Abstract
In yeast, septins form rings at the mother-bud neck and function as diffusion barriers. In animals, septins form filaments that can colocalize with other cytoskeletal elements. In the filamentous fungus Aspergillus nidulans there are five septin genes, aspA (an ortholog of Saccharomyces cerevisiae CDC11), aspB (an ortholog of S. cerevisiae CDC3), aspC (an ortholog of S. cerevisiae CDC12), aspD (an ortholog of S. cerevisiae CDC10), and aspE (found only in filamentous fungi). The aspB gene was previously reported to be the most highly expressed Aspergillus nidulans septin and to be essential. Using improved gene targeting techniques, we found that deletion of aspB is not lethal but results in delayed septation, increased emergence of germ tubes and branches, and greatly reduced conidiation. We also found that AspB-green fluorescent protein (GFP) localizes as rings and collars at septa, branches, and emerging layers of the conidiophore and as bars and filaments in conidia and hyphae. Bars are found in dormant and isotropically expanding conidia and in subapical nongrowing regions of hyphae and display fast movements. Filaments form as the germ tube emerges, localize to hyphal and branch tips, and display slower movements. All visible AspB-GFP structures are retained in ΔaspD and lost in ΔaspA and ΔaspC strains. Interestingly, in the ΔaspE mutant, AspB-GFP rings, bars, and filaments are visible in early growth, but AspB-GFP rods and filaments disappear after septum formation. AspE orthologs are only found in filamentous fungi, suggesting that this class of septins might be required for stability of septin bars and filaments in highly polar cells.
INTRODUCTION
Septins are evolutionarily conserved GTP binding proteins that form complexes and are increasingly viewed as cytoskeletal elements (62). Septins are found in all eukaryotes except higher plants (20, 52) and are involved in a variety of cellular processes including cytokinesis, vesicle trafficking, cytoskeleton organization, polarity, and formation of diffusion barriers (35, 39, 44, 53, 62). Perhaps, not surprisingly given their roles in such critical processes, septin defects have been associated with many human diseases (29).
The septins were first discovered in the budding yeast Saccharomyces cerevisiae as temperature-sensitive cell cycle mutants defective in cytokinesis (31, 41). Early in the yeast cell cycle, septins localize as a cap at the future site of bud emergence. As the bud emerges, septins localize first as a ring at the mother/bud neck and later as an hourglass structure as the bud develops (25, 26, 37). At cytokinesis, the hourglass structure converts into two rings. Septins at the neck form diffusion barriers that keep the cellular machinery necessary for growth and cytokinesis properly localized (16, 18, 22, 49, 50, 60). In addition, septins are part of the morphogenesis checkpoint that coordinates nuclear division with bud formation (37).
In S. cerevisiae septins organize into hetero-oligomeric octamers that associate end to end to form nonpolar filaments (6). Electron microscopy studies have shown that long filaments can align in pairs forming higher-order structures after the initial filament is formed, and FRAP (fluorescence recovery after photobleaching) experiments showed that septin turnover occurs prior to bud emergence, hourglass splitting, and ring disassembly, while there is no turnover during bud formation and cytokinesis (3, 4, 12, 17). Removal of septin members from the complex can result in abnormalities whose severity depends on which member is removed (43).
Septins have also been studied in other fungal systems. In the fission yeast Schizosaccharomyces pombe, septins assemble into a ring that splits and encloses the actin-myosin ring during cytokinesis, although they are not essential for the process (2). In the dimorphic pathogen Candida albicans, septin mutations result in defects in cell separation. During yeast phase growth, septins localize very similarly to those in S. cerevisiae. During hyphal growth, septins assemble and disassemble forming bands at the base of emerging germ tubes and septation sites, collars at hyphal tips, and filaments in mature chlamydospores concomitant with thickening of the cell wall (5, 27, 58, 63). In the filamentous fungus Ashbya gossypii septins appear to promote mitosis near new branches and septins form rings made of discrete thick septin bars in hyphae and thin filaments at hyphal tips. In the basidiomycete Cryptococcus neoformans, septins are involved in morphology, sporulation, clamp cell fusion, and nuclear dynamics and septins localize to emerging spores, septa, and clamp connections (36). In addition, Cdc10 and Cdc3 were occasionally observed to form filaments in hyphae. In the dimorphic basidiomycete Ustilago maydis, septins are not essential but are required for normal growth and morphology and localize to the yeast neck and as a band at the growing hyphal tip (1, 8). In addition, Sep4 (Cdc10) uniquely formed septin fibers along the cell cortex (1), and septins underwent dynamic rearrangements from hourglass collars into ring structures during septation (7, 10). In the rice blast fungus Magnaporthe oryzae, septins form rings that anticipate the site of the appressorium septum (54). In the fungal pathogen Aspergillus fumigatus, septins localized to the cytoplasm, septa, hyphal tips, as well as emerging branches and conidiophore layers, and AspD and AspE formed filaments along the hypha (32).
The filamentous fungus Aspergillus nidulans has five septins: AspA, AspB, AspC, AspD, and AspE (48). We previously showed that deletions of aspA, aspC, or both aspA and aspC result in early and increased germ tube and branch emergence, abnormal septation, and disorganized conidiophores and that AspA and AspC localization is codependent, suggesting that these septins interact (38). Both AspA and AspC localized as spots and bars in dormant and expanding conidia, rings at forming septa and the bases of emerging germ tubes, branches, and conidiophore layers, and as spots and filaments in the cytoplasm and cell cortex (38). aspB was previously reported to be an essential gene and immunofluorescence showed that AspB localizes to sites of septation and branching and to emerging conidiophore layers (47, 64). We show here that deletion of aspB is not lethal but causes aberrant morphology in several developmental stages and reduced conidiation. We also show that AspB-green fluorescent protein (GFP) localizes not only as dots, rings, and caps but also as bars and filaments throughout vegetative growth and asexual reproduction. By combining AspB-GFP strains with asp deletion strains, we also show that AspA, AspB, AspC, and AspD septins interact with each other and propose a model of septin heteropolymer formation.
MATERIALS AND METHODS
Strains and growth conditions.
Experiments were carried out using the A. nidulans strains listed in Table 1. Fungal cultures for microscopy, crosses, DNA isolation, and strain characterization studies were grown in minimal medium (MM; 1% glucose, nitrate salts, biotin, trace elements, 1% thiamine [pH 6.5]) or with complete medium (CM; 1% glucose, 2% peptone, 1% yeast extract, 1% Casamino Acids, 0.01% vitamins and supplements, nitrate salt solution, and trace elements [pH 6.5]); 1.8% agar was added for solid medium (33). Additional supplements were added depending on strains auxotrophic markers (i.e., pyridoxine HCl, p-aminobenzoate, riboflavin HCl, arginine, uridine, and uracil) (34). Strains were incubated at 30°C unless otherwise indicated.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotypea | Source or referenceb |
|---|---|---|
| pFNO3 | ga5-gfp AfpyrG kan | FGSC (67) |
| A1145 (TN02A7) | pyrG89; pyroA4; nkuA::argB; riboB2 | FGSC (67) |
| A1147 (TNO2A25) | pyrG89; argB2; pabaB22nku::argB; riboB2 | FGSC (67) |
| A850 | biA1; _argB::trpC_B; methG1; veA1; trpC801 | FGSC |
| A773 | pyrG89; wA3; pyroA4 | FGSC |
| ARL115 | pyrG89 aspB::aspB-gfp-AfpyrG; argB2; pabaB22; nku::argB; riboB2 | This study |
| ARL144 | pyrG89 aspB::AfpyrG; pyroA4; nkuA::argB; riboB2 | This study |
| ARL148 | pyrG89 aspD::AfpyrG; pyroA4; nkuA::argB; riboB2 | This study |
| ARL157 | aspC::AfpyrG; pyrG89; pyroA4 | 38 |
| ARL162 | aspC::AfpyrG; aspA::argB2; pyrG89; pyroA4; biA1_argB::trpC_B; veA1; trpC801 | 38 |
| AYR1 | pyrG89 aspB::AfpyrG; pyroA4; riboB2 | This study |
| AYR6 | pyrG89 aspB::aspB-gfp-AfpyrG; argB2 | This study |
| AYR10 | aspB::aspB-gfp-AfpyrG\\ | This study |
| AYR20 | aspB::aspB-gfp-AfpyrG; aspA::argB2 | This study |
| AYR21 | aspB::aspB-gfp-AfpyrG; aspA::argB2 | This study |
| AYR22 | aspB::aspB-gfp-AfpyrG; aspA::argB2 | This study |
| AYR23 | aspB::aspB-gfp-AfpyrG; aspC::AfpyrG | This study |
| AYR24 | aspB::aspB-gfp-AfpyrG; aspC::AfpyrG | This study |
| AYR25 | aspB::aspB-gfp-AfpyrG; aspC::AfpyrG; aspA::argB2 | This study |
| AYR26 | aspB::aspB-gfp-AfpyrG; aspE::AfpyrG | This study |
| AYR27 | aspB::aspB-gfp-AfpyrG; aspE::AfpyrG | This study |
| AYR32 | aspB::AfpyrG; pyroA4; argB2 | This study |
| AYR35 | aspB::aspB-gfp-AfpyrG; pyroA4 | This study |
| AYR43 | aspD::AfpyrG | This study |
| AYR45 | aspB::aspB-gfp-AfpyrG; nkuA::argB; aspD::Afpyr | This study |
| AYR64 | aspB::aspB-gfp-AfpyrG; pyroA4; aspD::pyrGAf | This study |
| AYR65 | aspB::aspB-gfp-AfpyrG; pyroA4; aspD::pyrGAf | This study |
| ASH26 | aspA::argB2 pyrG89 wa3argB::trpC_B methG1 pyroA4 | 38 |
| ASH41 | aspE::AfpyrG; riboB2 | This study |
The symbol “\\” indicates haploids fused to make a diploid strain.
