Abstract
Summary: Detection of viruses by the innate immune system involves the action of specialized pattern recognition receptors. Intracellular RIG-I receptors sense the presence of viral nucleic acids in infected cells and trigger signaling pathways that lead to the production of proinflammatory and antiviral proteins. Over the past few years, posttranslational modification of RIG-I and downstream signaling proteins by different types of ubiquitination has been found to be a key event in the regulation of RIG-I-induced NF-κB and interferon regulatory factor 3 (IRF3) activation. Multiple ubiquitin ligases, deubiquitinases, and ubiquitin binding scaffold proteins contribute to both positive and negative regulation of the RIG-I-induced antiviral immune response. A better understanding of the function and activity of these proteins might eventually lead to the development of novel therapeutic approaches for management of viral diseases.
INTRODUCTION
Ubiquitination is one of the most versatile posttranslational modifications and is indispensable for cellular homeostasis. Ubiquitin precursors are posttranslationally processed into peptides of 76 amino acids (5), and covalent attachment of these peptides to target proteins alters their functional properties. The transfer of ubiquitin to its substrate occurs in a three-step enzymological process. Ubiquitin is first activated by formation of a high-energy thioester bond with a ubiquitin-activating enzyme (E1) in an ATP-dependent manner. Ubiquitin is then transferred to a ubiquitin-conjugating enzyme (E2) (165), which, in association with a third enzyme called the ubiquitin ligase or E3, determines the substrate specificity and transfers a single activated ubiquitin molecule to the ε-amino group of a lysine residue on the target protein. Only two ubiquitin-specific E1 enzymes and 38 E2 enzymes have been identified in humans, but about 600 E3 ubiquitin ligases are encoded in the human genome. The E3 family can be divided according to the presence of conserved catalytic domains into three groups: HECT (homologous to E6-associated protein [E6AP] C terminus) (116) and those containing a U box (43) or RING (“really interesting new gene”) domain (19). Each E3 also harbors distinct protein interaction motifs that play a role in determining substrate specificity.
Ubiquitination can be monomeric, but the initial attachment of a single ubiquitin molecule to its substrate is typically followed by attachment of a ubiquitin peptide to start the formation of multimeric polyubiquitin chains. During this process, each of the seven lysine residues of ubiquitin (K6, K11, K27, K29, K33, K48, and K63) can be used to generate isopeptide bonds between sequential ubiquitin molecules. Additionally, ubiquitin can be connected head to tail by linking the carboxyl terminus of one ubiquitin molecule to the amino terminus of the next molecule to generate linear ubiquitin chains. Each chain takes on a distinct three-dimensional conformation that can be recognized by distinct ubiquitin binding domains (UBDs) present in a wide variety of proteins. The type of ubiquitination determines the fate of the ubiquitinated protein. For example, K48-linked polyubiquitination is required for proteosomal degradation of a protein, whereas K63-linked polyubiquitination is associated with nondegradative signaling events (51). Although K63-polyubiquitin chains have been shown to bind the proteasome in vitro, there is no evidence that K63-polyubiquitinated proteins are targeted for proteasomal degradation, possibly because these ubiquitin chains are bound to proteins containing certain UBDs and thus are not delivered to the proteasome. Protein ubiquitination is reversible. It is estimated that the human genome encodes approximately 100 ubiquitin-specific proteases (95) called deubiquitinating enzymes (DUBs). These enzymes can reverse ubiquitination by cleaving off ubiquitin chains at their base or by depolymerizing ubiquitin stretches. Based on domain homology, DUBs can be further classified into five families: four groups of cysteine proteases, namely, the ubiquitin C-terminal hydrolases (UCHs), ubiquitin-specific proteases (USPs), Josephin domain, and ovarian tumor proteases (OTU), and a group of metalloproteases called JAB1/MPN/MOV34 metalloenzymes or JAMMs (62). Most DUBs are modular and contain additional protein-protein interaction domains or ubiquitin binding domains, both of which help to determine DUB substrate and ubiquitin chain linkage specificity (115). Interestingly, DUBs associate in multimeric protein complexes with E3 ligases, and in this way they directly regulate ubiquitin ligase activity (151).
Over the past few years, ubiquitination has been implicated in the regulation of numerous cellular processes, such as protein turnover (7, 14), cell cycle regulation, cell death, endocytosis, autophagy, DNA repair (1), and initiation and regulation of the innate and adaptive immune responses (9, 80). The role of ubiquitin in the activation of NF-κB, an important transcription factor in innate immune regulation, has been extensively studied and reviewed, especially in the context of signaling by tumor necrosis factor (TNF) receptor I (TNFRI) and Toll-like receptors (s) (42, 133, 160). However, recent research also indicates that ubiquitination has an important role in signal transduction during the innate antiviral immune response. The most compelling evidence for this probably came from a study that generated a cell line for inducible replacement of the four endogenous ubiquitin-encoding genes with a K63R mutant form of ubiquitin, which prevents the formation of K63-linked polyubiquitin chains (164). When these cells were infected with Sendai virus, antiviral signal transduction could not proceed in the absence of K63-specific ubiquitin chain formation (175). Conversely, restoring the system with wild-type ubiquitin or a K48R ubiquitin mutant had no significant effect. Many viruses have developed strategies to downmodulate the cellular antiviral response in order to promote their own propagation and to evade the host immune response. These strategies include the action of virus-encoded E3 ligases or DUBs that modulate the ubiquitination status of host cell proteins (73, 110, 112, 156). These phenomena further highlight the importance of ubiquitin-mediated signaling in protection against these pathogens.
THE FAMILY OF RIG-I-LIKE RECEPTORS
Viral infection induces a strong antiviral immune response characterized by robust production of proinflammatory cytokines and antiviral type I interferons (type I IFN). Expression of type I IFN is directed mainly by the transcription factors nuclear factor-κB (NF-κB) and interferon regulatory factor (IRF) (47). The release of type I IFN promotes the transcription of a variety of genes that lead to limitation of further viral replication. Type I IFN secretion also contributes to the activation of several types of immune cells, including dendritic cells (DC), natural killer cells, monocytes, and macrophages, and it shapes the adaptive immune response by acting directly on T and B cells.
The innate immune response against invading pathogens is often initiated by detection of conserved molecular patterns by pathogen recognition receptors (PRRs) (139). Unlike bacteria, fungi, and parasites, viruses rarely contain signature molecules that can be detected by PRRs because they employ the cellular machinery to replicate, and so the viral proteins are indistinguishable from self proteins. Therefore, the host relies on detection of viral nucleic acid by two types of PRRs: nucleic acid-sensing TLRs (TLR3, -7, -8, and -9), which are localized in the endolysosomes, and RIG-I (retinoic acid-inducible gene I)-like receptors (RLRs) and viral DNA sensors, which reside in the cytoplasm (97). Although RLRs are generally considered to be the main PRRs for sensing RNA viruses, recent studies also implicate additional cytoplasmic RNA sensors, including members of the NOD-like receptor (NLR) family, such as NOD2 (119) and NLRC5 (94), and the DExD/H helicases DDX1, DDX21, and DDX36 (178). The signaling events that depart from these receptors are beyond the scope of this review. So far, three members of the RLR family have been described: RIG-I, MDA5 (melanoma differentiation-associated gene 5), and LGP2 (Laboratory of Genetics and Physiology 2), which bind to structurally different RNA molecules (92, 166). All three members of the RLR family are characterized by the presence of a central DExD/H box RNA helicase domain. RIG-I and MDA5, but not LGP2, also carry two N-terminal CARDs (caspase activation and recruitment domains) that are essential for initiating downstream signaling events. Ligand binding occurs at the carboxyl-terminal domain (CTD), which contains a positively charged RNA binding pocket (15, 138). Additionally, the C-terminal ends of both RIG-I and LGP2 contain a regulatory domain (RD) that keeps these RNA sensors in an inactive conformation in the absence of their respective ligands.
Most cell types rely on RLRs for sensing viral nucleic acids, but plasmacytoid dendritic cells (pDC) do not. pDC, formerly referred to as natural interferon-producing cells, are specialized type I IFN producers and rely solely on the nucleic acid sensing TLRs (mainly TLR7, -8, and -9) for the robust production of type I IFNs (56). RIG-I is indispensable for type I IFN responses to many single-stranded RNA viruses, including negative-stranded viruses of the families Orthomyxoviridae (including influenza A virus) and Paramyxoviridae (such as mumps virus, measles virus, and Sendai virus) and positive-stranded viruses, e.g., hepatitis C virus (78). RIG-I-deficient cells fail to induce an antiviral immune response against these viruses (56, 58). Similarly, MDA5 is essential for protection against a different set of viruses, including picornaviruses, such as poliovirus and encephalomyocarditis virus (35). Some viruses can be recognized by either RIG-I or MDA5. Little is known about which viruses are detected by LGP2 (98), but recent evidence suggests that LGP2 facilitates recognition of viral RNA by MDA5 (123).
Specificity toward viral RNA is maintained by the endolysosomal compartmentalization of TLRs that sense viral nucleic acids and their ligands, rather than by the structural properties of the nucleic acids (11). On the other hand, RLRs reside in the cytoplasm and thus encounter a broad range of potential RNA ligands, including self RNAs such as tRNA, rRNA, mRNAs, and microRNAs. Therefore, RLRs must discriminate rigorously between self and foreign RNAs to prevent an uncontrolled antiviral immune response. While most self RNAs are capped at their 5′ ends, viral RNA is generally not modified, and several biochemical studies have identified unmodified 5′-triphosphorylated single-stranded RNA (ssRNA) as the optimal RIG-I agonist. Further detailed analysis demonstrated that base pairing close to the 5′ terminus of RNA is also required for efficient binding to the RIG-I CTD (124, 125). Interestingly, these structures can be found at the termini of the genomes of many ssRNA viruses, such as the Paramyxoviridae, Orthomyxoviridae, and Flaviviridae, which are known to fold into double-stranded panhandle-like structures. Ligand specificity of MDA5 is not well understood, but MDA5 generally binds to longer (>2-kb) highly branched double-stranded RNA (dsRNA) (57, 108). Some of these molecules are part of the dsRNA found in the viral genomes of Picornaviridae (35, 58), but others are generated as by-products of viral replication (159). Interestingly, another aspect of MDA5 specificity is that proper signal transduction downstream of MDA5 depends on the presence of mRNA that is not 2′-O methylated (16, 184).
Apart from the rigorous control of RLR signaling at the level of ligand specificity, downstream signaling pathways are also controlled stringently. In part, the modulation of these signaling events, as discussed below, is performed by the ubiquitination machinery in the initial stage of ligand recognition, during the signaling events downstream of RLRs, or during the final stage by controlling the transcription factors involved in antiviral gene expression.
