Abstract
Most plastid proteins are encoded by their nuclear genomes and need to be targeted across multiple envelope membranes. In vascular plants, the translocons at the outer and inner envelope membranes of chloroplasts (TOC and TIC, respectively) facilitate transport across the two plastid membranes. In contrast, several algal groups harbor more complex plastids, the so-called secondary plastids, which are surrounded by three or four membranes, but the plastid protein import machinery (in particular, how proteins cross the membrane corresponding to the secondary endosymbiont plasma membrane) remains unexplored in many of these algae. To reconstruct the putative protein import machinery of a secondary plastid, we used the chlorarachniophyte alga Bigelowiella natans, whose plastid is bounded by four membranes and still possesses a relict nucleus of a green algal endosymbiont (the nucleomorph) in the intermembrane space. We identified nine homologs of plant-like TOC/TIC components in the recently sequenced B. natans nuclear genome, adding to the two that remain in the nucleomorph genome (B. natans TOC75 [BnTOC75] and BnTIC20). All of these proteins were predicted to be localized to the plastid and might function in the inner two membranes. We also show that the homologs of a protein, Der1, that is known to mediate transport across the second membrane in the several lineages with secondary plastids of red algal origin is not associated with plastid protein targeting in B. natans. How plastid proteins cross this membrane remains a mystery, but it is clear that the protein transport machinery of chlorarachniophyte plastids differs from that of red algal secondary plastids.
INTRODUCTION
Plastids have been acquired by multiple endosymbiotic events between a nonphotosynthetic eukaryote and a photosynthetic organism. Streptophytes and chlorophytes (land plants and their green algal relatives), rhodophytes (red algae), and glaucophytes have primary plastids that originated from a single endosymbiosis with a cyanobacterium (52). In contrast, many other lineages (apicomplexans, dinoflagellates, heterokonts, haptophytes, cryptophytes, euglenophytes, and chlorarachniophytes) possess secondary plastids that have been acquired by endosymbioses between either green or red algal endosymbionts and a nonphotosynthetic host (42, 43). Following these events, massive amounts of genes on the endosymbiont genomes were lost or transferred to host nuclear genomes (7, 49). Many transferred genes encode proteins that are targeted back to the plastid, and in most cases targeting is mediated by an N-terminal presequence (8, 51) that is recognized by a series of protein complexes that progressively move the protein toward its target.
Primary plastids are commonly surrounded by two envelope membranes. Plastid-targeted proteins translated in the cytoplasm must cross these two membranes and generally possess an N-terminal cleavable targeting sequence, called a transit peptide (TP), which mediates this process (9). The import pathways of plastid preproteins have been studied mainly in vascular plants, namely, Arabidopsis thaliana and Pisum sativum, and two protein complexes are known as TOC and TIC (translocons at the outer and inner envelope membranes of chloroplasts, respectively), which selectively recognize the preproteins and facilitate their transport across two plastid envelope membranes (3, 45). Three proteins, TOC34, TOC75, and TOC159, have been identified as the components of the TOC core complex (3). TOC75 is a member of the OMP85 family that forms a protein-conducting channel, and TOC34 and TOC159 are considered candidates for primary receptors of plastid-targeted preproteins. Additionally, TOC64 has been reported as a TOC component that is loosely associated with the core complex. Eight proteins have been detected as TIC components: TIC20, TIC21, TIC22, TIC32, TIC40, TIC55, TIC62, and TIC110 (45). Three of them, TIC20, TIC21, and TIC110, are supposed to form protein-conducting channels. A soluble intermembrane protein, TIC22, has been proposed to serve as an adaptor that facilitates preprotein transport between TOC and TIC complexes. Three proteins, TIC32, TIC55, and TIC62, are predicted to be involved in the redox regulation of preprotein transport. TIC40 was described as a cochaperone that is associated with TIC110 and Hsp93. Many homologs of plant TOC and TIC components have been reported from a few green algae (Chlamydomonas reinhardtii, Ostreococcus lucimarinus, and Ostreococcus tauri) and a red alga (Cyanidioschizon merolae) (38). This implies that primary plastids utilize a common import pathway of TOC and TIC complexes for nucleus-encoded plastid preproteins.
Plastids derived from secondary endosymbioses are more complex. They are bounded by three or four envelope membranes, and targeting sequences and transport pathways of plastid preproteins are quite different from those for primary plastids (8). Nucleus-encoded plastid preproteins commonly carry an N-terminal bipartite targeting sequence that consists of a signal peptide (SP) and a transit peptide-like (TPL) sequence (51). These preproteins are cotranslationally transported into the endoplasmic reticulum (ER) by the SP, and the TPL sequence is involved in delivering preproteins from the ER into the plastid stroma across three or four envelope membranes. A few homologs of TOC and TIC components (TIC20, TIC22, TIC110, and/or TOC75) have been detected by comparative genome searches in several organisms bearing secondary plastids of red algal origin: the heterokont Phaeodactylum tricornutum (10), the cryptophyte Guillardia theta (26), and the apicomplexans Plasmodium falciparum (39) and Toxoplasma gondii (65). This suggests that the outermost membrane is crossed via the general ER-targeting/secretion pathway and that the third and fourth membranes are crossed using some kind of TOC/TIC complexes, but for most algae this still leaves one membrane unaccounted for: the second outermost membrane, which is homologous to the plasma membrane of the endosymbiont. In several lineages with red algal secondary plastids, ER-associated protein degradation (ERAD)-derived proteins (e.g., Der1-like proteins) have been identified as the preprotein translocon that mediates the crossing of this membrane (2). However, there are currently no data on the nature of this process for any secondary plastid of green algal origin. This is unfortunate, because this is arguably the only step where the parallel evolution of a similar pathway is not highly likely. The symbiont would import already-functional TOC and TIC systems, and because all extant secondary plastids reside in the endomembrane system, it is hardly unexpected that the host ER-targeting pathway was independently coopted for this targeting step. Crossing the second outermost membrane is different because neither the host nor the endosymbiont would have had a single, obvious, and ready-made system in place to carry out this step (which might explain why two lineages of algae, dinoflagellates and euglenids, have lost this membrane).
