Abstract
We have analysed DNA from African cassava mosaic virus (ACMV)-infected Nicotiana benthamiana by two-dimensional agarose gel electrophoresis and detected ACMV-specific DNAs by blot-hybridisation. ACMV DNA forms including the previously characterised single-stranded, open-circular, linear and supercoiled DNAs along with five previously uncharacterised heterogeneous DNAs (H1-H5) were resolved. The heterogeneous DNAs were characterised by their chromatographic properties on BND-cellulose and their ability to hybridise to strand-specific and double-stranded probes. The data suggest a rolling circle mechanism of DNA replication, based on the sizes and strand specificity of the heterogeneous single-stranded DNA forms and their electrophoretic properties in relation to genome length single-stranded DNAs. Second-strand synthesis on a single-stranded virus-sense template is evident from the position of heterogeneous subgenomic complementary-sense DNA (H3) associated with genome-length virus-sense template (VT) DNA. The position of heterogeneous virus-sense DNA (H5), ranging in size from one to two genome lengths, is consistent with its association with genome-length complementary-sense template (CT) DNA, reflecting virus-sense strand displacement during replication from a double-stranded intermediate. The absence of subgenomic complementary-sense DNA associated with the displaced virus-sense strand suggests that replication proceeds via an obligate single-stranded intermediate. The other species of heterogeneous DNAs comprised concatemeric single-stranded virus-sense DNA (H4), and double-stranded or partially single-stranded DNA (H1 and H2).
Full text
PDF



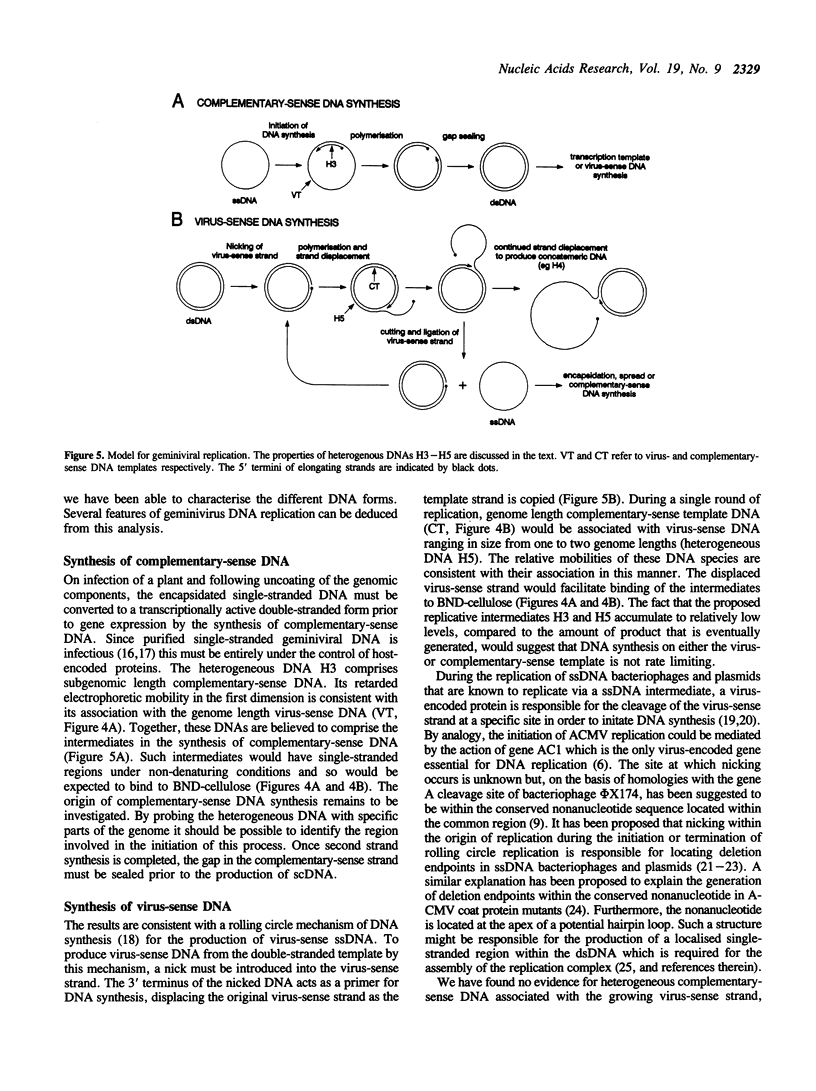

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Covey S. N., Hull R. Transcription of cauliflower mosaic virus DNA. Detection of transcripts, properties, and location of the gene encoding the virus inclusion body protein. Virology. 1981 Jun;111(2):463–474. doi: 10.1016/0042-6822(81)90349-4. [DOI] [PubMed] [Google Scholar]
- Etessami P., Callis R., Ellwood S., Stanley J. Delimitation of essential genes of cassava latent virus DNA 2. Nucleic Acids Res. 1988 Jun 10;16(11):4811–4829. doi: 10.1093/nar/16.11.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etessami P., Saunders K., Watts J., Stanley J. Mutational analysis of complementary-sense genes of African cassava mosaic virus DNA A. J Gen Virol. 1991 May;72(Pt 5):1005–1012. doi: 10.1099/0022-1317-72-5-1005. [DOI] [PubMed] [Google Scholar]
- Etessami P., Watts J., Stanley J. Size reversion of African cassava mosaic virus coat protein gene deletion mutants during infection of Nicotiana benthamiana. J Gen Virol. 1989 Feb;70(Pt 2):277–289. doi: 10.1099/0022-1317-70-2-277. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Gay N. J., Walker J. E. Homology between human bladder carcinoma oncogene product and mitochondrial ATP-synthase. Nature. 1983 Jan 20;301(5897):262–264. doi: 10.1038/301262a0. [DOI] [PubMed] [Google Scholar]
- Gilbert W., Dressler D. DNA replication: the rolling circle model. Cold Spring Harb Symp Quant Biol. 1968;33:473–484. doi: 10.1101/sqb.1968.033.01.055. [DOI] [PubMed] [Google Scholar]
- Goodman R. M. Single-stranded DNA genome in a whitefly-transmitted plant virus. Virology. 1977 Nov;83(1):171–179. doi: 10.1016/0042-6822(77)90220-3. [DOI] [PubMed] [Google Scholar]