FGSC, Fungal Genetics Stock Center, Department of Microbiology, University of Kansas Medical Center (Kansas City, KS).
Gene targeting and tagging.
Septin gene replacements with AfpyrG and septin proteins fused with gfp were constructed as described by Yang et al. (67). We amplified ∼2 kb upstream and downstream of aspB from A. nidulans genomic DNA strain A850 and amplified the AfpyrG marker from the plasmid pFNO3 and using the primers listed in Table 2. AccuPrime Pfx DNA polymerase was used to amplify and fuse DNA fragments as described by Szewczyk et al. (59). PCR products were separated on a 1% agarose gel and purified using the QIA quick gel extraction kit (Qiagen, Inc., Valencia, CA). A standard A. nidulans protoplast transformation protocol was used to transform ΔnkuA strains (Table 1) (59, 67–68). Transformants were selected by auxotrophic markers and verified by PCR as described in Yang et al. (67) and confirmed by Southern blot analyses (9). All PCR and Southern blot primers were designed to amplify products of different sizes for both positive and negative results, and the untagged wild-type strain A850 was used as a control. Genetic crosses were used to recover nkuA+ strains of interest (30). Strains were crossed to wild-type strains A773 and A850. aspB/aspB-gfp heterozygous diploids were constructed as previously described (30) using AYR6 (haploid aspB-gfp) and wild-type A850 as parents.
Table 2.
Primers for constructing aspB-gfp and ΔaspBstrains
| Primer | Sequence (5′–′3) |
|---|---|
| For aspB GFP tag | |
| AspB-GSP1 | CAGAAGGTGCAACCTGTTCAGGGGAACTTAC |
| AspB-GSP2 | ACGAAGAGAGAATCCCTTCCTCTTTCCCTTTTC |
| AspB-GFP1 | GGAAAGAGGAAGGGATTCTCTCTTCGTGGAGCTGGTGCAGGCGCTG |
| AspB-GFP2 | CGGGGTTTCCGACTAAGCGTCTGTCTGTCTGAGAGGAGGCACTGATGCG |
| AspB-GSP3 | ACAGACGCTTAGTCGGAAACCCCGACGGTC |
| AspB-GSP4 | GATACTGAACGTTCTCATCGCCCGCAAGC |
| AspB-SSP3 | GCGTCGATGCTAAGAATTAGCTTCCC |
| AspB-SSP4 | CGAGATCCATGCTAGCGTCATAGTAC |
| For aspB deletion | |
| AspB-Up-F | CTGTTCAATTGGATACTGCCGAG |
| AspB-Up-R | GAAGATGGAGTCAGCAGCTGTATAGG |
| AspB pyrG1 | GCCTATACAGCTGCTGACTCCATCTTCTGCCTCAAACAATGCTCTTCACCCTC |
| AspB pyrG2 | GTGGAGAATCAAACGTAGAAGTTCCAATAAGTGTCTGAGAGGAGGCACTGATGCG |
| AspB-Down-F | CTTCTACGTTTGATTCTCCACG |
| AspB-Down-R | CTACAGGATGACACCCAGTCAG |
| AspBKOck-up | GGTCATTCCTGGTGTGACAGTACC |
| pyrgAFcheck Rv | CAGAGCCCACAGAGCGCCTTGAG |
Microscopy.
Conidia were harvested from agar plates with sterile water. For fungal observation and characterization, 104 or 106 spores were grown in 10 ml of MM or CM liquid in a petri dish containing a glass coverslip and incubated at 30°C. To examine conidiophores, a flame-sterile coverslip was put over a water agar plate, followed by a block of MM or CM plus supplements. The block was inoculated with fungal spores in water and a flame-sterile coverslip was placed on top. Unsealed plates were incubated for 1, 2, and 3 days at 30°C. Calcofluor Blankophour BDH was used to stain septa, and Hoechst 33342 (for dormant conidia) and Hoechst 33258 bis-benzimide were used to stain nuclei (46). For in vivo GFP observations, strains were incubated as described above until the appropriate time point at 30°C, and coverslips with attached fungus were not fixed but mounted on slides in 8 to 10 μl of liquid media. The incubation times for examining fungal development at 30°C were as follows: isotropic growth, 4 to 5 h; germ tube emergence, 6 to 7 h; hyphal elongation and septation, 9 to 11 h; and branching, 12 to 16 h. Fungal hyphae were viewed by using a Zeiss Axioplan microscope with a Plan-Neofluar 100×/1.30 NA oil immersion objective lens and the X-Cite fluorescence illumination system (EXFO) and were digitally photographed using a Zeiss Axiocam MRc charge-coupled device (CCD) camera and software. Photoshop CS3 was used to combine light images with fluorescence images and for micrograph organization, improved contrast, and brightness and/or hue saturation coloration.
Confocal microscopy.
Strains were prepared as described above. A Leica SP2 spectral confocal scanning laser microscope was used with a 63× HCX PL APO 1.20 W CORR water immersion objective lens. The Ar/HeNe laser was used for GFP excitation with a wavelength of 488 nm (20%) and filter RT 30/70. Images were acquired with the AxioCam CCD (Zeiss) camera and Leica Confocal LCS Lite software. Image collection and Z-stacks were calculated by the software based on the observed fluorescence of each sample. The beam expander was set to 3, and the scanning setting was 1,024 × 1,024. Reference images were taken with white light microscopy. Line and frame average varied on the GFP background, but in general we used a line average of 1 to 4 and a frame average 2 to 6, and Q-Lut was used as a reference to avoid collection of saturated pixels. Maximum projection was used in the LS software to combine Z-stack images.
Time-lapse microscopy.
For in vivo GFP examination, strains were prepared as described above, but a Gene Frame (Thermo Scientific) adhesive was used prior to mounting. This adhesive was mounted onto slides making a small well to which 100 μl of MM liquid medium was added, and coverslips with adhering fungus were floated on top. An Olympus IX-71 inverted fluorescence microscope with the Olympus 100×/1.35 NA U Plan APO objective lens was used. Images were acquired with the Photometrix Cool Snap HQ CCD camera. Delta Vision Experiment Designer software was used to collect time-lapse data and movies. The time lapse was set for collection of GFP and DIC simultaneously with a time frame of 5 s, but due to the exposure and dual image collection time lapse frames varied from 5 to 10 s. We used flat-field calibration and when focus was lost it was adjusted manually. The data was analyzed using differential interference contrast (DIC) microscopy as reference and movies were edited with Windows Movie Maker to show data from the same focal plane.
RESULTS
aspB is nonessential but is required for normal growth and morphology.