IT ALL STARTS WITH UBIQUITINATION OF RIG-I
TRIM25
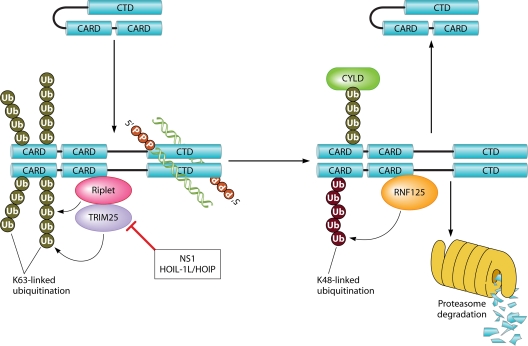
One of the first events following viral infection is robust ubiquitination of RIG-I. This is mediated partly by the K63-specific RING ubiquitin ligase TRIM25 (Fig. 1), a member of the tripartite motif (TRIM) protein family (29). TRIM proteins are characterized by the presence of a RING/Bbox/coiled-coil (RBCC) tripartite motif consisting of a RING finger domain with E3 ligase activity, one or two B box domains, and a coiled-coil domain, and they are involved in numerous cellular processes, including antiviral immune regulation (85, 89). Biochemical analysis revealed lysine residue 172 of RIG-I as the primary target of TRIM25-mediated ubiquitination. This amino acid proved to be crucial: mutating K172 almost completely abrogates downstream signaling because RIG-I can no longer bind to the MAVS (mitochondrial antiviral signaling protein) adaptor protein. Furthermore, a splice variant of RIG-I lacking the TRIM25 interaction domain that is strongly upregulated after viral infection acts as a naturally occurring dominant negative inhibitor of RIG-I signaling (28). Interestingly, the influenza A virus nonstructural protein 1 (NS1), which sequesters dsRNA, also efficiently inhibits TRIM25-induced RIG-I ubiquitination by blocking TRIM25 oligomerization and subsequent activation (Fig. 1) (27). This finding further underscores the importance of TRIM25 in the RIG-I signaling pathway.
Fig 1.
RIG-I (de)activation. 5′-triphosphorylated dsRNA binds to the C-terminal domain of RIG-I. This causes dimerization of RIG-I and induces it to undergo a conformational change that exposes the two N-terminal CARDs. This allows the binding of Riplet and TRIM25 ubiquitin ligases, which attach K63-specific polyubiquitin chains to RIG-I. The functions of TRIM25 are antagonized by the influenza virus protein NS1 and by the HOIL-1L/HOIP complex. Additional unanchored K63-linked polyubiquitin chains bind to the CARDs. These ubiquitin modifications facilitate the binding of the mitochondrial adaptor protein MAVS. After signal transduction, RNF125 polymerizes K48-linked ubiquitin chains onto RIG-I, which leads to its degradation by the proteasome. CYLD constitutively removes the activating K63-linked polyubiquitin chains from RIG-I.
Riplet
Using either the C-terminal region of RIG-I or the full-length protein as bait in a yeast two-hybrid screening, two groups independently identified the ring finger E3 ligase Riplet as a novel binding partner for RIG-I. Riplet stands for RING finger protein leading to RIG-I activation, and it is also named RNF135 or REUL. Like TRIM25, Riplet attaches K63-linked polyubiquitin chains to RIG-I and thus promotes downstream signaling (Fig. 1). However, in the case of Riplet, the RIG-I ubiquitination sites remain controversial. Whereas Oshiumi and colleagues identified K849 and K851, located on the CTD of RIG-I, as the crucial ubiquitin-anchoring residues (100), Gao et al. pinpointed K154, K164, and K172 on the N-terminal CARDs (30). This contradiction could have been due to the different cloning strategies used to generate RIG-I deletion mutants for protein interaction studies. The generation of RNF135-deficient mice established the essential role for Riplet in vivo, since these animals were more susceptible to vesicular stomatitis virus (VSV) infection and could not mount an effective antiviral immune response (101). Furthermore, both Riplet-deficient fibroblasts and conventional DC, but not pDC, showed defective early type I IFN production in response to VSV, influenza A virus, and hepatitis C Virus.
Unanchored Polyubiquitin Chains
Recently, Zeng and colleagues designed an elegant “infection-free” system for studying the RIG-I signaling pathway in vitro (174). RIG-I-induced IRF3 and NF-κB could be activated by incubating purified RIG-I with its ligand, 5′ppp-ssRNA, together with recombinant E1 and E2 (either Ubc5c/Ubc5b or Ubc13/Uev1A) and a RIG-I-specific E3 enzyme (TRIM25). Remarkably, replacing the whole ubiquitin machinery by in vitro-translated K63-linked ubiquitin chains fully restored IRF3 activation. The two sequential CARDs of RIG-I were crucial for polyubiquitin binding, and they had the strongest affinity for short tetrameric K63-linked polyubiquitin chains, which are also the most potent activators of IRF3. These observations led the authors to propose the following RIG-I activation model. When the CTD of RIG-I recognizes viral RNA molecules, RIG-I undergoes conformational changes that expose the N-terminal tandem CARDs, which then noncovalently bind free K63-linked polyubiquitin chains. The bound ubiquitin stretches then serve as scaffolds that promote the oligomerization of multiple activated RIG-I molecules and the subsequent binding of the adaptor protein MAVS. This assembly provides the basis for the antiviral signaling platform (Fig. 1). However, the source of the free polyubiquitin chains is not clear. In the study by Zeng et al. (174), the abundant unanchored polyubiquitin chains pulled down by RIG-I CARDs were most likely due to overexpression of the CARDs. In cells containing endogenous full-length RIG-I, it was proposed that viral RNA binding to the CTD of RIG-I exposes the N-terminal CARD, which then recruits TRIM25 to make unanchored K63 polyubiquitin chains. These unanchored polyubiquitin chains would be rapidly degraded unless they are bound to the RIG-I CARDs. Alternatively, it cannot be excluded that TRIM25 directly conjugates K63 polyubiquitin chains onto RIG-I, which would create a microenvironment in which abundant K63-linked polyubiquitin chains are available for cleavage by an unknown deubiquitinase and presented to the RIG-I CARD. So far, unanchored polyubiquitin chains have also been shown to be involved in the activation of both TAK1 and IKK protein kinase complexes (162) and in the regulation of p53 protein stability (17).
RIG-I-INDUCED SIGNAL TRANSDUCTION AND UBIQUITINATION GO HAND IN HAND
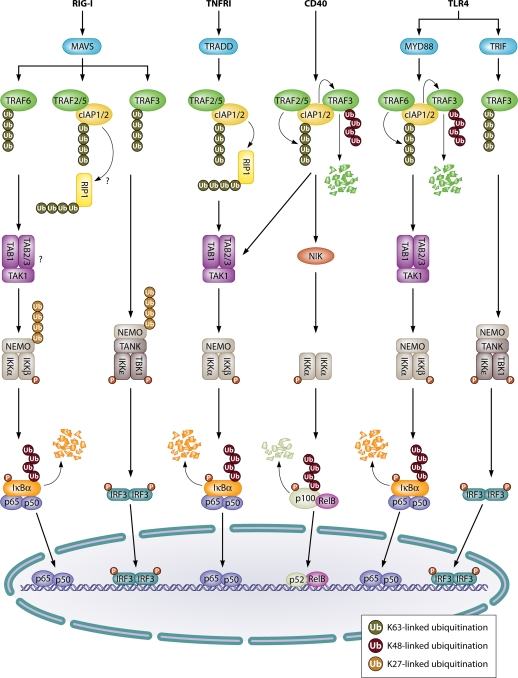
When RIG-I recognizes its ligand, it rapidly associates with MAVS, also called Cardif, IPS-1, or VISA. MAVS contains an N-terminal caspase activation and recruitment domain (CARD), which allows it to associate with the exposed CARD present on activated RIG-I and MDA5 molecules. MAVS harbors on its carboxyl terminus a transmembrane (TM) domain, which anchors the protein in the outer membranes of mitochondria, where it functions as the central adaptor protein in RLR signaling and controls downstream signaling events (60, 66, 86, 163). However, antiviral signaling is not restricted to mitochondria. MITA (mediator of IRF3 activation, or STING) facilitates MAVS complex formation and colocalizes with both mitochondria and the endoplasmic reticulum (ER) (54, 182). Furthermore, selective targeting of MAVS to peroxisomes can sustain the immediate-early expression of antiviral genes (20). Activated MAVS interacts with several different downstream signaling proteins to form an antiviral signaling platform. Among these molecules are TRADD (tumor necrosis factor receptor type 1-associated death domain) (87), RIP1 (receptor-interacting serine/threonine-protein kinase 1), FADD (Fas-associated protein with death domain) (6), and the proapoptotic proteins caspase-8 and caspase-10 (136), which were originally characterized in the context of signal transduction mediated by tumor necrosis factor receptor I (TNFRI). These proteins also mediate downstream signaling to NF-κB and IRF3/7. As discussed below, RIG-I signaling leading to the activation of these transcription factors shows many similarities with the TNFRI and TLR signaling pathways (Fig. 2).
Fig 2.
Similarities between signaling by RIG-I, TNFRI, CD40, and TLR4. Upon activation, RIG-I recruits the E3s TRAF6, TRAF2/5, cIAP1/2, and TRAF3 through the MAVS adaptor protein. When TRAF6 and RIP1 are K63 (auto)polyubiquitinated, they recruit the TAK1 and IKK complexes to the RIG-I receptor complex. TAK1 phosphorylates and activates the IKK complex, leading to activation of IKKβ and subsequent IκBα phosphorylation and degradation. This allows the translocation of NF-κB (p65/p50 dimer) to the nucleus. Similar mechanisms are involved in TNFRI-mediated NF-κB activation, which involves recruitment of TRAF2/5 via the TRADD adaptor protein. The RIG-I-induced TRAF3-mediated K63-linked autopolyubiquitination is involved in the activation of TBK1 and IKKε kinases, which phosphorylate IRF3, leading to its dimerization and nuclear translocation. Additionally, K27-linked polyubiquitination of NEMO by TRIM23 triggers NF-κB and IRF3 activation. The cIAP1/2-mediated K63-linked polyubiquitination of RIP1 and TRAF6/RIP1-mediated recruitment of the TAK1 complex (arrows) are hypothetical and are based on similarities between TNFRI and TLR4 signal transduction. Upon CD40 stimulation, TRAF2/5 targets cIAP1/2 for K63-linked polyubiquitination, which then mediate K48-linked polyubiquitination of TRAF3, which targets it for proteasomal degradation. This prevents the constitutive degradation of NIK by TRAF3, allowing NIK-mediated IKKα activation and IKKα-mediated phosphorylation of p100 NF-κB. This leads to partial processing of p100 into the p52 NF-κB subunit. The noncanonical NF-κB dimer p52/Rel-B then translocates to the nucleus. TLR4 stimulation induces the selective ubiquitination of TRAF3, which is either targeted for K48-linked polyubiquitination by cIAP1/2 (MyD88 dependent), leading to NF-κB activation, or subjected to K63-linked polyubiquitination, leading to IRF3 activation (TRIF dependent). It should be mentioned that specific signaling molecules have sometimes been reported to be modified by types of ubiquitination other than those that are indicated (e.g., linear polyubiquitination of NEMO in the case of TNF signaling or K63-linked polyubiquitination in case of TCR signaling), but for reasons of simplicity these have been omitted from the figure.
Role of Polyubiquitin Binding Scaffold Proteins
The IKK complex.
