Abstract
In the multinucleate filamentous fungus Ashbya gossypii, nuclei divide asynchronously in a common cytoplasm. We hypothesize that the division cycle machinery has a limited zone of influence in the cytoplasm to promote nuclear autonomy. Mitochondria in cultured mammalian cells undergo cell cycle-specific changes in morphology and membrane potential and therefore can serve as a reporter of the cell cycle state of the cytoplasm. To evaluate if the cell cycle state of nuclei in A. gossypii can influence the adjacent cytoplasm, we tested whether local mitochondrial morphology and membrane potential in A. gossypii are associated with the division state of a nearby nucleus. We found that mitochondria exhibit substantial heterogeneity in both morphology and membrane potential within a single multinucleated cell. Notably, differences in mitochondrial morphology or potential are not associated with a specific nuclear division state. Heterokaryon mutants with a mixture of nuclei with deletions of and wild type for the mitochondrial fusion/fission genes DNM1 and FZO1 exhibit altered mitochondrial morphology and severe growth and sporulation defects. This dominant effect suggests that the gene products may be required locally near their expression site rather than diffusing widely in the cell. Our results demonstrate that mitochondrial dynamics are essential in these large syncytial cells, yet morphology and membrane potential are independent of nuclear cycle state.
INTRODUCTION
Multinucleate cells exist naturally in diverse environments, including skeletal muscle, mammalian placenta, fruit fly embryos, pathogenic fungi, and malignant tumors. Despite their prevalence, relatively little is understood about the regulation of the cell cycle in multinucleated cells. Early cell cycle research demonstrated that nuclei of artificially fused mammalian cells synchronize their division cycles (33). Subsequent work over the past decades has led to a cell cycle paradigm in which oscillating expression and degradation of specific protein regulators controls the cycle of cell division. Because these protein regulators are diffusible and move between the cytoplasm and nucleus, it is thought that all nuclei in an artificially multinucleated cell experience the same concentrations of these regulators at all times and, therefore, synchronize their division cycles.
However, nuclei in a common cytoplasm do not always act in unison. DNA damage to just one nucleus in binucleate sea urchin embryos or Saccharomyces cerevisiae cells disrupts nuclear synchrony by delaying cell cycle progression of only the damaged nucleus (7, 18). In marsupial cells with two mitotic spindles, the anaphase transition of one spindle can be delayed by perturbing chromosome attachment (34). Asynchronous nuclear division has also been observed during normal growth in syncytial fungal systems, including Neurospora crassa and Ashbya gossypii (10, 12). A. gossypii grows as a vegetative mycelium with haploid nuclei and sporulates to produce haploid ascospores. Single A. gossypii cells contain dozens of asynchronously dividing nuclei linearly organized in hyphae, and nuclei come within 1 μm of neighbors when passing each other in the hypha (12). Sister nuclei, which result from a single mitosis event, immediately lose synchrony during the subsequent G1 phase, which has been observed to last from 10 to over 65 min in A. gossypii (28).
Throughout uninucleate cells, variable timing of the G1/S transition contributes substantially to intercellular variability in division timing. While cell size is a major factor influencing this variability in uninucleate cells, multiple nuclei inhabiting a common cytoplasm must establish such variability by a different mechanism, such as molecular noise in transcription (9). Deletion of components of the G1 transcriptional regulatory circuit in A. gossypii results in shorter, less variable division times and increased synchrony among nuclei, suggesting that variability in division timing is established at least in part by local fluctuations in the transcription of cell cycle regulator genes (28). However, the abilities of nuclei in a common cytoplasm to respond to local fluctuations in transcription and to undergo division cycles independently of each other also require nuclear autonomy of unknown molecular origin.