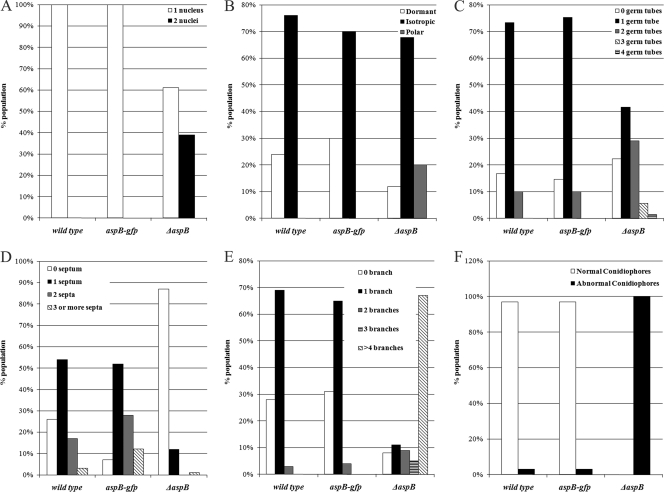
We previously reported that the A. nidulans septin aspB was essential based on failure to recover gene replacement strains in multiple experiments (47). Recently, using newly available fusion PCR techniques and A. nidulans ΔnkuA strains (67), we recovered ΔaspB::AfpyrG transformants. Using PCR and DNA hybridization, we verified that the aspB gene had been replaced with AfpyrG in these strains and that there were no extra insertions of the disruption cassette (data not shown). After crossing out the ΔnkuA mutation, we analyzed the early growth of a ΔaspB mutant by DIC microscopy and Hoechst 33342 staining of nuclei (the strains are described in Table 1). We used plate growth assays to test the ΔaspB mutant for sensitivity to the cell wall perturbing agent Calcofluor and to elevated temperature (42°C) but found no obvious difference compared to the wild-type strain (data not shown).
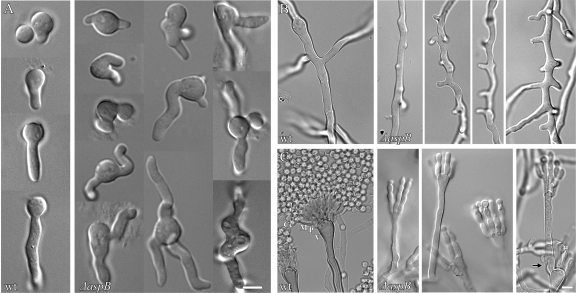
Freshly harvested wild-type conidia all measured ∼3 μm in diameter, as expected (45, 61). In contrast, ΔaspB conidia ranged from approximately 3 to 5 μm (data not shown). As expected, wild-type (wt) conidia were all uninucleate (n = 200). However, ΔaspB conidia were both uninucleate (61%) and binucleate (39%), and the binucleate conidia were generally larger (n = 200) (Fig. 1A and data not shown). To determine whether aspB has roles in the establishment of polarity and germ tube emergence, we compared early development of wt and ΔaspB strains. After 4 h of incubation at 30°C, all wt cells were still isotropic, showing no sign of polarity (0%, n = 300). In contrast, 20% of ΔaspB cells showed the tear-drop shape, indicating polarization just before germ tube emergence (n = 300) (Fig. 1B). After 6 h of incubation, 72% of wt cells formed a single, straight germ tube, with 10% forming two germ tubes (n = 200). In contrast, 29% of ΔaspB cells formed two bent or kinked germ tubes and another 5% of ΔaspB cells formed three or four bent or kinked germ tubes (n = 200) (Fig. 1C and Fig. 2A).
Fig 1.
aspB is required for normal growth and morphology. (A) The ΔaspB mutant forms uninucleate and binucleate spores. Dormant spores from wt, aspB-gfp, and ΔaspB strains were stained with Hoechst 33342, and the numbers of nuclei were counted (n = 200). (B) The ΔaspB mutant breaks dormancy earlier than the wild-type strain. Spores were incubated for 4 h at 30°C and categorized as dormant (∼2.5 μm), isotropic (∼5 μm), and polar (presence of a germ tube) (n = 200). (C) The ΔaspB mutant forms multiple germ tubes. Spores were incubated for 6 h at 30°C, and the numbers of germ tubes were counted (n = 300). (D) The ΔaspB mutant delays septation. Spores were incubated for 11 h at 30°C, and the number of septa were counted (n = 200). (E) The ΔaspB mutant hyperbranches. Spores were incubated for 14 h at 30°C, and the numbers of branches per compartment delineated by two septa were counted (n = 200). (F) The ΔaspB mutant forms abnormal conidiophores. Spores were incubated in agar between coverslips for 2 days at 30°C. Conidiophores were categorized as normal if all layers were present and abnormal if layers were absent or aberrant (n = 200).
Fig 2.
The ΔaspB mutant shows a hyper-emergence of growth and abnormal conidiophores. (A) The ΔaspB mutant forms multiple germ tubes, whereas the wild-type strain forms one germ tube. (B) The ΔaspB mutant forms multiple abnormal and stunted branches per compartment, whereas the wild-type strain forms one branch per compartment. (C) The ΔaspB mutant forms disorganized conidiophores. An arrow denotes a new aerial hypha arising from a vesicle. The wild-type strain forms many conidia, and the conidiophores are organized with different layers (V, vesicle; P, phialide; M, metulae; Cc, conidial chain). Conditions for growth were as described in Fig. 1. Scale bar, 5 μm.
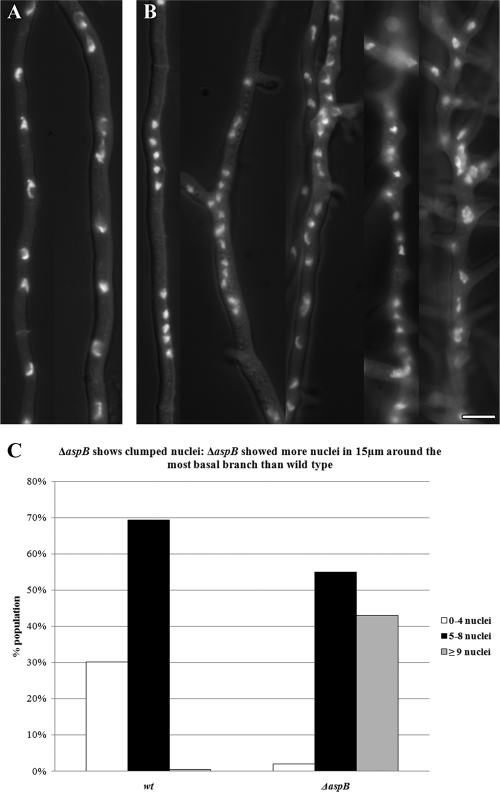
In wt A. nidulans, septation gives rise to an actively growing tip compartment and inactive subapical compartments that later initiate polar growth by branching, typically forming one branch per compartment (19, 21). After 11 h of incubation at 30°C, 71% of wt hyphae had 1 to 2 septa (n = 200), while only 12% of ΔaspB germlings had septa (Fig. 1D). However, at 14 h, virtually all ΔaspB cells had septa, indicating that septation was delayed but not absent in the deletion strain. As expected, after 11 to 14 h of incubation at 30°C, the wt strain showed branches that were uniform tubes of ∼5 μm in diameter with a single branch in most (69%) compartments (n = 200). In contrast, the ΔaspB mutant made branches that were often short, thin (2 to 2.5 μm in diameter) and hooked (67%) with two or more branches seen in 80% of compartments (Fig. 1E and Fig. 2B). Even after 16 to 18 h of incubation at 30°C, most ΔaspB branches remained thin, short, and hooked, indicating a defect in branching rather than a simple delay (data not shown). In wt A. nidulans, nuclei are distributed evenly along the hypha (66). To compare nuclear distribution in wt versus ΔaspB strains, we observed nuclei in a 15-μm zone bisected by the most basal branch. After 14 to 16 h of incubation at 30°C, nuclei in the wt strain were evenly spaced along the hypha and relatively uniform in size, with 70% of hyphae containing five to eight nuclei in the first basal branch region. In contrast, ΔaspB nuclei frequently clumped and appeared smaller and more irregular, with 42% showing more than eight nuclei in the first basal branch region (Fig. 3).
Fig 3.