In this study, we characterize the Der1-like proteins, as well as all recognizable TOC and TIC components, of the chlorarachniophyte Bigelowiella natans. Chlorarachniophytes possess four-membrane plastids derived from a green algal endosymbiont, and these plastids retain a highly reduced nucleus, the so-called nucleomorph, between the inner and outer pair of membranes (50). Plastid preproteins of chlorarachniophytes, like those of other secondary algal groups, possess an N-terminal bipartite targeting sequence consisting of an SP and a TPL sequence (54). Our previous studies demonstrated that the preproteins are transported into the plastids via the ER and that a net positive charge of TPL sequences is essential for passing through the inner two plastid membranes (31, 33). This suggests the existence of translocons that selectively recognize the TPL sequence and facilitate preprotein transport across the two inner plastid membranes. The complete sequence of the nucleomorph genome encodes one component of each of the TOC and TIC complexes, TOC75 and TIC20, but no other targeting gene products (25). We have therefore comprehensively searched the recently completed B. natans nuclear genome to develop a picture of what the plastid translocons in this organism might look like. We show that neither Der1-like protein (based on the localization of green fluorescent protein [GFP] fusion proteins) appears to be specifically associated with plastid protein targeting. Moreover, we show that both TOC and TIC complexes are apparently reduced and that the TOC in particular has lost all putative receptor proteins so that only the channel protein remains. Overall, these results support the conclusion that protein targeting to chlorarachniophyte plastids differs in several ways from that of secondary plastids of a red algal origin, at least one of which (transport across the second outermost membrane) is likely fundamentally different. So although many aspects of the evolutionary integration of red and green algal plastid symbionts and their hosts have proceeded in parallel in different lineages, for other steps, different solutions have also been found, as expected for a system assembled by “tinkering.”
MATERIALS AND METHODS
Identification of Der1, TOC, and TIC homologs in the Bigelowiella natans genome.
Homologs of the ERAD protein Der1 were identified by similarity with homologs from other eukaryotes using BLASTP searches against a library of predicted proteins from the B. natans genome sequencing project (sequenced at the DOE Joint Genome Institute). To identify the components of TOC and TIC homologs in B. natans, we used a collection of all proteins from both complexes from P. sativum, A. thaliana, C. reinhardtii, O. lucimarinus, and O. tauri, summarized in a previous study (38). These TOC and TIC sequences were used to search the predicted proteins from the B. natans genome using BLASTP. For a more sensitive manual survey to identify less-conserved potential homologs, we used the results of Smith-Waterman searches provided by JGI. Detected homologs to reciprocal BLASTP sequences were then used against P. sativum sequences in the NCBI server, and resulting E values are indicated (Table 1). Some predicted TOC and TIC homologs were absent from transcriptome surveys of B. natans (namely, B. natans TIC20 [BnTIC20], BnTIC21, BnTIC32, BnTIC40, and BnOMP85), and in these cases, we carried out 5′ and/or 3′ rapid amplification of cDNA ends (RACE) to confirm that they represent expressed genes. Total RNA was isolated from B. natans cells using TRIzol reagent (Invitrogen), and cDNA was synthesized using the FirstChoice RNA ligase-mediated (RLM)-RACE kit (Ambion) according to the manufacturer's protocols. Each target cDNA was amplified by PCR using Econo Taq DNA polymerase (Lucigen) with specific primer sets (shown in Table S1 in the supplemental material). Resulting PCR products were cloned into the pSC-A-amp/kan plasmid of a StrataClone PCR cloning kit and subsequently sequenced with an Applied Biosystems 3730 DNA analyzer and a BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems).
Table 1.
Putative TOC and TIC components in the Bigelowiella natans genomea
| Component | P. sativum accession no. | B. natans accession no.b | No. of aa | Presence of: |
TPL net charge (no. of aa) | E value | |
|---|---|---|---|---|---|---|---|
| SP sequence (probability score) | TPL sequence (probability score) | ||||||
| TOC34 | Q41009 | — | |||||
| TOC64 | AAF62870 | (JGI 89506) | 784 | Yes (0.998) | No (0.493) | 4 (50) | 7.00E−20 |
| TOC75 | Q43715 | ABA27321(Nm) | 818 | No | No (0.427) | −2.9 (50) | 9.00E−24 |
| OMP85 | Q43715 | JGI 79166 | 899 | Yes (0.786) | No (0.435) | 10 (50) | 0.002 |
| TOC159 | AF75761 | — | |||||
| TIC20 | AAC64607 | ABA27416 (Nm) | 207 | No | No (0.458) | 9 (50) | 5.00E−5 |
| JGI 115241 | 322 | Yes (1.000) | Yes (0.549) | 3.1 (32) | 1.00E−5 | ||
| TIC21 | ABG00264 | JGI 66755 | 272 | Yes (1.000) | Yes (0.518) | 7.1 (36) | 2.00E−4 |
| TIC22 | AAC64606 | JGI 127449 | 341 | Yes (0.996) | Yes (0.530) | 8 (32) | 0.001 |
| JGI 66754 | 330 | Yes (0.948) | Yes (0.538) | 9.1 (53) | 0.15 | ||
| TIC32 | AAS38575 | JGI 91380 | 527 | Yes (0.329) | Yes (0.521) | 0 (9) | 3.00E−32 |
| TIC40 | CAB50925 | JGI 82201 | 569 | Yes (0.574) | Yes (0.556) | 11.2 (58) | 2.00E−22 |
| TIC55 | CAA04157 | JGI 77479 | 648 | Yes (0.600) | Yes (0.514) | 6.2 (26) | 7.00E−59 |
| TIC62 | CAC87810 | JGI 86815 | 339 | Yes (0.835) | Yes (0.560) | 12.3 (53) | 3.00E−46 |
| TIC110 | CAA92823 | — | |||||
The GenBank accession or JGI protein identification number of each TOC/TIC component is recorded. “SP sequence” indicates the result of signal peptide prediction by the HMM method of SignalP, and the probability scores are shown in parentheses. “TPL sequence” is the result of transit peptide prediction by ChloroP. “TPL net charge” shows the net charge of the predicted TPL sequence, with the predicted length of the TPL sequence in parentheses (the N-terminal 50 aa of TOC64, TOC75, and TIC20 were used for the net charge calculation, since their TPL regions could not be predicted by ChloroP). The BLASTP E values against P. sativum TOC/TIC components are in the rightmost column.
Predictions of N-terminal SP sequences were performed by the Hidden Markov Models (HMM) method of the SignalP 3.0 server (6), and TPL sequences were predicted by the ChloroP 1.1 server (20). The probability of each prediction is indicated in Table 1. The overall charge of each predicted TPL sequence was calculated using the Peptide Property Calculator (Innovagen). Transmembrane α-helices were predicted with the TMHMM server, v. 2.0 (46), and SOSUI, v. 1.11 (34). Predictions of α-helices and β-strands in BnTOC75-nucleomorph (Nm) and BnOMP85 were made with PROFsec of the PredictProtein server (55). Protein motifs and domains were identified using two databases: PFAM (23) and SMART (48). We collected TOC and TIC homologs from plants (P. sativum, A. thaliana, Oryza sativa, and Physcomitrella patens) and green algae (C. reinhardtii, Chlorella variabilis, Micromonas pusilla, O. tauri, O. lucimarinus, and/or Volvox carteri) using Phytozome v. 7.0 and JGI websites, and alignments of homologs were created by the ClustalW program in MEGA 4 (63). Sequence conservations were shown as logo plots generating by WebLogo v. 2.8.2 (19).