Transcription factors of the NF-κB family regulate the expression of many genes and usually function as a p65-p50 heterodimer. Under normal conditions, NF-κB is associated with IκBα (inhibitor of NF-κB), which retains NF-κB in the cytoplasm by masking its nuclear localization signal sequence. Upon stimulation of TNFRI, the IKK (IκBα kinase) complex phosphorylates IκBα and thereby primes it for K48-linked polyubiquitination. This process is mediated by the cullin-RING SCFβTrCP (Skp1-Cul1-Fbox protein/β-transducin repeat-containing protein) and results in proteasomal degradation of IκBα and nuclear translocation of NF-κB (135). The IKK complex consists of three subunits: two enzymatically active kinases (IKKα and IKKβ; the latter is crucial for IκBα phosphorylation) and a regulatory subunit called NEMO (NF-κB essential modulator) or IKKγ. The mechanism by which RIG-I activates NF-κB is still unclear but most likely resembles TNFRI signaling to NF-κB (Fig. 2). TNF-induced activation of the IKK complex requires the TAK1 (transforming growth factor β [TGFβ]-activated kinase 1) complex, including TAK1 itself and the regulatory subunits TAB1 (TAK1 binding protein) and either TAB2 or TAB3 (75). A typical feature of the regulatory subunits NEMO, TAB2, and TAB3 is the presence of specific UBDs that recognize K63-linked and linear polyubiquitin chains and mediate the recruitment of IKK and TAK1 kinases to ubiquitinated upstream signaling proteins in the TNFRI signaling complex. NEMO and TAB2/3 are also ubiquitinated, which is believed to bring the TAK1 and IKK complexes close to each other. Interestingly, NEMO and TAB scaffold proteins can also be activated by unanchored K63-linked polyubiquitin chains (162). It will be worthwhile to analyze whether similar mechanisms contribute to RIG-I induced NF-κB activation.
IRF3 activation.
Activation and nuclear translocation of IRF3 require its phosphorylation by the IKK-related kinase TBK1 or IKKε and its subsequent dimerization (44, 84, 107). While TBK1 is constitutively expressed in most cell types, IKKε needs to be upregulated in many cell types and is believed to be involved in maintaining rather than initiating the antiviral immune response (144). Interestingly, NEMO is indispensable for RLR-mediated IRF3 activation: type I IFN production is almost completely abrogated in NEMO-deficient fibroblasts (181). Like IKKα and IKKβ in TNFRI signaling, both TBK1 and IKKε associate with NEMO upon viral infection, but these interactions are facilitated by the NEMO-like adaptor proteins TANK (39), NAP1 (26), and SINTBAD (118), which are constitutively bound to both TBK1 and IKKε (13). It has been reported that upon viral infection, TANK interacts with NEMO. Moreover, the binding of TANK and the C-terminal UBDs of NEMO is required for IRF3 activation (175, 181). This indicates that the mechanism is analogous to that of TNFRI signaling, with NEMO acting as a bridging molecule linking upstream polyubiquitinated signaling molecules to the IKK complex. A putative role for the TAK1 complex in IRF3 activation has not been properly addressed.
Linear ubiquitination.
A new role for linear ubiquitination in TNF-induced NF-κB signaling was reported recently. Linear polyubiquitin chains can be generated by the linear ubiquitin assembly complex (LUBAC), which is composed of the E3 enzymes HOIL-1L (heme-oxidized IRP2 ubiquitin ligase 1) and HOIP (HOIL-1L-interacting protein) and the ubiquitin binding protein SHARPIN (SHANK-associated RH domain interactor) (33, 52, 145). HOIL-1L and HOIP jointly catalyze the attachment of linear ubiquitin chains to NEMO, which generates a stable signaling platform that promotes the activation of the IKK complex, which results in NF-κB activation (40, 146, 154). Unlike TNFR signaling, however, RIG-I-mediated NF-κB activation seems independent of LUBAC (53).
Role of E3 Ubiquitin Ligases
The cIAP1/2-TRAF2-TRAF5 complex.
During TNFRI signaling, the E3 ligases cIAP1 and cIAP2 (cellular inhibitor of apoptosis 1 and 2), in conjunction with their E2 Ubc5 (ubiquitin-conjugating enzyme 5), mediate the K63-specific polyubiquitination of RIP1 at the receptor complex (8, 79). These K63-linked chains support the recruitment of the TAK1 and IKK complexes by binding to the UBDs of their respective regulatory subunits, TAB2/3 and NEMO (21). In addition, the RING finger E3 ligases TRAF2 and TRAF5 (TNF-receptor associated factor 2 and 5), both of which exhibit K63-specific autoubiquitinating activities, are recruited to the TNFRI signaling complex, and they are believed to have redundant functions in activating the cIAPs. Similarly, RIP1 is recruited to the MAVS complex by interacting directly with TRADD and MAVS. RIP1-deficient fibroblasts are highly susceptible to viral infection and fail to produce type I IFNs upon RIG-I stimulation (6, 87). Upon viral infection, RIP1 is targeted for K63-linked polyubiquitination, and this modification is linked to its ability to promote downstream signaling. Mutation of the crucial ubiquitin attachment site K377 of RIP1 impairs NEMO recruitment and decreases subsequent IRF3 activation (111). The E3 ligases cIAP1/2 (81), TRAF2 (88), and TRAF5 (142) are also recruited to the MAVS complex upon RIG-I stimulation (Fig. 2). Biochemical studies revealed TRAF2 and TRAF5 interaction motifs in the C-terminal region of MAVS. Interaction of TRAF5 with MAVS leads to TRAF5 autoubiquitination and subsequent activation of IRF3 (142). Although depletion of either TRAF2 or TRAF5 led to reduced IRF3 and NF-κB activation upon RIG-I stimulation, little information is known about the underlying mechanisms involved.
TRAF6 activates NF-κB and IRF7.
While TRAF2 and TRAF5 are crucial for TNFRI signaling, another member of the TRAF family of E3 enzymes, TRAF6, mediates the signaling induced by TLRs and interleukin-1 receptor (IL-1R) (Fig. 2). In cooperation with the heterodimeric E2 Ubc13/Uev1, TRAF6 promotes its own K63-specific autoubiquitination and the synthesis of unanchored K63-specific polyubiquitin chains (67). As in TNFRI signaling, the polyubiquitin chains synthesized by TRAF6 on K63 promote the recruitment of both TAK1 and IKK protein complexes to the receptor, ultimately leading to NF-κB activation. Mutational analysis identified two putative TRAF6 interaction motifs on MAVS (163), suggesting a role for TRAF6 in RLR signaling. Indeed, TRAF6-deficient fibroblasts are more susceptible than normal fibroblasts to infection with Sendai virus and vesicular stomatitis virus (VSV) (64, 168). Although the activation of IRF7 after viral infection resembles IRF3 activation and involves its direct phosphorylation of IRF7 by TBK1 and IKKε, activation of IRF7 and NF-κB is impaired in TRAF6-deficient fibroblasts but IRF3 activation is not. This suggests the existence of a unique TRAF6-dependent pathway diverging from MAVS and specifically sustaining the activation of IRF7 and NF-κB (64). This separate activation of IRF3 and IRF7 is also observed in pDC, in which TRAF6 is necessary for IRF7 activation after TLR7 and TLR9 activation and type I IFN production (59). Uncoupling IRF3 from the IRF7 activation pathway might be a way of avoiding their simultaneous inhibition by virus-encoded inhibitory proteins.
K63-linked polyubiquitination of TRAF3 promotes IRF3 activation.
RLR-mediated IRF3 activation requires the action of an additional atypical TRAF family member, TRAF3 (41, 96). Like for TRAF2, -5, and -6, two TRAF3 interaction motifs have been identified on MAVS (105, 120, 141). Humans expressing mutated forms of TRAF3 are highly susceptible to encephalitis caused by herpes simplex virus, and this demonstrates the importance of TRAF3 in antiviral immunity (106). Unlike TRAF2, TRAF5, and TRAF6, which activate the canonical NF-κB signaling pathway, TRAF3 was initially described as an inhibitor of noncanonical NF-κB activation (70). The noncanonical or alternative NF-κB pathway is activated by a subset of TNFR superfamily members, including receptors for BAFF, CD40 ligand, and LTβ, which are associated, respectively, with B cell development, activation, and the formation of secondary and tertiary lymphoid organs. Noncanonical NF-κB signaling is characterized by the nuclear translocation of p52-RelB NF-κB family members. Upon receptor activation of the noncanonical NF-κB pathway, p52 is generated by partial posttranslational proteasomal degradation of the p100 precursor protein following its phosphorylation by IKKα and the NF-κB-inducing kinase (NIK) (134). NIK is also responsible for the activation of IKKα and thus plays a key role in the activation of the alternative NF-κB pathway. In resting cells, NIK serves as a substrate for a cytoplasmic E3 ligase complex containing TRAF2, TRAF3, and the cIAPs. The cIAPs mark NIK for K48-specific degradative ubiquitination, which keeps NIK levels constitutively low and thereby prevents p100 processing (70). TRAF3 enzymatic activity is not required during this process, but TRAF3 serves as an adaptor protein linking the cIAPs with NIK. Upon CD40 stimulation, the whole complex translocates to the cytoplasmic domain of CD40. There, TRAF2 and the cIAPs are activated and attach K48-linked ubiquitin chains to TRAF3, resulting in its degradation and consequently in stabilization of NIK levels and activation of the noncanonical NF-κB pathway (Fig. 2) (150, 173). Upon RLR stimulation, TRAF3 is not modified exclusively with K48-linked ubiquitin chains but also undergoes nondegradative K63-linked polyubiquitination (104). Using a cell-free reconstitution system mimicking physiological viral infection, Zeng and coworkers identified the ubiquitin-conjugating enzyme Ubc5 as an essential component of IRF3 activation (175). In conjunction with TRAF3, Ubc5 promoted K63-linked TRAF3 autoubiquitination, which enhanced its ability to bind to NEMO and in this way induced TBK1 and IRF3 activation.
Selective ubiquitination of TRAF3 is not restricted to RLR signaling. For instance, stimulation of TLR4 can induce both K48- and K63-linked polyubiquitination of TRAF3, which lead to different outcomes. TLR4 engagement induces activation of TRAF6 and cIAP1/2, which leads to K48-linked ubiquitination of TRAF3 and subsequent proteasomal degradation. This process supports mitogen-activated protein kinase (MAPK) activation. On the other hand, TLR4 activation also supports the nondegradative K63-linked autoubiquitination of TRAF3, which is required to sustain IRF3 activation (Fig. 2). Indeed, selective degradation of cIAPs by using small-molecule Smac mimetics did not affect TLR4-induced IRF3-dependent gene induction, but it completely abolished MAPK activation (148). Following this line of evidence, Smac mimetics should not influence TRAF3-induced IRF3 activation after RIG-I stimulation.