The ΔaspB mutant shows clumped nuclei. (A) Wild-type nuclei positioned along hyphae. (B) The ΔaspB mutant results in clumped nuclei, particularly near branches. (C) The ΔaspB mutant shows clumped nuclei. The ΔaspB mutant showed more nuclei in 15 μm around the most basal branch than did the wild-type strain. To delineate the area for nuclear counts, the most basal branch was identified; 7.5 μm from the center of the branch to the left and right of the branch was measured (total area = 15 μm; n = 200). Spores were incubated for 14 to 16 h at 30°C, and nuclei were stained with Hoechst 33258. Scale bar, 5 μm.
In wt A. nidulans, asexual reproduction occurs by the production of chains of asexual spores (conidia) on specialized multilayered conidiophores, with each layer emerging in a process that is similar to yeast budding (45). As expected, after 2 days of incubation at 30°C virtually all wt conidiophores were morphologically normal with multiple discrete, uniform, organized layers and long chains of conidia (97%, n = 200). In contrast, conidiophores in the ΔaspB strain showed a wide range of phenotypes, although all were morphologically abnormal with disorganized layers that formed much shorter chains of conidia (n = 200), (Fig. 1F and Fig. 2C).
AspB forms rings, bars, and filaments throughout vegetative growth and conidiophore production.
To characterize AspB localization in live cells, we constructed a strain in which a single copy of aspB-gfp driven by the native aspB promoter replaced the native aspB gene. Because diploid cells are larger and better for microscopy and to further reduce the concentration of GFP-tagged AspB in the cell, we also constructed a heterozygous diploid strain carrying a single copy of aspB-gfp and a single copy of wt aspB. In both the haploid aspB-gfp strain and the heterozygous diploid aspB-gfp/aspB strain, conidial nuclear number, polarity establishment, germ tube emergence, septation, branching, and conidiophore formation were nearly identical to the wild type, showing that the GFP tag does not interfere with normal function (Fig. 1A to F). We visualized AspB-GFP localization in live cells throughout development using fluorescence microscopy.
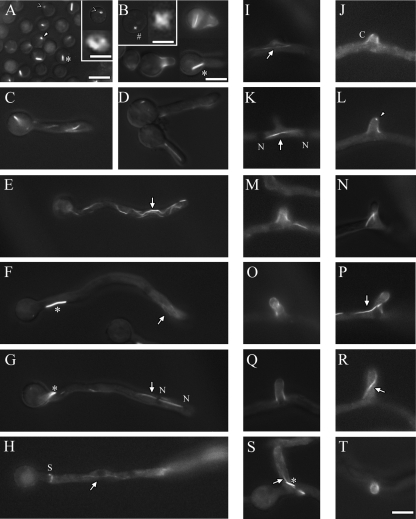
In freshly harvested conidia AspB-GFP localized as rings, dots and bars that appeared to be about 1 μm thick with variable lengths (Fig. 4A). As the spores began to swell, AspB-GFP also localized as small X-shaped structures (Fig. 4B, inset). AspB-GFP localized as a cap in emerging germ tubes and as a collar at the base of germ tubes (Fig. 4B). Bars were also seen in newly formed germ tubes (Fig. 4B). As germ tubes continued to extend, AspB-GFP filaments that appeared thinner and longer than bars were also observed (Fig. 4C to H). In older hyphae, tip localization was mostly as filaments, subapical localization was mostly as bars (Fig. 4F and G), and AspB-GFP localized as a ring at septa during septum formation (Fig. 4H) (64). During branch emergence AspB-GFP localized as filaments perpendicular to and just below nascent branches of <2.5 μm in length (Fig. 4I and K). As branches extended to ∼3 μm in length, the perpendicular filaments disappeared and AspB-GFP filaments were seen within branches (Fig. 4M, N, P, and R). In newly formed branches, AspB-GFP localized as a cap (Fig. 4J, L, M, O, Q, and T) or a single bright dot at branch tips (Fig. 4L). AspB-GFP bars and/or filaments were also seen in mature branches. A low level of cytoplasmic AspB-GFP was also seen in all developmental stages. This cytoplasmic signal was excluded from nuclei and the extreme apices (∼0.5 μm) of hyphae and branches.
Fig 4.
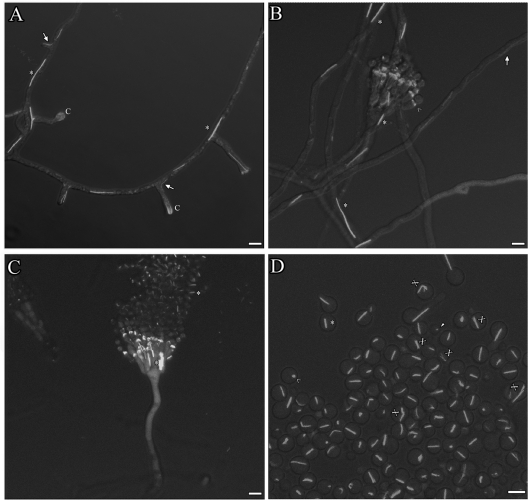
AspB forms rings, dots, bars, and filaments. The AspB-GFP strain was incubated at 30°C for 4 to 5 h to view isotropic growth, 6 to 7 h to view early germ tube emergence, 9 to 11 h to view hyphal elongation and septation, and 12 to 16 h to view branching. (A) AspB forms bars (*), rings (<), and dots (arrowhead) in dormant and germinating spores. The inset shows an enlarged view of a ring (scale bar, 0.5 μm). (B to D) AspB forms “X's” (#) in addition to bars, rings, and dots as conidia swell. The inset shows an enlarged view of an “X” (scale bar, 0.75 μm). AspB forms caps and collars as the germ tube emerges, bars that localize to conidia and filaments (arrows) that localize to tips. (E to G) AspB forms bars that localize subapically and filaments that localize to tips as the hypha extends. Cytoplasmic GFP fluorescence is excluded from nuclei (N). (H) AspB forms rings at septa. (I to R) AspB forms filaments in branching compartments, caps as the branch emerges, and dots at the tips of branches. Filaments localize to newly formed branches. (S) Filaments and bars localize to longer branches. (T) Top view of a branch cap. Scale bar, 5 μm.
During conidiogenesis, we observed AspB-GFP localization at several conidiophore layers by confocal microscopy (Fig. 5). AspB bars and filaments localized to the main hypha at the base of the aerial hypha (Fig. 5A). AspB-GFP localized as a cap while the vesicle was swelling and then as rings through which each layer emerged. Both the vesicle cap and the rings disappeared once new growth emergence was complete in that area (Fig. 5B). AspB-GFP bars were observed in conidia attached to the conidiophore as part of a chain and in single conidia that were no longer attached to chains (Fig. 5C and D). In these free conidia, the ends of some AspB-GFP bars appeared to be slightly out of register, giving the impression of being frayed (Fig. 5D).
Fig 5.
AspB forms bars, filaments, caps, and rings during conidiophore formation. (A) AspB-GFP localization at several stages of early conidiophore development. AspB-GFP forms bars (*) and filaments (arrow) in conidiophore stalks. AspB-GFP forms caps (C) in aerial hyphae. AspB-GFP caps are diffuse in swollen vesicles. (B) AspB-GFP remains localized at the phialide-conidium interface as conidia form. AspB-GFP forms rings (>) at the base of budding layers. (C) Conidial chains attached to conidiophores show AspB-GFP localization as dots and bars. (D) AspB-GFP forms bars on conidia freshly detached from conidiophores. Some bars looked “frayed” (X) at the ends. Spores were incubated in agar between coverslips for 1, 2, and 3 days at 30°C and viewed by confocal microscopy. Scale bar, 5 μm.
AspB-GFP bars and filaments show dynamic movement.
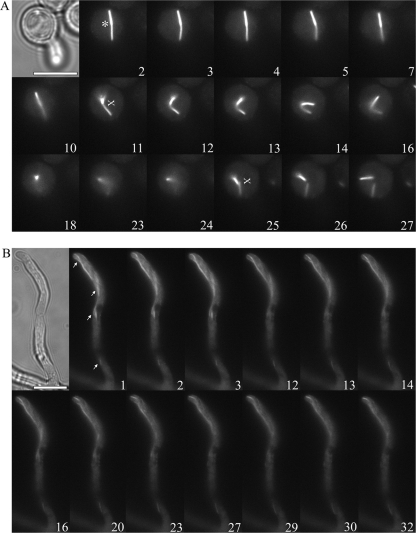
Septins have been reported to be dynamic, changing conformational arrangements, with subunits being recycled from one structure to another during development (13, 26, 35, 40). To determine whether AspB-GFP bars show dynamic movement in A. nidulans, we used time-lapse fluorescence microscopy (see Movies S1 and S2 in the supplemental material). During isotropic growth and early germ tube emergence, AspB-GFP bars moved freely within the conidium and/or bounced back and forth from the tips of germ tubes. Many bars appeared to have one end anchored in the region of the membrane, while the other end moved in and out of the plane of focus. For example, Fig. 6A shows a bar that appeared to have one end anchored and one end oscillating back and forth (Fig. 6A, time frames 2 to 7). Eventually, both ends began to move freely, followed by an apparent break into two pieces that moved independently in and out of the plane of focus (Fig. 6A, time frames 10 to 27). In contrast to the rapid large-scale movements of AspB bars in cells with young germ tubes, AspB filaments in longer hyphae showed more restricted movement. Figure 6B shows the slower gradual movement in and out of the plane of focus of a long filament that extends through the hypha into the branch tip.