Phylogenetic analyses.
Homologs of Der1, TIC20, and OMP85 (TOC75) were collected for apicomplexans, heterokonts, cryptophytes, red algae, green algae, and/or vascular plants using GenBank, JGI, or C. merolae genome project websites. Automatic alignment was performed by the L-INS-I method of the MAFFT package (41), and Gblocks (12) was used to automatically and reproducibly eliminate poorly aligned positions, with no gapped positions allowed, the minimum number of sequences for a conserved and a flank position set to 50% of the number of taxa plus one, the maximum of contiguous nonconserved positions set to 10, and the minimum length of a block set to 8; the resulting unambiguously aligned positions were manually inspected with SeaView (27). Maximum-likelihood phylogenetic analyses were performed with RAxML 7.2.8 (62), in combination with the rapid hill-climbing algorithm and the site-homogeneous LG + Γ + F model of evolution (-m PROTGAMMALGF, 4 discrete rate categories). Statistical support was evaluated with nonparametric bootstrapping using 100 replicates.
Subcellular localization of B. natans Der1-like proteins.
To construct two GFP expression vectors, cDNA fragments of two Der1-like genes (BnDer1-49642 and BnDer1-92850 genes) were amplified by PCR with specific primers shown in Table S2 in the supplemental material, and then each fragment was inserted between HindIII and NcoI sites of the pLaRGfp+mc vector (33). These plasmids were cloned in the DH5α strain of Escherichia coli and purified with a QIAprep Spin miniprep kit (Qiagen). To analyze the subcellular localization of GFP fusion proteins, we transformed a related chlorarachniophyte species, Amorphochlora amoebiformis, with these plasmids using a Biolistic PDS-1000/He particle delivery system (Bio-Rad) as described previously (32). Transiently transformed cells were observed under an Axioplan 2 fluorescence microscope (Carl Zeiss AG) with a 3CCD HD XL H1S video camera (Canon). GFP fluorescence and plastid autofluorescence were detected with filter 17 (excitation band-pass [BP], 485/20 nm; emission BP, 515 to 565 nm) and filter 15 (excitation BP, 546/12 nm; emission long-pass [LP], 590 nm), respectively.
Nucleotide sequence accession numbers.
cDNA sequences from 5′- and/or 3′-RACE analyses are deposited in GenBank under accession no. JQ088188 to JQ088192.
RESULTS AND DISCUSSION
To create a model of the plastid preprotein transport pathway in chlorarachniophytes, the B. natans genome sequence was surveyed for proteins potentially involved in preprotein translocons using reciprocal BLAST and Smith-Waterman searches. We can divide this transport process into three stages. The first is the targeting of plastid preproteins to the endomembrane system, resulting in the crossing of the outermost membrane. This is, according to all available evidence, mediated by the signal peptide in all secondary plastids and will not be discussed further. The second part of the pathway, and the most mysterious, is the crossing of the second outermost membrane. There is no direct evidence for how this interendomembrane sorting takes place in chlorarachniophytes, though two hypotheses have been proposed. In one, a duplicated ERAD complex controls this process, as is the case in at least some red secondary plastids (59), and in the other, a duplicated TOC complex carries it out (13). The third stage is the crossing of the inner two plastid membranes, and it is widely assumed that this is mediated by standard TOC and TIC complexes inherited from the primary algal endosymbiont, although few of the proteins making up this complex have been identified.
ERAD-derived proteins of the B. natans genome.
Several algal groups (heterokonts, haptophytes, and cryptophytes) and a parasitic group (apicomplexans) harbor four-membrane-bounded plastids derived from a red algal endosymbiont, and it was from these that the first direct evidence for how preproteins cross the second membrane was found. Sommer et al. (59) described a nucleomorph-encoded Der1-like protein as the symbiont ERAD-like machinery (SELMA) and speculated it to be involved in the crossing of the second outermost plastid membrane in cryptophytes. Several studies have subsequently proposed that symbiont-specific Der1-like proteins commonly act as the second outermost membrane translocon of secondary plastids in heterokonts (30, 59), haptophytes (22), and apicomplexans (1, 60).
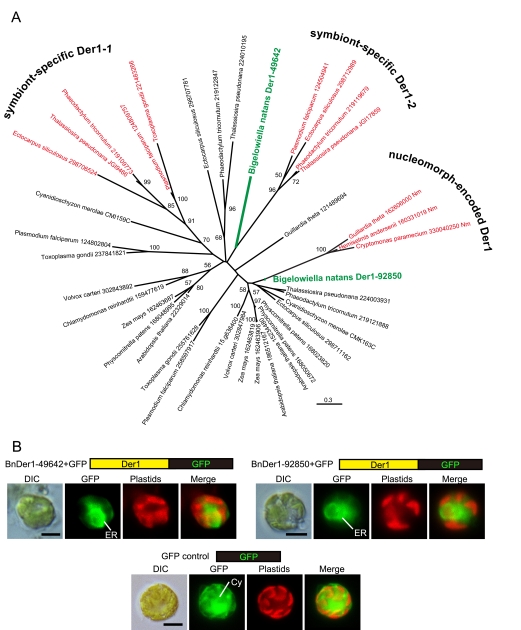
To determine whether Der1 is involved in crossing the second outermost membrane in chlorarachniophyte plastids, we searched for Der1 homologs in the B. natans genome. We detected two Der1-like proteins (BnDer1-49642 and BnDer1-92850) in the nuclear genome, and phylogenetic analyses suggested that these proteins were weakly related to nucleomorph-encoded Der1-like proteins of cryptophytes and symbiont-specific Der1-like proteins, respectively (Fig. 1A). This suggests that chlorarachniophytes might have independently coopted the same ERAD-derived system to target plastid preproteins as did red algal secondary plastids. However, the phylogenetic position of neither protein is statistically supported, so more-direct evidence for the functional roles of these proteins, as a plastid translocon or a general ERAD component, was sought. Specifically, we tested the localizations of both B. natans Der1-like proteins using a GFP fusion protein and the genetic transformation system of a related chlorarachniophyte, Amorphochlora amoebiformis. We created two plasmid constructs that express GFP fused with full-length BnDer1-49642 (217 amino acids [aa]) or BnDer1-92850 (251 aa). A. amoebiformis cells were transformed with each plasmid individually, and GFP localizations were examined. Both fusion proteins led to green fluorescence exclusively in the ER (Fig. 1B). This indicates that both B. natans Der1-like proteins function in the ER as general components of ERAD machinery and that neither of them is a translocon specifically at the second outermost membrane of the plastid. This result is not surprising, since organisms with red algal secondary plastids possess four homologs of Der1 proteins in their genomes: two of them are components of ERAD machinery and the other two are components of the SELMA, so there is a precedent for such algae using multiple ERAD-specific Der1 proteins. Although this does not reveal how proteins cross the second membrane in chlorarachniophytes, it does show that this lineage has evolved a system for transporting preproteins across the second plastid membrane that is distinct from the ERAD-derived system of red algal plastid-bearing organisms.