Despite the progress in our understanding of the signaling pathways leading to IRF3 activation, several questions remain. For example, how can NEMO promote both IKKα/IKKβ and TBK1/IKKε activation? Perhaps the differential recruitment of additional adaptor proteins, such as TANK, NAP1, and SINTBAD, causes the NEMO complex to specifically activate IRF3 rather than NF-κB. Likewise, the recruitment of additional E3 ligases that promote IRF3 activation might alter the ubiquitination status of NEMO and its interaction partners. Different types of ubiquitination might, for example, direct the activation of either NF-κB or IRF3. Interestingly, NEMO is K27 polyubiquitinated by the E3 ligase TRIM23 after viral infection. However, this modification is essential for both IRF3 and NF-κB activation in response to viral infection (Fig. 2) (2). Another important question concerns the signals that favor K63 linkage-induced (auto)activation of TRAF3 rather than its K48-specific polyubiquitination and degradation. At least for TLR4 signaling, excluding cIAP1/2 from the signaling complex is known to support IRF3 activation (148). It is possible that similar mechanisms specify RIG-I-induced activation of IRF3 and NF-κB. Finally, most studies have focused on RIG-I-mediated signal transduction, but the molecular mechanisms involved in MDA5 (and LGP2)-induced NF-κB and IRF3/7 activation have been somewhat neglected.
PUTTING A BRAKE ON RIG-I SIGNAL TRANSDUCTION
An uncontrolled and sustained innate immune response can result in tissue damage and chronic inflammatory diseases. Considering that about 20 viral RNA molecules can be sufficient for IRF3 activation (174), subtle control mechanisms are essential because of the abundance of cellular RNAs. RIG-I signaling is therefore negatively regulated at multiple levels and involves deubiquitination of specific proteins as well as their proteasomal degradation induced by K48-specific ubiquitin ligases.
Deubiquitinating Enzymes
CYLD controls the ubiquitination status of RIG-I.
CYLD is a deubiquitinase (DUB) of the USP family of DUBs and is specific for K63-linked and linear ubiquitin chains. Mutations in the CYLD gene are linked to familial cylindromatosis (10, 137), a condition marked by benign tumors of skin appendages. Apart from its tumor-suppressive capacities, CYLD is an essential modulator of both innate and adaptive immune responses. By deubiquitinating essential signaling molecules (TRAF2, TRAF6, RIP1, and NEMO), CYLD limits the NF-κB activation induced by TNFRI, CD40, or TLR stimulation (82). In T cells, CYLD is required for positive selection of developing double-positive thymocytes, resulting in fewer peripheral T cells in CYLD knockout mice (114, 147). However, CYLD-deficient peripheral T cells are hyperresponsive to T cell receptor (TCR) stimulation due to spontaneous TAK1 activity (113). This hyperresponsiveness leads to enhanced basal JNK and NF-κB activation and to the development of spontaneous intestinal inflammatory disease (113). Similarly, CYLD-deficient peripheral B cells proliferate excessively and spontaneously activate both canonical and noncanonical NF-κB pathways (48, 55). Expression profiling studies of several CARD-containing proteins and various DUBs showed that RIG-I and CYLD cluster closely together, suggesting a potential physiological role for CYLD in the regulation of the RIG-I pathway (25). Indeed, CYLD was shown to interact with the CARDs of RIG-I and to remove K63-linked polyubiquitin chains from RIG-I, which inhibits downstream signaling. DC lacking CYLD constitutively polyubiquitinate RIG-I and show enhanced activity of TBK1 and IKKε, suggesting that CYLD regulates basal RIG-I activity by modulating its K63-polyubiquitin status (177).
A20 disrupts the TRAF3-TBK1-IKKε complex.
A20 is a DUB that contains an OTU domain and has been implicated in the negative regulation of several proinflammatory signaling pathways, including TNFRI, TLR4, NOD2 (nucleotide binding oligomerization domain containing 2), B cell antigen receptor, T cell antigen receptor, and CD40 signaling (155). In humans, mutations in A20 have been linked to several autoimmune and inflammatory disorders, such as systemic lupus erythematosus (SLE), arthritis, Crohn's disease, psoriasis, and type I diabetes (152). These associations point to an important role for A20 in homeostatic immune regulation. Indeed, A20-deficient mice die prematurely from severe multiorgan inflammation (68). This phenotype could be reversed by intercrossing the mice with MyD88 knockout animals or treating the mice with broad-spectrum antibiotics, indicating that the severe inflammation seen in A20-deficient mice is induced by TLR-dependent signals triggered by commensal intestinal flora (149). More recently, studies on cell type-specific A20 knockout mice revealed the key role of A20 in innate immunity and inflammation (references 65, 74, 83, and 153 and our unpublished results). Reminiscent of the case for CYLD, A20 has also been described as a putative tumor suppressor gene that is mutated in several B cell lymphomas (152). Specific deletion of A20 in B cells led to enhanced proinflammatory cytokine production, elevated germinal center formation, autoantibody production, and B cell hyperresponsiveness associated with B cell hyperplasia. A20 has both deubiquitinating activity (mediated by the N-terminal OTU domain) and ubiquitin ligase activity (mediated by the C-terminal zinc finger-containing domain). Both activities contribute to the inhibition of TNF-induced signaling, in which A20 mediates the sequential depolymerization of K63-polyubiquitin chains on RIP1 and the subsequent attachment of K48-linked chains, leading to the proteasomal degradation of RIP1 (161). A20 also leads to the deubiquitination of TRAF6, NEMO, and RIP2, which interferes with their ability to promote NF-κB activation. It has been proposed that several ubiquitin binding adaptor proteins, such as TAX1BP1 (50, 126) and ABIN-1 (157), recruit A20 to its substrates. How A20 acts on such a broad range of substrates and interferes with several signaling cascades is not entirely clear. However, A20 was recently found to interact with and induce the degradation of the E2 enzymes Ubc13 and Ubc5, illustrating an alternative mechanism by which A20 might prevent ubiquitination of multiple proteins (128). A20 overexpression also restricts RIG-I-mediated signaling (104). Furthermore, A20 overexpression could inhibit IRF3 activation induced by Sendai virus (158), Newcastle disease virus (122), VSV (72), and influenza A virus (99), further indicating a role for A20 in the negative regulation of RIG-I signaling. A20 overexpression was found to disrupt the TRAF3-containing TBK1 and IKKε activating complex (104), but the mechanism by which A20 interferes with RIG-I signaling is still unclear. Although modulation of TNFRI signaling requires both the DUB and ubiquitin ligase activities of A20, DUB activity is not required for inhibiting RIG-I-mediated antiviral signaling. Moreover, the TAX binding protein 1 (TAX1BP1) of human T-lymphotropic virus, an adaptor protein that is crucial for the NF-κB inhibitory functions of A20 (50, 126), is also indispensable for A20-mediated inhibition of IRF3 activation. ABIN-1 cooperates with A20 in a similar fashion to disrupt the TRAF3-TBK1-IKKε signaling complex (31). Effective RIP1 degradation during TNF-induced signaling requires the cooperation of A20 with two other ubiquitin ligases, ITCH (Itchy, also called AIP4 [atrophin-1-interacting protein 4]) and RNF11 (127, 129). Whether these E3 ligases are also essential for A20-mediated RIG-I inhibition is not known.
TRAF3 deubiquitination by DUBA.
Deubiquitinating enzyme A (DUBA) (also known as OTUD5) is another DUB of the OTU subfamily. It was identified as a potent inhibitor of both RIG-I- and MDA5-mediated signaling during small interfering RNA (siRNA)-based screening for regulators of type I IFN production (61). DUBA was shown to physically interact with TRAF3 and attenuate its K63-specific autoubiquitination, interfering with its capacity to bind and activate TBK1. A ubiquitin interaction motif in the C-terminal part of DUBA facilitates binding to activated polyubiquitinated TRAF3. As expected, depletion of DUBA does not affect noncanonical NF-κB activation, which is entirely dependent on TRAF3 K48-specific degradative ubiquitination. Although indications for an in vivo role of DUBA are still lacking because DUBA-deficient animals have not been generated yet, indirect evidence came from studies with IL-1R knockout animals. Strikingly, IL-1R-deficient conventional DC are unable to produce IFN-β, possibly due to increased basal expression levels of DUBA (36).
OTUB1 and OTUB2 deubiquitinate TRAF3 and TRAF6.
Two other OTU domain-containing DUBs, OTUB1 and OTUB1, were identified in a protease cDNA array screen as inhibitors of RIG-I- and TLR3-induced IRF3 activation. Upon viral infection, recruitment of both OTUB1 and OTUB2 to the MAVS complex at the mitochondria promotes deubiquitination of TRAF3 and TRAF6 (69). In vitro studies have shown that OTUB1 is specific for K48-linked ubiquitin chains (22). It is therefore not clear how OTUB1 can modify K63 polyubiquitination of TRAF3 and TRAF6 in cells. Interestingly, a similar discrepancy in ubiquitin chain specificity has been noticed in the case of A20 (161). It is possible that certain cellular proteins change the specificity of OTUB1 and A20 from K48- to K63-specific deubiquitination. Alternatively, OTUB1, like A20, might affect K63-linked polyubiquitination independently of its catalytic activity by interfering with E2 enzymes. In this context, OTUB1 has been shown to interact with and inhibit the activity of the E2 enzyme Ubc13, which is the cognate enzyme for the E3 RNF168, during the DNA damage response. This inhibition of Ubc13 prevents the RNF168-mediated ubiquitination of chromatin binding proteins and consequently interferes with DNA repair (91). It is possible that OTUB1 modulates K63-linked polyubiquitination of TRAF3 and TRAF6 in a similar manner.
K48-Specific E3 Ligases Trigger Proteasomal Degradation of RIG-I and Downstream Signaling Molecules
RNF125 promotes degradation of RIG-I, MDA5, and MAVS.
The E3 ligase RNF125 was identified during attempts to isolate binding partners of UbcH8 (4), an E2 known to promote ISG15 conjugation to RIG-I (3). RNF125 (also known as TRAC-1 or T cell RING 1) belongs to a small subfamily of RING finger ubiquitin E3 ligases and is characterized by C-terminal Zn fingers and a K48-specific ubiquitin interaction motif (34). RNF125 positively regulates T cell receptor signaling (179) and inhibits HIV-1 replication (132). Together with the E2 ubiquitin-conjugating enzyme UbcH5, RNF125 mediates the attachment of K48-linked polyubiquitin chains to RIG-I, MDA5, and MAVS, targeting them for proteasomal degradation and thereby inhibiting the antiviral immune response (4).
Linear ubiquitination inhibits RIG-I signaling.
Linear polyubiquitin chains generated by the linear ubiquitin assembly complex (LUBAC), which is composed of the E3 enzymes HOIL-1L and HOIP, are involved in NF-κB activation after TNF stimulation (40, 146, 154). In contrast, LUBAC seems to be unnecessary for RIG-I-mediated NF-κB activation. In fact, LUBAC acts as a negative regulator of RIG-I-mediated activation of NF-κB and IRF3. HOIL-1L and HOIP seem to interfere with antiviral signaling at different levels during RIG-I activation. While HOIL-1L and HOIP act synergistically to polyubiquitinate the RIG-I-modifying E3 ligase TRIM25, leading to its degradation, HOIL-1L could act independently from HOIP by competing with TRIM25 for binding to RIG-I (53). Although linear ubiquitination has been shown to also mark proteins for proteasomal degradation (180), it is not clear which chain type is responsible for proteasomal degradation of TRIM25. LUBAC has also been shown to promote K48 polyubiquitination of TRIM25, but there is no evidence for direct conjugation of K48-polyubiquitin chains to TRIM25 by this complex (53).
