Abstract
Rapid clonal antigenic variation in Babesia bovis involves the variant erythrocyte surface antigen-1 (VESA1) protein expressed on the infected-erythrocyte surface. Because of the significance of this heterodimeric protein for demonstrated mechanisms of parasite survival and virulence, there is a need to understand how expression of the ves multigene family encoding this protein is controlled. As an initial step toward this goal, we present here initial characterization of the ves promoter driving transcription of VESA1a and -1b subunits. A series of transfection constructs containing various sequence elements from the in vivo locus of active ves transcription (LAT) were used to drive expression of the firefly luciferase gene in a dual luciferase-normalized assay. The results of this approach reveal the presence of two bidirectional promoter activities within the 434-bp intergenic region (IGr), influenced by putative regulatory sequences embedded within the flanking ves1α and ves1β genes. Repressor-like effects on the apposing gene were observed for intron 1 of both ves1α and ves1β. This effect is apparently not dependent upon intronic promoter activity and acts only in cis. The expression of genes within the ves family is likely modulated by local elements embedded within ves coding sequences outside the intergenic promoter region in concert with chromatin modifications. These results provide a framework to help us begin to understand gene regulation during antigenic variation in B. bovis.
INTRODUCTION
The protozoal bovine hemoparasite Babesia bovis sequesters mature developmental stages in the deep vasculature through the process of cytoadhesion, a phenomenon in which parasitized erythrocytes (PE) bind to the capillary and postcapillary venous endothelium. B. bovis cytoadhesion may occur in essentially any major organ (55–57), including the brain (1), and adhesive parasites may be selected which cytoadhere in vitro to cultured bovine brain capillary (39) and umbilical vein (34) endothelial cells. The sequestration of the mature developmental stages through cytoadhesion presumably precludes their passage through the spleen and thus mediates avoidance of splenic clearance (4).
To achieve cytoadhesion, B. bovis must export parasite-derived proteins to the surface of the infected erythrocyte (7, 39). Although the benefits of cytoadhesion have not yet been rigorously defined, the parasite-derived cytoadhesion ligands on the PE surface ultimately result in a host adaptive immune response to the exported proteins. As a result of this immune selection pressure, B. bovis undergoes rapid structural and antigenic variation of the surface proteins (6, 38). The parasite protein mediating both antigenic variation and cytoadhesion in B. bovis is the variant erythrocyte surface antigen-1 (VESA1) (6, 7, 37, 38). VESA1 is a heterodimeric protein whose subunits, VESA1a and -1b, are isolate specific and size polymorphic (7). This protein undergoes rapid structural and antigenic variation (6, 38) over time through a process of segmental gene conversion (2) that, when exposed to immune selection pressure, results in population-wide phenotypic changes (6). Variation may also occur during growth in in vitro culture in the absence of such selection pressure, although variants typically are slow to dominate the population under in vitro conditions (unpublished observations). Thus, antigenic variation would appear to be an ongoing, seemingly stochastic process. Such variation, and the function of the variant proteins as cytoadhesion ligands, is a trait shared between B. bovis and the human malarial parasite, Plasmodium falciparum (3).
The two subunits of VESA1 are encoded by members of the ves multigene family, with the 1a subunit encoded by ves1α genes (5) and the 1b subunit encoded by ves1β genes (58). There are approximately 150 members of the ves multigene family in the B. bovis genome (9). A majority of members are organized as head-to-head, divergent gene pairs and most often include one ves1α and one ves1β gene, although pairs containing two ves1α genes have also been observed (2, 9). Transcription of ves genes appears to be monoparalogous, with both ves1α and ves1β transcripts arising from a single locus (60). Although it is likely that many such loci are capable of driving transcription, to date only a single locus, termed the “locus of active ves transcription” (LAT) (2), has been observed to be transcriptionally active. The divergent, clustered organization of the actively transcribed genes could present the parasite with a mechanism by which to coordinately regulate the expression of the two genes and thus their encoded subunits. Given the adhesive function of this protein, the ability to arrive at a consistent subunit stoichiometry would seem to be beneficial, although this may not always be achieved (58). However, it also creates certain logistical problems and raises fundamental questions about how transcription may be organized to proceed bidirectionally, whether it involves simultaneous transcription of overlapping sequences or whether such overlap occurs within individual parasites. Preliminary mapping of transcription start sites for the ves1α gene by 5′ rapid amplification of cDNA ends (5′-RACE) revealed a beginning on the “ves1β” side of the intergenic region (IGr) in which ves promoter activities presumably would reside (5). A reanalysis of ves1α and ves1β transcripts by the use of a cap-dependent form of 5′-RACE suggests that transcription begins at several sites for each gene, including on the “distal half” of the IGr (this study). Given the quasipalindromic nature of the ves LAT, including a pair of inverted repeat sequences on each side of the IGr (2), this result is perhaps not surprising.
Another bidirectional promoter region in B. bovis, that which controls the EF1α genes, has previously been described (48). This region was shown to be composed of two nearly identical unidirectional promoters and identical EF1α genes. Given their nearly identical sequences, the two promoters and genes are likely the result of recent gene duplication and inversion events. However, ves genes and promoter regions are much more complex. Despite their overall similarities, ves genes bear distinct branch-associated asymmetries in sequence and organization, including within the IGr. We hypothesized that this complexity was key to regulation of the locus for the previously observed monoparalogous transcription (60).
To understand how ves genes are regulated requires first dissecting their structure and then identifying the relevant regulatory regions controlling gene expression, transcription factor binding sites, and local epigenetic chromatin modifications and remodeling. To initiate this process, we chose the approach of transiently transfecting parasites with plasmid constructs containing various regions of the LAT IGr to drive expression of a firefly luciferase reporter gene. These data were normalized by cotransfection with a second, Renilla luciferase-containing construct, enabling direct comparisons between the various constructs with regard to levels of gene expression. We report here that the quasipalindromic IGr of the LAT contains two bidirectional promoter activities whose levels of activity are affected by sequences embedded within the apposing flanking ves1α and ves1β genes. Furthermore, similar segments derived from transcriptionally silent ves loci were found to be capable of driving significant levels of marker gene expression, suggesting the capacity of such loci to accommodate epigenetic in situ transcriptional switching. This work should facilitate future investigation of the various influences involved in regulating this gene family and ultimately the development of strategies to ameliorate aspects of disease associated with this virulence trait.
MATERIALS AND METHODS
Parasite culture.
The B. bovis C9.1 and CE11 clonal lines were used in this study. Derivation of these lines by limiting dilution cloning and in vitro cultivation under microaerophilous stationary-phase conditions has been described previously (6, 27, 37).
Basic luciferase reporter constructs.
A number of different plasmids were constructed to assess promoter function (Fig. 1). A promoterless pGEM-Luc reporter vector (Promega Corporation, Madison, WI) was used as the basis for construction of reporter plasmids. A presumptive terminator sequence from the β-tubulin gene was amplified by PCR using B. bovis C9.1 clonal line genomic DNA (gDNA) and primers XW23 and XW24 (sequences for all primers used in this study are available upon request). The product was cloned into the SacI site of pGEM-Luc 3′ to luciferase coding sequences to yield the promoterless control plasmid, pLuc-T3. pLuc-T3 was used as a promoterless negative control in all assays and as the foundation for assembly of all firefly luciferase constructs. For the assembly of a construct using 5′ regulatory sequences of calmodulin, 1,397 bp of sequence upstream of the calmodulin gene start codon was amplified using B. bovis C9.1 gDNA and primers XW17 and XW18. The amplicon was cloned into the HindIII site of pLuc-T3 to yield pC5-Luc. A plasmid containing ves gene 5′ regulatory sequences was created by PCR amplification of the 678 bp upstream of the start codon of the LAT-associated ves1α gene (DQ267461) from phagemid 6-1 (DQ267460.1) (2) by the use of primers XW25 and XW26. The product was cloned into the HindIII site of pLuc-T3 to yield plasmid pLAT_Vα5-βI1. In addition to intergenic region sequences, this construct contained exon 1 and intron 1 of the apposing ves1β gene. Plasmids with reverse-oriented 5′ sequences served as reverse promoter negative controls. The pEF1α-IG plasmid, a kind gift from C. Suarez, was previously demonstrated to possess significant promoter activity (48) and in initial experiments was used as a positive control. An internal-control plasmid for normalization of firefly luciferase values among samples was constructed as follows: a 2,726-bp fragment containing 1,699 bp of the firefly luciferase gene plus 996 bp of β-tubulin 3′ sequences was removed from the pC5-Luc plasmid by digestion with BamHI and SacI. This was replaced by a fragment of 1,595 bp containing 933 bp of R. reniformis luciferase gene plus 662 bp of P. falciparum calmodulin polyadenylation sequences obtained by digestion of pPfrLuc plasmid DNA (a gift from D. Wirth [32, 33], provided to us by T. Bonilla) with BamHI and SacI. The resulting plasmid was named pC5Renilla.
