Abstract
Summary: The cytokine storm has captured the attention of the public and the scientific community alike, and while the general notion of an excessive or uncontrolled release of proinflammatory cytokines is well known, the concept of a cytokine storm and the biological consequences of cytokine overproduction are not clearly defined. Cytokine storms are associated with a wide variety of infectious and noninfectious diseases. The term was popularized largely in the context of avian H5N1 influenza virus infection, bringing the term into popular media. In this review, we focus on the cytokine storm in the context of virus infection, and we highlight how high-throughput genomic methods are revealing the importance of the kinetics of cytokine gene expression and the remarkable degree of redundancy and overlap in cytokine signaling. We also address evidence for and against the role of the cytokine storm in the pathology of clinical and infectious disease and discuss why it has been so difficult to use knowledge of the cytokine storm and immunomodulatory therapies to improve the clinical outcomes for patients with severe acute infections.
INTRODUCTION
The term “cytokine storm” calls up vivid images of an immune system gone awry and an inflammatory response flaring out of control (Fig. 1). The term has captured the attention of the public and the scientific community alike and is increasingly being used in both the popular media and the scientific literature. However, while the general concept of an excessive or uncontrolled release of proinflammatory cytokines is well known, an actual definition of what constitutes a cytokine storm is lacking. Furthermore, there is not a good understanding of the molecular events that precipitate a cytokine storm, of the contribution such a “storm” makes to pathogenesis, or of what therapeutic strategies might be used to prevent the storm or quell it once it has started.
Fig 1.
Imagery of a cytokine storm.
Although the concept certainly predates the coining of the term, the first use of “cytokine storm” appears to be in an article published in 1993 on graft-versus-host disease (47). The use of the term in infectious disease research began in early 2000 in reports on cytomegalovirus (6), Epstein-Barr virus-associated hemophagocytic lymphohistiocytosis (70), group A streptococcus (15), influenza virus (154), variola virus (71), and severe acute respiratory syndrome coronavirus (SARS-CoV) (63). The term appears to have first been applied in the context of avian H5N1 influenza virus infection in 2005 (155), after which it began to appear more frequently in the scientific literature. Public interest in “bird flu” also brought the term cytokine storm into the popular media. A recent Google search for cytokine storm yielded 323,000 hits; over 170,000 were within the past year alone.
Cytokine storms are associated with a wide variety of infectious and noninfectious diseases and have even been the unfortunate consequence of attempts at therapeutic intervention (136). Previous reviews have centered on the advent of the concept (29) or its role in graft-versus-host disease (46), multiple sclerosis (89), pancreatitis (94), or multiple organ dysfunction syndrome (145). Though the term was not explicitly stated, recent reviews have addressed potential cellular and molecular mechanisms contributing to the cytokine storm in viral disease (66, 81), some of which specifically focused on influenza (114, 115). In this review, we focus on the cytokine storm in the context of infection, with particular emphasis on respiratory viruses. We also highlight how high-throughput genomic methods are revealing new insights into the cytokine storm. These methods are especially useful for obtaining global views of the complex and intertwined molecular events that attend the upregulation of multiple cytokines.
Our goals in this review are to better define the concept of a cytokine storm and the biological consequences of cytokine overproduction. We will also look at the cytokine storm through the lens of genomics, which is revealing the importance of the kinetics of cytokine gene expression and the remarkable degree of redundancy and overlap in cytokine signaling. Finally, we will address evidence for and against the role of the cytokine storm in the pathology of clinical and infectious disease and discuss why it has been so difficult to use knowledge of the cytokine storm and immunomodulatory therapies to improve the clinical outcomes for patients with severe acute infections.
CYTOKINES
Cytokines are a diverse group of small proteins that are secreted by cells for the purpose of intercellular signaling and communication. Specific cytokines have autocrine, paracrine, and/or endocrine activity and, through receptor binding, can elicit a variety of responses, depending upon the cytokine and the target cell. Among the many functions of cytokines are the control of cell proliferation and differentiation and the regulation of angiogenesis and immune and inflammatory responses (Table 1).
Table 1.
Major types and actions of cytokines
| Type | Actions |
|---|---|
| Interferons | Regulation of innate immunity, activation of antiviral properties, antiproliferative effects |
| Interleukins | Growth and differentiation of leukocytes; many are proinflammatory |
| Chemokines | Control of chemotaxis, leukocyte recruitment; many are proinflammatory |
| Colony-stimulating factors | Stimulation of hematopoietic progenitor cell proliferation and differentiation |
| Tumor necrosis factor | Proinflammatory, activates cytotoxic T lymphocytes |
Many cytokines have multiple and sometimes unrelated functions that depend on the target cell or on the presence or absence of other cytokines. Some have limited sequence similarity and engage distinct receptors yet transduce signals through common intracellular pathways (e.g., type I and type III interferons [IFNs]). In part because of this diversity of structure and function, the classification and naming of cytokines have been a challenge. The complex network of the cytokine response is best considered a series of overlapping networks, each with a degree of redundancy and with alternate pathways. This combination of overlap and redundancy has important implications with respect to identifying the key steps in the cytokine response to infection and in targeting specific cytokines for therapeutic intervention. While many infections are characterized by broadly similar cytokine profiles, their clinical presentations can be quite different. Prior to delving into a discussion of cytokine storms, it is worth taking a brief look at the cytokines at the heart of the cytokine storm.
Cytokines Associated with the Cytokine Storm
Interferons.
The interferons (IFNs) are a family of cytokines that play a central role in innate immunity to viruses and other microbial pathogens (45, 75). They are classified into three major types (types I, II, and III) on the basis of their receptor specificity. Type I IFNs (IFN-α and IFN-β) signal through a heterodimeric receptor complex, IFNAR1/IFNAR2, whereas type II IFN (IFN-γ) signals through IFN-γR1/IFN-γR2. Lambda IFNs are a new class of IFN with antiviral properties (127), protecting mice against influenza A virus (106). IFN-λ1, -λ2, and -λ3 (also referred to as interleukin-29 [IL-29], IL-28a, and IL-28b) bind receptor complex IL-28R/IL-10Rβ yet are functionally similar to type I IFNs in that both transduce signals through the Jak-STAT signaling pathway. Receptor binding results in the initiation of downstream signaling cascades, the result of which is the activation of transcription factors and the induction of hundreds of IFN-stimulated genes. These genes encode protein products with antiviral, antiproliferative, or immunomodulatory properties. Such effects have led to the therapeutic use of IFNs (often in combination with other drugs) in the treatment of viral diseases such as hepatitis C and hepatitis B, certain types of leukemia and lymphoma, and multiple sclerosis (17, 51).
Interleukins.
In contrast to the IFNs, the interleukins are a diverse family of immune system regulators that function primarily in immune cell differentiation and activation. They may be either pro- or anti-inflammatory and, like all cytokines, elicit a wide variety of responses. The interleukin designation was originally coined to refer to cytokines produced by leukocytes that function in intercellular communication; however, interleukins are now known to be produced by a wide variety of cell types. Although a common nomenclature system has been adopted, the designation and naming of interleukins continue to be confusing (18). IL-1, for example, is actually a family of cytokines encoded by 11 genes (132). As new functions are elucidated for some IL-1 family members, new interleukin designations have been proposed (40).
IL-1α and IL-1β are proinflammatory cytokines that mediate the host response to infection through both direct and indirect mechanisms (41). Among their biological functions, these cytokines increase acute-phase signaling, trafficking of immune cells to the site of primary infection, epithelial cell activation, and secondary cytokine production. Inflammasomes are cytosolic macromolecular complexes comprised of members of the nucleotide-binding domain and leucine-rich-repeat-containing receptor (NLR) family (the NLRP3 inflammasome is one the best characterized) that produce IL-1β and IL-18 in defense against pathogens, and their activity is regulated by type I IFN (57). The acute-phase response to infection results in a wide range of local effects and systemic alterations that are evidenced in changes that are generally proinflammatory, such as the increase in specific cytokine production, and which can be linked to viral clearance, such as increases in complement (53). IL-1 receptor signaling is responsible for acute lung immunopathology but increases the survival of mice infected with influenza virus by enhancing IgM antibody responses and recruiting CD4+ T cells to the site of infection (124).
Chemokines.
The largest family of cytokines is the chemokines, with 44 members (and increasing) that bind to one or more of 21 G-protein-coupled receptors (33). These small secreted proteins are classified into four types (CXC, CC, C, and CX3C), depending upon the spacing of their first two cysteine residues. Chemokines function as chemoattractants to control the migration of cells, particularly those of the immune system, and contribute to such diverse processes as embryogenesis, innate and adaptive immune system development and function, and cancer metastasis (119). For example, differential expression of IFN-γ-regulated CXCR3 ligands, CXCL9, CXCL10, and CXCL11, appears to regulate T-cell function and infiltration in the periphery during inflammation (reviewed in reference 56). The majority of chemokines are considered to be proinflammatory, and they are released by a variety of cells in response to virus (or other microbial) infection. The release of proinflammatory chemokines results in the recruitment of immune system cells (neutrophils, monocytes/macrophages, and lymphocytes) to the site of infection. Whereas most cytokines have pleiotropic effects, chemokine recruitment of immune cells can be highly selective for specific cell types. For example, CXCL8 (IL-8), CCL2 (monocyte chemoattractant protein 1 [MCP-1]), and CCL11 (eotaxin) are major chemoattractant factors for neutrophil, monocyte, and eosinophil immune cells, respectively. Chemokines and their receptors have been heavily targeted by the pharmaceutical industry, but with limited success (54).
CSFs.
Colony-stimulating factors (CSFs), such as granulocyte-macrophage colony-stimulating factor (GM-CSF), macrophage colony-stimulating factor (M-CSF), and granulocyte colony-stimulating factor (G-CSF), stimulate hematopoietic progenitor cell proliferation and differentiation. Colony-stimulating factors are also associated with inflammation, and there is evidence that these factors may be part of a mutually dependent proinflammatory cytokine network that includes IL-1 and tumor necrosis factor (TNF) (58). It is thought that by functioning to increase the number of cytokine-producing macrophages at a site of inflammation, colony-stimulating factors may be part of an amplification cascade that serves to perpetuate inflammatory reactions.
TNFs.
Tumor necrosis factor (TNF) is perhaps the best known and most intensely studied of the proinflammatory cytokines, and it plays a prominent role in the cytokine storm literature. The name “tumor necrosis factor” was first used in 1975 for a cytotoxic serum factor capable of inducing tumor regression in mice (23), which soon thereafter was reported to play a role in the pathogenesis of malaria and sepsis (14, 30, 31). TNF is now considered a central cytokine in acute viral diseases, including those caused by influenza virus, dengue virus, and Ebola virus. TNF is expressed by a variety of immune cells, and its primary receptor, TNFR1, appears to be expressed by all cell types, ensuring widespread effects of this cytokine. The pleiotropic effects of TNF are further amplified by the existence of a superfamily of TNF proteins that consists of 19 members that signal through 29 receptors (2). Excess TNF production is associated with a number of chronic inflammatory and autoimmune diseases, and TNF inhibitors have been approved for the treatment of inflammatory bowel disease, psoriasis, and rheumatoid arthritis (79, 126). In contrast, the use of TNF inhibitors for the treatment of sepsis has not been successful (49), possibly due to the early release and short circulating half-life of the cytokine (30).