AIP4 promotes degradation of MAVS.
After initializing the antiviral immune response, MAVS is ubiquitinated on K48 by the HECT domain-containing E3 ligase AIP4 (or its murine ortholog ITCH); this leads to rapid degradation of MAVS (169). Typically, HECT E3 ligases contain N-terminal WW domains, which allow them to interact with phosphorylated serine or threonine residues that are followed by a proline in their target proteins. AIP4 cannot bind directly to MAVS, but it is sequestered to MAVS by PCBP2. The latter protein contains small multiple WW binding domains, and its expression is augmented upon viral infection. Overexpression of PCBP2 enhanced the interaction of AIP4 with MAVS and subsequently promoted its K48-specific ubiquitination. AIP4 has been implicated in the modulation of several other biological pathways, including TNFRI signaling. In this context, AIP4 associates with A20 and TAXBP1 to promote K48-linked polyubiquitination of RIP1, which inhibits the downstream signaling that leads to NF-κB activation (127). Itch-deficient mice develop several immune disorders, including dermatitis, lymphoid hyperplasia, and progressive pulmonary interstitial inflammation, and they die at the age of 6 to 8 months (49). The autoimmune phenotype of ITCH knockout mice could be attributed to changes in T lymphocytes (24, 77, 103). However, failure to inhibit RLR signaling in the absence of ITCH, which leads to enhanced type I IFN production, could contribute to the autoimmune disorders of these mice.
Degradation of MITA by RNF5.
The activity of MITA, a crucial adaptor protein promoting the recruitment of TBK1 and IKKε to MAVS, was recently shown to be modulated by the RING domain-containing E3 ligase RNF5, which specifically promotes MITA degradation at the mitochondria (183). RNF5 localized at the ER has been implicated in protein quality control (18, 143). Interestingly RNF5 translocates to the mitochondria following viral infection, where it induces proteasomal degradation of MITA by targeting it for K48-linked polyubiquitination, hence inhibiting downstream signaling.
Triad3A induces degradation of TRAF3.
The essential role of TRAF3 in antiviral signal transduction makes it an attractive target for proteins modulating RLR signaling. In addition to the removal of K63-linked polyubiquitin chains from TRAF3 by DUBs such as DUBA and OTUB1, TRAF3 is also subjected to K48-linked degradative polyubiquitination. Upon viral infection, Triad3A, the dominant splice variant of the RING finger type E3 ligase Triad, is upregulated; it functions as a K48-specific E3 that increases TRAF3 turnover (93).
K48-Specific Ubiquitin Ligases Targeting Transcription Factors
Regulating the stability of transcription factors is an important mechanism for controlling immune homeostasis. The transcription of IFN-β requires the cooperative binding of several transcription factors, such as ATF-2/c-Jun, NF-κB, and IRF3 or IRF7, to the IFN-β promoter (102). The activation of each of these transcription factors is well described, but the mechanisms involved in the disassembly of this complex are not entirely understood. When it binds its promoter, the NF-κB subunit p65 is degraded by nuclear proteasomes, a process that is controlled by the E3 ligases SOCS1 (suppressor of cytokine signaling 1) (117), PDLIM2 (PDZ and LIM domain protein 2) (140), and COMMD1 (COMM domain-containing protein 1) (32). Interestingly, IRF3 is also degraded after prolonged viral infection, and its degradation could be blocked by the proteasome inhibitor MG-132. One of the proteins regulating the stability of activated IRF3 is the peptidyl-prolyl isomerase Pin1. IRF3 activation depends on the immediate inducible phosphorylation of two C-terminal serine clusters (71). Pin1 contains an N-terminal WW domain that specifically interacts with a phosphorylated serine 339-proline 340 motif on IRF3, leading to its polyubiquitination and degradation. Phosphorylation of Ser339 is delayed compared to the activating C-terminal phosphorylation of IRF3 and coincides with decreased type I IFN production (121). The kinases and ubiquitin ligases involved in proteasome-mediated degradation of IRF3 are unknown. However, several E3 ligases have been shown to promote K48-specific polyubiquitination of IRF3. One of these is the ubiquitin ligase HOIL-1L (also called RBCK1) (176). Overexpression of HOIL-1L promotes the proteasome-mediated degradation of IRF3, but no biochemical studies have proved that IRF3 is a direct substrate for the ligase activity of HOIL-1L. Also, HOIL-1L deficiency in fibroblasts does not affect IRF3 stability, and this brings into question the proposed role of HOIL-1L as a predominant IRF3-specific E3 ligase (53). Another potential candidate for K48-linked polyubiquitination of IRF3 is the recently discovered E3 ligase RAUL, which can directly catalyze K48-linked polyubiquitination of both IRF3 and IRF7 (171). Nonetheless, degradation of IRF3 by RAUL occurs independently of the phosphorylation status of Ser339, suggesting that RAUL primarily regulates basal IRF3 and IRF7 levels. Another good candidate is the RING domain-containing E3 ligase TRIM21 (also known as Ro52), which is frequently found as an autoantigen in SLE and Sjörgen's syndrome. TRIM21 migrates to the nucleus upon IFN-β treatment and catalyzes the K48-linked polyubiquitination of IRF3 (46), IRF7 (45, 170), and the proinflammatory transcription factor IRF5 (23). In contrast, TRIM21 promotes the ubiquitination and stabilization of IRF8, a transcription factor involved in differentiation of macrophages, DC, and B cells. Stabilization of IRF8 leads to increased IL-12p40 expression (63). This demonstrates an additional level of complexity in the regulatory potential of TRIM21. The important immune regulatory role of TRIM21 is reflected in knockout mice, which develop IL-23/Th17-dependent inflammation and systemic autoimmunity in response to minor insults such as ear tagging (12, 23).
CONCLUSIONS AND PERSPECTIVES
Since the initial report in 2004 describing RIG-I as a cytosolic sensor of viral RNA (167), RIG-I and its family members MDA5 and LGP2 have been studied extensively. Substantial progress has been made in the characterization of the molecules involved in RLR-mediated signal transduction, and several studies have revealed a key role for various ubiquitin-modifying enzymes in the induction of the antiviral immune response. The importance of ubiquitination in RLR-induced antiviral signaling is also emphasized by the fact that several viruses encode proteins that interfere with ubiquitination as a defense against RLR-induced antiviral immune responses. For example, the influenza A virus protein NS1 prevents type I IFN production by interfering with the TRIM25-mediated activation of RIG-I (27). The immediate-early transcription factor RTA (replication and transcription activator) of Kaposi's sarcoma-associated herpesvirus promotes IRF3 and IRF7 degradation by stimulating their K48-specific polyubiquitination by RAUL (171, 172). Nevertheless, much work is required to understand how the infected cell senses viruses and how viruses fight back. For example, what signaling pathways besides IFN induction are regulated by RLRs? Can RLRs sense self molecules under certain conditions? What are the molecular patterns recognized by MDA5, and what is the function of LGP2? Do different cell types respond in different ways? How do different PRRs talk to each other? Is compartmentalization of ubiquitin networks during antiviral RIG-I signaling critical for cellular responses? Compartmentalization of E3 ligases, DUBs, and ubiquitin receptors is a general phenomenon in the ubiquitin-mediated signaling field, and various E3 ligases are recruited to the mitochondria, where they control mitochondrial function (37). It will therefore be of interest to study the spatial regulation of the ubiquitin network at the mitochondria following RLR stimulation. Furthermore, we do not know much about the dynamic regulation of immune cell activation, which is likely very complex. A combination of imaging technologies, systems biology, genetics, and immunology will be needed to uncover the details of pathogen-induced inflammatory responses and how they relates to pathogenesis of viral diseases.
Polymorphisms in RIG-I, MDA5, and MAVS have been linked to autoimmune disorders such as type I diabetes (131) and SLE (76, 109). Similarly, polymorphisms in TRIM21 (23) and A20 (38, 90) have been linked to SLE. This is somewhat expected because most of these conditions are associated with increased type I IFN production. In addition, aberrant expression of TBK1 and IKKε has been shown to contribute to breast cancer formation and lung cancer progression (130). It is still unknown if similar polymorphisms or aberrant gene expression of RLRs and RLR signaling proteins affect human susceptibility to viral infections. We anticipate that E3 ubiquitin ligases and DUBs will prove to be important new targets for mechanism-driven drug discovery. Furthermore, antisense targeting of specific negative regulatory proteins involved in RLR signaling (e.g., DUBs and ubiquitin ligases) by using synthetic 5′-triphosphate RNA oligonucleotides to stimulate RIG-I might be useful for treatment not only of infectious diseases but also of immune disorders and cancer. A more complete understanding of how RIG-I is regulated by ubiquitination will facilitate the difficult task of selectively interfering with harmful immune responses while enhancing beneficial ones.
ACKNOWLEDGMENTS
We thank Sandra Gardam and Amin Bredan for critical reading of the manuscript.
J. Maelfait was supported as a Ph.D. fellow by a grant from the Instituut voor Innovatie door Wetenschap en Technologie (IWT) and an Emmanuel van der Schueren award. Research in our lab is further supported by grants from the Fonds voor Wetenschappelijk Onderzoek-Vlaanderen (grants G.0619.10, G.0089.10, and 3G023611), the Interuniversity Attraction Poles program (IAP6/18), the Belgian Foundation against Cancer, the Strategic Basic Research program of the IWT, the Queen Elisabeth Medical Foundation, the Hercules Foundation, and the Group-ID MRP of Ghent University.
Biographies

Jonathan Maelfait obtained his Ph.D. in Sciences-Biotechnology in 2011 from Ghent University, Belgium. He currently works as a postdoctoral researcher in the Laboratory of Molecular Signal Transduction in Inflammation at the Flanders Institute for Biotechnology (VIB) and in the Department of Biomedical Molecular Biology of Ghent University. His research focuses on pathogen recognition receptors and the ubiquitin-mediated regulation of innate immune responses.

Rudi Beyaert obtained his Ph.D. in Sciences-Biotechnology in 1992 at Ghent University. In 1996 he started the Laboratory of Molecular Signal Transduction in Inflammation at the Flanders Institute for Biotechnology (VIB) in Ghent. In 2003 he was appointed as Full Professor in Molecular Biology at Ghent University, and in 2006 he became the Associate Director of the Department for Molecular Biomedical Research of the VIB. His research focuses on the molecular mechanisms that control inflammation and immunity. More specifically, his team is studying signal transduction pathways leading to NF-κB- and IRF3-dependent gene expression in response to proinflammatory cytokines (e.g., TNF and IL-1), antigen receptors, and infection, with a special focus on their regulation by ubiquitination and other posttranslational mechanisms. He has published over 160 papers on his research, which have received more than 8,000 citations.