Fig 1.
Unrooted maximum-likelihood phylogenetic tree of Der1-like proteins and subcellular localizations of B. natans Der1-like proteins. (A) B. natans sequences are shown in green, and Der1 sequences of putative translocons at the second plastid membrane are indicated in red. The numbers next to the species names indicate GenBank, JGI, or C. merolae genome identifiers. The values at nodes are the bootstrap support values and are indicated only when they are higher than 50%. The scale bar represents the estimated number of amino acid substitutions per site. (B) GFP localization of a transformed cell expressing BnDer1-49642-GFP, BnDer1-92850-GFP, and control GFP, respectively. The pictures labeled “GFP” and “Plastids” show GFP fluorescence (green) and chlorophyll autofluorescence (red), respectively. DIC, differential interference contrast image; SP, signal peptide; ER, endoplasmic reticulum; Cy, cytoplasm. Scale bar, 5 μm.
TOC and TIC complexes in B. natans.
TOC and TIC protein sequences of vascular plants and green algae were used for an initial BLASTP analysis against all predicted protein sequences of the B. natans genome, and Smith-Waterman searches were used for more sensitive detections. The predicted TOC and TIC homologs of B. natans were subsequently analyzed by BLASTP analysis of P. sativum protein sequences, and the resulting E values are shown in Table 1. We also predicted targeting sequences and functional domains/motifs using bioinformatic tools and manual curations. Overall, we identified 11 members of TOC/TIC components from the B. natans genomes, and all of them were predicted to be localized in the plastid.
TOC proteins.
The outer membrane translocon complex of vascular plants consists mainly of four TOC components. The B. natans nucleomorph genome encodes a copy of TOC75, and we found another homolog of OMP85 in the nuclear genome, but our searches did not clearly identify any additional component of the TOC complex.
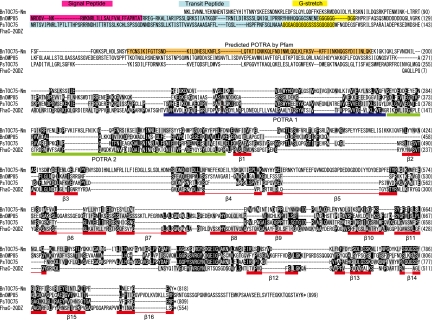
TOC75 is a member of outer membrane protein 85 (OMP85) family and forms a protein-conducting channel. This protein has two typical structural domains; the N-terminal region contains polypeptide transport-associated (POTRA) domains, and the C-terminal bacterial surface antigen domain forms a transmembrane β-barrel (24). POTRA domains share the same strand-helix-helix-strand-strand topology in their secondary structures (56), and the β-barrel domain contains 16 β-strands (21). A homolog of TOC75 in B. natans, BnTOC75-Nm, has previously been identified in the nucleomorph genome (25). This protein is predicted to possess an N-terminal POTRA domain and a bacterial surface antigen domain in its C-terminal region, according to PFAM searches (Fig. 2). Secondary-structure prediction based on PROFsec suggests that the putative POTRA domain consists of three β-strands, with the first and second strands separated by two α-helices, and that the C-terminal region contains 16 putative β-strands. However, it was difficult to predict regions of canonical POTRA1 and POTRA2 sequences and 16 β-strands from the alignment with FhaC, whose crystal structure has been reported (18) (Fig. 2). All together, these data suggest that BnTOC75-Nm probably forms a transmembrane β-barrel and functions as a protein-conducting channel. We also found a nucleus-encoded homolog of the OMP85 family, named BnOMP85, that was weakly predicted by reciprocal BLASTP searches (its E value against P. sativum TOC75 is 0.002 [Table 1]). Our phylogenetic analysis indicates that BnOMP85 is closely related with OMP85-like proteins of heterokonts and apicomplexans, but it is unclear whether these proteins are grouped with either TOC75 or OEP80 (Fig. S1). On the basis of the alignment with FhaC, this protein is predicted to include the POTRA2 domain and 16 β-strands in its C-terminal bacterial surface antigen domain (Fig. 2), which implies that BnOMP85 also functions as a protein-conducting channel. In vascular plants, TOC75 preproteins carry a cleavable transit peptide followed by a polyglycine stretch at their N termini, and these proteins should be inserted into the outer membrane from the stroma side. It has been reported that both of these motifs play an essential role in the outer plastid membrane localization of TOC75 in P. sativum (5) and A. thaliana (36). Interestingly, nucleus-encoded BnOMP85 possesses an N-terminal bipartite plastid targeting sequence followed by a polyglycine stretch containing 8 glycine residues, which implies that BnOMP85 may be inserted into the third membrane (counting from the outside) in a way similar to that of plant TOC75 (Fig. 2, 3). In contrast, BnTOC75-Nm lacks both a canonical transit peptide and a polyglycine stretch. There is no experimental evidence for the localization of BnTOC75-Nm, but if it occupies a location homologous to that in plants and green algae, it would be found in the third membrane (Fig. 3), although how it is targeted there is unknown and seems to be different from that of plant TOC75.
Fig 2.
Alignment of BnTOC75-Nm, BnOMP85, P. sativum TOC75, and Bordetella pertussis FhaC. Amino acids identical in at least two of the sequences are shaded in black, and conserved substitutions are shaded in gray. The putative signal peptide, transit peptide, and polyglycine stretch are highlighted with pink, blue, and yellow, respectively. POTRA1, POTRA2, and 16 β-strands (β-1 to -16) of FhaC that have been reported previously (18) are indicated below the sequence.
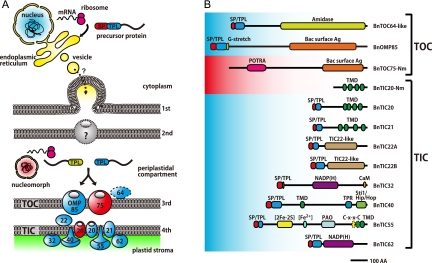
Fig 3.
Reconstructed translocons in a chlorarachniophyte plastid. (A) Predicted localization of B. natans TOC and TIC homologs. Nucleus-encoded components are highlighted with blue, and nucleomorph-encoded ones are red. Arrows indicate the transport pathway of plastid precursor proteins. (B) Schematic illustration of putative B. natans TOC and TIC proteins. Predicted signals, motifs, and domains are plotted in each sequence. Abbreviations: SP, signal peptide; Ag, antigen; TPL, transit peptide-like sequence; G-stretch, polyglycine stretch; TMD, transmembrane domain; POTRA, polypeptide transport-associated domain; CaM, calmodulin-binding site; TPR, tetratricopeptide repeat; PAO, pheophorbide a oxygenase; [2Fe-2S], 2Fe-2S binding site; [Fe2+], mononuclear-iron-binding site.