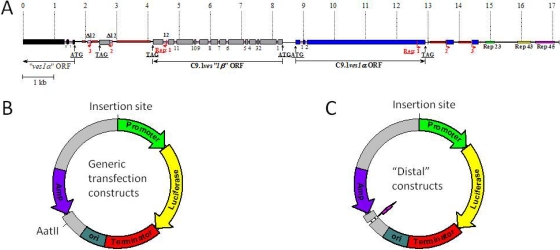
Fig 1.
Schematic diagrams of LAT organization and of constructs used in this study. (A) Schematic diagram showing the overall organization of the known, active B. bovis C9.1 line LAT in situ (adapted from reference 2 with permission). The ves1α gene exons are shown in blue, ves1β exons in light gray, and downstream ves1α gene exons in black. Introns are shown as thicker pink segments and are numbered. Rep 1, 2, and 3 and thicker red segments indicate repeated 3′ sequences found at the ends of the ves1α and ves1β genes. Rep23, Rep 43, and Rep 45 and thickened green, yellow, and bright pink segments indicate segments containing tandem repeats or a 23-, 43-, or 45-bp repeat length. A more complete description of the site and structure of the proteins is available in reference 2. (B) Schematic diagram of the overall organization of the transfection constructs used in this study. All were administered as supercoiled circular DNA. (C) Schematic diagram of constructs made to test for repressor-like activity (the reverse orientation is shown). The AatII insertion site is the region in which various sequences (offset, short pink arrow) were tested for repressor activity; these insertions destroyed the AatII site. All elements are drawn to scale except the promoter region. The promoter sequences differed in length among individual constructs; as drawn, the promoter would be 1 kbp in length.
Constructs with intergenic regions from alternative ves donor loci.
The 669 bp of 5′ sequences from the ves1αC gene was amplified from cosmid 1E10 (GenBank accession number AY279553) (2) by PCR, using primers XW77 and XW78, with creation of BamHI restriction sites on both ends. The amplicon was inserted into pLuc-T3, and the correctly oriented plasmid was named p1E10. Another 658 bp of 5′ sequences of ves1αA was amplified from cosmid S621 (GenBank accession number AY279554) (2) with XW92 and XW76. The amplicon was inserted into pLuc-T3 to yield construct pS621. All plasmid constructs were confirmed by restriction digestion and sequencing. Schematic diagrams of plasmid configuration are provided in Fig. 1B and C, and each panel indicates the region of LAT sequences (or corresponding regions) included in constructs used in the experiments represented by the figure.
Constructs with select ves intergenic region sequences.
The 434-bp LAT intergenic region was amplified from phagemid 6-1 (2) by the use of primers XW79 and XW80 and inserted into the BamHI site of pLuc-T3. As only plasmid with reversely inserted 5′ sequences was recovered (pLAT_IgVβ5), an ApaI restriction site was engineered into the 5′ end of the insertion by the use of XW95. This facilitated oriented insertion of the product into pLuc-T3, yielding pLAT_IgVα5. Fragments containing one-half of the Igr were amplified from pLAT_IgVα5 with creation of an ApaI site at the 5′ end and BamHI site at the 3′ end and were cloned by directional insertion. Sequences to create the 233 bp of pLAT_halfIG#1 were amplified from pLAT_IgVα5 by the use of XW106 and XW80. Sequences for the 200 bp of pLAT_halfIG#2 were amplified from pLAT_IgVα5 by the use of XW95 and XW107. The 200 bp of pLAT_halfIG#3 was amplified from pLAT_IgVα5 by the use of XW108 and XW79. Finally, the 233 bp of pLAT_halfIG#4 was amplified from pLAT_IgVα5 by the use of XW109 and XW110. PCR products were inserted into pLuc-T3 doubly digested with ApaI plus BamHI. The position of each relative to the full IGr, and its orientation relative to the luciferase gene upon insertion, is shown here (see Fig. 4).
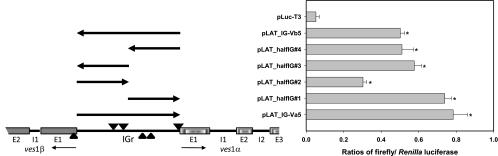
Fig 4.
The LAT IGr contains at least two bidirectional promoter activities. Complete or half-IGr segments were tested for promoter activity in B. bovis C9.1-parasitized erythrocytes by the dual-luciferase assay. Significant levels of promoter activity were observed with the full-length IGr and with both half-IGr constructs, regardless of the orientation of the sequences relative to the firefly luciferase gene. In vivo transcription start sites of ves1α and ves1β transcripts, observed by a cap-dependent 5′-RACE method, are indicated for comparison. Downward-pointing small triangles, ves1α transcripts; upward-pointing small triangles, ves1β transcripts. Arrows and asterisks are as defined in the Fig. 3 legend.
Constructs with additional ves gene sequences.
Sequences for the creation of constructs containing additional ves sequences were amplified from phagemid 3-2-1 (2). The 584-bp fragment used to create pLAT_Vα5-βE1 was amplified with primers XW124 and XW80. The 3′ antisense primer used in all ves1β5 constructs was XW79. It was paired with XW125 for the 557 bp of the pLAT_Vβ5-αE1 construct; with XW111 for the 670 bp of the pLAT_Vβ5-αI1 construct; with XW126 for the 734 bp of the Vβ5-E2 fragment; or with XW112 for the 807 bp of the pLAT_Vβ5-αI2 construct. Products were inserted into pLuc-T3 doubly digested with ApaI plus BamHI. These constructs are shown schematically (see Fig. 5). A control construct was assembled by amplifying a 238-bp segment of the msp1α gene from Anaplasma marginale Florida isolate genomic DNA by the use of primers DA129-Am5 and DA130-Am3. The resulting amplicon was inserted into the HindIII site of pLAT_IgVα5 to generate pMsp_IgVα5.
Fig 5.
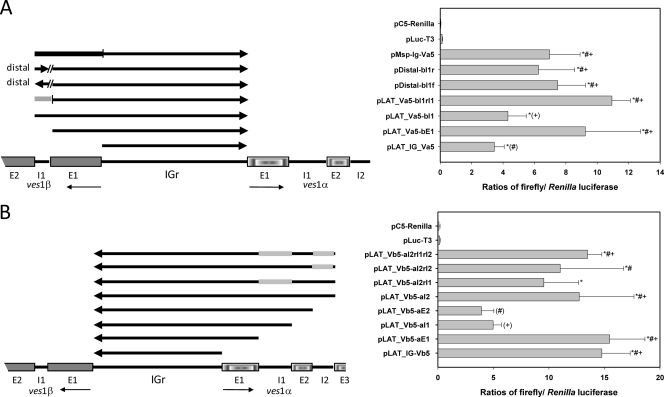
Flanking sequences influence IGr promoter activity. (A) “α orientation” full-length IGr and various lengths of additional flanking or distal ves1β gene sequences were inserted into the pLuc-T3 construct to test for promoter activity. The light gray bar indicates reversal of ves1β intron 1 orientation. The “distal” constructs contain ves1β intron 1 within the AatII site on the opposite side of the pGEM plasmid backbone, whereas pMsp_IgVα5 contained coding sequences from the A. marginale msp1α gene in place of ves flanking sequences (dark gray bar). Samples are marked to indicate which pairwise comparisons are statistically significantly different at P ≤ 0.05 as follows: #, those differing significantly from pLAT_IgVα5; (#), pLAT_IgVα5; +, those differing significantly from pLAT_Vα5-βI1; (+), pLAT_Vα5-βI1. No other pairwise comparisons yielded statistically significant differences. (B) “β orientation” full-length IGr and various lengths of additional flanking sequences from the ves1α gene were inserted into the pLuc-T3 construct to test for promoter activity. The thick gray bars indicate reversal of orientation of those sequences. Arrows and asterisks are as explained in the Fig. 3 legend. Samples are marked to indicate which pairwise comparisons are statistically significantly different at P ≤ 0.05 as follows: #, those differing significantly from pLAT_Vβ5-αE2; (#), pLAT_Vβ5-αE2; +, those differing significantly from pLAT_Vβ5-αI1; (+), pLAT_Vβ5-αI1. No other pairwise comparisons yielded statistically significant differences.