REFERENCES
- 1. Al-Hakim A, et al. 2010. The ubiquitous role of ubiquitin in the DNA damage response. DNA Repair (Amst.) 9:1229–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arimoto K, et al. 2010. Polyubiquitin conjugation to NEMO by triparite motif protein 23 (TRIM23) is critical in antiviral defense. Proc. Natl. Acad. Sci. U. S. A. 107:15856–15861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arimoto K, Konishi H, Shimotohno K. 2008. UbcH8 regulates ubiquitin and ISG15 conjugation to RIG-I. Mol. Immunol. 45:1078–1084 [DOI] [PubMed] [Google Scholar]
- 4. Arimoto K, et al. 2007. Negative regulation of the RIG-I signaling by the ubiquitin ligase RNF125. Proc. Natl. Acad. Sci. U. S. A. 104:7500–7505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baker RT, Board PG. 1987. The human ubiquitin gene family: structure of a gene and pseudogenes from the Ub B subfamily. Nucleic Acids Res. 15:443–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Balachandran S, Thomas E, Barber GN. 2004. A FADD-dependent innate immune mechanism in mammalian cells. Nature 432:401–405 [DOI] [PubMed] [Google Scholar]
- 7. Bedford L, Lowe J, Dick LR, Mayer RJ, Brownell JE. 2011. Ubiquitin-like protein conjugation and the ubiquitin-proteasome system as drug targets. Nat. Rev. Drug Discov. 10:29–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bertrand MJ, et al. 2008. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol. Cell 30:689–700 [DOI] [PubMed] [Google Scholar]
- 9. Bhoj VG, Chen ZJ. 2009. Ubiquitylation in innate and adaptive immunity. Nature 458:430–437 [DOI] [PubMed] [Google Scholar]
- 10. Bignell GR, et al. 2000. Identification of the familial cylindromatosis tumour-suppressor gene. Nat. Genet. 25:160–165 [DOI] [PubMed] [Google Scholar]
- 11. Blasius AL, Beutler B. 2010. Intracellular Toll-like receptors. Immunity 32:305–315 [DOI] [PubMed] [Google Scholar]
- 12. Bolland S, Garcia-Sastre A. 2009. Vicious circle: systemic autoreactivity in Ro52/TRIM21-deficient mice. J. Exp. Med. 206:1647–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chau TL, et al. 2008. Are the IKKs and IKK-related kinases TBK1 and IKK-epsilon similarly activated? Trends Biochem. Sci. 33:171–180 [DOI] [PubMed] [Google Scholar]
- 14. Clague MJ, Urbe S. 2010. Ubiquitin: same molecule, different degradation pathways. Cell 143:682–685 [DOI] [PubMed] [Google Scholar]
- 15. Cui S, et al. 2008. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol. Cell 29:169–179 [DOI] [PubMed] [Google Scholar]
- 16. Daffis S, et al. 2010. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 468:452–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dayal S, et al. 2009. Suppression of the deubiquitinating enzyme USP5 causes the accumulation of unanchored polyubiquitin and the activation of p53. J. Biol. Chem. 284:5030–5041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Delaunay A, et al. 2008. The ER-bound RING finger protein 5 (RNF5/RMA1) causes degenerative myopathy in transgenic mice and is deregulated in inclusion body myositis. PLoS One 3:e1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Deshaies RJ, Joazeiro CA. 2009. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78:399–434 [DOI] [PubMed] [Google Scholar]
- 20. Dixit E, et al. 2010. Peroxisomes are signaling platforms for antiviral innate immunity. Cell 141:668–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJ. 2006. Activation of IKK by TNFalpha requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol. Cell 22:245–257 [DOI] [PubMed] [Google Scholar]
- 22. Edelmann MJ, et al. 2009. Structural basis and specificity of human otubain 1-mediated deubiquitination. Biochem. J. 418:379–390 [DOI] [PubMed] [Google Scholar]
- 23. Espinosa A, et al. 2009. Loss of the lupus autoantigen Ro52/Trim21 induces tissue inflammation and systemic autoimmunity by disregulating the IL-23-Th17 pathway. J. Exp. Med. 206:1661–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fang D, et al. 2002. Dysregulation of T lymphocyte function in itchy mice: a role for Itch in TH2 differentiation. Nat. Immunol. 3:281–287 [DOI] [PubMed] [Google Scholar]
- 25. Friedman CS, et al. 2008. The tumour suppressor CYLD is a negative regulator of RIG-I-mediated antiviral response. EMBO Rep. 9:930–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fujita F, et al. 2003. Identification of NAP1, a regulatory subunit of IkappaB kinase-related kinases that potentiates NF-kappaB signaling. Mol. Cell. Biol. 23:7780–7793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gack MU, et al. 2009. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe 5:439–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gack MU, et al. 2008. Roles of RIG-I N-terminal tandem CARD and splice variant in TRIM25-mediated antiviral signal transduction. Proc. Natl. Acad. Sci. U. S. A. 105:16743–16748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gack MU, et al. 2007. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446:916–920 [DOI] [PubMed] [Google Scholar]
- 30. Gao D, et al. 2009. REUL is a novel E3 ubiquitin ligase and stimulator of retinoic-acid-inducible gene-I. PLoS One 4:e5760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gao L, et al. 2011. ABIN1 protein cooperates with TAX1BP1 and A20 proteins to inhibit antiviral signaling. J. Biol. Chem. 286:36592–36602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Geng H, Wittwer T, Dittrich-Breiholz O, Kracht M, Schmitz ML. 2009. Phosphorylation of NF-kappaB p65 at Ser468 controls its COMMD1-dependent ubiquitination and target gene-specific proteasomal elimination. EMBO Rep. 10:381–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gerlach B, et al. 2011. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature 471:591–596 [DOI] [PubMed] [Google Scholar]
- 34. Giannini AL, Gao Y, Bijlmakers MJ. 2008. T-cell regulator RNF125/TRAC-1 belongs to a novel family of ubiquitin ligases with zinc fingers and a ubiquitin-binding domain. Biochem. J. 410:101–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gitlin L, et al. 2006. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc. Natl. Acad. Sci. U. S. A. 103:8459–8464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gonzalez-Navajas JM, et al. 2010. Interleukin 1 receptor signaling regulates DUBA expression and facilitates Toll-like receptor 9-driven antiinflammatory cytokine production. J. Exp. Med. 207:2799–2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Grabbe C, Husnjak K, Dikic I. 2011. The spatial and temporal organization of ubiquitin networks. Nat. Rev. Mol. Cell Biol. 12:295–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Graham RR, et al. 2008. Genetic variants near TNFAIP3 on 6q23 are associated with systemic lupus erythematosus. Nat. Genet. 40:1059–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Guo B, Cheng G. 2007. Modulation of the interferon antiviral response by the TBK1/IKKi adaptor protein TANK. J. Biol. Chem. 282:11817–11826 [DOI] [PubMed] [Google Scholar]
- 40. Haas TL, et al. 2009. Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Mol. Cell 36:831–844 [DOI] [PubMed] [Google Scholar]
- 41. Hacker H, et al. 2006. Specificity in Toll-like receptor signalling through distinct effector functions of TRAF3 and TRAF6. Nature 439:204–207 [DOI] [PubMed] [Google Scholar]
- 42. Harhaj EW, Dixit VM. 2011. Deubiquitinases in the regulation of NF-kappaB signaling. Cell Res. 21:22–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hatakeyama S, Nakayama KI. 2003. U-box proteins as a new family of ubiquitin ligases. Biochem. Biophys. Res. Commun. 302:635–645 [DOI] [PubMed] [Google Scholar]
- 44. Hemmi H, et al. 2004. The roles of two IkappaB kinase-related kinases in lipopolysaccharide and double stranded RNA signaling and viral infection. J. Exp. Med. 199:1641–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Higgs R, et al. 2010. Self protection from anti-viral responses—Ro52 promotes degradation of the transcription factor IRF7 downstream of the viral Toll-like receptors. PLoS One 5:e11776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Higgs R, et al. 2008. The E3 ubiquitin ligase Ro52 negatively regulates IFN-beta production post-pathogen recognition by polyubiquitin-mediated degradation of IRF3. J. Immunol. 181:1780–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Honda K, et al. 2005. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 434:772–777 [DOI] [PubMed] [Google Scholar]
- 48. Hovelmeyer N, et al. 2007. Regulation of B cell homeostasis and activation by the tumor suppressor gene CYLD. J. Exp. Med. 204:2615–2627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hustad CM, et al. 1995. Molecular genetic characterization of six recessive viable alleles of the mouse agouti locus. Genetics 140:255–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Iha H, et al. 2008. Inflammatory cardiac valvulitis in TAX1BP1-deficient mice through selective NF-kappaB activation. EMBO J. 27:629–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ikeda F, Crosetto N, Dikic I. 2010. What determines the specificity and outcomes of ubiquitin signaling? Cell 143:677–681 [DOI] [PubMed] [Google Scholar]
- 52. Ikeda F, et al. 2011. SHARPIN forms a linear ubiquitin ligase complex regulating NF-kappaB activity and apoptosis. Nature 471:637–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Inn KS, et al. 2011. Linear ubiquitin assembly complex negatively regulates RIG-I- and TRIM25-mediated type I interferon induction. Mol. Cell 41:354–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ishikawa H, Barber GN. 2008. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455:674–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jin W, et al. 2007. Deubiquitinating enzyme CYLD regulates the peripheral development and naive phenotype maintenance of B cells. J. Biol. Chem. 282:15884–15893 [DOI] [PubMed] [Google Scholar]
- 56. Kato H, et al. 2005. Cell type-specific involvement of RIG-I in antiviral response. Immunity 23:19–28 [DOI] [PubMed] [Google Scholar]
- 57. Kato H, et al. 2008. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 205:1601–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kato H, et al. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101–105 [DOI] [PubMed] [Google Scholar]
- 59. Kawai T, et al. 2004. Interferon-alpha induction through Toll-like receptors involves a direct interaction of IRF7 with MyD88 and TRAF6. Nat. Immunol. 5:1061–1068 [DOI] [PubMed] [Google Scholar]
- 60. Kawai T, et al. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6:981–988 [DOI] [PubMed] [Google Scholar]
- 61. Kayagaki N, et al. 2007. DUBA: a deubiquitinase that regulates type I interferon production. Science 318:1628–1632 [DOI] [PubMed] [Google Scholar]
- 62. Komander D. 2010. Mechanism, specificity and structure of the deubiquitinases. Subcell. Biochem. 54:69–87 [DOI] [PubMed] [Google Scholar]
- 63. Kong HJ, et al. 2007. Autoantigen Ro52 is an interferon inducible E3 ligase that ubiquitinates IRF-8 and enhances cytokine expression in macrophages. J. Immunol. 179:26–30 [DOI] [PubMed] [Google Scholar]