In syncytial Drosophila melanogaster embryos, the endoplasmic reticulum (ER) and Golgi complex form compartmentalized structures surrounding individual nuclei, and these organized compartments of cytoplasm correspond to the regional heterogeneity of the plasma membrane (11, 25). It is possible that nuclear autonomy in A. gossypii is established by a similar organization of cytoplasm, which may create functionally distinct zones around each nucleus in a single cell. Such “islands of cytoplasm” may contribute to the establishment of both nuclear autonomy and variability in nuclear division by limiting the diffusion of signaling molecules between neighboring nuclei. We hypothesized that if cytoplasmic islands exist in A. gossypii, organelles may display intracellular heterogeneity dependent on the division state of a nearby nucleus.
In this study, we analyze mitochondria, which have been shown to undergo defined changes during the cell cycle in other systems. Mitochondria are highly dynamic organelles, whose morphology is regulated by the conserved GTPases Fzo1p and Dnm1p, responsible for mitochondrial fusion and fission, respectively (8). In cultured mammalian cells, mitochondria form a hyperfused network at the G1/S transition, with increased mitochondrial membrane potential and ATP output, and mitochondria fragment during mitosis (26, 38). Furthermore, disrupted mitochondrial function can initiate a G1/S arrest in Drosophila (23, 24). Although analysis of mitochondrial function and morphology and the cell cycle has been limited primarily to cultured mammalian cells and Drosophila, a hyperfused mitochondrial network at the G1/S transition was first described during meiosis and sporulation in budding yeast, suggesting the possibility of an evolutionarily conserved relationship between mitochondria and cell cycle progression throughout eukaryotes (27). Therefore, we set out to determine if the mitochondrial state could be a reporter of the cell cycle phase of the cytoplasm in multinucleate A. gossypii cells. In this study, we evaluate mitochondrial morphology and membrane potential through time and space within A. gossypii cells and investigate connections between the nuclear cycle and mitochondrial dynamics.
MATERIALS AND METHODS
Strain construction.
A. gossypii was cultured and transformed as previously described (43). PCR was performed using standard methods with DNA polymerase from Roche Diagnostics (Indianapolis, IN) and Invitrogen (Carlsbad, CA). Restriction enzymes were from New England BioLabs (Beverly, MA), and oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, IA). DNA manipulations were performed according to the protocol of Sambrook and Russell (36). All DNA sequencing was performed by Dartmouth College Core Facilities (Hanover, NH).
All A. gossypii strains, plasmids, and oligonucleotide primers used in this study are listed in Tables 1, 2, and 3. To generate plasmid AGB131, a 2,456-bp product containing CFP-NAT1 was PCR amplified from AGB011 using AGO184 and AGO187. This fragment was cotransformed into DHD5 yeast with AGB028 to yield AGB131, which was verified by sequencing with AGO186. To generate the plasmid AGB133, the plasmid AGB131 was digested with HindIII and XhoI, and a 3,389-bp segment containing A. gossypii HHF1 (AgHHF1)-CFP-NAT1 was isolated by gel extraction. This segment was ligated into pUC19 and verified by digestion with NruI and NdeI. A. gossypii strain AG261 was generated by digestion of AGB133 with NheI and HindIII and transformation of this fragment into the wild type (WT). To generate a stable homokaryon strain, single spores were isolated from initial transformants and grown. Transformation was confirmed by fluorescence microscopy.
Table 1.
A. gossypii strains used in this study
| Strain | Relevant genotypea | Reference or source |
|---|---|---|
| WT | leu2Δ thr4Δ | 2 |
| AG261 | AgHHF1-CFP-NAT1 leu2Δ thr4Δ | This study |
| AG270 | AgTUB4-mCherry-NAT1leu2Δ thr4Δ | 28 |
| AG469 | [HISpr-mitoGFP-GEN3] AgTUB4-mCherry-NAT1leu2Δ thr4Δ | This study |
| AG510 | (HISpr-mitoGFP-GEN3 +WT) AgTUB4-mCherry-NAT1leu2Δ thr4Δ | This study |
| AG594 | (dnm1Δ-HISpr-mitoGFP-GEN3 +WT) AgTUB4-mCherry-NAT1leu2Δ thr4Δ | This study |
| AG595 | (fzo1Δ-HISpr-mitoGFP-GEN3 +WT) AgTUB4-mCherry-NAT1leu2Δ thr4Δ | This study |
Genes expressed from a plasmid are indicated by brackets, and heterokaryon alleles are indicated by parentheses.