TOC64 is an integral membrane protein composed of an N-terminal transmembrane domain, an inactive amidase domain, and C-terminal tetratricopeptide repeat (TPR) motifs (57). The B. natans nuclear genome encodes a single homolog of amidase with a canonical N-terminal bipartite targeting sequence consisting of an SP and a weakly predicted TPL sequence (Fig. 3; Table 1). However, this protein lacks the TPR motifs, which are proposed to be a chaperone-docking site, as well as the N-terminal transmembrane domain. These suggest that the B. natans amidase homolog is probably targeted to the plastid but that it possibly lacks the function of an import receptor, so it is not certain that the TOC64 component is strictly required for protein transport into the chlorarachniophyte plastids. This view is supported by a few additional observations: the genomes of both a green alga (C. reinhardtii) and a red alga (C. merolae) also lack TOC64 (38), and it has been demonstrated that the A. thaliana TOC64 protein is not essential for efficient preprotein transport into plastids (4).
Both TOC34 and TOC159 are GTPase proteins that function as receptors of plastid preproteins and, together with the protein-conducting channel TOC75, constitute the core TOC components (3). Plastid preproteins are initially identified by TOC34 and TOC159 by the direct binding of transit peptides or an indirect binding to transit peptide associated with Hsp70 and 14-3-3 proteins. Bound proteins are then threaded into the TOC75 cannel. In our survey, no homolog of either TOC34 or TOC159 was found in the B. natans genome. Although it is possible that the sensitivity of our survey was not sufficient to detect these proteins, overall, the evidence suggests that the B. natans genome lacks canonical plant-like receptors of plastid preproteins. We propose two possible implications for the absence of TOC34 and TOC159 receptors in B. natans. First, the TPL sequence of plastid preproteins might be directly identified by BnTOC75-Nm or BnOMP85 without receptors, since it has been reported that P. sativum TOC75 contains a transit peptide-binding site in its N-terminal region, including POTRA domains (21). Alternatively, B. natans might use novel preprotein receptors which are divergent from plant TOC components. Our previous studies have indicated that a net positive charge of TPL sequences is essential for protein transport across the two inner plastid membranes (31, 33). This implies that novel receptors might recognize the TPL sequence net charge on the third outermost plastid membrane.
TIC proteins.
In vascular plants, eight TIC components have been identified so far (45). In this study, we found 7 homologs of TIC components in the B. natans nuclear genome (TIC20, TIC21, TIC22, TIC32, TIC40, TIC55, and TIC62 were identified, and TIC110 was not identified). A homolog of TIC20 is also encoded in the nucleomorph genome (Table 1).
TIC110, TIC20, and TIC21 are proposed to be components of protein-conducting channels at the inner plastid membrane. TIC110 is the most abundant component of the TIC complex, acting as the main channel for protein transport (29). TIC20 is an alternative candidate for the inner membrane channel, which is an integral membrane protein with four α-helical transmembrane domains (14). TIC21 also contains four α-helices and has been postulated to be a putative channel (64). We detected single TIC20 and TIC21 homologs (BnTIC20 and BnTIC21) but no TIC110 in the B. natans nuclear genome (Table 1). BnTIC20 and BnTIC21 were predicted to be integral membrane proteins with four transmembrane α-helices (Fig. 3). Both of them possess an N-terminal bipartite targeting sequence, and their TPL sequences carry a net positive charge (Table 1). These data suggest that BnTIC20 and BnTIC21 might be targeted to the plastid stroma and inserted in the innermost plastid membrane via multiple α-helices. Since the B. natans genome lacks a TIC110 homolog, BnTIC20 and/or BnTIC21 presumably functions as a protein-conducting channel at the innermost membrane. Interestingly, the nucleomorph genomes of two chlorarachniophyte species (B. natans and Gymnochlora stellata) also encode TIC20 homologs, which are each predicted to possess four transmembrane α-helices (Fig. 3). To date, two to four TIC20 paralogs have been described in plants and green algae, and a phylogenetic analysis has demonstrated that TIC20 homologs form two distinct clades (38). Reconstruction of the phylogeny analysis, including the phylogeny of both chlorarachniophyte TIC20 homologs (see Fig. S2 in the supplemental material), indicates that nucleus-encoded TIC20 branches in one of these clades and that nucleomorph-encoded TIC20 branches within the other. On the basis of this phylogeny, the nucleomorph-encoded TIC20 protein is the more likely candidate for being the channel-forming protein, because it is comparatively closely related to two TIC20 homologs (A. thaliana TIC20-I [AtTIC20-I] and AtTIC20-IV) that have been reported to be the major functional isoforms in A. thaliana (40).
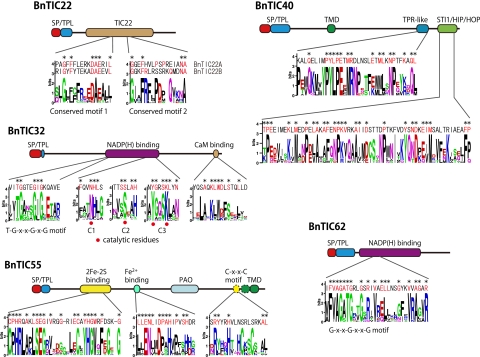
TIC22 is a soluble intermembrane protein that is peripherally bound to the outer face of the inner plastid membrane (44). This protein has been postulated to serve as an adaptor between TOC and TIC complexes. In the B. natans genome, two homologs of TIC22 were weakly predicted by reciprocal BLASTP searches against P. sativum TIC22 (E values of 0.15 and 0.001 [Table 1]). Although the accuracy of BLASTP predictions is not high, these proteins share two conserved motifs with TIC22 homologs of plants and green algae (Fig. 4) and carry a canonical N-terminal bipartite targeting sequence consisting of an SP and a pronounced net positively charged TPL sequence (Table 1). Therefore, we postulate that these two proteins are TIC22 homologs of B. natans (BnTIC22A and BnTIC22B) and that they are most likely localized in the intermembrane space between the inner two plastid membranes. However, it remains unclear how these proteins are inserted into the intermembrane space.
Fig 4.
Protein structures of BnTIC22, BnTIC40, BnTIC32, BnTIC55, and BnTIC62. Sequence conservations among TIC homologs of land plants (Pisum sativum, Arabidopsis thaliana, Oryza sativa, and Physcomitrella patens) and green algae (Chlamydomonas reinhardtii, Chlorella variabilis, Micromonas pusilla, Ostreococcus tauri, O. lucimarinus, and/or Volvox carteri) are shown as logo plots generating by WebLogo v. 2.8.2. B. natans TIC sequences are shown above the logo plots, and conserved amino acids are indicated by asterisks.