Fig 6.
Motion of AspB bars and filaments. (A) AspB-GFP bar (*) in a germinating conidium moves away from the cell periphery, oscillates in and out of the plane of focus, and breaks in two. “Frayed” ends (X) can be observed. (B) AspB-GFP filaments (arrow) show slower movement than bars at the hyphal tips. Cytoplasmic GFP fluorescence is excluded from nuclei. Time lapse fluorescence microscopy was used. Numbers represent time lapse frames from Movies S1 and S2 in the supplemental material. Frames are 5 to 10 s apart. Conditions for growth as in Fig. 4. Scale bar, 5 μm.
All AspB rings, bars, and filaments are lost in the absence of AspA and AspC, and AspB bars and filaments are lost in post-septation hyphae in the absence of AspE.
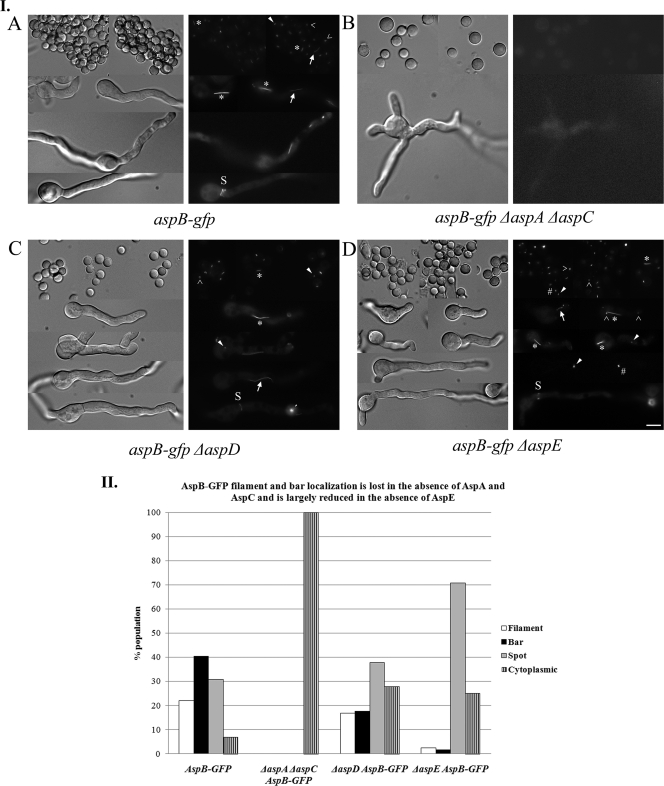
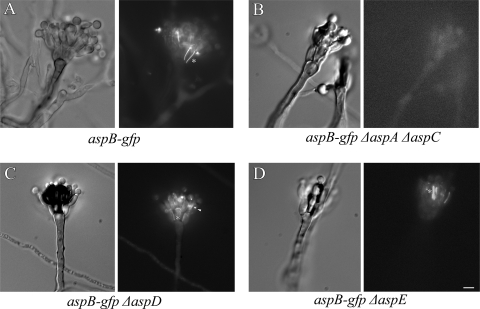
We recently showed that in A. nidulans septins AspA and AspC interact as AspA-GFP forms small, abnormal structures in ΔaspC strains, while AspC-GFP does not localize in ΔaspA strains (38). To determine whether AspB interacts with other septins, we crossed the aspB-gfp strain to ΔaspA, ΔaspC, ΔaspA ΔaspC, ΔaspD, and ΔaspE strains and selected progeny carrying aspB-gfp and the appropriate septin deletion. Progeny were checked by PCR to verify the aspB-gfp cassette was present, and the appropriate septin was deleted. Strains were characterized throughout vegetative growth and asexual reproduction. AspB-GFP rings, bars, and filaments were lost in all stages of development and reproduction in ΔaspA, ΔaspC, and ΔaspA ΔaspC strains, with only cytoplasmic localization detected (Fig. 7IB and II, Fig. 8B, and data not shown). In the ΔaspD strain, AspB-GFP localization to septa, conidiophores, and conidia was normal (Fig. 7IC and Fig. 8C), while AspB-GFP bars were present only half as often, and filaments were slightly reduced relative to the wt strain (Fig. 7IC and II). In the ΔaspE strain, AspB-GFP bars, rings, and filaments were present through early germ tube emergence as in the wt strain, and localization to septa and conidiophores appeared to be normal (Fig. 7ID and Fig. 8D). However, in post-septation ΔaspE hyphae the localization was mostly as dots and/or X's with very few of the bars and filaments that are seen in the wt strain at this stage (Fig. 7ID and II).
Fig 7.
AspB structures are lost in the absence of AspA and AspC and filaments and bars are lost in hyphae in the absence of AspE. (I) AspB-GFP structures in septin deletion backgrounds. (A) AspB-GFP structures. Rings (>), dots (arrowhead), bars (*) filaments (arrow), and at septa (S) are indicated. (B) All AspB-GFP structures are lost in the ΔaspA ΔaspC mutant. (C) All AspB-GFP structures are found in ΔaspD strains. (D) All AspB-GFP structures are found in early ΔaspE cells but are mostly seen as dots and/or “X's” in hyphae. Conditions for growth were as described in Fig. 4. Scale bar, 5 μm. (II) AspB-GFP structures found in septin deletion backgrounds. Spores were incubated for 10 h at 30°C, and AspB structures were classified as filament, bar, and spot (rings, dots, and X's) and cytoplasmic (n = 100). Scale bar, 5 μm.
Fig 8.
AspB bars are lost in conidiophores in the absence of AspA and AspC. (A) AspB-GFP bars (*) in conidiophores. (B) AspB-GFP bars are lost in conidiophores in the ΔaspA ΔaspC mutant. (C) AspB-GFP bars and dots (arrowhead) in the ΔaspD mutant. (D) AspB-GFP bars and dots in the ΔaspE mutant. Conditions for growth were as described as in Fig. 5. Scale bar, 5 μm.
DISCUSSION
The Cdc3 ortholog AspB is not essential and plays a role in restricting emergence of new growth foci.
The budding yeast S. cerevisiae has four core septins, Cdc3, Cdc10, Cdc11, and Cdc12, along with three other septins that play roles in different developmental stages and mutant backgrounds. The core septins typically form rings at the mother-daughter neck, and their functions include organizing the division site and preventing diffusion of mRNAs and proteins. Although there is still some controversy about exactly how the core septins are arranged at the mother-daughter neck, their ability to form continuous filaments appears to be essential for localization to the membrane as visible rings and for their function as diffusion barriers (43). Recently, septins have been characterized in several filamentous fungi where they play a variety of roles and take on a range of morphologies and localizations (11, 24, 49). The filamentous fungus A. nidulans has five septins (AspA to AspE), one representative of each of the S. cerevisiae core septins and a single septin found only in filamentous fungi (AspA is orthologous to Cdc11, AspB is orthologous to Cdc3, AspC is orthologous to Cdc12, and AspD is orthologous to Cdc10; AspE is found only in filamentous fungi.) (48, 52).
We previously reported that aspB, the CDC3 ortholog, was essential in A. nidulans (47) and that a temperature-sensitive mutant of aspB (aspB ts) made septa that weakly labeled with the chitin-binding reagent Calcofluor and had a slightly higher number of branches at restrictive temperature (64). In the present study we created an aspB-null allele (ΔaspB) taking advantage of the recent development of fusion PCR and ΔnkuA strains to enhance gene replacement by homologous integration (59, 67). In addition to less effective methods for gene replacement, our earlier efforts to isolate the ΔaspB mutant were likely confounded by a strategy that depended on isolating conidiating colony sectors from an aspB/ΔaspB::argB diploid since our present results show that ΔaspB strains have greatly reduced conidiation.