Table 2.
Plasmids used in this study
| Plasmid | Name | Vector | Insert | Reference or source |
|---|---|---|---|---|
| pUC19 | ||||
| pRS416 | 39 | |||
| AGB011 | pAGT102 | pUC19 | CFP-NAT1 | 20 |
| AGB028 | pAG1552 | pRS416 | AgHHF1-GEN3 | 1 |
| AGB096 | ScHIS3pr-SWE1-6HA | pRS416 | ScHIS3pr-SWE1-6HA | 16 |
| AGB131 | pRS416 | AgHHF1-CFP-NAT1 | This study | |
| AGB133 | pUC19 | AgHHF1-CFP-NAT1 | This study | |
| AGB280 | B1220 | pYX142 | Su9(1-69)-GFP+ | 10 |
| AGB303 | pGEN::ScHIS3pr | pRS416 | GEN3-ScHIS3pr | This study |
| AGB319 | pGEN::ScHIS3pr-mitoGFP | pRS416 | GEN3-ScHIS3pr-mitoGFP | This study |
Table 3.
Oligonucleotide primers used in this study
| Primer | Sequence |
|---|---|
| AGO098 | CTTGCCATCCTATGGAACTG |
| AGO184 | GCAAAAGCAACCGTGACACACGATATGGCAGATTCACATACATATATGCCTGCAGCCAAACAGTGTTC |
| AGO186 | GACAACATCCAGGGCATCAC |
| AGO187 | TACGCGCTCAAGCGCCAGGGCCGCACCTTGTACGGCTTCGGCGGCAAAACGACGGCCAGTGAATTCG |
| AGO200 | AGACACGTGCTGAAGTCAAG |
| AGO202 | GTCTTGTAGTTCCCGTCATC |
| AGO471 | TTACTCACCACTGCGATCCC |
| AGO510 | TGGCCTCCTCTAGTACACTC |
| AGO512 | TCGCGAGCCCATTTATACCC |
| AGO717 | GATGCCTTATACCGGCATAC |
| AGO718 | ATTGCGCAGTTCCGGACATGCC |
| AGO751 | CCTACACCGAACTGAGATAC |
| AGO802 | GATTACTAGTATGGCCTCCACTCGTGTCCTC |
| AGO803 | GCATCCGCGGGCTGAGCTCTTATTTGTAGAG |
| AGO815 | AGCGCGCAATTAACCCTCAC |
| AGO839 | TCCCTCCATGAGTTCCCATCGCGTAGAGCATCATATGCGCGGACAGCAGGCATGCAAGCTTAGATC |
| AGO840 | CGCGTAAAGAATGGTCGCTCCGTGCTGCCGTAGACTGCGCTGTCGTGAGCTCTTATTTGTAGAGTTC |
| AGO908 | GAGGTTTGGAGGAGCACGCGCGGAAGATTGGTAAAGTTGGACACAATTCAGGCATGCAAGCTTAG |
| AGO909 | AGTGGATACTAGAGTGTGAAAGGTTCTGAGGCATTGTCATAAATGGCGGGCTGAGCTCTTATTTG |
| AGO910 | CTATTTACATAACGTAGACGGGTCGCCAAAGAACCAAGAAGCCCTGCGGGCTGAGCTCTTATTTG |
| AGO911 | CGAACACAAGGATACTACGGCAGGTTGCCTCAAAGACAGTGACTAATTCAGGCATGCAAGCTTAG |
The plasmid AGB303 was generated by digestion of AGB096 with BamHI and EcoRI and ligation of the fragment containing Saccharomyces cerevisiae HIS3 promoter (ScHIS3pr) into pRS416, which had also been digested by BamHI and EcoRI. The resulting plasmid was sequenced with AGO471, AGO512, and AGO751 for verification. To generate the plasmid ABG319, a 965-bp product containing the green fluorescent protein with a mitochondrial targeting sequence (mitoGFP) was amplified from AGB280 using PCR primers AGO802 and AGO803 (10). This product was digested with SpeI and SacII and ligated into the plasmid AGB303, which had also been digested with SpeI and SacII. AGB319 was verified by digestion with EcoRI and NdeI and sequencing using primers AGO200, AGO202, AGO510, and AGO815. AGB319 was transformed into A. gossypii to generate strain AG469. Transformation was verified by fluorescence microscopy.