Cytokine Dynamics
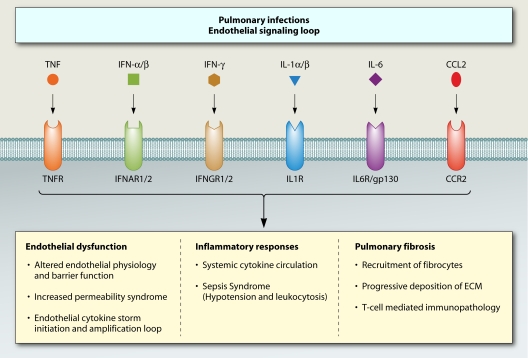
We still do not understand the complex nature of the immune response and have probably underestimated its dynamic nature during acute infection. For example, while TNF promotes IL-1 generation, inducing changes in endothelial cell physiology within the local microenvironment (109), increased TNF can also exert broad systemic effects beyond the site of infection. An experimental study of rabbits with severe Pseudomonas pneumonia and systemic sepsis provided definitive evidence that TNF instilled into the lungs could pass into the systemic circulation, providing direct communication between the lungs and the bloodstream (80). Nevertheless, we have relied for too long on clinical studies using intermittent sampling from one compartment (typically the peripheral blood), although many of the critical responses are likely to be far more localized in tissue. For instance, in severe infections affecting the deep tissues of the respiratory tract (as has been described in avian influenza or severe primary influenza virus infections), it is probable that it is the immunological cascade that takes place directly in the deep tissues that is crucial to immunopathology rather than what can be measured as a “spillover” in the peripheral blood (28, 82, 128). It is important to also consider the compartmentalization of tissue-specific microenvironments. For example, within the lung, the respiratory epithelium and alveolar macrophages normally maintain homeostasis by limiting activation of innate immunity in airways and alveolar spaces (61). In mice, lymphoid tissue around airways also can dampen initial responses to viral challenge but can be induced to provide efficient stimulation of cellular and humoral immune responses with more severe influenza virus infection (107). Similar compartmentalization is likely to be important in infections of the central nervous system (i.e., bacterial and tuberculous meningitis, encephalitis, and fungal infections) and perhaps in more subtle ways in infections such as dengue, in which the clinical syndrome is dominated by capillary permeability and plasma leakage (130, 131).
To date, most studies have focused on direct measurements of a few cytokines and chemokines in the peripheral blood compartment and have failed to interrogate the whole of the immune cascade in the context of the infecting pathogen and the rapidly changing immune environment in tissues. Interestingly, while the peripheral blood may not provide an accurate picture of the cytokine profiles in a tissue, in the lungs, the location of the initial infection does not seem to be a determinant of the severity of local and systemic cytokine storms. For example, influenza viruses infect and destroy the ciliated epithelial cells of the conducting airways, whereas SARS-CoV infects type II pneumocytes in the alveolar walls and hantavirus particles infect microvascular endothelial cells in the alveolar walls, yet all can lead to indistinguishable clinical syndromes of acute lung injury (ALI) with respiratory failure, sepsis, and a cytokine storm. Bacterial exotoxins, such as the Pseudomonas type III exotoxin and staphylococcal enterotoxins, are directly lytic to alveolar cells and destroy the gas exchange parenchyma of the lungs (48, 80, 112). Although bacterial, viral, and fungal products drive inflammation in the lungs via interactions with surface Toll-like receptors (TLR) and intracellular receptors for DNA and RNA (NOD-like and RIG-I-like receptors), endogenous products such as oxidized phospholipids and matrix breakdown products also drive inflammation through TLR4 and probably other TLRs, providing a mechanism for perpetuating inflammation when microbial products are being cleared from the lungs (68, 72). Mitochondrial membrane proteins and cellular ATP also have been implicated in driving endogenous inflammatory responses (156).
Regulation of Pro- and Anti-Inflammatory Cytokines
The intensity of the inflammatory response in the lungs reflects a balance between proinflammatory cytokines (e.g., TNF and IL-1β) and their cognate soluble receptors or inhibitors (TNFR1, TNFR2, and IL-1RA), which inhibit the activity of these inflammatory cytokines in the aqueous phase of alveolar fluid (113). One mechanism to dampen lung inflammation is by regulating the activation of specific cell types. For example, CD200R expression on alveolar macrophages helps resolve lung inflammation during influenza virus infection by restraining macrophage activity (133). Negative regulators, such as IL-1 receptor-associated kinase (IRAK-M), suppressor of cytokine signaling 1 (SOCS1), phosphoinositide-3-OH kinase (PI3K), Toll-interacting protein (TOLLIP), and zinc finger protein A20 (34), also help maintain innate immune processes by preventing aberrant TLR activation. The production of anti-inflammatory cytokines, mainly IL-10 by macrophages and certain types of T cells (Th2 and regulatory T cells) and B cells (105), represents another mechanism involved in regulating proinflammatory responses. Although IL-10 is most commonly recognized as an anti-inflammatory cytokine, recent evidence has linked IL-10 to a potential role in fibrosis, where increased IL-10 expression was reported to induce collagen production and fibrocyte recruitment into the lung (135). In contrast, interactions between IL-6 and its soluble receptor enhance the activity of IL-6 on target cells, providing a mechanism for enhancing the activity of TNF and IL-1β when the concentrations of soluble TNF receptors and IL-1RA are very high (113). As the balance of pro- and anti-inflammatory mechanisms is critical to maintaining lung immune homeostasis, it is conceivable that if one or more of these regulatory mechanisms are absent or aberrantly regulated, then the outcome may contribute toward a cytokine storm.
THE CYTOKINE STORM
Cytokine Storm Pathology
Inflammation associated with a cytokine storm begins at a local site and spreads throughout the body via the systemic circulation. Rubor (redness), tumor (swelling or edema), calor (heat), dolor (pain), and “functio laesa” (loss of function) are the hallmarks of acute inflammation. When localized in skin or other tissue, these responses increase blood flow, enable vascular leukocytes and plasma proteins to reach extravascular sites of injury, increase local temperatures (which is advantageous for host defense against bacterial infections), and generate pain, thereby warning the host of the local responses. These responses often occur at the expense of local organ function, particularly when tissue edema causes a rise in extravascular pressures and a reduction in tissue perfusion. Compensatory repair processes are initiated soon after inflammation begins, and in many cases the repair process completely restores tissue and organ function. When severe inflammation or the primary etiological agent triggering inflammation damages local tissue structures, healing occurs with fibrosis, which can result in persistent organ dysfunction.
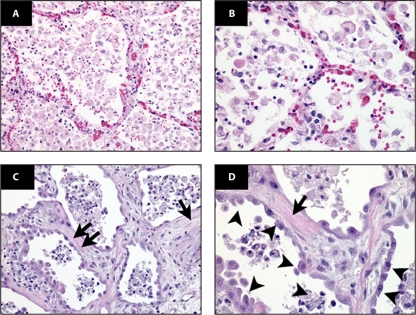
Acute lung injury (ALI) is a common consequence of a cytokine storm in the lung alveolar environment and systemic circulation and is most commonly associated with suspected or proven infections in the lungs or other organs (121). In humans, ALI is characterized by an acute mononuclear/neutrophilic inflammatory response followed by a chronic fibroproliferative phase marked by progressive collagen deposition in the lung (Fig. 2) (reviewed in reference 96). Pathogen-induced lung injury can progress into ALI or its more severe form, acute respiratory distress syndrome (ARDS), as seen with SARS-CoV and influenza virus infections. IL-1β is a key cytokine driving proinflammatory activity in bronchoalveolar lavage fluid of patients with lung injury (118). Intense inflammation in the lungs also can have other systemic effects on other organs, as the combination of severe HCl injury in the lungs and mechanical ventilation in rabbits leads to renal dysfunction and evidence of apoptosis in renal tubular epithelial cells (69).
Fig 2.
Human ARDS. Photomicrographs from the lungs of 2 different patients with ARDS, stained with hematoxylin and eosin (H&E) are shown. The alveolar spaces are filled with a mixed mononuclear/neutrophilic infiltrate, the alveolar walls are thickened, and the septae are edematous. Note the presence of cellular debris and proteinaceous material in the air spaces (A [magnification, ×200] and B [magnification, ×400]). In later stages (C [magnification, ×200] and D [magnification, ×400]), there is a fibroproliferative response with collagen deposition in the alveolar walls (arrows). Note that the alveolar epithelium has been replaced with cuboidal cells (arrowheads). (Figure and legend reprinted from reference 96 with permission of the publisher.)
The cytokine storm is best exemplified by severe lung infections, in which local inflammation spills over into the systemic circulation, producing systemic sepsis, as defined by persistent hypotension, hyper- or hypothermia, leukocytosis or leukopenia, and often thrombocytopenia (84). Viral, bacterial, and fungal pulmonary infections all cause the sepsis syndrome, and these etiological agents are difficult to differentiate on clinical grounds. In some cases, persistent tissue damage without severe microbial infection in the lungs also is associated with a cytokine storm and clinical manifestations that mimic sepsis syndrome. In addition to lung infections, the cytokine storm is a consequence of severe infections in the gastrointestinal tract, urinary tract, central nervous system, skin, joint spaces, and other sites.
Studies of patients with severe sepsis due to pulmonary or nonpulmonary infections show characteristic plasma cytokine profiles, which change over time. The acute-response cytokines TNF and IL-1β and the chemotactic cytokines IL-8 and MCP-1 appear in the early minutes to hours after infection, followed by a more sustained increase in IL-6. The anti-inflammatory cytokine IL-10 appears somewhat later, as the body attempts to control the acute systemic inflammatory response. Plasma samples from a laboratory worker who developed septic shock following the deliberate injection of a large amount of bacterial endotoxin (in an attempt to treat a recently diagnosed tumor) provided insight into this sequence of cytokines following entry of bacterial products into the systemic circulation (138). A similar picture appeared in six healthy volunteers treated with an activating antibody against CD28 during a phase 1 clinical trial (136). IL-6 concentrations in peripheral blood have been used to assess the intensity of systemic cytokine responses in patients with sepsis, because IL-6 production is stimulated by TNF and IL-1β, providing an integrated signal of these two early-response cytokines (1).
Systemic production of IL-10 following the onset of a cytokine storm is a marker of a counter-anti-inflammatory response that has been termed “immunoparalysis,” in that it is associated with downregulation of neutrophil and monocyte function in the systemic circulation (32, 42, 49). Downregulation of systemic inflammation might be conceptually beneficial in controlling systemic responses to local infections (108). However, it has been suggested that patients who survive the initial cytokine storm but subsequently die may be those who do not recover from immunoparalysis. Patients with persistent downregulation of HLA-DR (a marker of immunosuppression) on monocytes 3 to 4 days after the onset of severe sepsis and cytokine storm have a high mortality rate, suggesting a rationale for therapy to reverse immunosuppression under such circumstances (104).