Constructs with inverted intronic sequences.
The 43 bp of ves1β intron 1 sequence was inverted by a crossover PCR approach (41). The intron was amplified from pLAT_Vα5-βI1 with primers DA119(ApaI) and XW120. The amplicon was connected with ApaI-linearized pLAT_Vα5-βE1 by crossover PCR with primers DA119 (ApaI) and XW80. The fragment was cut with BamHI and ApaI and ligated into similarly cut pLuc-T3 plasmid to yield pLAT_Vα5-βI1rI1. To reverse ves1α introns 1 and/or 2, introns were first amplified with Phusion polymerase (New England BioLabs, Beverley, MA). ves1α introns 1 and 2 were amplified from pLAT_Vβ5-αI2 with primers DA121 and DA122 and primers DA123 and DA124, respectively. To prepare vector for ves1α intron 1, all other plasmid sequences were amplified from pLAT_vβ5-αI2 by inverse PCR with primers DA125 and DA126. Similarly, for ves1α intron 2, all other plasmid sequences were amplified by inverse PCR with DA127 and DA128. Introns 1 and 2 were inserted into the relevant vectors by the use of InFusion reagents as recommended by the manufacturer (Clontech, Mountain View, CA) to make pLAT_Vβ5-αI2rI1 and pLAT_Vβ5-αI2rI2, respectively. To reverse both introns, pLAT_Vβ5-αI2rI2 was amplified with DA125 and DA126 and the amplified reversed intron 1 was inserted, using InFusion reagents, to create pLAT_Vβ5-αI2rI1rI2. These constructs are shown (see Fig. 5).
Constructs employing only intronic sequences as promoter.
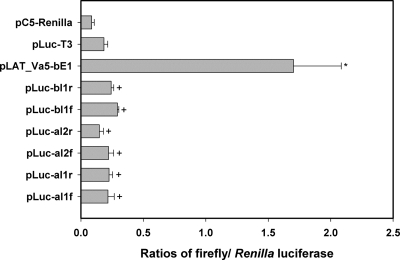
Intron 2 (72 bp) of the C9.1 line-expressed ves1α gene was amplified from pgC3c (5), in both polarities, with primers DA111 and DA112 and primers DA113 and DA114. The amplified sequences were inserted into pLuc-T3 plasmid linearized using HindIII and In-Fusion reagents (Clontech, Mt. View, CA) to yield pLuc_aI2F and pLuc_aI2R. Intron 1 of the ves1α gene was amplified from construct pLAT_vβ5-αI2 with primers DA133 and DA134, whereas intron 1 of the ves1β gene was amplified from pLAT_vα5-βI1 with primers DA131 and DA132. Amplicons were inserted into the BamHI site of pLuc-T3 to yield constructs pLuc-αI1f, pLuc-αI1r, pLuc-αI2f, pLuc-αI2r, pLuc-βI1f, and pLuc-βI1r. These constructs are shown (see Fig. 6).
Fig 6.
ves1α and ves1β introns have no apparent promoter-like activity. ves1α introns 1 and 2 and ves1β intron 1 were tested directly for promoter activity by insertion into the pLuc-T3 construct to drive luciferase expression. None of these sequences demonstrated promoter activity significantly greater than that of the pLuc-T3 promoterless control. This experiment was repeated two times, using B. bovis CE11-parasitized erythrocytes. The asterisk indicates a statistically significant difference from pLuc-T3 at P ≤ 0.05; the plus signs indicate those samples whose results are statistically significantly different from those of pLAT_Vα5-βE1 at P ≤ 0.05. No other pairwise comparisons revealed significant differences.
Constructs employing intronic sequences as distal elements.
The pLAT_vα5-βI1 plasmid was opened with AatII at a location 2,204 nucleotides (nt) distal to the HindIII site containing ves promoter sequences. ves1β intron 1 sequences were amplified with primers DA107 and DA108 and primers DA109 and DA110. Amplicons were inserted into the AatII site of pLAT_Vα5-βE1, in both polarities, by the use of In-Fusion reagents (Clontech) to yield pDistal_βI1f and pDistal_βI1r (see Fig. 5).
Nomenclature of construct names.
Constructs containing IGr sequences from the LAT ves locus are designated “pLAT_V” followed by an “α5” or a “β5.” These designate whether the “α side” or “β side” of the IGr is directly apposed to the luciferase reporter as the 5′ promoter sequence. Sequences beyond the IGr (i.e., apposing the luciferase gene) are indicated by an “-E#” or “-I#” to indicate that they extend to the end of an exon or intron, respectively, with the “#” referring to the number of the exon or intron. A superscripted “rI#” indicates reversal of that specific intron within the construct. For example, “pLAT_Vβ5-αI2rI1” contains the IGr oriented with its β side facing the luciferase gene and includes ves1α gene sequences through the end of intron 2 but with intron 1 sequences in a reversed orientation. Schematic drawings are provided in each figure to clarify the sequences contained within each individual construct and their orientations.
Transient transfection of constructs.
Plasmid DNAs were prepared using EndoFree plasmid purification reagents (Qiagen, Inc., La Jolla, CA) or PureYield plasmid (Promega Corporation, Madison, WI) or Zyppe Plasmid Max (Zymo Research, Irvine, CA) preparation reagents, following the instructions of the manufacturers. DNAs were suspended in cytomix buffer before use. DNA concentrations were determined by UV spectrometry and by comparison with known amounts of standard markers on agarose gels. Before transfections, plasmid DNAs were diluted into cytomix (120 mM KCl, 0.15 mM CaCl2, 10 mM K2HPO4/KH2PO4, 25 mM HEPES, 2 mM EGTA, 5 mM MgCl2, pH 7.6). Parasites were used when cultures reached 5% to 15% parasitized erythrocytes. For transfecting infected red blood cells (IRBCs), parasite cultures were centrifuged at 3,300 × g for 10 min at 4°C. The cell pellet was washed two times with cytomix, under the same conditions. For each transfection, 100 μl of packed IRBCs was electroporated with 11.5 pmol of transfection construct DNA and either 3.8 or 7.7 pmol of pC5 Renilla mixed in an approximately 450-μl total volume (the amount was always consistent within an experimental series). Electroporation was performed with a Bio-Rad Gene Pulser II electroporation apparatus (Bio-Rad Laboratories, Hercules, CA) under conditions of 1.25 kV, 25 μF capacitance, and 200 Ω resistance, in 2-mm-gap cuvettes (Fisher Biotech, Atlanta, GA). Transfected cells were then transferred into 6-well plates containing 3 ml of a 2.5% packed cell volume suspension of uninfected erythrocytes in complete medium. For periods longer than 24 h, the medium was changed and 50 μl of packed RBCs was added into each well on the day following transfection. All wells were treated identically. At the desired times posttransfection (p.t.), RBCs were collected into 2-ml microcentrifuge tubes, sedimented by centrifugation at 3,300 × g for 10 min, and washed two times with 1× Vega y Martinez (VYM) buffer (51) under the same conditions. Following initial experiments to define the time course of marker expression (Fig. 2), subsequent experiments involved a standard 24-h incubation. Infected erythrocytes were selectively lysed by treatment with NH4Cl-Tris, essentially as described previously (31), and then centrifuged at 12,000 × g for 10 min. The unlysed parasite pellet was washed two times with 1× VYM buffer under the same conditions and then lysed with 100 μl of freshly prepared passive lysis buffer (Promega Corporation, Madison, WI) for 25 min.