To generate dnm1Δ and fzo1Δ heterokaryon strains (AG594 and AG595), a deletion cassette containing GEN3 and mitoGFP with 45 bp of homology to genomic regions bordering the gene of interest was generated by PCR using oligonucleotide primer pairs AGO-908/AGO-909 and AGO-910/AGO-911, respectively. The template for the PCR was a 6.5-kb gel-purified segment of AGB319, obtained from double digestion with EcoRI and StuI. The deletion cassette was transformed into strain AG270. Two independent transformations with the dnm1Δ cassette yielded two independent transformants with similarly disrupted growth and mitochondrial morphology. One transformation with the fzo1Δ cassette yielded two independent transformants with disrupted growth and mitochondrial morphology, distinct from that of the dnm1Δ strain. Strain AG510 was generated by transformation of AG270 with a deletion cassette containing the GEN3 gene and mitoGFP with 45 bp of homology to an intergenic region of chromosome I. The cassette was generated by PCR using a 3-kb gel-purified segment of AGB319 resulting from digestion with PvuII and XmnI as a template and oligonucleotide primers AGO839 and AGO840. Genomic DNA was purified from transformants, and integration of the cassette was verified by analytical PCR with primers AGO200, AGO98, AGO717, and AGO718.
Throughout, the word “hypha” describes a single, linear branch of a cell, “cell” describes a network of hyphae with a continuous cytoplasm, and “mycelium” describes the macroscopic growth on solid media, which can be composed of many individual cells.
Growth assay.
Approximately 5 mm3 of live mycelium was inoculated onto the center of Ashbya full medium (AFM) agar plates containing 200 μg/ml G418 for strain AG510, the dnm1Δ mutant, and the fzo1Δ mutant or containing no G418 for the WT (strain AG270). Plates were imaged daily for 7 days using a Bio-Rad ChemiDoc XR5 molecular imager with ImageLab software. Colony area and perimeter were measured using a macro written in ImageJ (NIH), which “autothresholded” images and determined colony area and perimeter.
Fixation and DNA staining.
Cells were fixed in 2% paraformaldehyde (Fisher Scientific, Fair Lawn, NJ) for 8 min at 30°C with occasional gentle inversion. They were washed 3 times in 1× phosphate-buffered saline (PBS) and stained with 2 μg/ml Hoechst dye (Invitrogen) for 10 min at room temperature. Cells were again washed in PBS, resuspended in ProLong Gold antifade reagent (Invitrogen), and plated on glass slides, which were stored at −20°C.
Nocodazole treatment.
Cells were grown in AFM containing 100 μg/ml ampicillin and 300 μg/ml G418 to select for the plasmid from which mitoGFP was expressed. After 12 h, an additional 300 μg/ml G418 was added, and 15 μg/ml nocodazole (Sigma-Aldrich, St. Louis, MO) was added to half of the culture. Treated cells were imaged after 30 or 180 min, and untreated cells were imaged after 180 min. Before live imaging, cells were collected by low-speed centrifugation, washed in low-fluorescence minimal medium, and plated on a 2% agarose minimal medium gel pad.