Host Susceptibility to the Cytokine Storm
One of the challenging clinical questions about the cytokine storm is why some individuals seem particularly susceptible yet others seem relatively resistant, and there has been a great deal of interest in identifying underlying genetic mechanisms (149). Recent studies have shown a vast amount of variability in the innate immune responses of healthy humans, as reflected by the intermediate phenotype of whole-blood cytokine responses to bacterial products (151). Hyper- and hyporesponders to bacterial products are identifiable in the healthy population, which is explainable in part by genetically determined differences in the structure and function of TLR receptors, particularly TLR1 (150). In a large population of septic patients, those with a single nucleotide polymorphism (SNP) marking a hyperfunctioning variant of TLR1 had increased organ dysfunction and morbidity from Gram-positive bacteremia (150). Other genetic polymorphisms also contribute to the severity of the host response in sepsis and the cytokine storm, but the TLR1 polymorphism has a particularly strong relationship to Gram-positive infections (149).
Variants of TLR4, the principal receptor for lipopolysaccharide (LPS), can predispose individuals to sepsis, as evidenced by increased markers of systemic inflammation, including C-reactive protein (CRP), LPS-binding protein, and peripheral leukocytes in healthy humans exposed to endotoxin (99). Genome-wide association studies (GWAS) have associated TLR4 polymorphisms with increased susceptibility to pathogens and severity of disease. For example, TLR4 Asp299Gly occurred at a high frequency in Ghanaian children with severe malaria (100), and the polymorphism was also associated with manifestation of malaria during pregnancy in Ghanaian women (101). A recent GWAS identified multiple polymorphisms in cytokine-inducible SRC homology 2 (SH2) domain protein (CISH), a SOCS family member that controls IL-2 signaling, that were associated with increased susceptibility to bacteremia, tuberculosis, and severe malaria in persons in Gambia, Hong Kong, Kenya, Malawi, and Vietnam (77). Several GWAS have also identified variants of IFN-λ3 that were associated with spontaneous resolution and successful treatment of hepatitis C virus (HCV) infection (55, 140). The potential of polyethylene glycol (PEG)–IFN-λ1 as a novel therapeutic in the treatment of HCV is currently being investigated, though we will likely become less reliant on PEG-IFN therapies with the development of new HCV drugs such as the protease inhibitor telaprevir.
Because genetics play an important role in differential disease phenotypes, new mouse resources such as the Collaborative Cross are being utilized to further investigate host genetics and the biological pathways involved in microbial control. The Collaborative Cross is a recombinant inbred mouse resource (5) designed to capture the genetic heterogeneity of the human population, supporting systems genetics and predictive biology. Studies investigating host responses to influenza among genetically distinct recombinant inbred mice, such as BXD RI lines derived from crosses between DBA/2J and C57BL/6J mice, have demonstrated increased transcriptional responses associated with inflammation and antiviral immunity in mice that are more highly susceptible to infection (3, 16). Future investigations will likely reveal underlying genetic variants influencing host responses that contribute toward a cytokine storm during infection.
GENOMIC VIEWS OF THE CYTOKINE STORM
Functional genomics lends itself to a deeper understanding of infectious disease by encompassing both the pathogen and the host response. Microarray technologies provide a global view of gene expression changes induced by a variety of stimuli, enabling us to simultaneously profile tens of thousands of transcriptional changes from an organ or tissue compartment. The functional associations among these gene expression patterns show perturbations in cellular signaling pathways and cellular networks, with the implication that their differential regulation may contribute toward the resolution of infection or, alternatively, have detrimental consequences leading to a fatal outcome. The compilation of cytokine and chemokine genomic data from influenza, SARS-CoV, and dengue studies provides important insight into our understanding of the cytokine storm. In particular, the dynamic transcriptional responses among the molecular components involved in cytokine and chemokine gene expression, including their kinetic properties and the timing of gene activation, are beginning to detail the events surrounding the cytokine storm.
Cytokine Gene Expression Kinetics
The paradigm of “hit hard and hit early” popularized for the treatment of AIDS also appears to apply to the way H5N1 avian influenza virus causes tissue damage in human infections. The rapid and intense nature of the host inflammatory response is the suspected cause of severe lung damage (116). In a case study conducted in Ho Chi Minh City, Vietnam, comparing 18 people infected with H5N1 virus in 2004 and 2005 to 8 people infected with seasonal H1N1 influenza virus, elevated levels of MCP-1 (known as CCL2), IFN-γ-inducible IP-10 (CXCL10), MIG protein (CXCL9), and IL-8 were observed in H5N1 virus-infected patients who progressed to severe lung injury (38). Over the past decade, compelling genomic evidence from animal model systems indicates that highly pathogenic influenza viruses aberrantly regulate cytokine and chemokine transcriptional responses, leading to a cytokine storm.
The first genomic study to analyze host transcriptional responses to the 1918 pandemic influenza virus was led by Kash and colleagues. Extensive lung damage in mice infected with 1918 virus was accompanied by highly upregulated cytokine and chemokine gene expression (73). Transcriptional activation of innate immune genes was observed as early as 1 day postinfection and remained sustained through the course of infection. These genomic data suggested that the host response enhances 1918 virus pathogenesis by initiating a cytokine storm that contributes to increased disease severity. An overly aggressive innate immune response marked by early expression of proinflammatory cytokine genes was also observed in 1918 virus-infected macaques (28). For example, strong upregulation of IL-6, IL-8, CCL2, and CCL5 cytokine and chemokine gene expression in the lungs was mirrored by elevated levels of IL-8, CCL2, and CCL5 in the sera of infected animals (78). Microarray analysis of lung tissue from macaques infected with H5N1 virus revealed prolonged expression of CCL2, CXCL10, and CXCL9 genes among a gene set of 45 significantly differentially expressed cytokine and chemokine genes (7). The strong interferon and inflammatory transcriptional responses early in infection combined with histopathologic findings for H5N1-infected type II pneumocytes likely account for irreversible lung damage caused by inflammation. IFN signaling appears to play a critical role in restricting highly pathogenic influenza viruses to the lung microenvironment, as IFNAR deficiency in mice results in dissemination of the 1918 virus to brain and spleen (27).
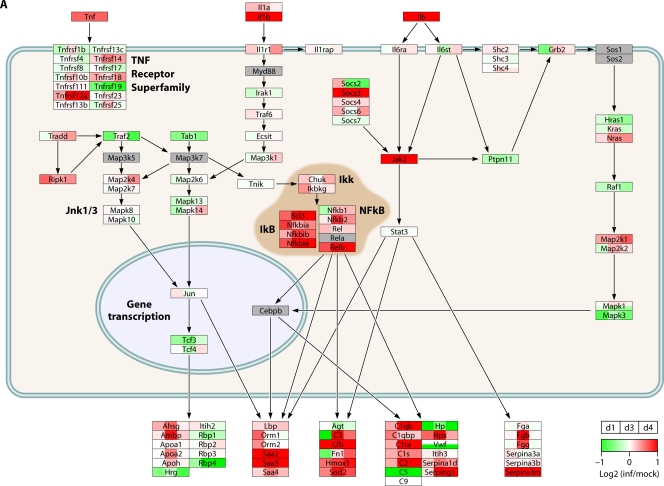
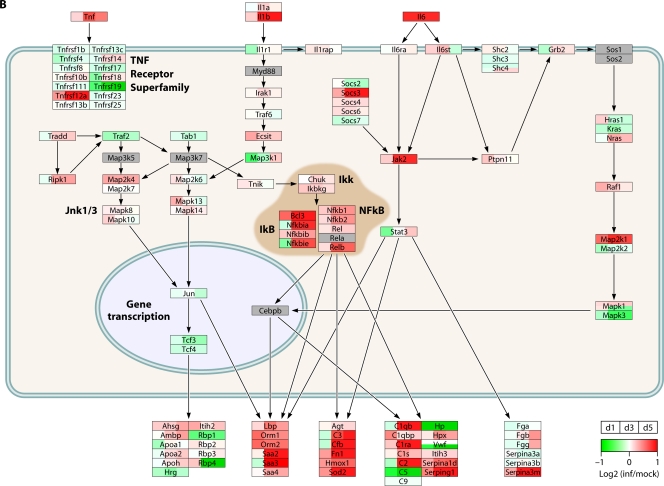
Direct comparison of transcriptional responses in infected macaque bronchi showed that these viruses induce similar IFN-regulated gene expression patterns, while genes associated with inflammation were significantly differentially regulated by 1918 and H5N1 viruses. Among the inflammatory response genes differentially regulated by 1918 and H5N1 viruses were those for colony-stimulating factor 3 (CSF3R) and IFN-γ. A large portion of the differentially expressed inflammatory response genes were anticorrelated in gene expression (upregulated in 1918 virus-infected bronchial tissue but downregulated in H5N1 virus-infected bronchial tissue), including inflammasome genes NLRP3 and IL1B, cytokine genes TNF and IFNB1, and cytokine receptor genes TNFRSF1B and IL4R. These findings suggest that an accelerated or excessive activation of the inflammasome is detrimental rather than protective in macaques infected with 1918 virus and that dysregulation of type I IFN signaling genes may impact regulation of inflammasome activity. The network architecture for acute-phase molecular interactions and transcriptional responses to 1918, H5N1, and seasonal H1N1 virus infections is displayed in Fig. 3.
Fig 3.
H5N1 and H1N1 influenza viruses differentially regulate the acute-phase response during infection. The diagrams show transcriptional changes for cytokine signaling pathway molecules in mouse lungs infected with either H5N1 virus (27) (A), 1918 virus (27) (B), or seasonal H1N1 virus (73) (C). Microarray data were background corrected and normalized by LOESS and quantile normalization. Expression values were represented as log2 ratios of infected to respective mock-infected samples, where biological and technical replicates for a given condition were averaged. PathVisio was used for data visualization (143) of the acute-phase response signaling pathway from IPA (Ingenuity Systems). Official gene symbols are shown. Serum amyloid (Saa2 and Saa3) and serine (or cysteine) peptidase inhibitor (Serpina1d and Serpina3a) represent acute-phase response proteins. Red indicates that the gene expression is increased relative to that for the uninfected reference. Green indicates that the gene expression is lower than that for the uninfected reference. White indicates no change in gene expression, and gray indicates that a molecule was not detected by microarray analysis. inf, infected; d, days postinfection.
TNF is one of the most prominent cytokines upregulated during H5N1 infection. It is highly expressed across infection models, including primary human respiratory epithelial cells (24, 125) and human monocyte-derived macrophages (26, 59, 62), compared with during seasonal H1N1 influenza virus and swine-origin H1N1 influenza virus (SOIV) infection (111). While a range of proinflammatory cytokines are induced in respiratory epithelial cells, TNF is not directly induced in respiratory epithelial cells by influenza virus infection (24) but rather is induced by secondary mediator cascades distinct from the primary effects of the virus (83). The concept of secondary effects of cytokine cascades, as opposed to the direct effect of the virus, in amplifying and broadening the proinflammatory response may be important in the escalation of cytokine storm (24, 111, 125).
Hyperresponsiveness of PRRs
The magnitude and range of cytokine and chemokine gene expression profiles are related to the recognition of pathogen-associated molecular patterns (PAMPs) by a variety of host pathogen recognition receptors (PRRs). As discussed above, there are several classes of cellular PRRs that activate and modulate innate immunity during infection, the specificity of which is determined by the recognized microbial motif, including TLRs that engage LPS, molecules derived from protozoan parasites, and viral RNA products (110, 152, 153), RLRs such as RIG-I, LGP2, and MDA5 (reviewed in reference 92), and NLRs such as NOD2 (122). RIG-I is critical for type I IFN transcription during viral infection (76). RIG-I-deficient cells show reduced expression of IFN-α and -β genes as well as IFN-stimulated genes during influenza virus infection (91). In the absence of RIG-I signaling, mice fail to produce type I IFN in response to vesicular stomatitis virus (VSV), Newcastle disease virus (NDV), and Sendai virus infection (74).