In our previous work we postulated that the ability of the aspB ts mutant to make septa at restrictive temperature indicated that this mutant allele might have partial function. To our surprise, the ΔaspB-null allele described here was also able to complete septation, although more slowly than the wild type. The more rapid loss of Calcofluor label at septa in the ΔaspB mutant versus the wild type was also similar to that observed in the aspB ts mutant. Our results suggest that AspB is not essential for septation, but that its absence results in abnormal septal organization, though ultrastructural studies are needed to clarify this point.
We observed early polarization and a more dramatic increase in the emergence of germ tubes and branches in the ΔaspB strain compared to our earlier observations of the aspB ts mutant. In the ΔaspB mutant, as well as ΔaspA and ΔaspC mutants, we also observed abnormal binucleate conidia and abnormally spaced clusters of nuclei in hyphae (Fig. 1 and 3) (57, 65). In S. cerevisiae, septin mutations result in an aberrant bud emergence pattern, chains of elongated buds, and multinucleated buds from continued nuclear division (25, 31). The early and increased emergence of growth foci in the form of germ tubes and branches in the ΔaspB mutant suggests that A. nidulans septins might play a role in selecting new growth sites similar to the role of S. cerevisiae septins in bud site selection (25). However, since yeast septin mutants do not display the simultaneous emergence of multiple buds, AspB also appears to have a role in restricting new growth foci not seen in S. cerevisiae. A. nidulans ΔaspA and ΔaspC strains also show the emergence of extra germ tubes and branches, as does a septin deletion mutant in another filamentous fungus, U. maydis (1, 38). Thus, the restriction of new growth foci might be a common function for septins in other filamentous fungi.
AspB forms prominent bars and filaments in addition to rings.
We previously reported that AspB localized as rings to forming septa, branches and emerging layers of conidiophores based on immunofluorescence using polyclonal anti-AspB antibodies (64). We report here that in addition to the previously observed rings, A. nidulans AspB also forms prominent bars and filaments based on localization of AspB-GFP. The most obvious explanation for the additional septin structures seen here is that the AspB bars and filaments are artifacts resulting from perturbations in expression and/or the addition of the GFP tag. We do not think this explanation is correct for several reasons. First, we constructed strains so that the aspB-gfp fusion replaces the native aspB gene at the endogenous locus and is driven by the native aspB promoter. Thus, our aspB-gfp fusion is expressed at levels as close to wild-type aspB as possible. Second, all mutant phenotypes of the ΔaspB mutant were rescued in the aspB-gfp strain and in a heterozygous aspB-gfp/aspB diploid (Fig. 1). Thus, AspB-GFP is fully functional. Third, the absence of the Cdc11 ortholog AspA or the Cdc12 ortholog AspC eliminated all AspB-GFP localization, suggesting the formation of a multi-septin core complex similar to that found in S. cerevisiae. Fourth, AspB-GFP bars and filaments were not found in post-septation hyphae of the ΔaspE mutant, and bars were absent from regions undergoing branching (data not shown), suggesting developmental regulation of AspB bars and filaments. Such developmental regulation seems unlikely for artifactual structures. Finally, there have been recent reports of similar septin bar and filament structures in other filamentous fungi. In C. albicans Cdc10 septin filaments were observed during the final stage of chlamydospore morphogenesis (42), and Cdc3 and Cdc10 filaments were observed in a small percentage of C. neoformans dikaryotic hyphae (36). In A. gossypii, a series of short bars has been observed to form the mature septin ring (15). In U. maydis the Cdc10 filaments extend from pole to pole in the budding stage and in A. fumigatus AspD and AspE septins form visible filaments (1, 32).
There are several reasons that we might have failed to observe AspB bars and filaments in our earlier immunofluorescence work. In our previous study, the affinity-purified polyclonal antibodies were of such low titer that they had to be used undiluted for immunofluorescence, and we used a less intense light source and a less sensitive camera than in the present study. A more interesting possibility than these trivial explanations is that the experimental manipulations required for immunofluorescence might have caused the bar and filament subclasses of septins to be disrupted. Immunofluorescence of cytoplasmic proteins in fungi requires enzymatic digestion of the fungal cell wall for the antibodies to reach their target epitopes. If septin bars and filaments depend on the cell wall to help stabilize them in some way, but septin rings do not, then cell wall digestion would be expected to eliminate only these subclasses of septin structures. Indeed, a connection between the fungal cell wall and the septins would be consistent with a recently postulated role for septins in cortical stability in animal cells (23).
Because septin bars are absent from yeast and yeast septin mutants do not display the emergence of extra growth foci, we speculate that septin bars might have a role in suppressing new growth. All freshly harvested, dormant A. nidulans conidia contained a prominent AspB bar. Thus, the AspB bar is present before growth initiation, as would be expected if it functions to prevent germ tube emergence. Interestingly, in post-septation hyphae AspB bars generally were seen in quiescent subapical regions, whereas AspB filaments were observed in actively growing areas, including branching compartments and emerging branches. We only rarely observed bars and filaments within the same compartment of post-septation hyphae (data not shown). Similarly, during conidiophore development, AspB bars were only seen in quiescent regions where conidiophore layers were not actively budding. The presence of septin bars in dormant spores and quiescent regions of hyphae and conidiophores is consistent with these bars playing a role in restricting the emergence of new growth foci. It seems plausible that cortical rigidity maintained via some sort of septin-cell wall connection might be needed to prevent the inappropriate protrusion of new growth foci from the cell.
Model of A. nidulans septin interactions.
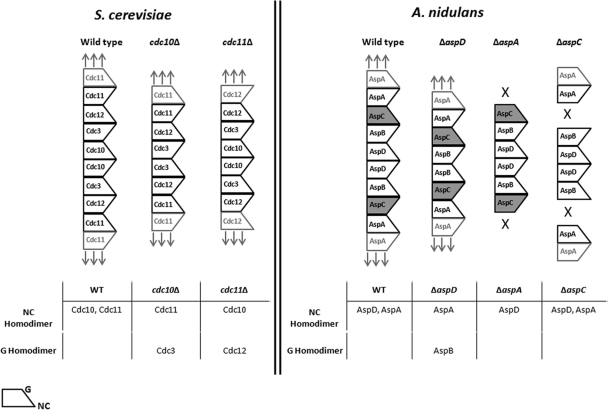
Structural studies of human septins have shown that individual septins interact with each other via two different interfaces termed the G interface and the NC interface (55, 56). In S. cerevisiae the core septins assemble into a nonpolar hetero-octamer rod in the order: Cdc11-Cdc12-Cdc3-Cdc10-Cdc10-Cdc3-Cdc12-Cdc11 (6) (Fig. 9). At the center of the octamer rod Cdc10 forms a homodimer through its NC interface. At the ends of the octamer rod Cdc11 forms homodimers through its NC interfaces to allow end to end association of septin rods into longer structures (Fig. 9). Using an elegant combination of deletion and site-directed mutants, McMurray et al. (43) demonstrated that in the absence of the central Cdc10 septin, the newly exposed Cdc3 septin homodimerizes by its G interface, allowing an alternative septin structure to be made. Similarly, in the absence of the terminal Cdc11, the newly exposed Cdc12 homodimerizes by its G interface allowing a different alternative septin linear structure to be made.
Fig 9.
Model of septin interactions in Aspergillus nidulans. (Left panel) Order of septin subunits in S. cerevisiae (adapted with permission from reference 6; additional data are from reference 43). (Right panel) Postulated order of septin subunits in A. nidulans. The wide end of septin subunit represents the NC interface; the narrow end represents the G interface. Boldface text denotes a single heteropolymer septin rod. The lighter text represents neighboring rods. Arrows indicate that septin polymer can be extended. X's indicate that septin polymer cannot be extended. Dark shading of AspC indicates that it differs from its S. cerevisiae ortholog Cdc12 because it is unable to form homopolymers via its G interface. See the text for further details.
A. nidulans has a single ortholog of each of the core S. cerevisiae septins along with a septin found only in filamentous fungi (AspE) (52). Based on orthology with S. cerevisiae septins and the AspB-GFP localization we observed in septin deletion strains, we propose the following model (Fig. 9).