- 64. Konno H, et al. 2009. TRAF6 establishes innate immune responses by activating NF-kappaB and IRF7 upon sensing cytosolic viral RNA and DNA. PLoS One 4:e5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kool M, et al. 2011. The ubiquitin-editing protein A20 prevents dendritic cell activation, recognition of apoptotic cells, and systemic autoimmunity. Immunity 35:82–96 [DOI] [PubMed] [Google Scholar]
- 66. Kumar H, et al. 2006. Essential role of IPS-1 in innate immune responses against RNA viruses. J. Exp. Med. 203:1795–1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lamothe B, et al. 2007. TRAF6 ubiquitin ligase is essential for RANKL signaling and osteoclast differentiation. Biochem. Biophys. Res. Commun. 359:1044–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lee EG, et al. 2000. Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice. Science 289:2350–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Li S, et al. 2010. Regulation of virus-triggered signaling by OTUB1- and OTUB2-mediated deubiquitination of TRAF3 and TRAF6. J. Biol. Chem. 285:4291–4297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Liao G, Zhang M, Harhaj EW, Sun SC. 2004. Regulation of the NF-kappaB-inducing kinase by tumor necrosis factor receptor-associated factor 3-induced degradation. J. Biol. Chem. 279:26243–26250 [DOI] [PubMed] [Google Scholar]
- 71. Lin R, Heylbroeck C, Pitha PM, Hiscott J. 1998. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol. Cell. Biol. 18:2986–2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Lin R, et al. 2006. Negative regulation of the retinoic acid-inducible gene I-induced antiviral state by the ubiquitin-editing protein A20. J. Biol. Chem. 281:2095–2103 [DOI] [PubMed] [Google Scholar]
- 73. Lindner HA. 2007. Deubiquitination in virus infection. Virology 362:245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lippens S, et al. 2011. Keratinocyte-specific ablation of the NF-kappaB regulatory protein A20 (TNFAIP3) reveals a role in the control of epidermal homeostasis. Cell Death Differ. 18:1845–1853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Liu S, Chen ZJ. 2011. Expanding role of ubiquitination in NF-kappaB signaling. Cell Res. 21:6–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Liu X, et al. 2011. Possible association of VISA gene polymorphisms with susceptibility to systemic lupus erythematosus in Chinese population. Mol. Biol. Rep. 38:4583–4588 [DOI] [PubMed] [Google Scholar]
- 77. Liu YC. 2007. The E3 ubiquitin ligase Itch in T cell activation, differentiation, and tolerance. Semin. Immunol. 19:197–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Loo YM, et al. 2008. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J. Virol. 82:335–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Mahoney DJ, et al. 2008. Both cIAP1 and cIAP2 regulate TNFalpha-mediated NF-kappaB activation. Proc. Natl. Acad. Sci. U. S. A. 105:11778–11783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Malynn BA, Ma A. 2010. Ubiquitin makes its mark on immune regulation. Immunity 33:843–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Mao AP, et al. 2010. Virus-triggered ubiquitination of TRAF3/6 by cIAP1/2 is essential for induction of interferon-beta (IFN-beta) and cellular antiviral response. J. Biol. Chem. 285:9470–9476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Massoumi R. 2010. Ubiquitin chain cleavage: CYLD at work. Trends Biochem. Sci. 35:392–399 [DOI] [PubMed] [Google Scholar]
- 83. Matmati M, et al. 2011. A20 (TNFAIP3) deficiency in myeloid cells triggers erosive polyarthritis resembling rheumatoid arthritis. Nat. Genet. 43:908–912 [DOI] [PubMed] [Google Scholar]
- 84. Matsui K, et al. 2006. Role of TANK-binding kinase 1 and inducible IkappaB kinase in IFN responses against viruses in innate immune cells. J. Immunol. 177:5785–5789 [DOI] [PubMed] [Google Scholar]
- 85. McNab FW, Rajsbaum R, Stoye JP, O'Garra A. 2011. Tripartite-motif proteins and innate immune regulation. Curr. Opin. Immunol. 23:46–56 [DOI] [PubMed] [Google Scholar]
- 86. Meylan E, et al. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167–1172 [DOI] [PubMed] [Google Scholar]
- 87. Michallet MC, et al. 2008. TRADD protein is an essential component of the RIG-like helicase antiviral pathway. Immunity 28:651–661 [DOI] [PubMed] [Google Scholar]
- 88. Mikkelsen SS, et al. 2009. RIG-I-mediated activation of p38 MAPK is essential for viral induction of interferon and activation of dendritic cells: dependence on TRAF2 and TAK1. J. Biol. Chem. 284:10774–10782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Munir M. 2010. TRIM proteins: another class of viral victims. Sci. Signal. 3:jc2. [DOI] [PubMed] [Google Scholar]
- 90. Musone SL, et al. 2008. Multiple polymorphisms in the TNFAIP3 region are independently associated with systemic lupus erythematosus. Nat. Genet. 40:1062–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Nakada S, et al. 2010. Non-canonical inhibition of DNA damage-dependent ubiquitination by OTUB1. Nature 466:941–946 [DOI] [PubMed] [Google Scholar]
- 92. Nakhaei P, Genin P, Civas A, Hiscott J. 2009. RIG-I-like receptors: sensing and responding to RNA virus infection. Semin. Immunol. 21:215–222 [DOI] [PubMed] [Google Scholar]
- 93. Nakhaei P, et al. 2009. The E3 ubiquitin ligase Triad3A negatively regulates the RIG-I/MAVS signaling pathway by targeting TRAF3 for degradation. PLoS Pathog. 5:e1000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Neerincx A, et al. 2010. A role for the human nucleotide-binding domain, leucine-rich repeat-containing family member NLRC5 in antiviral responses. J. Biol. Chem. 285:26223–26232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Nijman SM, et al. 2005. A genomic and functional inventory of deubiquitinating enzymes. Cell 123:773–786 [DOI] [PubMed] [Google Scholar]
- 96. Oganesyan G, et al. 2006. Critical role of TRAF3 in the Toll-like receptor-dependent and -independent antiviral response. Nature 439:208–211 [DOI] [PubMed] [Google Scholar]
- 97. O'Neill LA, Bowie AG. 2010. Sensing and signaling in antiviral innate immunity. Curr. Biol. 20:R328–R333 [DOI] [PubMed] [Google Scholar]
- 98. Onoguchi K, Yoneyama M, Fujita T. 2011. Retinoic acid-inducible gene-I-like receptors. J. Interferon Cytokine Res. 31:27–31 [DOI] [PubMed] [Google Scholar]
- 99. Onose A, et al. 2006. An inhibitory effect of A20 on NF-kappaB activation in airway epithelium upon influenza virus infection. Eur. J. Pharmacol. 541:198–204 [DOI] [PubMed] [Google Scholar]
- 100. Oshiumi H, Matsumoto M, Hatakeyama S, Seya T. 2009. Riplet/RNF135, a RING finger protein, ubiquitinates RIG-I to promote interferon-beta induction during the early phase of viral infection. J. Biol. Chem. 284:807–817 [DOI] [PubMed] [Google Scholar]
- 101. Oshiumi H, et al. 2010. The ubiquitin ligase Riplet is essential for RIG-I-dependent innate immune responses to RNA virus infection. Cell Host Microbe 8:496–509 [DOI] [PubMed] [Google Scholar]
- 102. Panne D. 2008. The enhanceosome. Curr. Opin. Struct. Biol. 18:236–242 [DOI] [PubMed] [Google Scholar]
- 103. Parravicini V, Field AC, Tomlinson PD, Basson MA, Zamoyska R. 2008. Itch−/− alphabeta and gammadelta T cells independently contribute to autoimmunity in Itchy mice. Blood 111:4273–7282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Parvatiyar K, Barber GN, Harhaj EW. 2010. TAX1BP1 and A20 inhibit antiviral signaling by targeting TBK1-IKKi kinases. J. Biol. Chem. 285:14999–15009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Paz S, et al. 2011. A functional C-terminal TRAF3-binding site in MAVS participates in positive and negative regulation of the IFN antiviral response. Cell Res. 21:895–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Perez de Diego R, et al. 2010. Human TRAF3 adaptor molecule deficiency leads to impaired Toll-like receptor 3 response and susceptibility to herpes simplex encephalitis. Immunity 33:400–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Perry AK, Chow EK, Goodnough JB, Yeh WC, Cheng G. 2004. Differential requirement for TANK-binding kinase-1 in type I interferon responses to toll-like receptor activation and viral infection. J. Exp. Med. 199:1651–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Pichlmair A, et al. 2009. Activation of MDA5 requires higher-order RNA structures generated during virus infection. J. Virol. 83:10761–10769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Pothlichet J, et al. 2011. A loss-of-function variant of the antiviral molecule MAVS is associated with a subset of systemic lupus patients. EMBO Mol. Med. 3:142–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Rahman MM, McFadden G. 2011. Modulation of NF-kappaB signalling by microbial pathogens. Nat. Rev. Microbiol. 9:291–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Rajput A, et al. 2011. RIG-I RNA helicase activation of IRF3 transcription factor is negatively regulated by caspase-8-mediated cleavage of the RIP1 protein. Immunity 34:340–351 [DOI] [PubMed] [Google Scholar]
- 112. Randow F, Lehner PJ. 2009. Viral avoidance and exploitation of the ubiquitin system. Nat. Cell Biol. 11:527–534 [DOI] [PubMed] [Google Scholar]
- 113. Reiley WW, et al. 2007. Deubiquitinating enzyme CYLD negatively regulates the ubiquitin-dependent kinase Tak1 and prevents abnormal T cell responses. J. Exp. Med. 204:1475–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Reiley WW, et al. 2006. Regulation of T cell development by the deubiquitinating enzyme CYLD. Nat. Immunol. 7:411–417 [DOI] [PubMed] [Google Scholar]
- 115. Reyes-Turcu FE, Shanks JR, Komander D, Wilkinson KD. 2008. Recognition of polyubiquitin isoforms by the multiple ubiquitin binding modules of isopeptidase T. J. Biol. Chem. 283:19581–19592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Rotin D, Kumar S. 2009. Physiological functions of the HECT family of ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 10:398–409 [DOI] [PubMed] [Google Scholar]
- 117. Ryo A, et al. 2003. Regulation of NF-kappaB signaling by Pin1-dependent prolyl isomerization and ubiquitin-mediated proteolysis of p65/RelA. Mol. Cell 12:1413–1426 [DOI] [PubMed] [Google Scholar]
- 118. Ryzhakov G, Randow F. 2007. SINTBAD, a novel component of innate antiviral immunity, shares a TBK1-binding domain with NAP1 and TANK. EMBO J. 26:3180–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Sabbah A, et al. 2009. Activation of innate immune antiviral responses by Nod2. Nat. Immunol. 10:1073–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Saha SK, et al. 2006. Regulation of antiviral responses by a direct and specific interaction between TRAF3 and Cardif. EMBO J. 25:3257–3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Saitoh T, et al. 2006. Negative regulation of interferon-regulatory factor 3-dependent innate antiviral response by the prolyl isomerase Pin1. Nat. Immunol. 7:598–605 [DOI] [PubMed] [Google Scholar]
- 122. Saitoh T, et al. 2005. A20 is a negative regulator of IFN regulatory factor 3 signaling. J. Immunol. 174:1507–1512 [DOI] [PubMed] [Google Scholar]