Mitochondrion visualization of dnm1Δ and fzo1Δ heterokaryons and AG510.
A small amount of mycelium was collected from the edge of a colony growing on an AFM agar plate with 200 μg/ml G418 at 30°C. Mycelium was suspended in AFM and incubated with shaking in a baffled flask at 30°C for 3 h. One hundred nanomolar MitoTracker Green FM (1 mM stock in dimethyl sulfoxide [DMSO]; Invitrogen) was added for 30 min, and cells were collected by centrifugation, washed with minimal medium, and imaged on a 2% agarose minimal medium gel pad.
Simultaneous MitoTracker and TMRE staining.
WT cells (see Fig. 3B) were grown for 10.5 h in filter-sterilized AFM containing 100 μg/ml ampicillin. One hundred nanomolar MitoTracker Green FM (1 mM stock in DMSO; Invitrogen) was added, and cells were stained at 30°C for 30 min. Cells were collected by centrifugation and plated on a gel pad made with 2% agarose dissolved in low-fluorescence minimal medium containing 15 nM tetramethylrhodamine ethyl ester (TMRE) (1 mM stock in DMSO; Sigma-Aldrich, St. Louis, MO). Cells were placed in a humid chamber at 30°C for 30 min to stain and settle before being imaged. For carbonyl cyanide m-chlorophenyl hydrazone (CCCP) and oligomycin experiments (see Fig. 3A), WT cells were grown for 10 h in filter-sterilized AFM, stained with 100 nM MitoTracker Green FM for 30 min at 30°C, collected by centrifugation, and resuspended in liquid minimal medium. Cells were then stained with 15 nM TMRE for 20 min at 30°C, treated with 5 μg/ml oligomycin (5 mg/ml stock in DMSO; Sigma-Aldrich, St. Louis, MO), 10 μM CCCP (100 mM stock in DMSO; Sigma-Aldrich, St. Louis, MO), or nothing, collected by centrifugation, and imaged immediately without being washed. H4-CFP-expressing cells (strain AG261 [see Fig. 4 and 6]) were treated in a similar manner but grown for 10 h and stained with 125 nM MitoTracker Green FM and 5 nM TMRE.
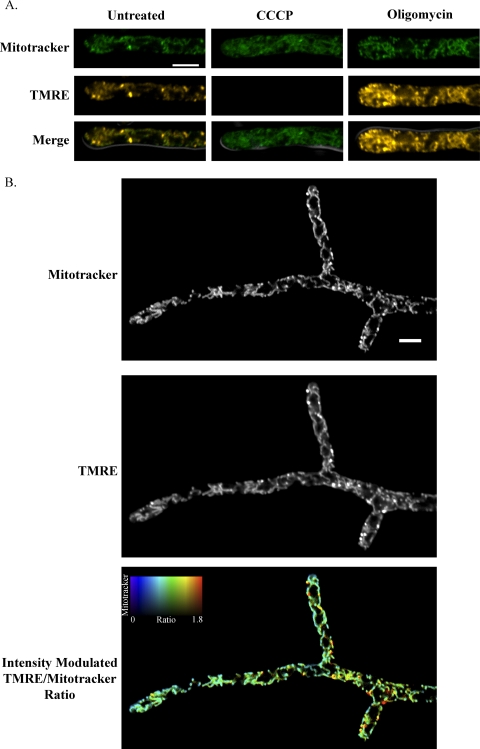
Fig 3.