Infection with malarial parasites causes proinflammatory priming of TLR responses via TLR9 activation. Plasmodium infection initiates IL-12 and IFN-γ production, which in turn enhances TLR expression and innate immune signaling pathways. The increased TLR expression is thought to lead to a stage of hyperresponsiveness to TLR agonists. This was observed in humans acutely infected with Plasmodium falciparum, resulting in enhanced activation of innate immune cells to TLR agonists. Genes involved in TLR signaling pathways for which enhanced expression was observed during P. falciparum infection included the CD39, TLR4, TLR1, TLR8, and MYD88 genes. This phenomenon was also observed, in a TLR9/IL-12/IFN-γ-dependent manner, in mice infected with Plasmodium chabaudi (50). Similarly, respiratory syncytial virus (RSV) infection results in increased TLR4 expression on airway epithelial cells that sensitizes cells to endotoxin by enhancing LPS-receptor engagement (103), and TLR4 variants may attenuate innate immune responses by disrupting RSV triggering of TLR4 surface expression (142). A hyperresponsive TLR state represents one mechanism by which a cytokine storm can be perpetuated.
Differential Host Proinflammatory Responses
Swine are an important reservoir for influenza virus and have been linked to the emergence of some of the most notable historic and contemporary influenza pandemics, including the most recent swine-origin pandemic virus arising in 2009. It is not clearly understood why influenza viruses that are associated with severe or fatal infection in humans and nonhuman primates are generally nonfatal and cause less severe disease in swine (146). To understand the host response to influenza in pigs, Ma and colleagues recently profiled the gene expression in the lungs of pigs infected with recent H1N1 influenza viruses, a 2009 pandemic H1N1 human isolate, and a 2009 swine H1N1 virus and compared transcriptional patterns with those of a historical isolate, A/swine/Iowa/1930, that causes mild clinical symptoms (93). Pigs infected with the contemporary viruses experienced a transient, early increase in immune response concurrent with greater clinical illness. This included greater expression of cytokines and chemokines such as TNF, TNFR, CXCL10, IFN-β, IL-12α, and IFN-related signaling molecules. In contrast to the case for lethal influenza virus infection in other animal models, the increased cytokine and chemokine gene expression resolved and all pigs recovered. The host lung environment regulating inflammatory responses to infection may be dependent on the species, with swine having a more controlled environment for otherwise pathogenic influenza viruses.
It is not clear why even though most human SOIV infections were mild, some individuals, particularly young, healthy adults, presented severe clinical symptoms. In a macaque infection model, clinically distinct Mexican isolates of SOIV showed an increase in IL-6, TNF, and IL-1β gene expression in the lungs and pronounced levels of IL-6 and MCP-1 in the plasma (123). The upregulation of these cytokines associated with the cytokine storm likely varies between pigs and macaques, potentially in the kinetics of PRR stimulation, allowing the pig to serve as a reservoir for influenza viruses with pathogenic potential. It appears that pigs mount a protective proinflammatory response without tipping the scale to promote an adverse cytokine storm. Many of the proinflammatory cytokines contributing to the cytokine storm were observed to play a protective role against 2009 H1N1 pandemic influenza virus infection. Different influenza viruses may present agonists accounting for differential host proinflammatory responses to infection.
Key Cytokine Storm Mediators
The functional role of cytokines during influenza virus infection has been pursued largely using single cytokine gene and cytokine receptor gene knockout (KO) mice, including those lacking IL-6 (137), IL-17RA (35), or CCR2 (39), or using KO mice lacking multiple cytokine receptor genes, such as IL-1R and TNFR (117). Transgenic mice with humanized immune systems present a greater opportunity to study cytokines in a more clinically relevant animal model. For example, IL-3 and GM-CSF knock-in mice (hIL-3/GM-CSF KI mice) have been used to study human innate immune responses in the lung and may provide a good resource in future cytokine storm investigations (148). The pivotal contribution of some cytokine storm mediators has been identified using KO mice, but the complete absence of a cytokine or its cognate receptor has also revealed compensation among cytokine responses to infection. In a study led by Belisle and colleagues, IL-1R and TNFR KO mice infected with the 1918 virus showed a protective role for IL-1α, while TNF exerted adverse cytokine storm effects. In the absence of TNF, mice showed an increase in survival that was associated with a decrease in IFN-signaling-related antiviral gene expression. The transcriptional response in IL1R KO mice during acute infection is strongly associated with an increase in TNF gene expression, revealing the interplay between IL-1 and TNF proinflammatory mediators (11). Cytokine signaling pathway redundancy is also observed with respect to type I IFN signaling, as there is strong upregulation of IFN-stimulated gene expression in IFN-α/βR KO mice infected with the 1918 virus (27). A combination of knockout mouse models and pharmacological intervention (administration of chemokine receptor small-molecule antagonists and/or cytokine antibodies) will likely provide greater insight into the cytokine storm and the redundancy that may exacerbate disease (Fig. 4). However, dampening an overactive proinflammatory response is not as straightforward as targeting the key cytokine mediators, since most chemokine receptors, for example, bind several ligands and contain small ligand binding pockets that have proved difficult to identify effective antagonists.
Fig 4.
Mediators of the cytokine storm and the associated phenotypes with infection outcome. The diagram shows the functional roles of key cytokines and chemokines and their cognate receptors in the development of the main clinical outcomes associated with the cytokine storm. The redundancy of the cytokine and chemokine signaling pathways is emphasized.
Strength of Chemoattractant Responses
SARS provides one example of chemokine responses that lead to a strong proinflammatory response in the lungs. One characteristic feature of SARS is the development of pulmonary fibrosis, observed with inflammation and hypercytokinemia. Atypical IFN-mediated immune responses suggest that type I IFN signaling may be important in the pathogenesis of SARS-CoV (21). Cameron and colleagues (21) assessed gene signatures and protein profiles associated with nonsevere and severe clinical courses of SARS in humans with acute SARS-CoV infection. Gene expression signatures for type I and type II IFN responses in the lungs of SARS patients revealed persistent expression of CXCL10, CCL2, IFNAR1, IFNGR1, and CD58. High plasma levels of the IFN-stimulated chemokines CXCL10 and CCL2 were found in infected patients, and those who died from infection exhibited significantly higher levels of CXCL10 than those who recovered. In addition to playing a distinct role in the early clinical course and persistence of SARS in infected patients, differential chemokine expression is observed in viral lung disease caused by respiratory syncytial virus (RSV) (36). Genomic studies have revealed chemokine gene expression and regulation patterns that can impact Th1- and Th2-biased immunopathology.
Genomic studies of SARS-CoV infection in ferrets showed enhanced expression of chemokine ligands, in particular, CCL2, CCL4, CCL14, CCL19, and CCL25 (37). Upregulation of the fibrosis-associated chemokine CCL2 was shown to be mediated by ERK1/2 in human lung A549 epithelial cells incubated with SARS-CoV spike (S) protein or SARS virus-like particles (VLPs) (25). Characterization of a recombinant mouse-adapted SARS-CoV strain (rMA15) in STAT1 knockout mice revealed increased pathology in the absence of STAT1 signaling, including evidence of pulmonary fibrotic lesions (52). In particular, microarray profiling showed a skewing toward Th2 responses that was influenced by the absence of STAT1 signaling and likely accounted for the protein deposition in the lungs (158). Enhanced antiviral gene expression to SAR-CoV in IFNAR KO mice suggested that alternate signaling pathways were compensating for the lack of type I IFN signaling through IFNAR, possibly Jak-STAT signaling activated by type III IFNs.
Increased Capillary Permeability Syndrome
We have been discussing how cytokines are associated with increased inflammation. Another consequence of the cytokine storm in dengue virus infection is an increased capillary permeability syndrome. Dengue is an acute systemic viral illness caused by one of four serotypes of dengue virus. There are no animal models that mimic the clinical syndrome of severe dengue in humans, and therefore observational studies of patient cohorts are the main source of understanding the immunopathogenesis of dengue virus infections. Intriguingly, severe dengue occurs most commonly in individuals experiencing a second infection with a serotype distinct from that of a past exposure. In these so-called heterotypic infections, the host anamnestic immune response is postulated to mechanistically contribute to the syndrome of increased capillary permeability that characterizes severe dengue.
In the first few days of clinically apparent infection, there is an innate immune response in all patients. Gene expression studies have shown that type I IFN-inducible transcripts are the most prominent overabundant family of transcripts in blood leukocytes in the early acute phase of dengue relative to the late convalescence phase (60). Transcripts from pathways associated with signaling through TLRs, IL-27, IL-12, IFN-γ, and the Jak-STAT pathway are also overabundant in the acute phase. Interconnecting these overabundant transcripts were transcription factors involved in IFN/NF-κB signaling responses, particularly STAT1, STAT2, STAT3, IRF7, IRF9, IRF1, CEBPB, and SP1 (60, 90). Serum concentrations of IFN-α, IFN-γ, IL-10, and TNF receptors are elevated in this early phase and remain elevated until defervescence occurs (86). Elevated concentrations of TNF-RII predict severe dengue in children (13). Gene expression analysis of primary human cells infected with dengue virus revealed strong upregulation of CCL8 and CXCL10, and these proteins were increased in infected patients during the febrile period of the disease (10).
The mechanism through which dengue virus elicited cytokines such as TNF might mediate endothelial dysfunction are not clear, though changes in the integrity of inter-endothelial cell junctions is a possible cause. Most permeability-inducing factors bind to endothelial cell plasma membrane receptors, activate heterotrimeric G proteins, and cause an increase in intracellular Ca2+. This results in myosin-driven endothelial contraction and opening of tight junctions. Other studies have suggested a role for vascular endothelial growth factor (VEGF) and its regulatory receptors in mediating capillary permeability. In vivo, plasma levels of free but not total VEGF-A are elevated in severely ill patients at the time of plasma leakage, while VEGFR2 levels are diminished (134). This, together with dengue virus-induced changes to endothelial cell VEGFR2 surface expression and responsiveness to VEGF stimulation, has led to the suggestion that the VEGF axis may be an important mechanism of plasma leakage in severe dengue.
Increased capillary permeability in dengue usually occurs between days 4 and 6 of illness, is transient, and only occasionally results in hypovolemic shock. At this stage of the illness, viremia is in steep decline and serum cytokine concentrations of IFN-γ and IL-10 are at or near their peak levels (86). One hypothesis is that the functional phenotype of dengue virus-responsive CD8+ T cells, together with the magnitude of the T-cell response, may play a role in the development of severe disease. In particular, T-cell responses in severely ill patients are limited, in that they produce IFN-γ and/or TNF and rarely CD107a, a marker of cytotoxic degranulation. Conversely, in patients with uncomplicated dengue, relatively more CD8+ T cells display CD107a and few express only IFN-γ and/or TNF. These and other data (102) suggest that T cells are potent contributors of cytokines to the inflammatory milieu in infected tissues and that this contributes to vasodilation during this critical phase of the illness, although not all studies have found evidence in support of this hypothesis (44).