- 123. Satoh T, et al. 2010. LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc. Natl. Acad. Sci. U. S. A. 107:1512–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Schlee M, et al. 2009. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity 31:25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Schmidt A, et al. 2009. 5′-triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc. Natl. Acad. Sci. U. S. A. 106:12067–12072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Shembade N, Harhaj NS, Liebl DJ, Harhaj EW. 2007. Essential role for TAX1BP1 in the termination of TNF-alpha-, IL-1- and LPS-mediated NF-kappaB and JNK signaling. EMBO J. 26:3910–3922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Shembade N, et al. 2008. The E3 ligase Itch negatively regulates inflammatory signaling pathways by controlling the function of the ubiquitin-editing enzyme A20. Nat. Immunol. 9:254–262 [DOI] [PubMed] [Google Scholar]
- 128. Shembade N, Ma A, Harhaj EW. 2010. Inhibition of NF-kappaB signaling by A20 through disruption of ubiquitin enzyme complexes. Science 327:1135–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Shembade N, Parvatiyar K, Harhaj NS, Harhaj EW. 2009. The ubiquitin-editing enzyme A20 requires RNF11 to downregulate NF-kappaB signalling. EMBO J. 28:513–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Shen RR, Hahn WC. 2011. Emerging roles for the non-canonical IKKs in cancer. Oncogene 30:631–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Shigemoto T, et al. 2009. Identification of loss of function mutations in human genes encoding RIG-I and MDA5: implications for resistance to type I diabetes. J. Biol. Chem. 284:13348–13354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Shoji-Kawata S, et al. 2007. The RING finger ubiquitin ligase RNF125/TRAC-1 down-modulates HIV-1 replication in primary human peripheral blood mononuclear cells. Virology 368:191–204 [DOI] [PubMed] [Google Scholar]
- 133. Skaug B, Jiang X, Chen ZJ. 2009. The role of ubiquitin in NF-kappaB regulatory pathways. Annu. Rev. Biochem. 78:769–796 [DOI] [PubMed] [Google Scholar]
- 134. Sun SC. 2011. Non-canonical NF-kappaB signaling pathway. Cell Res. 21:71–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Suzuki H, et al. 1999. IkappaBalpha ubiquitination is catalyzed by an SCF-like complex containing Skp1, cullin-1, and two F-box/WD40-repeat proteins, betaTrCP1 and betaTrCP2. Biochem. Biophys. Res. Commun. 256:127–132 [DOI] [PubMed] [Google Scholar]
- 136. Takahashi K, et al. 2006. Roles of caspase-8 and caspase-10 in innate immune responses to double-stranded RNA. J. Immunol. 176:4520–4524 [DOI] [PubMed] [Google Scholar]
- 137. Takahashi M, et al. 2000. Linkage and LOH studies in 19 cylindromatosis families show no evidence of genetic heterogeneity and refine the CYLD locus on chromosome 16q12-q13. Hum. Genet. 106:58–65 [DOI] [PubMed] [Google Scholar]
- 138. Takahasi K, et al. 2008. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol. Cell 29:428–440 [DOI] [PubMed] [Google Scholar]
- 139. Takeuchi O, Akira S. 2010. Pattern recognition receptors and inflammation. Cell 140:805–820 [DOI] [PubMed] [Google Scholar]
- 140. Tanaka T, Grusby MJ, Kaisho T. 2007. PDLIM2-mediated termination of transcription factor NF-kappaB activation by intranuclear sequestration and degradation of the p65 subunit. Nat. Immunol. 8:584–591 [DOI] [PubMed] [Google Scholar]
- 141. Tang ED, Wang CY. 2009. MAVS self-association mediates antiviral innate immune signaling. J. Virol. 83:3420–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Tang ED, Wang CY. 2010. TRAF5 is a downstream target of MAVS in antiviral innate immune signaling. PLoS One 5:e9172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Tcherpakov M, et al. 2009. Regulation of endoplasmic reticulum-associated degradation by RNF5-dependent ubiquitination of JNK-associated membrane protein (JAMP). J. Biol. Chem. 284:12099–12109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Tenoever BR, et al. 2007. Multiple functions of the IKK-related kinase IKKepsilon in interferon-mediated antiviral immunity. Science 315:1274–1278 [DOI] [PubMed] [Google Scholar]
- 145. Tokunaga F, et al. 2011. SHARPIN is a component of the NF-kappaB-activating linear ubiquitin chain assembly complex. Nature 471:633–636 [DOI] [PubMed] [Google Scholar]
- 146. Tokunaga F, et al. 2009. Involvement of linear polyubiquitylation of NEMO in NF-kappaB activation. Nat. Cell Biol. 11:123–132 [DOI] [PubMed] [Google Scholar]
- 147. Tsagaratou A, Trompouki E, Grammenoudi S, Kontoyiannis DL, Mosialos G. 2010. Thymocyte-specific truncation of the deubiquitinating domain of CYLD impairs positive selection in a NF-kappaB essential modulator-dependent manner. J. Immunol. 185:2032–2043 [DOI] [PubMed] [Google Scholar]
- 148. Tseng PH, et al. 2010. Different modes of ubiquitination of the adaptor TRAF3 selectively activate the expression of type I interferons and proinflammatory cytokines. Nat. Immunol. 11:70–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Turer EE, et al. 2008. Homeostatic MyD88-dependent signals cause lethal inflammation in the absence of A20. J. Exp. Med. 205:451–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Vallabhapurapu S, et al. 2008. Nonredundant and complementary functions of TRAF2 and TRAF3 in a ubiquitination cascade that activates NIK-dependent alternative NF-kappaB signaling. Nat. Immunol. 9:1364–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Ventii KH, Wilkinson KD. 2008. Protein partners of deubiquitinating enzymes. Biochem. J. 414:161–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Vereecke L, Beyaert R, van Loo G. 2009. The ubiquitin-editing enzyme A20 (TNFAIP3) is a central regulator of immunopathology. Trends Immunol. 30:383–391 [DOI] [PubMed] [Google Scholar]
- 153. Vereecke L, et al. 2010. Enterocyte-specific A20 deficiency sensitizes to tumor necrosis factor-induced toxicity and experimental colitis. J. Exp. Med. 207:1513–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Verhelst K, Verstrepen L, Carpentier I, Beyaert R. 2011. Linear ubiquitination in NF-kappaB signaling and inflammation: what we do understand and what we do not. Biochem. Pharmacol. 82:1057–1065 [DOI] [PubMed] [Google Scholar]
- 155. Verstrepen L, et al. 2010. Expression, biological activities and mechanisms of action of A20 (TNFAIP3). Biochem. Pharmacol. 80:2009–2020 [DOI] [PubMed] [Google Scholar]
- 156. Viswanathan K, Fruh K, DeFilippis V. 2010. Viral hijacking of the host ubiquitin system to evade interferon responses. Curr. Opin. Microbiol. 13:517–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Wagner S, et al. 2008. Ubiquitin binding mediates the NF-kappaB inhibitory potential of ABIN proteins. Oncogene 27:3739–3745 [DOI] [PubMed] [Google Scholar]
- 158. Wang YY, Li L, Han KJ, Zhai Z, Shu HB. 2004. A20 is a potent inhibitor of TLR3- and Sendai virus-induced activation of NF-kappaB and ISRE and IFN-beta promoter. FEBS Lett. 576:86–90 [DOI] [PubMed] [Google Scholar]
- 159. Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR. 2006. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J. Virol. 80:5059–5064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Wertz IE, Dixit VM. 2010. Signaling to NF-kappaB: regulation by ubiquitination. Cold Spring Harbor Perspect. Biol. 2:a003350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Wertz IE, et al. 2004. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature 430:694–699 [DOI] [PubMed] [Google Scholar]
- 162. Xia ZP, et al. 2009. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature 461:114–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Xu LG, et al. 2005. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell 19:727–740 [DOI] [PubMed] [Google Scholar]
- 164. Xu M, Skaug B, Zeng W, Chen ZJ. 2009. A ubiquitin replacement strategy in human cells reveals distinct mechanisms of IKK activation by TNFalpha and IL-1beta. Mol. Cell 36:302–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Ye Y, Rape M. 2009. Building ubiquitin chains: E2 enzymes at work. Nat. Rev. Mol. Cell Biol. 10:755–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Yoneyama M, Fujita T. 2009. RNA recognition and signal transduction by RIG-I-like receptors. Immunol. Rev. 227:54–65 [DOI] [PubMed] [Google Scholar]
- 167. Yoneyama M, et al. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730–737 [DOI] [PubMed] [Google Scholar]
- 168. Yoshida R, et al. 2008. TRAF6 and MEKK1 play a pivotal role in the RIG-I-like helicase antiviral pathway. J. Biol. Chem. 283:36211–36220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. You F, et al. 2009. PCBP2 mediates degradation of the adaptor MAVS via the HECT ubiquitin ligase AIP4. Nat. Immunol. 10:1300–1308 [DOI] [PubMed] [Google Scholar]
- 170. Young JA, et al. 2011. Fas-associated death domain (FADD) and the E3 ubiquitin-protein ligase TRIM21 interact to negatively regulate virus-induced interferon production. J. Biol. Chem. 286:6521–6531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Yu Y, Hayward GS. 2010. The ubiquitin E3 ligase RAUL negatively regulates type I interferon through ubiquitination of the transcription factors IRF7 and IRF3. Immunity 33:863–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Yu Y, Wang SE, Hayward GS. 2005. The KSHV immediate-early transcription factor RTA encodes ubiquitin E3 ligase activity that targets IRF7 for proteosome-mediated degradation. Immunity 22:59–70 [DOI] [PubMed] [Google Scholar]
- 173. Zarnegar BJ, et al. 2008. Noncanonical NF-kappaB activation requires coordinated assembly of a regulatory complex of the adaptors cIAP1, cIAP2, TRAF2 and TRAF3 and the kinase NIK. Nat. Immunol. 9:1371–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Zeng W, et al. 2010. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell 141:315–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Zeng W, Xu M, Liu S, Sun L, Chen ZJ. 2009. Key role of Ubc5 and lysine-63 polyubiquitination in viral activation of IRF3. Mol. Cell 36:315–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Zhang M, et al. 2008. Negative feedback regulation of cellular antiviral signaling by RBCK1-mediated degradation of IRF3. Cell Res. 18:1096–1104 [DOI] [PubMed] [Google Scholar]
- 177. Zhang M, et al. 2008. Regulation of IkappaB kinase-related kinases and antiviral responses by tumor suppressor CYLD. J. Biol. Chem. 283:18621–18626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Zhang Z, et al. 2011. DDX1, DDX21, and DHX36 helicases form a complex with the adaptor molecule TRIF to sense dsRNA in dendritic cells. Immunity 34:866–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Zhao H, et al. 2005. A novel E3 ubiquitin ligase TRAC-1 positively regulates T cell activation. J. Immunol. 174:5288–5297 [DOI] [PubMed] [Google Scholar]
- 180. Zhao S, Ulrich HD. 2010. Distinct consequences of posttranslational modification by linear versus K63-linked polyubiquitin chains. Proc. Natl. Acad. Sci. U. S. A. 107:7704–7709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Zhao T, et al. 2007. The NEMO adaptor bridges the nuclear factor-kappaB and interferon regulatory factor signaling pathways. Nat. Immunol. 8:592–600 [DOI] [PubMed] [Google Scholar]
- 182. Zhong B, et al. 2008. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 29:538–550 [DOI] [PubMed] [Google Scholar]
- 183. Zhong B, et al. 2009. The ubiquitin ligase RNF5 regulates antiviral responses by mediating degradation of the adaptor protein MITA. Immunity 30:397–407 [DOI] [PubMed] [Google Scholar]
- 184. Zust R, et al. 2011. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 12:137–143 [DOI] [PMC free article] [PubMed] [Google Scholar]