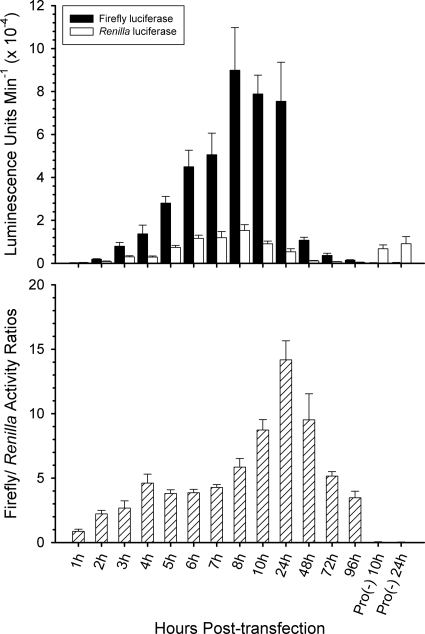
Fig 2.
Time course of luciferase expression. B. bovis C9.1-parasitized erythrocytes were cotransfected with pLAT_Vα5-βI1 (see Fig. 3 for included promoter sequences) and pC5 Renilla and placed back into culture as described in Materials and Methods. Timed aliquots were then collected at the various time points and processed for the dual-luciferase assay. The times are given as hours posttransfection. Pro(−), pLuc-T3 promoterless negative control. The top plot indicates the empirical amounts of firefly and Renilla luciferase activities; the bottom shows the ratios of the two activities for each time point. The error bars represent ±1 standard deviation (s.d.) of the results determined with triplicate samples. The experiment was repeated two times.
Dual-luciferase reporter assay.
The dual-luciferase assay was performed following the instructions of the manufacturer (Promega Corp.), with lysates analyzed in triplicate. Both firefly and Renilla luciferase activities were quantified with a MicroBeta Jet scintillation spectrometer (PerkinElmer, San Jose, CA), taking 10-s measurements for each reporter. To enhance accuracy and minimize background, lysates were centrifuged for 2 min at 12,000 × g at room temperature prior to analysis. A 100-μl volume of extract was dispensed into the wells, and three consecutive recordings were taken. The average reading of each well was subtracted from luciferase activity measurements taken from the same well. Readings from mock-transfected parasite lysates were subtracted from all luciferase readings, and the levels of firefly luciferase activity were normalized to the level of Renilla reniformis luciferase activity. Samples were prepared in triplicate, and each experiment was repeated at least three times or as indicated in the figure legends. Firefly/Renilla luciferase ratios differed between experiments, being affected by several factors, including percent parasitized erythrocytes and the age and history of the substrate. Therefore, direct comparisons among ratios are made only for samples within the same experiment, although relative expression patterns were qualitatively reproducible among experiments.
Statistical analysis of signals.
Normalized luciferase activities were plotted using Sigmaplot v. 11 (Systat Software, Inc., San Jose, CA) as the mean ± 1 standard deviation, calculated from triplicate samples. Differences in promoter activity values were determined by the Student-Newman-Keuls one-way analysis of variance (ANOVA) test (21), as implemented in SigmaPlot. This was performed on raw data when a Shapiro-Wilk test for normality was passed or on ranks when this test failed. A value of P ≤ 0.05 was accepted as the threshold at which differences between the values of two samples were considered statistically different.
Cap-dependent 5′-RACE.
Total RNA was isolated from B. bovis C9.1 line parasites by the use of Totally RNA reagents. The RNA was treated with Turbo DNase followed by DNase inactivation reagent (all from Ambion, Inc., Austin, TX) per the manufacturer's instructions. Transcription start sites were mapped by a 5′ cap-dependent RNA ligase-mediated rapid amplification of cDNA ends method (First Choice RLM-RACE; Ambion). The procedure was performed essentially as described by the manufacturer, except that each enzymatic step was preceded by heating of the RNA to 80°C for 3 min, followed by snap chilling on ice to reduce secondary structures. cDNA was made by reverse transcription at 65 to 70°C with Thermo-X reverse transcriptase (RT) (Ambion), by the use of BAK172 and BAK171 as gene-specific primers for ves1α and ves1β genes, respectively. Nested PCRs were performed using 5′-RACE Outer Primer in combination with BAK80H as the primary gene-specific primer for ves1β transcripts or BAK169 for ves1α transcripts. BAK166H and BAK167H, along with a modified form of the manufacturer's nested 5′-end adapter primers (InnerL primer), were used as nested primers for ves1α and ves1β transcripts, respectively. All PCRs were performed using iProof (Bio-Rad Laboratories, Hercules, CA) or Phusion polymerase. PCR conditions were optimized for each reaction due to the difficulty in amplifying highly structured DNA. Due to cloning difficulties, amplicons were purified from agarose gels and sequenced directly by dideoxynucleotide inhibitor chemistry. Sequence reactions were analyzed by the University of Florida Interdisciplinary Center for Biotechnology Research DNA Sequencing Laboratory.
Identification of ves-associated RNA fragment ends.
Total RNA was decapped with tobacco acid pyrophosphatase and then circularized by ligation in dilute solution with T4 RNA ligase, essentially as described by Mandl et al. (30). cDNA was made by reverse transcription with SuperScript II reverse transcriptase and either PD2Fa,b or PD1R primers to detect antisense or sense transcripts, respectively. Primers DA59 and DA35 were used for assessing the 5′ end of the ves1α gene for local ncRNAs and primers BAK18 plus DA35 as controls to capture the ends of full-length ves1α mRNAs. PCRs were amplified with Phusion polymerase.
Circular chromatin conformation capture (4C).
In vitro-cultured B. bovis parasites at a level of 5% to 10% parasitized erythrocytes were centrifuged and washed with 1× VYM buffer (51) by centrifugation at 3,300 × g for 10 min at 4°C. The parasitized erythrocytes were then lysed with 0.06% saponin (Sigma Chemical Co. St. Louis, MO) in 1× VYM on ice, sedimented, and washed with phosphate-buffered saline (PBS) at 6,700 × g for 10 min at 4°C. They were then cross-linked with 2% (vol/vol) formaldehyde in 10% (vol/vol) defibrinated adult bovine serum–PBS at 20°C for 10 min before cross-linking was quenched on ice with 0.125 M glycine (final concentration) (22). Supernatants were removed following centrifugation at 225 × g for 8 min at 4°C. Cells were then washed one time with ice-cold “solution A” [20 mM piperazine-1,4-bis(2-ethanesulfonic acid) (pH 7.5), 15 mM NaCl, 60 mM KCl, 14 mM 2-mercaptoethanol, 0.5 mM ethyleneglycol-bis(β-aminoethylether)-N,N,N′,N′-tetraacetic acid, 4 mM EDTA, 0.15 mM spermine, 0.5 mM spermidine, 0.125 mM phenylmethylsulfonyl fluoride] (25) at 16,000 × g for 10 min at 4°C and resuspended in solution A on ice. NP-40 was added at 0.67% (vol/vol), and the parasites were disrupted by Dounce homogenization. Nuclei were sedimented at 4,500 × g for 10 min at 4°C and washed once with solution A. Restriction digestion of chromatin with EcoRI, ligation with T4 DNA ligase, reversal of cross-links, and DNA purification were subsequently performed as described previously (22). Inverse PCR of the circularized products was performed using iProof DNA polymerase (Bio-Rad) and primer pair AB1 and BAK40R, AB1 and BAK23, AB2 and BAK40R, AB2 and BAK23, AB3 and BAK40R, or AB3 and BAK23. Control reactions included nonfixation and nonligation of samples and single-primer PCRs. Only products which were dependent upon fixation, ligation, and the presence of both primers for amplification were recovered from agarose gels and cloned by Topo-TA Cloning (Invitrogen, Carlsbad, CA). Products were sequenced, and nucleotide BLAST analyses were run against all B. bovis sequences (taxid 5865) and the B. bovis T2Bo isolate genome (taxid 484906) to identify captured sequences.