TARGETING THE CYTOKINE STORM
Across much of the world, infectious diseases remain a very real threat, accounting for approximately half of all deaths. Malaria, tuberculosis, HIV disease, influenza, dengue, and endemic and emerging infections all contribute to morbidity and mortality. As economies develop, urbanization and environmental degradation gather pace and the structures of societies change, creating many new challenges in the 21st century. In addition to the emergence of new diseases, the continued rise of drug resistance among all the major infections is outpacing the rate of discovery of new antibiotics. Against this backdrop of antimicrobial resistance and the emergence of new pathogens, increasing interest has focused on the development of drugs that target the immune response to infection. As we have discussed, many acute infections are characterized by a powerful and potentially destructive immune response, and it would seem logical to target this response in order to reduce the self-inflicted damage initiated by the host in response to infection (129). Yet to date, successful targeting of the immune system during acute infections has proved to be extraordinarily difficult and largely unsuccessful.
Challenges of Current Immunotherapies
A great deal of effort has been devoted to targeting the host response with a variety of anti-inflammatory drugs and adjunct approaches in a range of acute severe infections, including treatment with corticosteroids, aspirin, monoclonal antibodies (MAbs), anti-cytokine and anti-chemokine agents, plasma exchange, and statins. Despite these efforts, none has been proved to be effective (98), and some have worsened the outcome (19). For example, in severe malaria, an infection characterized by a powerful immunological cascade, a dozen interventions targeting the host response to infection have been tried over the last 20 years, including artemisinin-based combinations and treatment with chloroquine. The majority of antimalarial drugs sold in Africa are either ineffective or accelerate malarial parasite resistance owing to the distribution of counterfeit drugs (9, 43). The capillary permeability syndrome that is characteristic of severe dengue is a result of endothelial dysfunction and compromised integrity of endothelial cell junctions. Clinical trials to test the ability of early corticosteroid therapy to attenuate the host inflammatory response in dengue, and in turn capillary permeability, are ongoing (ClinicalTrials.gov, ISRCTN39575233). Previous trials of corticosteroids have involved patients with hypovolemic shock and did not show benefit, though this could have reflected the late timing of therapy. The availability of generic anticytokine MAb therapies might open new avenues for therapy in the future. Nevertheless, despite strong evidence of a cytokine storm in many severe acute infections, it has proven difficult to intervene and improve clinical outcomes, for reasons that remain elusive.
Implications for Therapeutic Strategies
Immunomodulatory drugs that diminish inflammation during infection with drug treatment show therapeutic benefit (Table 2). Sphingosine receptors play an important role in innate immune responses (120), and sphingosine analogs have shown potential for controlling the cytokine storm caused by influenza virus (95). Sphingosine-1-phosphate (S1P) receptor 1 suppresses immune cell recruitment through downregulation of cytokine and chemokine production by respiratory endothelial cells in the presence of S1P1-selective agonists, CYM-5442 and RP-002, including production of IFN-α, CCL2, IL-6, TNF-α, and IFN-γ, (139). Teijaro and colleagues (139) demonstrated that blunted innate chemokine and cytokine responses mediated by S1P1-selective agonists protected mice from lethal infection with pandemic H1N1 influenza virus. Agonism of S1P1 receptor has no effect on viral replication; however, some strategies dampening the immune response can correlate with increased susceptibility to infection. This was observed in mice treated with DNA encoding transforming growth factor β1 (TGF-β1), which inhibited pulmonary eosinophilic responses yet increased susceptibility to Cryptococcus neoformans, influenza virus, and respiratory syncytial virus (147).
Table 2.
Anti-inflammatory properties of immunomodulatory drugs
| Type of therapeutic | Drug(s) | Immunomodulatory effect(s) (reference[s]) |
|---|---|---|
| COX inhibitors | Mesalamine, celecoxib | Coadministration of COX inhibitors with zanamivir diminished cellular infiltrate and improved survival of H5N1 virus-infected mice compared to antiviral treatment alone (22, 157) |
| CCR2 inhibitor | PF-04178903 | Increased survival of mice infected with influenza virus and reduced lung immunopathology (87, 88) |
| Sphingosine receptor agonists | Suppresses cytokine and chemokine production; sphingosine receptors have been shown to play an important role in innate immune responses (120) | |
| Anti-TNF agents | Mediator of pulmonary inflammation during influenza A viral pneumonia; decreased severity of pulmonary immunopathology and prolonged survival of A/PR/8-infected mice (67) | |
| Statins | Simvastatin | Statins were not found to reduce the risk of developing severe disease in patients with pandemic influenza (H1N1) 2009 (144) |
| OX40 | OX40-Ig fusion proteins | OX40 plays a critical role in T-cell-mediated immunopathology in the lung during viral infection (65); ligation on activated T cells reduces pulmonary eosinophilia during Cryptococcus neoformans infection (64) |
| PPARα/PPARγ agonists | Gemfibrozil, pioglitazone, rosiglitazone, 15d-PGJ2, ciglitazone, troglitazone | 15d-PGJ2, ciglitazone, and troglitazone decreased production of IL-1α, IL-6, and TNF cytokines, CXCL8 and CCL5 chemokines, and ICAM-1 in RSV-infected lung epithelial cells (4); administration of gemfibrozil (intraperitoneally) on days 4 to 10 after exposure to H2N2 influenza virus and following the onset of illness significantly increased survival in mice with severe influenza (20) |
Peroxisome proliferator-activated receptors (PPARs) are lipid-activated transcription factors that act as key regulators of lipid metabolism and inflammation (reviewed in reference 12). PPAR-γ agonists downregulate the inflammatory response to virus-induced lung inflammation (8). For example, gemfibrozil targets PPARα, dampening the inflammatory response and improving the survival of mice infected with influenza H2N2 virus (20). Combination therapy consisting of zanamvir, an inhibitor of the influenza virus neuraminidase, and celecoxib and mesalazine, inhibitors of inflammation, increased the survival of mice infected with a highly pathogenic strain of H5N1 influenza virus (157). The cyclooxygenase-2 (COX-2) inhibitors celecoxib and mesalazine also reduced the mortality rate of mice infected with H5N1 influenza virus when administered in combination with zanamivir (22). These studies demonstrated that antiviral treatment was less effective alone than in combination with immunomodulatory therapies that suppressed inflammation. Once initiated, the proinflammatory cytokines and chemokines can continue to drive the progression of immunopathologic events in the absence of continued viral replication.
In simple terms, it is likely that during a severe acute infection, certain elements of the immune response need enhancing at times and need suppressing at other times. What is required of the immune system early in the infection, when the pathogen is dividing rapidly and reaching high infectious loads, may be very different from what is needed later, possibly only a few hours later, when either the pathogen is at steady state or the pathogen load is falling due to either the effects of appropriate antimicrobial therapy or the clearance of the pathogen by the immune response. Treating patients with the “right” immunomodulating drug but at the “wrong” time could worsen the clinical outcome. We still do not understand the delicate nature of this rapidly changing immune response, and until we do, it is unlikely that we will develop rational therapies that target the exact phase of the immune cascade and administer those therapies at the time they are needed. It might be that within the long list of adjunct interventions that have been tried in acute severe infections, therapies exist that would be successful if only the immune dynamic were understood so that they could be used at the right time and their pharmacological profile fit the need for a potentially short-acting drug. We know that individuals respond differently to infection and that individual responses to antibiotics and adjunct therapies also vary (141). These variations in host response are likely to be complex and controlled by host and pathogen genetic determinants as well as by the immune memory of each individual.
CONCLUDING REMARKS
We are in a potentially golden age of scientific discovery led in part by recent technological advances in next-generation sequencing (NGS). For example, NGS has unveiled a more comprehensive view of the transcriptome, enabling identification of large and small RNAs, splice isoforms, and novel transcripts from unannotated genes. Yet the mind-numbing weight of data we can now accrue brings its own challenges. Overall, the need for better methods for NGS data analysis, including RNA-Seq bias correction and improved isoform quantification, is just one of the challenges faced with using this technology. There is no agreement on what should be measured and how it should be analyzed. An international consensus on this would help define how the field develops in the coming years. Moreover, we have split interdependent systems into silos of research such that studies of the immune cascade rarely integrate the pathogen or the host genetic makeup or attempt to interrogate the whole delicate and rapidly changing immune environment. The world of host and pathogen genetics has been revolutionized in the last decade, and through scale and increasingly sophisticated biostatistical analysis, what associations mean and how they should be interpreted will continue to be developed. The development of transcriptional signatures and proteomics to complement host and pathogen genetic studies is providing great steps forward; however, we need to standardize these integrated studies with much tighter definitions of the clinical and phenotypic data.
In this review we have focused on the biological consequences of a cytokine storm, namely, immunopathogenesis caused by SARS-CoV, influenza virus, and dengue virus infections. We have highlighted lessons learned from genomic studies within the past decade, in particular how the kinetics of cytokine and chemokine gene expression can influence cytokine overproduction, the redundancy that exists within these signaling systems that can affect the landscape of transcriptional responses, and how these early molecular events can influence later stages of disease phenotypes associated with the cytokine storm. Moving forward, the application of systems biology approaches to infectious disease research will offer further insight into the key cellular processes that either sustain the cytokine storm or contribute to its resolution. As part of a systems biology effort, the use of high-throughput technologies and analytical and computational modeling methods to identify and model the pathogen and host determinants and interactions underlying infectious disease will aid in the identification of molecular signatures of infection. For example, weighted gene correlation network analysis has been used to model the dynamic H5N1 virus-mediated transcriptional response in infected human Calu-3 bronchial epithelial cells (85), and transcriptional network inference has predicted gene behavior in mice and macaques, including key components of the NLRP3 inflammasome (CASP1, NLRP3, and PYCARD) and cytokine and chemokine genes (IL-6, CXCL10, and CCL4) relevant to cytokine storm that are conserved across species (97). Within this framework, we will be better able to predict the various facets of the circuit operation and coordination and the overall system in the resulting phenotypic outcome. This will impact how potential immunotherapies will perturb the circuitry and machinery related to the cytokine storm and likely will present key targets for therapeutic intervention and diagnostics.
ACKNOWLEDGMENTS
We thank Stewart Chang and Sean Proll for discussions and assistance with the original figures and Patrick Lane for the final figures and creative illustration of a cytokine storm.
Research in M.G.K.'s laboratory is supported by Public Health Service grants P01AI058113, P30DA015625, P51RR00166, R24RR016354, and U54AI081680 from the National Institutes of Health and by contract HHSN272200800060C from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services.
Biographies

Jennifer R. Tisoncik has a Ph.D. in microbiology and immunology from the University of Illinois at Chicago. She is currently a postdoctoral research fellow at the University of Washington in the laboratory of Michael Katze, where she is applying genomic and systems biology approaches to define and model virus-host interactions. Jennifer's research interest is focused on understanding innate immune responses to influenza. Her research has involved a comprehensive molecular analysis of H5N1 NA to characterize residues critical in its role in viral particle release and to identify mutations leading to antiviral resistance. She is currently studying differential transcriptional responses that can contribute to lung inflammation during influenza pathogenesis.