ΔΨM values are heterogeneous in single A. gossypii cells. (A) WT cells were stained with MitoTracker Green FM (MitoTracker), which accumulates in mitochondria based on mass, and tetramethylrhodamine ethyl ester (TMRE), which accumulates in mitochondria based on ΔΨM. Cells were treated with either oligomycin, which increases ΔΨM by inhibiting ATP synthase, or CCCP, which reduces ΔΨM by facilitating proton diffusion through the mitochondrial membrane. MitoTracker and TMRE fluorescence signals were acquired simultaneously for a single Z plane using a scanning confocal microscope. The merged image shows the cell outline by differential inference contrast (DIC) in gray, and the scale bar represents 5 μm. (B) WT cells were stained with MitoTracker and TMRE. In the intensity-modulated-ratio image, the TMRE/MitoTracker ratio is displayed using a colorimetric scale, with the brightness of each pixel modulated by the intensity of the MitoTracker signal.
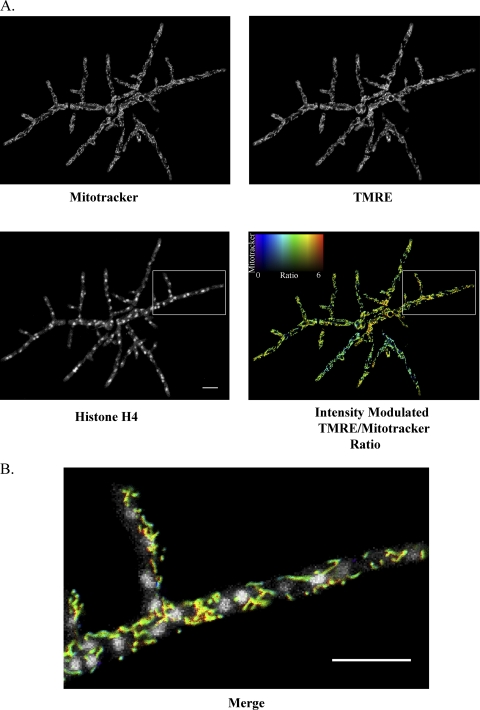
Fig 4.
Nuclei experience different ΔΨM environments within a single A. gossypii cell. (A) Cells expressing a CFP-tagged histone, H4 (AG261), were stained with MitoTracker and TMRE. MitoTracker and TMRE signals were acquired simultaneously for a single Z plane, and CFP signal was acquired in a Z stack. In the intensity-modulated-ratio image, the TMRE/MitoTracker ratio of each pixel is indicated by the hue and the intensity of the MitoTracker signal is indicated by the brightness. (B) The region indicated by the box is shown at higher magnification as a merge of the histone H4 signal and the intensity-modulated TMRE/MitoTracker ratio. Scale bars represent 10 μm.
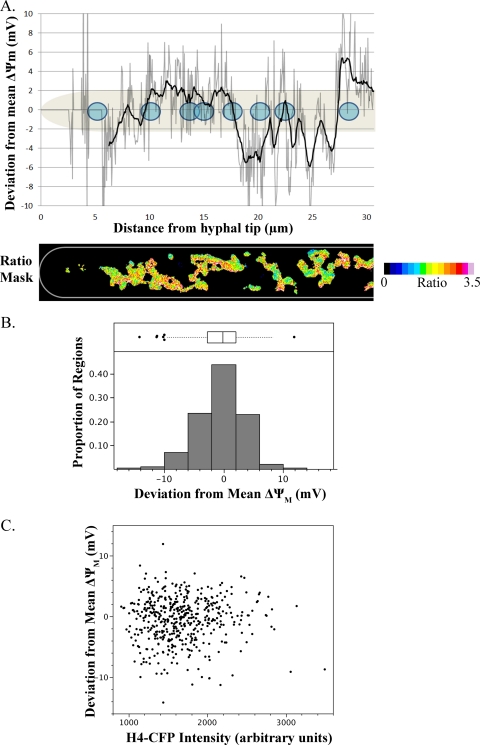
Fig 6.