- Gros M. F., te Riele H., Ehrlich S. D. Replication origin of a single-stranded DNA plasmid pC194. EMBO J. 1989 Sep;8(9):2711–2716. doi: 10.1002/j.1460-2075.1989.tb08412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros M. F., te Riele H., Ehrlich S. D. Rolling circle replication of single-stranded DNA plasmid pC194. EMBO J. 1987 Dec 1;6(12):3863–3869. doi: 10.1002/j.1460-2075.1987.tb02724.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss A., Ehrlich S. D. The family of highly interrelated single-stranded deoxyribonucleic acid plasmids. Microbiol Rev. 1989 Jun;53(2):231–241. doi: 10.1128/mr.53.2.231-241.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiger J. A., Jr, Sinsheimer R. L. Vegetative lambda DNA. IV. Fractionation of replicating lambda DNA on benzoylated-naphthoylated DEAE cellulose. J Mol Biol. 1969 Mar 28;40(3):467–490. doi: 10.1016/0022-2836(69)90166-1. [DOI] [PubMed] [Google Scholar]
- Michel B., Ehrlich S. D. Illegitimate recombination at the replication origin of bacteriophage M13. Proc Natl Acad Sci U S A. 1986 May;83(10):3386–3390. doi: 10.1073/pnas.83.10.3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel B., Ehrlich S. D. Illegitimate recombination occurs between the replication origin of the plasmid pC194 and a progressing replication fork. EMBO J. 1986 Dec 20;5(13):3691–3696. doi: 10.1002/j.1460-2075.1986.tb04701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noirot P., Bargonetti J., Novick R. P. Initiation of rolling-circle replication in pT181 plasmid: initiator protein enhances cruciform extrusion at the origin. Proc Natl Acad Sci U S A. 1990 Nov;87(21):8560–8564. doi: 10.1073/pnas.87.21.8560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers S. G., Bisaro D. M., Horsch R. B., Fraley R. T., Hoffmann N. L., Brand L., Elmer J. S., Lloyd A. M. Tomato golden mosaic virus A component DNA replicates autonomously in transgenic plants. Cell. 1986 May 23;45(4):593–600. doi: 10.1016/0092-8674(86)90291-6. [DOI] [PubMed] [Google Scholar]
- Saunders K., Lucy A. P., Covey S. N. Susceptibility of Brassica species to cauliflower mosaic virus infection is related to a specific stage in the virus multiplication cycle. J Gen Virol. 1990 Aug;71(Pt 8):1641–1647. doi: 10.1099/0022-1317-71-8-1641. [DOI] [PubMed] [Google Scholar]
- Stanley J., Townsend R. Characterisation of DNA forms associated with cassava latent virus infection. Nucleic Acids Res. 1985 Apr 11;13(7):2189–2206. doi: 10.1093/nar/13.7.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunter G., Bisaro D. M. Transactivation in a geminivirus: AL2 gene product is needed for coat protein expression. Virology. 1991 Jan;180(1):416–419. doi: 10.1016/0042-6822(91)90049-h. [DOI] [PubMed] [Google Scholar]
- Sunter G., Hartitz M. D., Hormuzdi S. G., Brough C. L., Bisaro D. M. Genetic analysis of tomato golden mosaic virus: ORF AL2 is required for coat protein accumulation while ORF AL3 is necessary for efficient DNA replication. Virology. 1990 Nov;179(1):69–77. doi: 10.1016/0042-6822(90)90275-v. [DOI] [PubMed] [Google Scholar]
- Townsend R., Stanley J., Curson S. J., Short M. N. Major polyadenylated transcripts of cassava latent virus and location of the gene encoding coat protein. EMBO J. 1985 Jan;4(1):33–37. doi: 10.1002/j.1460-2075.1985.tb02313.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend R., Watts J., Stanley J. Synthesis of viral DNA forms in Nicotiana plumbaginifolia protoplasts inoculated with cassava latent virus (CLV); evidence for the independent replication of one component of the CLV genome. Nucleic Acids Res. 1986 Feb 11;14(3):1253–1265. doi: 10.1093/nar/14.3.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]