RESULTS AND DISCUSSION
The ability to genetically manipulate B. bovis has been developed only very recently and remains in its rudimentary stages (47, 54). Because of its potential to expand our understanding of antigenic variation in B. bovis, we chose to employ transient transfection with luciferase reporter constructs (Fig. 1) to perform an initial characterization of promoter structure in the ves multigene family. We first established the time course of luciferase expression by the pC5Renilla positive-control and pLAT_Vα5-βI1 constructs in order to determine the optimal timing of sample assays employing ves promoter sequences. As seen in Fig. 2, both luciferases were detectable by 2 h posttransfection (p.t.) and rose steadily through 8 h p.t., at which point firefly luciferase expression by the pLAT_Vα5-βI1 construct effectively reached a plateau and Renilla luciferase expression by pC5Renilla began to slowly but progressively decline. When firefly values were normalized relative to luciferase values, a peak in ratios was observed 24 h p.t., yielding a very sensitive indicator of test promoter activity relative to the internal control. Signals from the promoterless control, pLuc-T3, were consistently extremely low. These dynamics were reproducible, and the 24 h p.t. time point was selected for subsequent experiments in which putative ves promoter activities were compared.
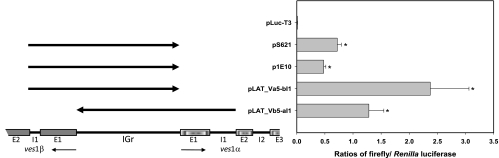
Transcription of ves genes in B. bovis occurs in a monoparalogous fashion at the locus of active ves transcription (LAT) (60). This site is organized as a pair of divergently oriented ves1α and ves1β genes (2), as is the case for the majority of this gene family (9). The preservation of this quasipalindromic organization at many ves loci throughout the B. bovis genome prompted us to ask whether nontranscribed ves loci possessed similar promoter activity. Constructs were therefore made containing the intergenic region (IGr) sequences, plus exon 1 and intron 1 of the apposing ves1α and ves1β genes of the LAT, and from ves1α genes from two identically organized, nontranscribed loci, previously cloned from the genome as cosmid 1E10 and phagemid 6-2-1 (2). When these constructs were tested for expression of firefly luciferase in a dual-luciferase assay, each demonstrated significant promoter activity relative to the promoterless control, pLuc-T3 (Fig. 3). This finding is important, because it supports the possibility that ves loci other than the LAT are competent to drive expression of VESA1 polypeptides in situ. This situation implies that selective activation of the LAT or some form of selective silencing of other ves loci or both occurs. Silencing may occur through local or generalized chromatin silencing or other phenomena; this remains to be determined. In P. falciparum, sense and antisense transcripts derived from intronic promoters, coupled with histone H3 lysine 9 trimethylation, are associated with silencing of the 5′ structural promoter of nonexpressed var genes (12, 16, 20, 29, 52). In Giardia lamblia, in contrast, an RNA interference (RNAi)-like mechanism appears to result in rapid posttranscriptional degradation of transcripts from the many nonexpressed members of the vsp gene family rather than selective transcription (42). The latter mechanism is unlikely in B. bovis due to the demonstrated lack of RNAi machinery in Plasmodium spp. (8) and the inability to identify orthologs of Dicer, Argonaute, and other components of RNAi in most other Apicomplexans (50), including B. bovis (9). However, noncoding RNAs (ncRNAs) are gaining recognition for playing a variety of roles in many systems, including gene silencing (43) and enhancement (40) and establishment of chromatin structure (53). Recently, ncRNAs associated with chromatin structure at telomeres have been identified in the related Apicomplexan malaria parasite Plasmodium falciparum (10). No evidence is yet available demonstrating actual mechanisms of ves gene silencing. Given the use by other Apicomplexans of histone modification-based epigenetic markings commonly associated with activation and silencing (23), such modifications are likely to play some role, but this remains to be established.
Fig 3.
Promoter activities of IGr sequences from various ves loci. B. bovis C9.1-parasitized erythrocytes were cotransfected with luciferase constructs containing the IGr and apposing gene sequences to the 3′ end of intron 1 from various ves loci. p1E10 and pS621 IGr sequences are derived from ves loci that are nontranscribed in the B. bovis C9.1 clonal line (13). Note that the S621 construct contains such sequences from a ves1α-ves1α pair. In this and subsequent figures, arrows above the LAT schematic drawing indicate the portion(s) of the locus (or comparable regions of other loci) included in luciferase constructs. Unless indicated otherwise, these sequences were included as putative promoters, and arrowheads indicate the 3′ end of each sequence adjacent to the luciferase gene. Asterisks indicate samples with values statistically significantly different from those determined with the pLuc-T3 promoterless control (P ≤ 0.05).
In order to begin dissection of the ves promoter, we chose to first include only IGr sequences, either as a complete IGr or as “half-IGr” elements. In keeping with the ves IGr's bidirectional function, IGr sequences demonstrated significant promoter activity in either orientation (Fig. 4). We anticipated that bidirectional transcriptional activity would arise either through a single, central promoter activity or two independent unidirectional promoter activities, in similarity to the EF1α locus (48). In contrast to expectations, each half IGr was found to possess significant promoter activity in both orientations (Fig. 4). Thus, at least two distinct bidirectional promoters, or as many as four unidirectional core promoter activities, reside within the 434-bp LAT IGr for the expression of the two polypeptides. The purpose of this unusually complex architecture is not clear but seems likely to serve some role in the regulation of ves gene expression. To determine whether each half of the IGr functions in vivo in driving transcription in either direction, we mapped the 5′ ends of ves1α and ves1β transcripts by a cap-dependent 5′-RACE method. As shown in Table 1 and Fig. 4, transcripts of both polarities originate from each side of the IGr, suggesting that each bidirectional promoter drives transcription in both directions in vivo. The longest transcripts thus have heavily overlapping 5′ untranslated regions, bringing into question whether individual parasites could simultaneously transcribe both ves1α and ves1β transcripts from bidirectional promoters in apposing half IGrs. Such a situation would seem more likely to result in transcription of either the ves1α or ves1β gene but not simultaneous transcription of both. This scenario is consistent with the recently reported independent and apparently temporally separated appearance of VESA1a and -1b polypeptides on the IRBC surface during parasite development (58). For the parasite to express VESA1 holoprotein in this case would require a switch in the transcriptional activity from one side of the IGr to the other, perhaps subject to local chromatin remodeling. In preliminary experiments, an apparent higher-order structure has been observed in presumably nucleosome-free IGr DNA within B. bovis chromatin (24). However, the presence of transcripts with 5′ ends originating from the same side of the IGr as the gene being transcribed suggests that the two genes could be transcribed simultaneously, depending upon where the RNA polymerase complex is assembled and how much of the IGr is involved spatially. These possibilities require further exploration.
Table 1.
ves transcription start sitesa
| Gene | TSS | TSS sequence |
|---|---|---|
| ves1α | −275 | CACC/ACTA |
| ves1α | −236 | ATCC/AGTA |
| ves1α | −7 | AAGC/ACTG |
| ves1β | −314 | CGAC/AGTA |
| ves1β | −276 | GGGA/TTCC |
| ves1β | +8 | TTTC/AGGG |
Positions are given relative to the ves1α or ves1β methionine start codon and shown schematically in Fig. 4. Transcription was usually found to begin with the A nucleotide of a CA dinucleotide (underlined).
TSS, transcription start site.