ΔΨM heterogeneity in single A. gossypii cells is independent of nuclear division state. (A) For the hypha displayed, the ratio of TMRE/MitoTracker signal was calculated for pixels defined as mitochondrial. For each mitochondrial pixel, the mean TMRE/MitoTracker ratio was calculated and compared to the mean TMRE/MitoTracker ratio of mitochondrial pixels in the entire hypha to determine each pixel's deviation from the mean ΔΨM of the hypha. This deviation was plotted versus distance from the hyphal tip (gray line) as the mean deviation of mitochondrial pixels within each column of pixels in the image (0.05-μm width). To reduce noise in the plotted trace, another curve was plotted (black line) in which each point represents the mean deviation of mitochondrial pixels within 0.5 μm of a given column. Positions of nuclei (shown in blue) were determined by visualization of CFP-tagged H4. (B) For 6 cells, the mean ratio value of all mitochondrial pixels within a 4-μm-diameter circular region centered on each nucleus was calculated and compared to the mean ratio of mitochondrial pixels in the entire cell to determine each region's deviation from the mean ΔΨM of the cell. Regions for which fewer than 5% of pixels were mitochondrial were excluded from analysis due to the contribution of noise, so data from 468 perinuclear regions are represented. (C) For each nucleus, division state was analyzed by fluorescence intensity of CFP-tagged histone H4, which increases during the nuclear division cycle. The deviation from the mean ΔΨM of each perinuclear region was plotted versus the H4-CFP intensity of the nucleus.
Wide-field fluorescent imaging and processing.
Wide-field images were acquired on a Zeiss AxioImager-M1 light microscope with a Plan-Apochromat 63×/1.4-numerical-aperture (NA) objective (see Fig. 1, 2, 7, and 8A) or a Plan-Neofluar 40×/1.3-NA objective (see Fig. 8B). GFP was visualized with Zeiss filter set 38E, Hoechst dye was visualized with Zeiss filter set 49, and mCherry was visualized with Chroma filter set 41043. The fluorescent light source was an Exfo X-Cite 120 lamp, and the camera was a Hamamatsu Orca-ER (model no. C4742-8012AG). Volocity 5 (Improvision-PerkinElmer) was used for all acquisitions. GFP signals were processed by 50 iterations of deconvolution using a point spread function generated in Volocity 4. Except where noted, wide-field images are presented as maximum projections of 0.3-μm Z-step images acquired through the depth of the cell.
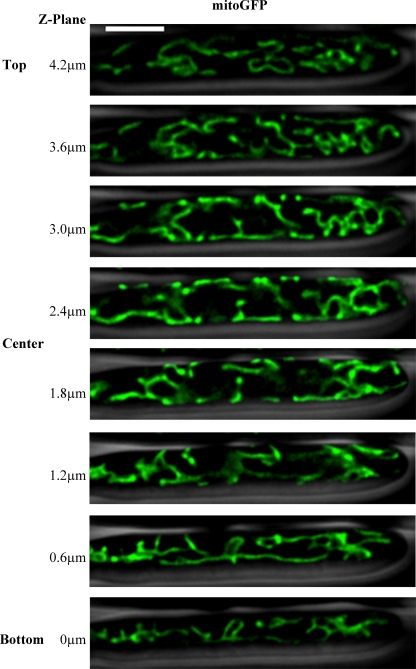
Fig 1.
Mitochondria form tubular networks in A. gossypii. Cells expressing mitoGFP (AG469) were visualized using wide-field fluorescence microscopy. Images were captured at 0.3-μm sections throughout the Z dimension of the cell, and the stack of images was deconvolved. Individual Z slices are displayed in 0.6-μm increments, with the cell outline from phase-contrast microscopy shown in gray. The scale bar represents 5 μm.