We propose that the A. nidulans core septins assemble with each septin in the same position and interacting via the same NC or G interface as its S. cerevisiae ortholog. The resulting hetero-octamer rod would have the order AspA-AspC-AspB-AspD-AspD-AspB-AspC-AspA, with AspD in the central position and AspA in the terminal position, both forming homopolymers through their NC interfaces. We also propose that in the absence of AspD, AspB forms a homopolymer through its G interface directly analogous to the ability of Cdc3 to form homopolymers via its G interface in the absence of Cdc10. This is consistent with our finding that all AspB-GFP structures are present in the ΔaspD strain (Fig. 7C). We further propose that, in contrast to its S. cerevisiae ortholog Cdc12, AspC is not able to dimerize via its G interface. Thus, in the ΔaspA mutant, no AspB-GFP structures are seen because the AspC-AspB-AspD-AspD-AspB-AspC heterohexamer rods cannot associate end to end to form larger visible structures, and in the ΔaspC mutant no AspB-GFP structures are seen because there is no connection formed between AspB and AspA (Fig. 7B and data not shown).
Unlike the core septins, AspE does not appear to be involved in the formation of septin higher-order structures until after septum formation. In the ΔaspE mutant, AspB-GFP localized to septin rings, filaments, and bars before septation; however, after septation, septin bars disappeared, although rings and filaments were still seen. In filamentous fungi, septation marks the transition from a unicellular to a multicellular state, and AspE only has orthologs in multicellular filamentous fungi and not in the unicellular yeasts. It is possible that AspE interacts with core septins to stabilize the longer bars seen post-septation. It is also possible that AspE acts to modify the cell cortex or some other cellular component after septation to allow the longer bars to form. Differences in the individual septins composing heteropolymers in different developmental states is not without precedent. In S. cerevisiae the non-core septins Spr3 and Spr28 are expressed only during sporulation (14, 51), and in C. albicans the non-core septin Sep7 has been shown to be required for hyphal but not yeast growth (28). Although it is not yet clear what biological role septin bars play in A. nidulans, it seems likely that they are involved in the transition from unicellular to multicellular growth.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the Robert Watkins American Society of Microbiology Fellowship, a Plant Biology Department Palfrey Grant, a Graduate Recruitment Opportunity Fellowship, and a Dissertation Completion Award from the University of Georgia to Y.H.-R. and by NSF grants MCB0211787 and IOS1051730 and support from the UGA Office of the Vice President for Research to M.M.
We thank Rebecca Lindsey for the construction of the aspB::aspB-gfp and aspB::AfpyrG strains.
Footnotes
Published ahead of print 13 January 2012
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1. Alvarez-Tabares I, Perez-Martin J. 2010. Septins from the phytopathogenic fungus Ustilago maydis are required for proper morphogenesis but dispensable for virulence. PLoS One 5:e12933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. An H, Morrell JL, Jennings JL, Link AJ, Gould KL. 2004. Requirements of fission yeast septins for complex formation, localization, and function. Mol. Biol. Cell 15:5551–5564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barral Y. 2008. Yeast septins: a cortical organizer, p 101–124 In Hall P, Russell H, Pringle J. (ed), The septins. John Wiley & Sons, Ltd, London, United Kingdom [Google Scholar]
- 4. Barral Y, Mermall V, Mooseker MS, Snyder M. 2000. Compartmentalization of the cell cortex by septins is required for maintenance of cell polarity in yeast. Mol. Cell 5:841–851 [DOI] [PubMed] [Google Scholar]
- 5. Berman J, Sudbery PE. 2002. Candida albicans: a molecular revolution built on lessons from budding yeast. Nat. Rev. Genet. 3:918–930 [DOI] [PubMed] [Google Scholar]
- 6. Bertin A, et al. 2008. Saccharomyces cerevisiae septins: supramolecular organization of hetero-oligomers and the mechanism of filament assembly. Proc. Natl. Acad. Sci. U. S. A. 105:8274–8279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bohmer C, Ripp C, Bolker M. 2009. The germinal centre kinase Don3 triggers the dynamic rearrangement of higher-order septin structures during cytokinesis in Ustilago maydis. Mol. Microbiol. 74:1484–1496 [DOI] [PubMed] [Google Scholar]
- 8. Boyce KJ, Chang H, D'Souza CA, Kronstad JW. 2005. An Ustilago maydis septin is required for filamentous growth in culture and for full symptom development on maize. Eukaryot. Cell 4:2044–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brown T. 1993. Analysis of DNA sequences by blotting and hybridization, p 1–2.9 In Current protocols in molecular biology, vol 2 John Wiley & Sons, Inc, New York, NY [Google Scholar]
- 10. Canovas D, Perez-Martin J. 2009. Sphingolipid biosynthesis is required for polar growth in the dimorphic phytopathogen Ustilago maydis. Fungal Genet. Biol. 46:190–200 [DOI] [PubMed] [Google Scholar]
- 11. Cao L, Yu W, Wu Y, Yu L. 2009. The evolution, complex structures and function of septin proteins. Cell. Mol. Life Sci. 66:3309–3323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Caviston JP, Longtine M, Pringle JR, Bi E. 2003. The role of Cdc42p GTPase-activating proteins in assembly of the septin ring in yeast. Mol. Biol. Cell 14:4051–4066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cid VJ, Adamikova L, Sanchez M, Molina M, Nombela C. 2001. Cell cycle control of septin ring dynamics in the budding yeast. Microbiology 147:1437–1450 [DOI] [PubMed] [Google Scholar]
- 14. De Virgilio C, DeMarini DJ, Pringle JR. 1996. SPR28, a sixth member of the septin gene family in Saccharomyces cerevisiae that is expressed specifically in sporulating cells. Microbiology 142(Pt 10):2897–2905 [DOI] [PubMed] [Google Scholar]
- 15. DeMay BS, Meseroll RA, Occhipinti P, Gladfelter AS. 2009. Regulation of distinct septin rings in a single cell by Elm1p and Gin4p kinases. Mol. Biol. Cell 20:2311–2326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dobbelaere J, Barral Y. 2004. Spatial coordination of cytokinetic events by compartmentalization of the cell cortex. Science 305:393–396 [DOI] [PubMed] [Google Scholar]
- 17. Dobbelaere J, Gentry MS, Hallberg RL, Barral Y. 2003. Phosphorylation-dependent regulation of septin dynamics during the cell cycle. Dev. Cell 4:345–357 [DOI] [PubMed] [Google Scholar]
- 18. Douglas LM, Alvarez FJ, McCreary C, Konopka JB. 2005. Septin function in yeast model systems and pathogenic fungi. Eukaryot. Cell 4:1503–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dynesen J, Nielsen J. 2003. Branching is coordinated with mitosis in growing hyphae of Aspergillus nidulans. Fungal Genet. Biol. 40:15–24 [DOI] [PubMed] [Google Scholar]
- 20. Estey MP, Kim MS, Trimble WS. 2011. Septins. Curr. Biol. 21:R384–R387 [DOI] [PubMed] [Google Scholar]
- 21. Fiddy C, Trinci AP. 1976. Mitosis, septation, branching, and the duplication cycle in Aspergillus nidulans. J. Gen. Microbiol. 97:169–184 [DOI] [PubMed] [Google Scholar]
- 22. Finger FP. 2005. Reining in cytokinesis with a septin corral. Bioessays 27:5–8 [DOI] [PubMed] [Google Scholar]
- 23. Gilden J, Krummel MF. 2010. Control of cortical rigidity by the cytoskeleton: emerging roles for septins. Cytoskeleton (Hoboken) 67:477–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gladfelter AS. 2010. Guides to the final frontier of the cytoskeleton: septins in filamentous fungi. Curr. Opin. Microbiol. 13:720–726 [DOI] [PubMed] [Google Scholar]
- 25. Gladfelter AS, Kozubowski L, Zyla TR, Lew DJ. 2005. Interplay between septin organization, cell cycle and cell shape in yeast. J. Cell Sci. 118:1617–1628 [DOI] [PubMed] [Google Scholar]
- 26. Gladfelter AS, Pringle JR, Lew DJ. 2001. The septin cortex at the yeast mother-bud neck. Curr. Opin. Microbiol. 4:681–689 [DOI] [PubMed] [Google Scholar]