The consistency of divergent ves gene organization led us to question whether this pattern of organization might exist to contribute to ves gene regulation. This possibility was queried by the construction of luciferase constructs containing progressively more apposing sequence on the opposite side of the IGr to assay for the influence of these sequences on expression levels. As seen in Fig. 5A, the inclusion of the IGr with the “α side” directing luciferase transcription, apposed by ves1β sequences, resulted in increased luciferase expression levels upon addition of exon 1. Addition of intron 1, however, reduced expression of luciferase by ≥50%, an effect that was abrogated by reversing the orientation of the intron. To control for the possibility that this was an artifact due solely to construct size, a plasmid containing a comparable length of coding sequence from the A. marginale Florida isolate msp1α gene in place of ves1β sequences, pMsp_IgVα5 (672 bp versus 678 bp for pLAT_Vα5-βI1), was constructed. Creation of a control construct of this length yielded levels of luciferase expression which were indistinguishable statistically from those generated in the presence of exon 1 but significantly higher than those seen with IGr alone or in the presence of intron 1 (Fig. 5A). Because of the apparent lack of sequence specificity revealed by inclusion of msp1α sequences, the enhancing effect of the addition of ves1β exon 1 to α-oriented IGr sequences may be nonspecific, perhaps due to local length-dependent structural characteristics of the plasmid construct. However, this result also indicates that the repressive effect of the ves1β intron 1 addition, which is lost upon inversion of this sequence, is not simply a length-effect artifact but rather illustrates the significance and apparent specificity of this repression. To test the possibility that ves1β intron 1 might act as a distal repressor, it was placed on the opposite side of plasmid pLAT_Vα5-βE1 in both orientations (Fig. 5A). When moved distally, no significant repression was observed, and there was no orientation dependence with respect to the results. These results suggest that the “default” activity of exogenously introduced ves promoters is “active” and that ves1β intron 1 possesses an orientation-dependent cis-acting repressive effect on expression of genes juxtaposed to the ves1α side of the IGr. The mechanism by which that might occur remains to be elucidated. Similarly, the ves1β side of the IGr was oriented next to the luciferase gene and additional lengths of the ves1α gene were added at the opposite side. The “β side” of the IGr behaved overall similarly but slightly differently (Fig. 5B). In this orientation, the IGr already had high activity, which did not change with addition of ves1α exon 1. The orientation dependence observed for the promoter activities of IGr-only constructs suggests that the clear asymmetries existing within the quasipalindromic IGrs (2), including those studied here, may underlie these differences. Importantly, the addition of ves1α intron 1 sequences again resulted in a highly significant reduction of luciferase expression driven by the ves1β side of the IGr. Promoter activity of the IGr was unchanged by further inclusion of ves1α exon 2, but inclusion of ves1α intron 2 resulted in the recovery of higher levels of luciferase expression. Unlike the situation with ves1β intron 1, reversing the orientation of ves1α intron 1 had only a minimal effect, and reversing intron 2, or both introns, had no effect (Fig. 5B). However, it should be noted that ves1α intron 1 was reversed in the context of being flanked by exon 2 and intron 2 downstream, whereas ves1β intron 1 was reversed without these downstream sequences.
The orientation dependence of ves1β intron 1-mediated repression led us to question whether this behavior might be due to the presence of a unidirectional promoter embedded in that region of both genes. ves1α intron 1 shares very similar overall structures among different loci and even shares patches of significant sequence homology with the IGr itself (alignment available upon request). Precedence for this possibility certainly exists in other systems. In P. falciparum, for example, the single intron present within each var gene contains a bidirectional promoter (11), the activity of which appears essential for maintenance of var gene silencing (17, 18, 20, 35). To test this possibility directly, we made a series of constructs in pLuc-T3 in which ves1α introns 1 and 2 and ves1β intron 1 were each placed directly next to luciferase in both orientations. No significant promoter activity was observed for any of the introns in either orientation (Fig. 6). Based on these results, we conclude that the diminished luciferase expression in the presence of intron 1 is unlikely to be due to intronic promoter activities having direct effects on the IGr, as has been suggested for var gene regulation in P. falciparum (14, 18). However, it must be recognized that these sequences were asked to act in isolation without any of the potential influences being from within the chromatin environment; when present within chromatin, they may behave differently. Whether these sequences influence transcription of the genes within which they are embedded is also not clear from these results, as these experiments demonstrate effects only on genes apposing them across the IGr sequences. It would be instructive from a mechanistic point of view to investigate the influence of the introns on the transcription start sites used within the IGr during expression of luciferase. Clearly, the interactions of intronic sequences with the ves IGr promoter region are complex and warrant further investigation.
One matter of note is that all the constructs presented in this study which used ves promoter sequences were potentially subjected to epigenetic silencing upon insertion into the parasite. Previously, reverse transcriptase-PCR analysis of steady-state RNA isolated from bulk cultured parasites strongly suggested that under normal in vivo conditions, a single ves locus is active at any one time (60). The standard conditions used in this study involved analysis of transfectants following a 24-h incubation period. Given that the reproductive cycle of B. bovis following release from erythrocytes by electroporation is approximately 8 to 10 h (19), most parasites would have gone through at least two and perhaps three cell cycles—long enough to have silenced episomal ves promoters through epigenetic markers laid down following replication. Furthermore, there is no selection and presumably no reproductive benefit gained from the presence of the episomal constructs. Therefore, silencing of the ves gene family likely depends upon more than just the presence of an active ves promoter and may, at a minimum, require integration within the chromatin environment. Regardless, these results imply that whatever mechanism underlies silencing of the ves gene family does not depend upon a strict, ves-specific promoter-counting mechanism, such as the exclusionary “transcription body” mechanism proposed to mediate monoallelic transcription of VSG genes in Trypanosoma brucei (36). Indeed, this effect is consistent with the sequential expression of VESA1a and -1b subunits on the IRBC surface (58). Active transcription of ves1β may suppress transcription from ves1α and vice versa. This possibility remains to be fully explored. The observation of 5′-capped ves1α transcripts spanning from approximately the AUG start codon to within the 5′ end of exon 3 is consistent with this possibility (Fig. 7). These transcripts were captured by the circularization-dependent RT-PCR approach of Mandl et al. (30) following treatment with tobacco acid pyrophosphatase by the use of primers DA59 and DA35. The transcripts were properly spliced at both introns 1 and 2, indicating an original sense orientation. However, each was recovered from antisense-targeted samples and did not include sequences used to reverse transcribe sense transcripts, suggesting a possible double-stranded RNA (dsRNA) origin. Whether they represent a bizarre artifact, a novel form of regulatory ncRNA, short transcripts resulting from paused transcription, or simply incompletely degraded transcripts undergoing turnover is not yet clear. However, their similar sizes and sequence limits are striking. Moreover, control reactions performed with primers DA35 and BAK18 captured bona fide 5′ and 3′ ends of full-length ves1α mRNAs (not shown). The possibility of regulation via ncRNAs is reminiscent of the “sterile transcripts” arising from intron 1 of P. falciparum var genes (44), which may play a role in silencing of the var family (13, 15, 17), and warrants further exploration using a nontargeted, unbiased approach.
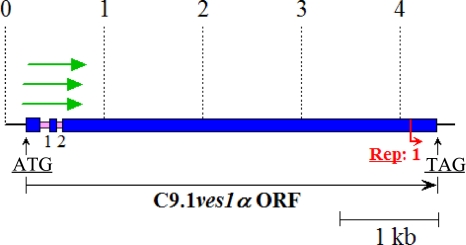
Fig 7.
Potential ncRNA fragments are found at the 5′ end of the ves1α gene. The schematic diagram illustrates the sizes and positions of potential ncRNAs (green arrows) recovered by RT-PCR of circularized RNAs relative to the ves1α gene. These RNAs spanned from approximately the 5′ end of the open reading frame for exon 1 (−38 to +16 nt range, relative to the AUG start codon) to within the 5′ end of exon 3 and were properly spliced. Labeling of the schematic matches Fig. 1.
