A., Wang A. H., Rich A. A bifurcated hydrogen-bonded conformation in the d(A.T) base pairs of the DNA dodecamer d(CGCAAATTTGCG) and its complex with distamycin. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8385–8389. doi: 10.1073/pnas.84.23.8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooney M., Czernuszewicz G., Postel E. H., Flint S. J., Hogan M. E. Site-specific oligonucleotide binding represses transcription of the human c-myc gene in vitro. Science. 1988 Jul 22;241(4864):456–459. doi: 10.1126/science.3293213. [DOI] [PubMed] [Google Scholar]
- Gray D. M., Johnson K. H., Vaughan M. R., Morris P. A., Sutherland J. C., Ratliff R. L. The vacuum UV CD bands of repeating DNA sequences are dependent on sequence and conformation. Biopolymers. 1990 Feb 5;29(2):317–323. doi: 10.1002/bip.360290204. [DOI] [PubMed] [Google Scholar]
- Gray D. M., Lang D., Kuner E., Vaughan M., Sutherland J. A thin quartz cell suitable for vacuum ultraviolet absorption and circular dichroism measurements. Anal Biochem. 1984 Jan;136(1):247–250. doi: 10.1016/0003-2697(84)90331-2. [DOI] [PubMed] [Google Scholar]
- Gray D. M., Ratliff R. L. Circular dichroism spectra of poly[d(AC):d(GT)], poly[r(AC):r(GU)], and hybrids poly[d(AC):r(GU)] and poly[r(AC):d(GT)] in the presence of ethanol. Biopolymers. 1975 Mar;14(3):487–498. doi: 10.1002/bip.1975.360140305. [DOI] [PubMed] [Google Scholar]
- Johnson K. H., Gray D. M., Morris P. A., Sutherland J. C. A.U and G.C base pairs in synthetic RNAS have characteristic vacuum UV CD bands. Biopolymers. 1990 Feb 5;29(2):325–333. doi: 10.1002/bip.360290205. [DOI] [PubMed] [Google Scholar]
- Krueger W. C., Li L. H., Moscowitz A., Prairie M. D., Petzold G., Swenson D. H. Binding of CC-1065 to poly- and oligonucleotides. Biopolymers. 1985 Aug;24(8):1549–1572. doi: 10.1002/bip.360240811. [DOI] [PubMed] [Google Scholar]
- Manzini G., Xodo L. E., Gasparotto D., Quadrifoglio F., van der Marel G. A., van Boom J. H. Triple helix formation by oligopurine-oligopyrimidine DNA fragments. Electrophoretic and thermodynamic behavior. J Mol Biol. 1990 Jun 20;213(4):833–843. doi: 10.1016/S0022-2836(05)80267-0. [DOI] [PubMed] [Google Scholar]
- Moser H. E., Dervan P. B. Sequence-specific cleavage of double helical DNA by triple helix formation. Science. 1987 Oct 30;238(4827):645–650. doi: 10.1126/science.3118463. [DOI] [PubMed] [Google Scholar]
- Nelson H. C., Finch J. T., Luisi B. F., Klug A. The structure of an oligo(dA).oligo(dT) tract and its biological implications. Nature. 1987 Nov 19;330(6145):221–226. doi: 10.1038/330221a0. [DOI] [PubMed] [Google Scholar]
- O'Connor T., Bina M. The structure of triple helical poly(U).poly(A).poly(U) studied by Raman spectroscopy. J Biomol Struct Dyn. 1984 Dec;2(3):615–625. doi: 10.1080/07391102.1984.10507595. [DOI] [PubMed] [Google Scholar]
- Sprecher C. A., Johnson W. C., Jr Circular dichroism of the nucleic acid monomers. Biopolymers. 1977 Oct;16(10):2243–2264. doi: 10.1002/bip.1977.360161012. [DOI] [PubMed] [Google Scholar]
- Steely H. T., Jr, Gray D. M., Ratliff R. L. CD of homopolymer DNA-RNA hybrid duplexes and triplexes containing A-T or A-U base pairs. Nucleic Acids Res. 1986 Dec 22;14(24):10071–10090. doi: 10.1093/nar/14.24.10071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel S. A., Dervan P. B. Site-specific cleavage of a yeast chromosome by oligonucleotide-directed triple-helix formation. Science. 1990 Jul 6;249(4964):73–75. doi: 10.1126/science.2195655. [DOI] [PubMed] [Google Scholar]