TIC40 has been described as a cochaperone protein with an N-terminal α-helical transmembrane domain, followed by a TPR motif that interacts with TIC110 and a Sti1/Hip/Hop domain that medicates the interaction with Hsp93 (16, 17, 61). We identified a TIC40 homolog in B. natans (BnTIC40) that is predicted to possess these three characteristic domains as well as an N-terminal bipartite targeting sequence (Fig. 3; Table 1). Notably, the C-terminal sequence contains a TPR-like motif and a Sti1/Hip/Hop domain sharing a high degree of similarity with TIC40 proteins of plants and green algae (Fig. 4). BnTIC40 appears to function as a translocon at the innermost membrane but likely does not interact with TIC110 as in plants, since this protein is absent in B. natans.
Three TIC components, TIC32 (15, 35), TIC55 (11), and TIC62 (47), contain either NADP(H)-binding sites or Rieske-type iron-sulfur clusters, and these are hypothesized to be involved in preprotein transport regulation depending on the metabolic state of the plastid. We found one homolog of each of these TIC components, each predicted to carry an N-terminal targeting sequence, suggesting plastid localization (Fig. 3, Table 1). B. natans TIC55 (BnTIC55) contains a 2Fe-2S binding site and a mononuclear iron binding site in its N-terminal region, followed by a domain with similarities to pheophorbide a oxygenase (PAO), and two transmembrane helices were predicted to fall in the C-terminal region based on the alignment with TOC55 homologs of plants and green algae (the TMHMM server predicted a single transmembrane helix) (Fig. 4). BnTIC55 closely resembled TIC55 homologs in these features, but there is a striking difference; although TOC55 homologs possess a C-terminal conserved domain, including an R-Y-X2-H-X3-C-X2-C motif of unknown function (28), two cysteine residues of this motif have been replaced by serine residues in BnTIC55 (Fig. 4). Both homologs of TIC32 and TIC62 in the B. natans genome (BnTIC32 and BnTIC62) were predicted to belong to the (extended) family of short-chain dehydrogenases (SDRs) containing conserved NADP(H)-binding sites (Fig. 3). BnTIC32 possesses the signature motif, T-G-X3-G-X-G, for NADP(H) binding and catalytic tetrad residues consisting of asparagine, serine, tyrosine, and lysine (35) (Fig. 4). The C-terminal region of BnTIC32 is loosely defined as a calmodulin-binding site that is present in plant TIC32 homologs (15) (Fig. 4). BnTIC62 also contains the G-X2-G-X2-G motif within its predicted NADP(H)-binding domain but lacks a C-terminal ferredoxin-NADPH reductase (FNR)-binding domain that has been reported for TIC62 of P. sativum and A. thaliana (47) (Fig. 4). These three TIC homologs, BnTIC55, BnTIC32, and BnTIC62, possess several domains with high resemblance to plant TIC components. However, a few domains having essential/unknown functions are weakly predicted or not detected in B. natans TIC homologs. We propose these three homologs to be putative TIC components, but it is unclear whether they have the same functions as plant TIC components.
Reconstructing chlorarachniophyte plastid-targeting translocons in B. natans.
Collectively, the B. natans nuclear and nucleomorph genomes encode two homologs of TOC75 and seven members of the TIC complex. These plant-like TOC and TIC components seem to be derived from the green algal endosymbiont via secondary endosymbiosis. It is noteworthy that only two components, TOC75 and TIC20, remain encoded in the nucleomorph genome and that these two proteins are also the putative protein-conducting channels. It is also noteworthy the way in which the B. natans TOC complex appears to have been reduced. The conserved pore is retained, but none of the receptors are evident. These proteins are present in the genome of C. reinhardtii, which is comparatively closely related to the green algal endosymbiont of chlorarachniophytes (38, 53), but were apparently lost in the B. natans lineage after the secondary endosymbiosis. Because primary plastids reside in the cytoplasm, their TOC complex must be able to distinguish plastid proteins from all other cytoplasmic and mitochondrial proteins. In contrast, secondary plastids reside within the endomembrane system and in chlorarachniophytes within a small residual volume of cytoplasm that is discontinuous with the rest of the host endomembrane system. Accordingly, the receptors at the third plastid membrane must distinguish plastid proteins only from the pool of nonplastid proteins that would reside in this compartment. Since this compartment is massively reduced compared to ordinary green algal cytoplasm, we hypothesize the receptors might require much less information to accurately recognize plastid proteins, which might in turn lead to a reduced demand for receptors.
An even more pressing problem is how plastid-targeted preproteins in chlorarachniophytes cross the second outermost membrane. We found no evidence to support the conclusion that ERAD-derived components, in particular, symbiont-specific Der1 proteins, are utilized to cross this membrane, as has been shown in all other algae that have retained it (2). While this may have been the most obvious possibility, we have shown that the localization patterns of B. natans Der1 proteins are consistent with a generalized ER function. However, the hypothesis that a duplicate TOC complex is located in the second membrane and mediates import is consistent with our findings, since two homologs of the OMP85 family (BnTOC75-Nm and BnOMP85) are encoded in the nuclear and the nucleomorph genome, respectively. It is possible that both putative channel proteins localize in the third membrane (counting from the outside). One paralogue of A. thaliana TOC75, the so-called AtOEP80 protein, is directly inserted into the outer plastid membrane from the cytoplasm without an N-terminal transit peptide (37), and both AtTOC75 and AtOEP80 have the same topology in the plastid outer membrane despite the opposite directions of their insertions (AtTOC75 and AtOEP80 are delivered from the stroma side and the cytoplasm, respectively) (58). If BnTOC75-Nm is targeted in the same way, both it and BnOMP85 may localize to the third membrane. However, it is also possible that BnTOC75-Nm might be integrated into the second outermost membrane, because this protein lacks a canonical plastid membrane targeting sequence that BnOMP85 and plant TOC75 preproteins possess. In this case, BnOMP85 would fulfill the canonical role of TOC75 in the third membrane, while BnTOC75-Nm would serve to translocate proteins across the second membrane, as the SELMA system does in red algal secondary plastids. The detailed localization of these putative channels should be confirmed in the future, but currently there is no method available in chlorarachniophytes that would allow us to unambiguously distinguish between localizations in any of the four plastid membranes (which might be demonstrated by a self-assembling split GFP system, for example). Moreover, we currently have no means to transform the nucleomorph genome, so even if the location could be distinguished, it would not yet be possible to determine how BnTOC75-Nm is targeted. Nevertheless, these two putative channels are fascinating candidates to solve this interesting puzzle in protein targeting.