A preliminary search for unique sequences interacting in trans as potential activating signals was attempted. This search was performed by analysis of sequences interacting with the LAT through a targeted circular chromatin conformation capture (4C) approach (59), using inverse PCR with primer pairs flanking the IGr to capture sequences interacting with the promoter region. We had hypothesized that a ves-specific enhancer, analogous to the “H enhancer” element implicated in controlling transcription of at least some mouse odorant receptor genes, might be uncovered (28). Rather, the sequences captured were almost solely from a variety of other ves (or ves-like) genes (data not shown). One reason for this may be simple proximity. Not only is the LAT flanked by ves sequences (2), but ves sequences are abundant in the genome, being present in approximately 150 copies (9) and accounting for perhaps 5% of the genome. Moreover, during portions of the cell cycle, the LAT may be silenced and remodeled into condensed chromatin, further exacerbating this effect. It was not generally possible to determine empirical distribution of captured ves sequences around the genome. The only B. bovis genome available at the time this work was done was from the T2Bo isolate (9) rather than the Mexico isolate used here, and the ves family is far too diverse to allow identification of specific members across isolates. Recently, five additional genomes from virulent and attenuated B. bovis were published (26), but those genomes also shed no light on the organization of this family in the Mexico isolate. However, it is plausible that the transcriptionally active ves genes within the LAT might preferentially associate with other ves loci through some specific functional interaction. Conserved functional roles such as those conjectured here are suggested by the conservation of internal sequences within ves1α introns 1 and 2 and ves1β intron 1 (60) (additional data not shown; alignments available upon request) among members of this multigene family (2). This remains to be further investigated, the physical association of distant ves genes remains to be critically demonstrated (requiring acquisition of the genome of the Mexico isolate), and an unbiased assessment of ncRNAs needs to be made and the functional significance of such sequences and interactions determined.
We suggest that transcriptional regulation of ves genes depends upon a variety of influences. Among these, the putative repressor-like effect on the apposing gene exhibited by ves1α and ves1β introns 1 and perhaps additional, as-yet-undetected regulatory functions embedded within ves genes are likely to contribute to the overall control of any specific locus and perhaps the gene family as a whole. Moreover, preliminary evidence consistent with chromatin remodeling that enables the LAT (whichever ves locus is serving as the LAT at that moment) IGr to adopt an open structure with highly ordered nucleosomes has been obtained (reference 24 and manuscript in preparation). While ultimately illuminating, in the near term these observations place the issue of what enables one site to become activated while others remain silenced only one step further removed from our current point of understanding. The issue becomes what triggers chromatin remodeling of a given locus for transcription and how regulatory controls are exerted in trans. We propose that beginning the process of dissecting ves promoter structure and regulation with the newly available transfection and stable transformation technology (45, 46, 48, 49) would facilitate explicating this important issue.
ACKNOWLEDGMENTS
We thank Carlos Suarez for sharing pEF1α-IG to facilitate initiation of this work, Jorg Bungert for helpful suggestions, and Alexia Berg, Allison Vansickle, Christie Smesko, Brady Pratt, and Jimmy Kidwell for their assistance in animal handling.
This project was supported by NIH grant R01 AI055864.
Footnotes
Published ahead of print 27 January 2012
REFERENCES
- 1. Aikawa M, et al. 1992. A study on the pathogenesis of human cerebral malaria and cerebral babesiosis. Mem. Inst. Oswaldo Cruz 87(Suppl. 3): 297–301 [DOI] [PubMed] [Google Scholar]
- 2. Al-Khedery B, Allred DR. 2006. Antigenic variation in Babesia bovis occurs through segmental gene conversion of the ves multigene family, within a bidirectional site of transcription. Mol. Microbiol. 59:402–414 [DOI] [PubMed] [Google Scholar]
- 3. Allred DR, Al-Khedery B. 2004. Antigenic variation and cytoadhesion in Babesia bovis and Plasmodium falciparum: different logics achieve the same goal. Mol. Biochem. Parasitol. 134:27–35 [DOI] [PubMed] [Google Scholar]
- 4. Allred DR, Al-Khedery B, O'Connor RM. 2003. Antigenic variation and its significance to Babesia, p 273–290 In Craig A, Scherf A. (ed), Antigenic variation. Elsevier Science Ltd., Oxford, United Kingdom [Google Scholar]
- 5. Allred DR, et al. 2000. The ves multigene family of B. bovis encodes components of rapid antigenic variation at the infected erythrocyte surface. Mol. Cell 5:153–162 [DOI] [PubMed] [Google Scholar]
- 6. Allred DR, Cinque RM, Lane TJ, Ahrens KP. 1994. Antigenic variation of parasite-derived antigens on the surface of Babesia bovis-infected erythrocytes. Infect. Immun. 62:91–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Allred DR, Hines SA, Ahrens KP. 1993. Isolate-specific parasite antigens of the Babesia bovis-infected erythrocyte surface. Mol. Biochem. Parasitol. 60:121–132 [DOI] [PubMed] [Google Scholar]
- 8. Baum J, et al. 2009. Molecular genetics and comparative genomics reveal RNAi is not functional in malaria parasites. Nucleic Acids Res. 37:3788–3798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brayton KA, et al. 2007. Genome sequence of Babesia bovis and comparative analysis of Apicomplexan hemoprotozoa. PLoS Pathog. 3:e148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Broadbent KM, et al. 2011. A global transcriptional analysis of Plasmodium falciparum malaria reveals a novel family of telomere-associated lncRNAs. Genome Biol. 12:R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Calderwood MS, Gannoun-Zaki L, Wellems TE, Deitsch KW. 2003. Plasmodium falciparum var genes are regulated by two regions with separate promoters, one upstream of the coding region and a second within the intron. J. Biol. Chem. 278:34125–34132 [DOI] [PubMed] [Google Scholar]
- 12. Chookajorn T, et al. 2007. Epigenetic memory at malaria virulence genes. Proc. Natl. Acad. Sci. U. S. A. 104:899–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dzikowski R, Deitsch KW. 2008. Active transcription is required for maintenance of epigenetic memory in the malaria parasite Plasmodium falciparum. J. Mol. Biol. 382:288–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dzikowski R, Frank M, Deitsch K. 2006. Mutually exclusive expression of virulence genes by malaria parasites is only dependent on non-coding regulatory elements and is independent of chromosomal location or antigen production. PLoS Pathogens 2:e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dzikowski R, et al. 2007. Mechanisms underlying mutually exclusive expression of virulence genes by malaria parasites. EMBO Rep. 8:959–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Epp C, Li F, Howitt CA, Chookajorn T, Deitsch KW. 2009. Chromatin associated sense and antisense noncoding RNAs are transcribed from the var gene family of virulence genes of the malaria parasite Plasmoidum falciparum. RNA 15:116–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Frank M, Deitsch K. 2006. Activation, silencing and mutually exclusive expression within the var gene family of Plasmodium falciparum. Int. J. Parasitol. 36:975–985 [DOI] [PubMed] [Google Scholar]
- 18. Frank M, et al. 2006. Strict pairing of var promoters and introns is required for var gene silencing in the malaria parasite Plasmodium falciparum. J. Biol. Chem. 281:9942–9952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Franssen FFJ, Gaffar FR, Yatsuda AP, de Vries E. 2003. Characterisation of erythrocyte invasion by Babesia bovis merozoites efficiently released from their host cell after high-voltage pulsing. Microbes Infect. 5:365–372 [DOI] [PubMed] [Google Scholar]