- 27. Gladfelter AS, Sudbery P. 2008. Septins in four model fungal systems: diversity in form and function, p 125–146 In Hall P, Russell H, Pringle J. (ed), The septins. John Wiley, Ltd, London, United Kingdom [Google Scholar]
- 28. Gonzalez-Novo A, et al. 2008. Sep7 is essential to modify septin ring dynamics and inhibit cell separation during Candida albicans hyphal growth. Mol. Biol. Cell 19:1509–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hall PA, Finger FP. 2008. Septins and human disease, p 295–317 In Hall P, Russell H, Pringle J. (ed), The septins. John Wiley, Ltd, London, United Kingdom [Google Scholar]
- 30. Harris SD. 2001. Genetic analysis of ascomycete fungi, p 47–58 In Talbot NJ. (ed), Molecular and cellular biology of filamentous fungi: a practical approach. Oxford University Press, New York, NY [Google Scholar]
- 31. Hartwell LH. 1971. Genetic control of the cell division cycle in yeast. IV. Genes controlling bud emergence and cytokinesis. Exp. Cell Res. 69:265–276 [DOI] [PubMed] [Google Scholar]
- 32. Juvvadi PR, Fortwendel JR, Rogg LE, Steinbach WJ. 2011. Differential localization patterns of septins during growth of the human fungal pathogen Aspergillus fumigatus reveal novel functions. Biochem. Biophys. Res. Commun. 405:238–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kafer E. 1977. Meiotic and mitotic recombination in Aspergillus and its chromosomal aberrations. Adv. Genet. 19:33–131 [DOI] [PubMed] [Google Scholar]
- 34. Kaminskyj S. 2001. Fundamentals of growth, storage, genetics, and microscopy of Aspergillus nidulans. Fungal Genet. Newsl. 2001:25–31 [Google Scholar]
- 35. Kinoshita M. 2006. Diversity of septin scaffolds. Curr. Opin. Cell Biol. 18:54–60 [DOI] [PubMed] [Google Scholar]
- 36. Kozubowski L, Heitman J. 2010. Septins enforce morphogenetic events during sexual reproduction and contribute to virulence of Cryptococcus neoformans. Mol. Microbiol. 75:658–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lew DJ. 2003. The morphogenesis checkpoint: how yeast cells watch their figures. Curr. Opin. Cell Biol. 15:648–653 [DOI] [PubMed] [Google Scholar]
- 38. Lindsey R, Cowden S, Hernandez-Rodriguez Y, Momany M. 2010. Septins AspA and AspC are important for normal development and limit the emergence of new growth foci in the multicellular fungus Aspergillus nidulans. Eukaryot. Cell 9:155–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lindsey R, Momany M. 2006. Septin localization across kingdoms: three themes with variations. Curr. Opin. Microbiol. 9:559–565 [DOI] [PubMed] [Google Scholar]
- 40. Longtine MS, Bi E. 2003. Regulation of septin organization and function in yeast. Trends Cell Biol. 13:403–409 [DOI] [PubMed] [Google Scholar]
- 41. Longtine MS, et al. 1996. The septins: roles in cytokinesis and other processes. Curr. Opin. Cell Biol. 8:106–119 [DOI] [PubMed] [Google Scholar]
- 42. Martin SW, Douglas LM, Konopka JB. 2005. Cell cycle dynamics and quorum sensing in Candida albicans chlamydospores are distinct from budding and hyphal growth. Eukaryot. Cell 4:1191–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. McMurray MA, et al. 2011. Septin filament formation is essential in budding yeast. Dev. Cell 20:540–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. McMurray MA, Thorner J. 2008. Biochemical properties and supramolecular architecture of septin hetero oligomers and septin filaments, p 47–100 In Hall P, Russell H, Pringle J. (ed), The septins. John Wiley, Ltd, London, United Kingdom [Google Scholar]
- 45. Mims C, Richardson E, Timberlake W. 1988. Ultrastructural analysis of conidiophore development in the fungus Aspergillus nidulans using freeze-substitution. Protoplasma 144:132–141 [Google Scholar]
- 46. Momany M. 2001. Cell biology of the duplication cycle in fungi, p 119–125 In Talbot NJ. (ed), Molecular and cellular biology of filamentous fungi: a practical approach. Oxford University Press, London, United Kingdom [Google Scholar]
- 47. Momany M, Hamer JE. 1997. The Aspergillus nidulans septin encoding gene, aspB, is essential for growth. Fungal Genet. Biol. 21:92–100 [DOI] [PubMed] [Google Scholar]
- 48. Momany M, Zhao J, Lindsey R, Westfall PJ. 2001. Characterization of the Aspergillus nidulans septin (asp) gene family. Genetics 157:969–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Oh Y, Bi E. 2011. Septin structure and function in yeast and beyond. Trends Cell Biol. 21:141–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Orlando K, et al. 2011. Exo-endocytic trafficking and the septin-based diffusion barrier are required for the maintenance of Cdc42p polarization during budding yeast asymmetric growth. Mol. Biol. Cell 22:624–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ozsarac N, Bhattacharyya M, Dawes IW, Clancy MJ. 1995. The SPR3 gene encodes a sporulation-specific homologue of the yeast CDC3/10/11/12 family of bud neck microfilaments and is regulated by ABFI. Gene 164:157–162 [DOI] [PubMed] [Google Scholar]
- 52. Pan F, Malmberg RL, Momany M. 2007. Analysis of septins across kingdoms reveals orthology and new motifs. BMC Evol. Biol. 7:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Russell SE, Hall PA. 2005. Do septins have a role in cancer? Br. J. Cancer 93:499–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Saunders DG, Dagdas YF, Talbot NJ. 2010. Spatial uncoupling of mitosis and cytokinesis during appressorium-mediated plant infection by the rice blast fungus Magnaporthe oryzae. Plant Cell 22:2417–2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sirajuddin M, et al. 2007. Structural insight into filament formation by mammalian septins. Nature 449:311–315 [DOI] [PubMed] [Google Scholar]
- 56. Sirajuddin M, Farkasovsky M, Zent E, Wittinghofer A. 2009. GTP-induced conformational changes in septins and implications for function. Proc. Natl. Acad. Sci. U. S. A. 106:16592–16597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Spiliotis ET, Kinoshita M, Nelson WJ. 2005. A mitotic septin scaffold required for mammalian chromosome congression and segregation. Science 307:1781–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sudbery PE. 2001. The germ tubes of Candida albicans hyphae and pseudohyphae show different patterns of septin ring localization. Mol. Microbiol. 41:19–31 [DOI] [PubMed] [Google Scholar]
- 59. Szewczyk E, et al. 2006. Fusion PCR and gene targeting in Aspergillus nidulans. Nat. Protoc. 1:3111–3120 [DOI] [PubMed] [Google Scholar]
- 60. Takizawa PA, DeRisi JL, Wilhelm JE, Vale RD. 2000. Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science 290:341–344 [DOI] [PubMed] [Google Scholar]
- 61. Trinci AP. 1969. A kinetic study of the growth of Aspergillus nidulans and other fungi. J. Gen. Microbiol. 57:11–24 [DOI] [PubMed] [Google Scholar]
- 62. Versele M, Thorner J. 2005. Some assembly required: yeast septins provide the instruction manual. Trends Cell Biol. 15:414–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Warenda AJ, Konopka JB. 2002. Septin function in Candida albicans morphogenesis. Mol. Biol. Cell 13:2732–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Westfall PJ, Momany M. 2002. Aspergillus nidulans septin AspB plays pre- and postmitotic roles in septum, branch, and conidiophore development. Mol. Biol. Cell 13:110–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Xiang X, Beckwith SM, Morris NR. 1994. Cytoplasmic dynein is involved in nuclear migration in Aspergillus nidulans. Proc. Natl. Acad. Sci. U. S. A. 91:2100–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Xiang X, Fischer R. 2004. Nuclear migration and positioning in filamentous fungi. Fungal Genet. Biol. 41:411–419 [DOI] [PubMed] [Google Scholar]
- 67. Yang L, et al. 2004. Rapid production of gene replacement constructs and generation of a green fluorescent protein-tagged centromeric marker in Aspergillus nidulans. Eukaryot. Cell 3:1359–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yelton MM, Hamer JE, Timberlake WE. 1984. Transformation of Aspergillus nidulans by using a trpC plasmid. Proc. Natl. Acad. Sci. U. S. A. 81:1470–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.