Marcus J. Korth received his Ph.D. in microbiology from the University of Washington in 1993 for work on the role of fimbrial adhesins in Escherichia coli pathogenesis. After completing a postdoctoral fellowship with Dr. Michael Katze at the University of Washington, where he focused on characterizing the PKR inhibitor and endoplasmic reticulum molecular chaperone P58IPK, he has remained in the laboratory, where he is currently a Senior Research Scientist. He is interested in applying high-throughput technologies to the study of virus-host interaction.

Cameron Simmons has a Ph.D. from the University of Melbourne, Australia. Cam is currently a Professor of Infectious Diseases, a Wellcome Trust Senior Fellow, and Vice-Director of the Oxford University Clinical Research Unit in Ho Chi Minh City, Vietnam. Cam has spent the last decade working in Vietnam on tuberculosis, influenza, and, most recently, dengue. Cam's research group conducts clinical and basic research on therapeutic interventions for dengue, novel mosquito-based dengue control methods, and basic mechanisms of disease pathogenesis.

Jeremy Farrar trained in medicine at the University of London and received his Ph.D. in immunology from Oxford University. He is a Professor of Medicine at Oxford University, a Global Scholar of Princeton University, and an Adjunct Professor of Medicine at National University of Singapore. Since 1996 he has been the Director of the Oxford University Clinical Research Unit at the Hospital for Tropical Diseases Vietnam. His interests are in integrated clinical research across a range of infectious diseases, including influenza, central nervous system infections, dengue, typhoid, malaria, tuberculosis, opportunistic infections related to HIV, and emerging infectious diseases. He serves on a number of international advisory positions, including for dengue, influenza, typhoid, and emerging infections. His research focus is on the most important infectious diseases of the hospital and region in southeast Asia, and he seeks to help facilitate the best clinical research in infectious diseases, promoting the integration of evidence generated from research work into practice and policy.

Thomas R. Martin, M.D., is Emeritus Professor of Medicine and a former member of the Division of Pulmonary and Critical Care Medicine at the University of Washington. He received the M.D. degree from the University of Pennsylvania and then completed training in internal medicine and pulmonary and critical care medicine at the University of Washington in Seattle. He has had a longstanding interest in mechanisms of host defense, inflammation, and injury in the lungs and has served as the Director of the University of Washington Specialized Center in Acute Lung Injury Research. In June 2011, he became the Director of Clinical Sciences in the Respiratory Franchise at Novartis Pharmaceuticals in East Hanover, NJ.

Michael G. Katze has a Ph.D. in microbiology from Hahnemann University, PA. He is currently a Professor of Microbiology at the University of Washington, where he serves as Associate Director at the Washington National Primate Research Center and is Head of the Center's Division of Functional Genomics and Infectious Disease, Co-Director of the Pacific Northwest Regional Center of Excellence, Director of the Center for Systems and Translational Research on Infectious Disease (STRIDE), Program Director of an NIAID contract to use systems biology approaches to develop computational models of the host response to respiratory virus infection, and Program Director of a NIDA P30 Center focused on using genomic and proteomic technologies to study hepatitis C virus (HCV) and HCV-associated liver disease. His research interests are focused on applying genomic and proteomic technologies to the study of virus-host interactions and the innate immune response.
REFERENCES
- 1. Abraham E, et al. 1997. p55 Tumor necrosis factor receptor fusion protein in the treatment of patients with severe sepsis and septic shock. A randomized controlled multicenter trial. Ro 45-2081 Study Group. JAMA 277:1531–1538 [PubMed] [Google Scholar]
- 2. Aggarwal BB. 2003. Signalling pathways of the TNF superfamily: a double-edged sword. Nat. Rev. Immunol. 3:745–756 [DOI] [PubMed] [Google Scholar]
- 3. Alberts R, et al. 2010. Gene expression changes in the host response between resistant and susceptible inbred mouse strains after influenza A infection. Microbes Infect. 12:309–318 [DOI] [PubMed] [Google Scholar]
- 4. Arnold R, Neumann M, Konig W. 2007. Peroxisome proliferator-activated receptor-gamma agonists inhibit respiratory syncytial virus-induced expression of intercellular adhesion molecule-1 in human lung epithelial cells. Immunology 121:71–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aylor DL, et al. 2011. Genetic analysis of complex traits in the emerging collaborative cross. Genome Res. 21:1213–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barry SM, Johnson MA, Janossy G. 2000. Cytopathology or immunopathology? The puzzle of cytomegalovirus pneumonitis revisited. Bone Marrow Transplant. 26:591–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Baskin CR, et al. 2009. Early and sustained innate immune response defines pathology and death in nonhuman primates infected by highly pathogenic influenza virus. Proc. Natl. Acad. Sci. U. S. A. 106:3455–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bassaganya-Riera J, Song R, Roberts PC, Hontecillas R. 2010. PPAR-gamma activation as an anti-inflammatory therapy for respiratory virus infections. Viral Immunol. 23:343–352 [DOI] [PubMed] [Google Scholar]
- 9. Bate R, Coticelli P, Tren R, Attaran A. 2008. Antimalarial drug quality in the most severely malarious parts of Africa—a six country study. PLoS One 3:e2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Becerra A, et al. 2009. Gene expression profiling of dengue infected human primary cells identifies secreted mediators in vivo. J. Med. Virol. 81:1403–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Belisle SE, et al. 2010. Genomic profiling of tumor necrosis factor alpha (TNF-alpha) receptor and interleukin-1 receptor knockout mice reveals a link between TNF-alpha signaling and increased severity of 1918 pandemic influenza virus infection. J. Virol. 84:12576–12588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bensinger SJ, Tontonoz P. 2008. Integration of metabolism and inflammation by lipid-activated nuclear receptors. Nature 454:470–477 [DOI] [PubMed] [Google Scholar]
- 13. Bethell DB, et al. 1998. Pathophysiologic and prognostic role of cytokines in dengue hemorrhagic fever. J. Infect. Dis. 177:778–782 [DOI] [PubMed] [Google Scholar]
- 14. Beutler B, Milsark IW, Cerami AC. 1985. Passive immunization against cachectin/tumor necrosis factor protects mice from lethal effect of endotoxin. Science 229:869–871 [DOI] [PubMed] [Google Scholar]
- 15. Bisno AL, Brito MO, Collins CM. 2003. Molecular basis of group A streptococcal virulence. Lancet Infect. Dis. 3:191–200 [DOI] [PubMed] [Google Scholar]
- 16. Boon AC, et al. 2011. H5N1 influenza virus pathogenesis in genetically diverse mice is mediated at the level of viral load. mBio 2(5):e00171–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Borden EC, et al. 2007. Interferons at age 50: past, current and future impact on biomedicine. Nat. Rev. Drug Discov. 6:975–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brocker C, Thompson D, Matsumoto A, Nebert DW, Vasiliou V. 2010. Evolutionary divergence and functions of the human interleukin (IL) gene family. Hum. Genomics 5:30–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Brun-Buisson C, Richard JC, Mercat A, Thiebaut AC, Brochard L. 2011. Early corticosteroids in severe influenza A/H1N1 pneumonia and acute respiratory distress syndrome. Am. J. Respir. Crit. Care Med. 183:1200–1206 [DOI] [PubMed] [Google Scholar]
- 20. Budd A, et al. 2007. Increased survival after gemfibrozil treatment of severe mouse influenza. Antimicrob. Agents Chemother. 51:2965–2968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cameron MJ, et al. 2007. Interferon-mediated immunopathological events are associated with atypical innate and adaptive immune responses in patients with severe acute respiratory syndrome. J. Virol. 81:8692–8706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Carey MA, et al. 2010. Pharmacologic inhibition of COX-1 and COX-2 in influenza A viral infection in mice. PLoS One 5:e11610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Carswell EA, et al. 1975. An endotoxin-induced serum factor that causes necrosis of tumors. Proc. Natl. Acad. Sci. U. S. A. 72:3666–3670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chan MC, et al. 2005. Proinflammatory cytokine responses induced by influenza A (H5N1) viruses in primary human alveolar and bronchial epithelial cells. Respir. Res. 6:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen IY, et al. 2010. Upregulation of the chemokine (C-C motif) ligand 2 via a severe acute respiratory syndrome coronavirus spike-ACE2 signaling pathway. J. Virol. 84:7703–7712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cheung CY, et al. 2002. Induction of proinflammatory cytokines in human macrophages by influenza A (H5N1) viruses: a mechanism for the unusual severity of human disease? Lancet 360:1831–1837 [DOI] [PubMed] [Google Scholar]
- 27. Cilloniz C, et al. 2010. Lethal dissemination of H5N1 influenza virus is associated with dysregulation of inflammation and lipoxin signaling in a mouse model of infection. J. Virol. 84:7613–7624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cilloniz C, et al. 2009. Lethal influenza virus infection in macaques is associated with early dysregulation of inflammatory related genes. PLoS Pathog. 5:e1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Clark IA. 2007. The advent of the cytokine storm. Immunol. Cell Biol. 85:271–273 [DOI] [PubMed] [Google Scholar]
- 30. Clark IA. 2007. How TNF was recognized as a key mechanism of disease. Cytokine Growth Factor Rev. 18:335–343 [DOI] [PubMed] [Google Scholar]
- 31. Clark IA, Virelizier JL, Carswell EA, Wood PR. 1981. Possible importance of macrophage-derived mediators in acute malaria. Infect. Immun. 32:1058–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cohen J. 2002. The immunopathogenesis of sepsis. Nature 420:885–891 [DOI] [PubMed] [Google Scholar]
- 33. Comerford I, McColl SR. 2011. Focus on chemokines. Immunol. Cell Biol. 89:183–184 [DOI] [PubMed] [Google Scholar]
- 34. Coornaert B, Carpentier I, Beyaert R. 2009. A20: central gatekeeper in inflammation and immunity. J. Biol. Chem. 284:8217–8221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Crowe CR, et al. 2009. Critical role of IL-17RA in immunopathology of influenza infection. J. Immunol. 183:5301–5310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Culley FJ, et al. 2006. Role of CCL5 (RANTES) in viral lung disease. J. Virol. 80:8151–8157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Danesh A, et al. 2011. Early gene expression events in ferrets in response to SARS coronavirus infection versus direct interferon-alpha2b stimulation. Virology 409:102–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. de Jong MD, et al. 2006. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat. Med. 12:1203–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dessing MC, van der Sluijs KF, Florquin S, van der Poll T. 2007. Monocyte chemoattractant protein 1 contributes to an adequate immune response in influenza pneumonia. Clin. Immunol. 125:328–336 [DOI] [PubMed] [Google Scholar]