- 20. Gannoun-Zaki L, Jost A, Mu J, Deitsch KW, Wellems TE. 2005. A silenced Plasmodium falciparum var promoter can be activated in vivo through spontaneous deletion of a silencing element in the intron. Eukaryot. Cell 4:490–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Glantz SA. 1992. Primer of biostatistics, 3rd ed McGraw-Hill, Inc., New York, NY [Google Scholar]
- 22. Hagège H, et al. 2007. Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nat. Protoc. 2:1722–1733 [DOI] [PubMed] [Google Scholar]
- 23. Hakimi M-A, Deitsch KW. 2007. Epigenetics in Apicomplexa: control of gene expression during cell cycle progression, differentiation and antigenic variation. Curr. Opin. Microbiol. 10:357–362 [DOI] [PubMed] [Google Scholar]
- 24. Huang Y. 2009. Ph.D. thesis Higher-order structure in the Babesia bovis locus of active ves transcription. University of Florida, Gainesville, FL [Google Scholar]
- 25. Lanzer M, de Bruin D, Ravetch JV. 1992. Transcription mapping of a 100 kb locus of Plasmodium falciparum identifies an intergenic region in which transcription terminates and reinitiates. EMBO J. 11:1949–1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lau AOT, et al. 2011. Attenuation of virulence in an apicomplexan hemoparasite results in reduced genome diversity at the population level. BMC Genomics 12:410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Levy MG, Ristic M. 1980. Babesia bovis: continuous cultivation in a microaerophilous stationary phase culture. Science 207:1218–1220 [DOI] [PubMed] [Google Scholar]
- 28. Lomvardas S, et al. 2006. Interchromosomal interactions and olfactory receptor choice. Cell 126:403–413 [DOI] [PubMed] [Google Scholar]
- 29. Lopez-Rubio JJ, Mancio-Silva L, Scherf A. 2009. Genome-wide analysis of heterochromatin associates clonally variant gene regulation with perinuclear repressive centers in malaria parasites. Cell Host Microbe 5:179–190 [DOI] [PubMed] [Google Scholar]
- 30. Mandl CW, Heinz FX, Puchhammer-Stöckl E, Kunz C. 1991. Sequencing the termini of capped viral RNA by 5′-3′ ligation and PCR. Biotechniques 10:485–486 [PubMed] [Google Scholar]
- 31. Martin WJ, Finerty J, Rosenthal A. 1971. Isolation of Plasmodium berghei (malaria) parasites by ammonium chloride lysis of infected erythrocytes. Nat. New Biol. 233:260–261 [DOI] [PubMed] [Google Scholar]
- 32. Militello K, Dodge M, Bethke L, Wirth DF. 2004. Identification of regulatory elements in the Plasmodium falciparum genome. Mol. Biochem. Parasitol. 134:75–88 [DOI] [PubMed] [Google Scholar]
- 33. Militello KT, Wirth DF. 2003. A new reporter gene for transient transfection of Plasmodium falciparum. Parasitol. Res. 89:154–157 [DOI] [PubMed] [Google Scholar]
- 34. Molloy JB, Bowles PM, Jorgensen WK, Cooke BM. 2003. Babesia bovis: adhesion of parasitized red blood cells to bovine umbilical vein endothelial cells in vitro does not select for virulence. Exp. Parasitol. 103:182–184 [DOI] [PubMed] [Google Scholar]
- 35. Muhle RA, et al. 2009. A var gene promoter implicated in severe malaria nucleates silencing and is regulated by 3′ untranslated region and intronic cis-elements. Int. J. Parasitol. 39:1425–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Navarro M, Gull K. 2001. A polI transcriptional body associated with VSG mono-allelic expression in Trypanosoma brucei. Nature 414:759–762 [DOI] [PubMed] [Google Scholar]
- 37. O'Connor RM, Allred DR. 2000. Selection of Babesia bovis-infected erythrocytes for adhesion to endothelial cells co-selects for altered variant erythrocyte surface antigen isoforms. J. Immunol. 164:2037–2045 [DOI] [PubMed] [Google Scholar]
- 38. O'Connor RM, Lane TJ, Stroup SE, Allred DR. 1997. Characterization of a variant erythrocyte surface antigen (VESA1) expressed by Babesia bovis during antigenic variation. Mol. Biochem. Parasitol. 89:259–270 [DOI] [PubMed] [Google Scholar]
- 39. O'Connor RM, Long JA, Allred DR. 1999. Cytoadherence of Babesia bovis-infected erythrocytes to bovine brain capillary endothelial cells provides an in vitro model for sequestration. Infect. Immun. 67:3921–3928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ørom UA, et al. 2010. Long noncoding RNAs with enhancer-like function in human cells. Cell 143:46–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pont-Kingdon G. 1994. Construction of chimeric molecules by a two-step recombinant PCR method. Biotechniques 16:1010–1011 [PubMed] [Google Scholar]
- 42. Prucca CG, et al. 2008. Antigenic variation in Giardia lamblia is regulated by RNA interference. Nature 456:750–754 [DOI] [PubMed] [Google Scholar]
- 43. Schmitz K-M, Mayer C, Postepska A, Grummt I. 2010. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 24:2264–2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Su X, et al. 1995. The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes. Cell 82:89–100 [DOI] [PubMed] [Google Scholar]
- 45. Suarez CE, Lacy P, Laughery J, Gonzalez MG, McElwain T. 2007. Optimization of Babesia bovis transfection methods. Parassitologia 49:67–70 [PubMed] [Google Scholar]
- 46. Suarez CE, McElwain TF. 2008. Stable expression of a GFP-BSD fusion protein in Babesia bovis merozoites. Int. J. Parasitol. 39:289–297 [DOI] [PubMed] [Google Scholar]
- 47. Suarez CE, McElwain TF. 2010. Transfection systems for Babesia bovis: a review of methods for the transient and stable expression of exogenous genes. Vet. Parasitol. 167:205–215 [DOI] [PubMed] [Google Scholar]
- 48. Suarez CE, Norimine J, Lacy P, McElwain TF. 2006. Characterization and gene expression of Babesia bovis elongation factor-1α. Int. J. Parasitol. 36:965–973 [DOI] [PubMed] [Google Scholar]
- 49. Suarez CE, et al. 2004. Intergenic regions in the rhoptry associated protein-1 (rap-1) locus promote exogenous gene expression in Babesia bovis. Int. J. Parasitol. 34:1177–1184 [DOI] [PubMed] [Google Scholar]
- 50. Ullu E, Tschudi C, Chakraborty T. 2004. RNA interference in protozoan parasites. Cell. Microbiol. 6:509–519 [DOI] [PubMed] [Google Scholar]
- 51. Vega CA, Buening GM, Rodriguez SD, Carson CA. 1986. Concentration and enzyme content of in vitro-cultured Babesia bigemina-infected erythrocytes. J. Protozool. 33:514–518 [DOI] [PubMed] [Google Scholar]
- 52. Voss TS, et al. 2006. A var gene promoter controls allelic exclusion of virulence genes in Plasmodium falciparum malaria. Nature 439:1004–1008 [DOI] [PubMed] [Google Scholar]
- 53. Wang KC, et al. 2011. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472:120–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang X. 2010. Ph.D. thesis Development of a transfection system for genetic manipulation of Babesia bovis. University of Florida, Gainesville, FL [Google Scholar]
- 55. Wright IG. 1972. An electron microscopic study of intravascular agglutination in the cerebral cortex due to Babesia argentina infection. Int. J. Parasitol. 2:209–215 [DOI] [PubMed] [Google Scholar]
- 56. Wright IG. 1973. Ultrastructural changes in Babesia argentina-infected erythrocytes in kidney capillaries. J. Parasitol. 59:735–736 [PubMed] [Google Scholar]
- 57. Wright IG, Goodger BV, McKenna RV, Mahoney DF. 1979. Acute Babesia bovis infection: a study of the vascular lesions in kidney and lung. Z. Parasitenk. 60:19–27 [Google Scholar]
- 58. Xiao Y-P, Al-Khedery B, Allred DR. 2010. The Babesia bovis VESA1 virulence factor subunit 1b is encoded by the 1β branch of the ves multigene family. Mol. Biochem. Parasitol. 171:81–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhao Z, et al. 2006. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat. Genet. 38:1341–1347 [DOI] [PubMed] [Google Scholar]
- 60. Zupańska AK, Drummond PB, Swetnam DM, Al-Khedery B, Allred DR. 2009. Universal primers suitable to assess population dynamics reveal apparent mutually exclusive transcription of the Babesia bovis ves1α gene. Mol. Biochem. Parasitol. 166:47–53 [DOI] [PMC free article] [PubMed] [Google Scholar]