Further characterization of this important step in plastid targeting will be of great interest, not only from the perspective of chlorarachniophyte biology, but for helping to define the limits of the integration of endosymbionts, since this is arguably the only step where one might realistically expect to see a genuinely nonparallel development in green and red algal secondary plastids. The absence of a duplicated plastid-specific Der1 protein in chlorarachniophytes shows that they do use a nonhomologous system, and the onus is now on defining what that system is.
Supplementary Material
ACKNOWLEDGMENTS
We thank Joyce Wang and the Kronstad lab for granting access to the Particle Delivery System. We thank the U.S. Department of Energy Joint Genome Institute for sequencing the nuclear genome of B. natans and J. M. Archibald, M. W. Gray, G. I. McFadden, and C. E. Lane for project contributions.
This work was funded by a grant from the Natural Sciences and Engineering Research Council of Canada (227301) and by a grant to the Centre for Microbial Diversity and Evolution from the Tula Foundation. The work conducted by the U.S. Department of Energy Joint Genome Institute is supported by the Office of Science of the U.S. Department of Energy under contract no. DE-AC02-05CH11231. F.B. is supported by a prospective researcher postdoctoral fellowship from the Swiss National Science Foundation. P.J.K. is a Fellow of the Canadian Institute for Advanced Research and a Senior Scholar of the Michael Smith Foundation for Health Research.
Footnotes
Published ahead of print 20 January 2012
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1. Agrawal S, van Dooren GG, Beatty WL, Striepen B. 2009. Genetic evidence that an endosymbiont-derived endoplasmic reticulum-associated protein degradation (ERAD) system functions in import of apicoplast proteins. J. Boil. Chem. 284:33683–33691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Agrawal S, Striepen B. 2010. More membranes, more proteins: complex protein import mechanisms into secondary plastids. Protist 161:672–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Andrès C, Agne B, Kessler F. 2010. The TOC complex: preprotein gateway to the chloroplast. Biochim. Biophys. Acta 1803:715–723 [DOI] [PubMed] [Google Scholar]
- 4. Aronsson H, et al. 2007. Toc64/OEP64 is not essential for the efficient import of proteins into chloroplasts in Arabidopsis thaliana. Plant J. 52:53–68 [DOI] [PubMed] [Google Scholar]
- 5. Baldwin AJ, Inoue K. 2006. The most C-terminal tri-glycine segment within the polyglycine stretch of the pea Toc75 transit peptide plays a critical role for targeting the protein to the chloroplast outer envelope membrane. FEBS J. 273:1547–1555 [DOI] [PubMed] [Google Scholar]
- 6. Bendtsen JD, Nielsen H, von Heijne G, Brunak S. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783–795 [DOI] [PubMed] [Google Scholar]
- 7. Bock R, Timmis JN. 2008. Reconstructing evolution: gene transfer from plastids to the nucleus. Bioessays 30:556–566 [DOI] [PubMed] [Google Scholar]
- 8. Bolte K, et al. 2009. Protein targeting into secondary plastids. J. Eukaryot. Microbiol. 56:9–15 [DOI] [PubMed] [Google Scholar]
- 9. Bruce BD. 2000. Chloroplast transit peptides: structure, function and evolution. Trends Cell Biol. 10:440–447 [DOI] [PubMed] [Google Scholar]
- 10. Bullmann L, et al. 2010. Filling the gap, evolutionarily conserved Omp85 in plastids of chromalveolates. J. Biol. Chem. 285:6848–6856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Caliebe A, et al. 1997. The chloroplastic protein import machinery contains a Rieske-type iron-sulfur cluster and a mononuclear iron-binding protein. EMBO J. 16:7342–7350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17:540–552 [DOI] [PubMed] [Google Scholar]
- 13. Cavalier-Smith T. 2003. Genomic reduction and evolution of novel genetic membranes and protein-targeting machinery in eukaryote-eukaryote chimaeras (meta-algae). Philos. Trans. R. Soc. Lond. B Biol. Sci. 358:109–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen X, Smith MD, Fitzpatrick L, Schnell DJ. 2002. In vivo analysis of the role of atTic20 in protein import into chloroplasts. Plant Cell 14:641–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chigri F, et al. 2006. Calcium regulation of chloroplast protein translocation is mediated by calmodulin binding to Tic32. Proc. Natl. Acad. Sci. U. S. A. 103:16051–16056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chou ML, et al. 2003. Tic40, a membrane-anchored co-chaperone homolog in the chloroplast protein translocon. EMBO J. 22:2970–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chou ML, Chu CC, Chen LJ, Akita M, Li HM. 2006. Stimulation of transit-peptide release and ATP hydrolysis by a cochaperone during protein import into chloroplasts. J. Cell Biol. 175:893–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Clantin B, et al. 2007. Structure of the membrane protein FhaC: a member of the Omp85-TpsB transporter superfamily. Science 317:957–961 [DOI] [PubMed] [Google Scholar]
- 19. Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Emanuelsson O, Nielsen H, von Heijne G. 1999. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8:978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ertel F, et al. 2005. The evolutionarily related beta-barrel polypeptide transporters from Pisum sativum and Nostoc PCC7120 contain two distinct functional domains. J. Biol. Chem. 280:28281–28289 [DOI] [PubMed] [Google Scholar]
- 22. Felsner G, et al. 2011. ERAD components in organisms with complex red plastids suggest recruitment of a preexisting protein transport pathway for the periplastid membrane. Genome Biol. Evol. 3:140–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Finn RD, et al. 2010. The Pfam protein families database. Nucleic Acids Res. 38:D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gentle IE, Burri L, Lithgow T. 2005. Molecular architecture and function of the Omp85 family of proteins. Mol. Microbiol. 58:1216–1225 [DOI] [PubMed] [Google Scholar]
- 25. Gilson PR, et al. 2006. Complete nucleotide sequence of the chlorarachniophyte nucleomorph: nature's smallest nucleus. Proc. Natl. Acad. Sci. U. S. A. 103:9566–9571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gould SB, et al. 2006. Nucleus-to-nucleus gene transfer and protein retargeting into a remnant cytoplasm of cryptophytes and diatoms. Mol. Biol. Evol. 23:2413–2422 [DOI] [PubMed] [Google Scholar]
- 27. Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27:221–224 [DOI] [PubMed] [Google Scholar]
- 28. Gray J, et al. 2004. A small family of LLS1-related non-heme oxygenases in plants with an origin amongst oxygenic photosynthesizers. Plant Mol. Biol. 54:39–54 [DOI] [PubMed] [Google Scholar]