- 40. Dinarello C, et al. 2010. IL-1 family nomenclature. Nat. Immunol. 11:973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dinarello CA. 2009. Immunological and inflammatory functions of the interleukin-1 family. Annu. Rev. Immunol. 27:519–550 [DOI] [PubMed] [Google Scholar]
- 42. Docke WD, et al. 1997. Monocyte deactivation in septic patients: restoration by IFN-gamma treatment. Nat. Med. 3:678–681 [DOI] [PubMed] [Google Scholar]
- 43. Dondorp AM, et al. 2009. Artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 361:455–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dung NT, et al. 2010. Timing of CD8+ T cell responses in relation to commencement of capillary leakage in children with dengue. J. Immunol. 184:7281–7287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fensterl V, Sen GC. 2009. Interferons and viral infections. Biofactors 35:14–20 [DOI] [PubMed] [Google Scholar]
- 46. Ferrara JL. 1993. Cytokine dysregulation as a mechanism of graft versus host disease. Curr. Opin. Immunol. 5:794–799 [DOI] [PubMed] [Google Scholar]
- 47. Ferrara JL, Abhyankar S, Gilliland DG. 1993. Cytokine storm of graft-versus-host disease: a critical effector role for interleukin-1. Transplant. Proc. 25:1216–1217 [PubMed] [Google Scholar]
- 48. Finck-Barbancon V, et al. 1997. ExoU expression by Pseudomonas aeruginosa correlates with acute cytotoxicity and epithelial injury. Mol. Microbiol. 25:547–557 [DOI] [PubMed] [Google Scholar]
- 49. Fowler AA, Fisher BJ, Centor RM, Carchman RA. 1984. Development of the adult respiratory distress syndrome: progressive alteration of neutrophil chemotactic and secretory processes. Am. J. Pathol. 116:427–435 [PMC free article] [PubMed] [Google Scholar]
- 50. Franklin BS, et al. 2009. Malaria primes the innate immune response due to interferon-gamma induced enhancement of toll-like receptor expression and function. Proc. Natl. Acad. Sci. U. S. A. 106:5789–5794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Friedman RM. 2008. Clinical uses of interferons. Br. J. Clin. Pharmacol. 65:158–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Frieman MB, et al. 2010. SARS-CoV pathogenesis is regulated by a STAT1 dependent but a type I, II and III interferon receptor independent mechanism. PLoS Pathog. 6:e1000849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gabay C, Kushner I. 1999. Acute-phase proteins and other systemic responses to inflammation. N. Engl. J. Med. 340:448–454 [DOI] [PubMed] [Google Scholar]
- 54. Garin A, Proudfoot AE. 2011. Chemokines as targets for therapy. Exp. Cell Res. 317:602–612 [DOI] [PubMed] [Google Scholar]
- 55. Ge D, et al. 2009. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature 461:399–401 [DOI] [PubMed] [Google Scholar]
- 56. Groom JR, Luster AD. 2011. CXCR3 ligands: redundant, collaborative and antagonistic functions. Immunol. Cell Biol. 89:207–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Guarda G, et al. 2011. Type I interferon inhibits interleukin-1 production and inflammasome activation. Immunity 34:213–223 [DOI] [PubMed] [Google Scholar]
- 58. Hamilton JA. 2008. Colony-stimulating factors in inflammation and autoimmunity. Nat. Rev. Immunol. 8:533–544 [DOI] [PubMed] [Google Scholar]
- 59. Hinder F, et al. 1991. Influenza A virus infects macrophages and stimulates release of tumor necrosis factor-alpha. Pathobiology 59:227–231 [DOI] [PubMed] [Google Scholar]
- 60. Hoang LT, et al. 2010. The early whole-blood transcriptional signature of dengue virus and features associated with progression to dengue shock syndrome in Vietnamese children and young adults. J. Virol. 84:12982–12994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Holt PG, Strickland DH, Wikstrom ME, Jahnsen FL. 2008. Regulation of immunological homeostasis in the respiratory tract. Nat. Rev. Immunol. 8:142–152 [DOI] [PubMed] [Google Scholar]
- 62. Houde M, Arora DJ. 1990. Stimulation of tumor necrosis factor secretion by purified influenza virus neuraminidase. Cell. Immunol. 129:104–111 [DOI] [PubMed] [Google Scholar]
- 63. Huang KJ, et al. 2005. An interferon-gamma-related cytokine storm in SARS patients. J. Med. Virol. 75:185–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Humphreys IR, et al. 2003. OX40 ligation on activated T cells enhances the control of Cryptococcus neoformans and reduces pulmonary eosinophilia. J. Immunol. 170:6125–6132 [DOI] [PubMed] [Google Scholar]
- 65. Humphreys IR, et al. 2003. A critical role for OX40 in T cell-mediated immunopathology during lung viral infection. J. Exp. Med. 198:1237–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hussell T, Goulding J. 2010. Structured regulation of inflammation during respiratory viral infection. Lancet Infect. Dis. 10:360–366 [DOI] [PubMed] [Google Scholar]
- 67. Hussell T, Pennycook A, Openshaw PJ. 2001. Inhibition of tumor necrosis factor reduces the severity of virus-specific lung immunopathology. Eur. J. Immunol. 31:2566–2573 [DOI] [PubMed] [Google Scholar]
- 68. Imai Y, et al. 2008. Identification of oxidative stress and Toll-like receptor 4 signaling as a key pathway of acute lung injury. Cell 133:235–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Imai Y, et al. 2003. Injurious mechanical ventilation and end-organ epithelial cell apoptosis and organ dysfunction in an experimental model of acute respiratory distress syndrome. JAMA 289:2104–2112 [DOI] [PubMed] [Google Scholar]
- 70. Imashuku S. 2002. Clinical features and treatment strategies of Epstein-Barr virus-associated hemophagocytic lymphohistiocytosis. Crit. Rev. Oncol. Hematol. 44:259–272 [DOI] [PubMed] [Google Scholar]
- 71. Jahrling PB, et al. 2004. Exploring the potential of variola virus infection of cynomolgus macaques as a model for human smallpox. Proc. Natl. Acad. Sci. U. S. A. 101:15196–15200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Jiang D, et al. 2005. Regulation of lung injury and repair by Toll-like receptors and hyaluronan. Nat. Med. 11:1173–1179 [DOI] [PubMed] [Google Scholar]
- 73. Kash JC, et al. 2006. Genomic analysis of increased host immune and cell death responses induced by 1918 influenza virus. Nature 443:578–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kato H, et al. 2005. Cell type-specific involvement of RIG-I in antiviral response. Immunity 23:19–28 [DOI] [PubMed] [Google Scholar]
- 75. Katze MG, Fornek JL, Palermo RE, Walters KA, Korth MJ. 2008. Innate immune modulation by RNA viruses: emerging insights from functional genomics. Nat. Rev. Immunol. 8:644–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kawai T, et al. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6:981–988 [DOI] [PubMed] [Google Scholar]
- 77. Khor CC, et al. 2010. CISH and susceptibility to infectious diseases. N. Engl. J. Med. 362:2092–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kobasa D, et al. 2007. Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature 445:319–323 [DOI] [PubMed] [Google Scholar]
- 79. Kopf M, Bachmann MF, Marsland BJ. 2010. Averting inflammation by targeting the cytokine environment. Nat. Rev. Drug Discov. 9:703–718 [DOI] [PubMed] [Google Scholar]
- 80. Kurahashi K, et al. 1999. Pathogenesis of septic shock in Pseudomonas aeruginosa pneumonia. J. Clin. Invest. 104:743–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. La Gruta NL, Kedzierska K, Stambas J, Doherty PC. 2007. A question of self-preservation: immunopathology in influenza virus infection. Immunol. Cell Biol. 85:85–92 [DOI] [PubMed] [Google Scholar]
- 82. Lee SM, et al. 2010. Systems-level comparison of host responses induced by pandemic and seasonal influenza A H1N1 viruses in primary human type I-like alveolar epithelial cells in vitro. Respir. Res. 11:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Lee SM, et al. 2008. Hyperinduction of cyclooxygenase-2-mediated proinflammatory cascade: a mechanism for the pathogenesis of avian influenza H5N1 infection. J. Infect. Dis. 198:525–535 [DOI] [PubMed] [Google Scholar]
- 84. Levy MM, et al. 2003. 2001 SCCM/ESICM/ACCP/ATS/SIS International Sepsis Definitions Conference. Crit. Care Med. 31:1250–1256 [DOI] [PubMed] [Google Scholar]
- 85. Li C, et al. 2011. Host regulatory network response to infection with highly pathogenic H5N1 avian influenza virus. J. Virol. 85:10955–10967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Libraty DH, et al. 2002. Differing influences of virus burden and immune activation on disease severity in secondary dengue-3 virus infections. J. Infect. Dis. 185:1213–1221 [DOI] [PubMed] [Google Scholar]
- 87. Lin KL, Suzuki Y, Nakano H, Ramsburg E, Gunn MD. 2008. CCR2+ monocyte-derived dendritic cells and exudate macrophages produce influenza-induced pulmonary immune pathology and mortality. J. Immunol. 180:2562–2572 [DOI] [PubMed] [Google Scholar]
- 88. Lin KL, Sweeney S, Kang BD, Ramsburg E, Gunn MD. 2011. CCR2-antagonist prophylaxis reduces pulmonary immune pathology and markedly improves survival during influenza infection. J. Immunol. 186:508–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Link H. 1998. The cytokine storm in multiple sclerosis. Mult. Scler. 4:12–15 [DOI] [PubMed] [Google Scholar]
- 90. Long HT, et al. 2009. Patterns of gene transcript abundance in the blood of children with severe or uncomplicated dengue highlight differences in disease evolution and host response to dengue virus infection. J. Infect. Dis. 199:537–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Loo YM, et al. 2008. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J. Virol. 82:335–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Loo YM, Gale M., Jr 2011. Immune signaling by RIG-I-like receptors. Immunity 34:680–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Ma W, et al. 2011. 2009 pandemic H1N1 virus causes disease and upregulation of genes related to inflammatory and immune response, cell death and lipid metabolism in pigs. J. Virol. 85:11626–11637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Makhija R, Kingsnorth AN. 2002. Cytokine storm in acute pancreatitis. J. Hepatobil. Pancreat. Surg. 9:401–410 [DOI] [PubMed] [Google Scholar]
- 95. Marsolais D, et al. 2009. A critical role for the sphingosine analog AAL-R in dampening the cytokine response during influenza virus infection. Proc. Natl. Acad. Sci. U. S. A. 106:1560–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Matute-Bello G, Frevert CW, Martin TR. 2008. Animal models of acute lung injury. Am. J. Physiol. Lung Cell. Mol. Physiol. 295:L379–L399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. McDermott JE, et al. 2011. Conserved host response to highly pathogenic avian influenza virus infection in human cell culture, mouse and macaque model systems. BMC Syst. Biol. 5:190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Meijvis SC, et al. 2011. Dexamethasone and length of hospital stay in patients with community-acquired pneumonia: a randomised, double-blind, placebo-controlled trial. Lancet 377:2023–2030 [DOI] [PubMed] [Google Scholar]
- 99. Michel O, et al. 2003. Systemic responsiveness to lipopolysaccharide and polymorphisms in the Toll-like receptor 4 gene in human beings. J. Allergy Clin. Immunol. 112:923–929 [DOI] [PubMed] [Google Scholar]
- 100. Mockenhaupt FP, et al. 2006. Toll-like receptor (TLR) polymorphisms in African children: common TLR-4 variants predispose to severe malaria. Proc. Natl. Acad. Sci. U. S. A. 103:177–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Mockenhaupt FP, et al. 2006. Common polymorphisms of Toll-like receptors 4 and 9 are associated with the clinical manifestation of malaria during pregnancy. J. Infect. Dis. 194:184–188 [DOI] [PubMed] [Google Scholar]
- 102. Mongkolsapaya J, et al. 2003. Original antigenic sin and apoptosis in the pathogenesis of dengue hemorrhagic fever. Nat. Med. 9:921–927 [DOI] [PubMed] [Google Scholar]
- 103. Monick MM, et al. 2003. Respiratory syncytial virus up-regulates TLR4 and sensitizes airway epithelial cells to endotoxin. J. Biol. Chem. 278:53035–53044 [DOI] [PubMed] [Google Scholar]
- 104. Monneret G, et al. 2006. Persisting low monocyte human leukocyte antigen-DR expression predicts mortality in septic shock. Intensive Care Med. 32:1175–1183 [DOI] [PubMed] [Google Scholar]
- 105. Moore KW, de Waal Malefyt R, Coffman RL, O'Garra A. 2001. Interleukin-10 and the interleukin-10 receptor. Annu. Rev. Immunol. 19:683–765 [DOI] [PubMed] [Google Scholar]
- 106. Mordstein M, et al. 2008. Interferon-lambda contributes to innate immunity of mice against influenza A virus but not against hepatotropic viruses. PLoS Pathog. 4:e1000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Moyron-Quiroz JE, et al. 2004. Role of inducible bronchus associated lymphoid tissue (iBALT) in respiratory immunity. Nat. Med. 10:927–934 [DOI] [PubMed] [Google Scholar]
- 108. Munford RS, Pugin J. 2001. Normal responses to injury prevent systemic inflammation and can be immunosuppressive. Am. J. Respir. Crit. Care Med. 163:316–321 [DOI] [PubMed] [Google Scholar]
- 109. Nawroth PP, et al. 1986. Tumor necrosis factor/cachectin interacts with endothelial cell receptors to induce release of interleukin 1. J. Exp. Med. 163:1363–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Oshiumi H, Matsumoto M, Funami K, Akazawa T, Seya T. 2003. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-beta induction. Nat. Immunol. 4:161–167 [DOI] [PubMed] [Google Scholar]
- 111. Osterlund P, et al. 2010. Pandemic H1N1 2009 influenza A virus induces weak cytokine responses in human macrophages and dendritic cells and is highly sensitive to the antiviral actions of interferons. J. Virol. 84:1414–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Pankhaniya RR, et al. 2004. Pseudomonas aeruginosa causes acute lung injury via the catalytic activity of the patatin-like phospholipase domain of ExoU. Crit. Care Med. 32:2293–2299 [DOI] [PubMed] [Google Scholar]
- 113. Park WY, et al. 2001. Cytokine balance in the lungs of patients with acute respiratory distress syndrome. Am. J. Respir. Crit. Care Med. 164:1896–1903 [DOI] [PubMed] [Google Scholar]
- 114. Peiris JS, Cheung CY, Leung CY, Nicholls JM. 2009. Innate immune responses to influenza A H5N1: friend or foe? Trends Immunol. 30:574–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Peiris JS, Hui KP, Yen HL. 2010. Host response to influenza virus: protection versus immunopathology. Curr. Opin. Immunol. 22:475–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Peiris JS, et al. 2004. Re-emergence of fatal human influenza A subtype H5N1 disease. Lancet 363:617–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Perrone LA, Szretter KJ, Katz JM, Mizgerd JP, Tumpey TM. 2010. Mice lacking both TNF and IL-1 receptors exhibit reduced lung inflammation and delay in onset of death following infection with a highly virulent H5N1 virus. J. Infect. Dis. 202:1161–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Pugin J, Ricou B, Steinberg KP, Suter PM, Martin TR. 1996. Proinflammatory activity in bronchoalveolar lavage fluids from patients with ARDS, a prominent role for interleukin-1. Am. J. Respir. Crit. Care Med. 153:1850–1856 [DOI] [PubMed] [Google Scholar]
- 119. Raman D, Sobolik-Delmaire T, Richmond A. 2011. Chemokines in health and disease. Exp. Cell Res. 317:575–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Rivera J, Proia RL, Olivera A. 2008. The alliance of sphingosine-1-phosphate and its receptors in immunity. Nat. Rev. Immunol. 8:753–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Rubenfeld GD, et al. 2005. Incidence and outcomes of acute lung injury. N. Engl. J. Med. 353:1685–1693 [DOI] [PubMed] [Google Scholar]
- 122. Sabbah A, et al. 2009. Activation of innate immune antiviral responses by Nod2. Nat. Immunol. 10:1073–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Safronetz D, et al. 2011. Pandemic swine-origin H1N1 influenza A virus isolates show heterogeneous virulence in macaques. J. Virol. 85:1214–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Schmitz N, Kurrer M, Bachmann MF, Kopf M. 2005. Interleukin-1 is responsible for acute lung immunopathology but increases survival of respiratory influenza virus infection. J. Virol. 79:6441–6448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Seo SH, Webster RG. 2002. Tumor necrosis factor alpha exerts powerful anti-influenza virus effects in lung epithelial cells. J. Virol. 76:1071–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Sethi G, Sung B, Kunnumakkara AB, Aggarwal BB. 2009. Targeting TNF for treatment of cancer and autoimmunity. Adv. Exp. Med. Biol. 647:37–51 [DOI] [PubMed] [Google Scholar]
- 127. Sheppard P, et al. 2003. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat. Immunol. 4:63–68 [DOI] [PubMed] [Google Scholar]
- 128. Shinya K, et al. 2006. Avian flu: influenza virus receptors in the human airway. Nature 440:435–436 [DOI] [PubMed] [Google Scholar]
- 129. Simmons C, Farrar J. 2008. Insights into inflammation and influenza. N. Engl. J. Med. 359:1621–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Simmons CP, et al. 2007. Patterns of host genome-wide gene transcript abundance in the peripheral blood of patients with acute dengue hemorrhagic fever. J. Infect. Dis. 195:1097–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Simmons CP, et al. 2006. Pretreatment intracerebral and peripheral blood immune responses in Vietnamese adults with tuberculous meningitis: diagnostic value and relationship to disease severity and outcome. J. Immunol. 176:2007–2014 [DOI] [PubMed] [Google Scholar]
- 132. Sims JE, Smith DE. 2010. The IL-1 family: regulators of immunity. Nat. Rev. Immunol. 10:89–102 [DOI] [PubMed] [Google Scholar]
- 133. Snelgrove RJ, et al. 2008. A critical function for CD200 in lung immune homeostasis and the severity of influenza infection. Nat. Immunol. 9:1074–1083 [DOI] [PubMed] [Google Scholar]
- 134. Srikiatkhachorn A, et al. 2007. Virus-induced decline in soluble vascular endothelial growth receptor 2 is associated with plasma leakage in dengue hemorrhagic Fever. J. Virol. 81:1592–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Sun L, et al. 2011. New concepts of IL-10-induced lung fibrosis: fibrocyte recruitment and M2 activation in a CCL2/CCR2 axis. Am. J. Physiol. Lung Cell. Mol. Physiol. 300:L341–L353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Suntharalingam G, et al. 2006. Cytokine storm in a phase 1 trial of the anti-CD28 monoclonal antibody TGN1412. N. Engl. J. Med. 355:1018–1028 [DOI] [PubMed] [Google Scholar]
- 137. Szretter KJ, et al. 2007. Role of host cytokine responses in the pathogenesis of avian H5N1 influenza viruses in mice. J. Virol. 81:2736–2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Taveira da Silva AM, et al. 1993. Shock and multiple-organ dysfunction after self-administration of Salmonella endotoxin. N. Engl. J. Med. 328:1457–1460 [DOI] [PubMed] [Google Scholar]
- 139. Teijaro JR, et al. 2011. Endothelial cells are central orchestrators of cytokine amplification during influenza virus infection. Cell 146:980–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Thomas DL, et al. 2009. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature 461:798–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Tobin DM, et al. 2010. The lta4h locus modulates susceptibility to mycobacterial infection in zebrafish and humans. Cell 140:717–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Tulic MK, et al. 2007. TLR4 polymorphisms mediate impaired responses to respiratory syncytial virus and lipopolysaccharide. J. Immunol. 179:132–140 [DOI] [PubMed] [Google Scholar]
- 143. van Iersel MP, et al. 2008. Presenting and exploring biological pathways with PathVisio. BMC Bioinformatics 9:399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Viasus D, et al. 2011. Effect of immunomodulatory therapies in patients with pandemic influenza A (H1N1) 2009 complicated by pneumonia. J. Infect. 62:193–199 [DOI] [PubMed] [Google Scholar]
- 145. Wang H, Ma S. 2008. The cytokine storm and factors determining the sequence and severity of organ dysfunction in multiple organ dysfunction syndrome. Am. J. Emerg. Med. 26:711–715 [DOI] [PubMed] [Google Scholar]
- 146. Weingartl HM, et al. 2009. Experimental infection of pigs with the human 1918 pandemic influenza virus. J. Virol. 83:4287–4296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Williams AE, et al. 2005. TGF-beta prevents eosinophilic lung disease but impairs pathogen clearance. Microbes Infect. 7:365–374 [DOI] [PubMed] [Google Scholar]
- 148. Willinger T, et al. 2011. Human IL-3/GM-CSF knock-in mice support human alveolar macrophage development and human immune responses in the lung. Proc. Natl. Acad. Sci. U. S. A. 108:2390–2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Wurfel MM. 2008. Genetic insights into sepsis: what have we learned and how will it help? Curr. Pharm. Des. 14:1900–1911 [DOI] [PubMed] [Google Scholar]
- 150. Wurfel MM, et al. 2008. Toll-like receptor 1 polymorphisms affect innate immune responses and outcomes in sepsis. Am. J. Respir. Crit. Care Med. 178:710–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Wurfel MM, et al. 2005. Identification of high and low responders to lipopolysaccharide in normal subjects: an unbiased approach to identify modulators of innate immunity. J. Immunol. 175:2570–2578 [DOI] [PubMed] [Google Scholar]
- 152. Yamamoto M, et al. 2003. Role of adaptor TRIF in the MyD88-independent Toll-like receptor signaling pathway. Science 301:640–643 [DOI] [PubMed] [Google Scholar]
- 153. Yamamoto M, et al. 2002. A novel Toll/IL-1 receptor domain-containing adapter that preferentially activates the IFN-beta promoter in the Toll-like receptor signaling. J. Immunol. 169:6668–6672 [DOI] [PubMed] [Google Scholar]
- 154. Yokota S. 2003. Influenza-associated encephalopathy—pathophysiology and disease mechanisms. Nippon Rinsho. 61:1953–1958 [PubMed] [Google Scholar]
- 155. Yuen KY, Wong SS. 2005. Human infection by avian influenza A H5N1. Hong Kong Med. J. 11:189–199 [PubMed] [Google Scholar]
- 156. Zhang Q, et al. 2010. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature 464:104–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Zheng BJ, et al. 2008. Delayed antiviral plus immunomodulator treatment still reduces mortality in mice infected by high inoculum of influenza A/H5N1 virus. Proc. Natl. Acad. Sci. U. S. A. 105:8091–8096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Zornetzer GA, et al. 2010. Transcriptomic analysis reveals a mechanism for a prefibrotic phenotype in STAT1 knockout mice during severe acute respiratory syndrome coronavirus infection. J. Virol. 84:11297–11309 [DOI] [PMC free article] [PubMed] [Google Scholar]