- 29. Heins L, et al. 2002. The preprotein conducting channel at the inner envelope membrane of plastids. EMBO J. 21:2616–2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hempel F, Bullmann L, Lau J, Zauner S, Maier UG. 2009. ERAD-derived preprotein transport across the second outermost plastid membrane of diatoms. Mol. Biol. Evol. 26:1781–1790 [DOI] [PubMed] [Google Scholar]
- 31. Hirakawa Y, Gile GH, Ota S, Keeling PJ, Ishida K. 2010. Characterization of periplastidal compartment-targeting signals in chlorarachniophytes. Mol. Biol. Evol. 27:1538–1545 [DOI] [PubMed] [Google Scholar]
- 32. Hirakawa Y, Kofuji R, Ishida K. 2008. Transient transformation of a chlorarachniophyte alga, Lotharella amoebiformis (chlorarachniophyceae), with uidA and egfp reporter genes. J. Phycol. 44:814–820 [DOI] [PubMed] [Google Scholar]
- 33. Hirakawa Y, Nagamune K, Ishida K. 2009. Protein targeting into secondary plastids of chlorarachniophytes. Proc. Natl. Acad. Sci. U. S. A. 106:12820–12825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hirokawa T, Boon-Chieng S, Mitaku S. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14:378–379 [DOI] [PubMed] [Google Scholar]
- 35. Hörmann F, et al. 2004. Tic32, an essential component in chloroplast biogenesis. J. Biol. Chem. 279:34756–34762 [DOI] [PubMed] [Google Scholar]
- 36. Inoue K, Keegstra K. 2003. A polyglycine stretch is necessary for proper targeting of the protein translocation channel precursor to the outer envelope membrane of chloroplasts. Plant J. 34:661–669 [DOI] [PubMed] [Google Scholar]
- 37. Inoue K, Potter D. 2004. The chloroplastic protein translocation channel Toc75 and its paralog OEP80 represent two distinct protein families and are targeted to the chloroplastic outer envelope by different mechanisms. Plant J. 39:354–365 [DOI] [PubMed] [Google Scholar]
- 38. Kalanon M, McFadden GI. 2008. The chloroplast protein translocation complexes of Chlamydomonas reinhardtii: a bioinformatic comparison of Toc and Tic components in plants, green algae and red algae. Genetics 179:95–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kalanon M, Tonkin CJ, McFadden GI. 2009. Characterization of two putative protein translocation components in the apicoplast of Plasmodium falciparum. Eukaryot. Cell 8:1146–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kasmati AR, Töpel M, Patel R, Murtaza G, Jarvis P. 2011. Molecular and genetic analyses of Tic20 homologues in Arabidopsis thaliana chloroplasts. Plant J. 66:877–889 [DOI] [PubMed] [Google Scholar]
- 41. Katoh K, Kuma K, Toh H, Miyata T. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33:511–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Keeling PJ. 2009. Chromalveolates and the evolution of plastids by secondary endosymbiosis. J. Eukaryot. Microbiol. 56:1–8 [DOI] [PubMed] [Google Scholar]
- 43. Keeling PJ. 2010. The endosymbiotic origin, diversification and fate of plastids. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365:729–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kouranov A, Chen X, Fuks B, Schnell DJ. 1998. Tic20 and Tic22 are new components of the protein import apparatus at the chloroplast inner envelope membrane. J. Cell Biol. 143:991–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kovács-Bogdán E, Soll J, Bölter B. 2010. Protein import into chloroplasts: the Tic complex and its regulation. Biochim. Biophys. Acta 1803:740–747 [DOI] [PubMed] [Google Scholar]
- 46. Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567–580 [DOI] [PubMed] [Google Scholar]
- 47. Küchler M, Decker S, Hörmann F, Soll J, Heins L. 2002. Protein import into chloroplasts involves redox-regulated proteins. EMBO J. 21:6136–6145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Letunic I, Doerks T, Bork P. 2009. SMART 6: recent updates and new developments. Nucleic Acids Res. 37:D229–D232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Martin W, et al. 1998. Gene transfer to the nucleus and the evolution of chloroplasts. Nature 393:162–165 [DOI] [PubMed] [Google Scholar]
- 50. McFadden GI, Gilson PR, Hofmann CJ, Adcock GJ, Maier UG. 1994. Evidence that an amoeba acquired a chloroplast by retaining part of an engulfed eukaryotic alga. Proc. Natl. Acad. Sci. U. S. A. 91:3690–3694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Patron NJ, Waller RF. 2007. Transit peptide diversity and divergence: a global analysis of plastid targeting signals. Bioessays 29:1048–1058 [DOI] [PubMed] [Google Scholar]
- 52. Rodríguez-Ezpeleta N, et al. 2005. Monophyly of primary photosynthetic eukaryotes: green plants, red algae, and glaucophytes. Curr. Biol. 15:1325–1330 [DOI] [PubMed] [Google Scholar]
- 53. Rogers MB, Gilson PR, Su V, McFadden GI, Keeling PJ. 2007. The complete chloroplast genome of the chlorarachniophyte Bigelowiella natans: evidence for independent origins of chlorarachniophyte and euglenid secondary endosymbionts. Mol. Biol. Evol. 24:54–62 [DOI] [PubMed] [Google Scholar]
- 54. Rogers MB, et al. 2004. Plastid-targeting peptides from the chlorarachniophyte Bigelowiella natans. J. Eukaryot. Microbiol. 51:529–535 [DOI] [PubMed] [Google Scholar]
- 55. Rost B, Yachdav G, Liu J. 2004. The PredictProtein server. Nucleic Acids Res. 32:W321–W326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sánchez-Pulido L, Devos D, Genevrois S, Vicente MVA. 2003. POTRA: a conserved domain in the FtsQ family and a class of beta-barrel outer membrane proteins. Trends Biochem. Sci. 28:523–526 [DOI] [PubMed] [Google Scholar]
- 57. Sohrt K, Soll J. 2000. Toc64, a new component of the protein translocon of chloroplasts. J. Cell Biol. 148:1213–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sommer MS, et al. 2011. Chloroplast Omp85 proteins change orientation during evolution. Proc. Natl. Acad. Sci. U. S. A. 108:13841–13846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sommer MS, et al. 2007. Der1-mediated preprotein import into the periplastid compartment of chromalveolates? Mol. Biol. Evol. 24:918–928 [DOI] [PubMed] [Google Scholar]
- 60. Spork S, et al. 2009. An unusual ERAD-like complex is targeted to the apicoplast of Plasmodium falciparum. Eukaryot. Cell 8:1134–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Stahl T, Glockmann C, Soll J, Heins L. 1999. Tic40, a new “old” subunit of the chloroplast protein import translocon. J. Biol. Chem. 274:37467–37472 [DOI] [PubMed] [Google Scholar]
- 62. Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690 [DOI] [PubMed] [Google Scholar]
- 63. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 64. Teng YS, et al. 2006. Tic21 is an essential translocon component for protein translocation across the chloroplast inner envelope membrane. Plant Cell 18:2247–2257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. van Dooren GG, Tomova C, Agrawal S, Humbel BM, Striepen B. 2008. Toxoplasma gondii Tic20 is essential for apicoplast protein import. Proc. Natl. Acad. Sci. U. S. A. 105:13574–13579 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.