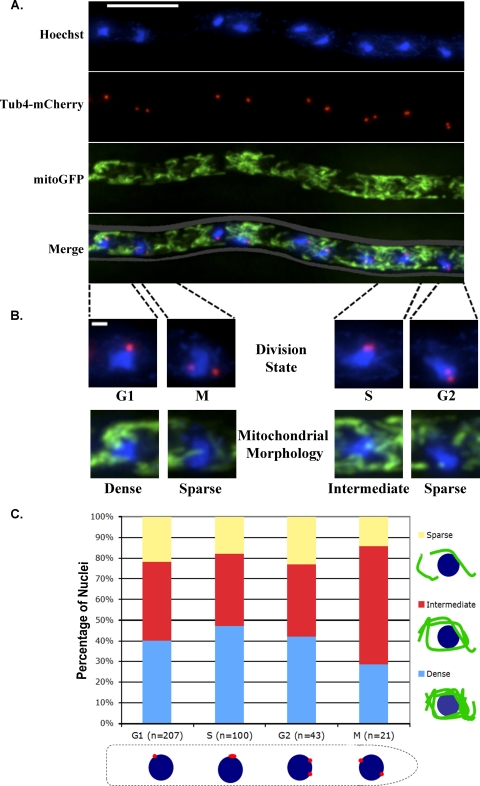
Fig 2.
Heterogeneity of mitochondrial morphology is independent of nuclear division. (A) Cells expressing mitoGFP to visualize mitochondria and Tub4-mCherry to visualize spindle pole bodies (SPBs) (AG469) were fixed, and DNA was stained with Hoechst dye. mitoGFP signal is shown in green, SPBs are shown in red, and nuclei are shown in blue. The scale bar represents 10 μm. (B) Nuclei were scored for division state using SPBs as a marker and for the morphology of mitochondria immediately surrounding a nucleus. Individual nuclei are shown to demonstrate scoring categories. The scale bar represents 1 μm. (C) Three hundred seventy-one nuclei were scored for division state and the morphology of surrounding mitochondria.
Fig 7.
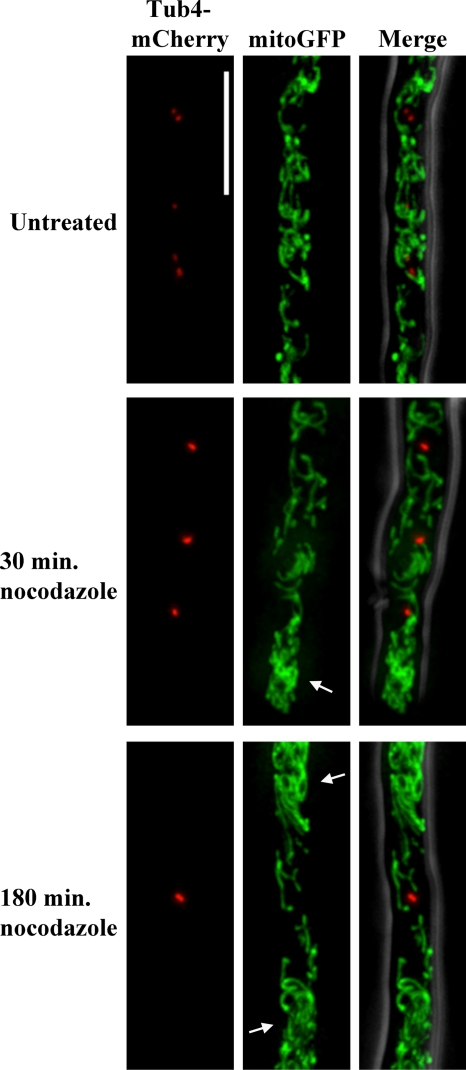
Microtubule depolymerization disrupts mitochondrial morphology, but G2/M arrest does not induce mitochondrial fragmentation. Cells expressing mitoGFP and Tub4-mCherry (AG469) were treated with the microtubule polymerization inhibitor nocodazole for 30 min to depolymerize cytoplasmic microtubules without widespread cell cycle arrest or 180 min to arrest nuclei at G2/M and imaged live. SPBs confirm G2/M arrest and increased internuclear distance due to inhibition of nuclear division. Arrows indicate particularly densely clumped mitochondria in nocodazole-treated cells. The scale bar represents 5 μm.
Fig 8.
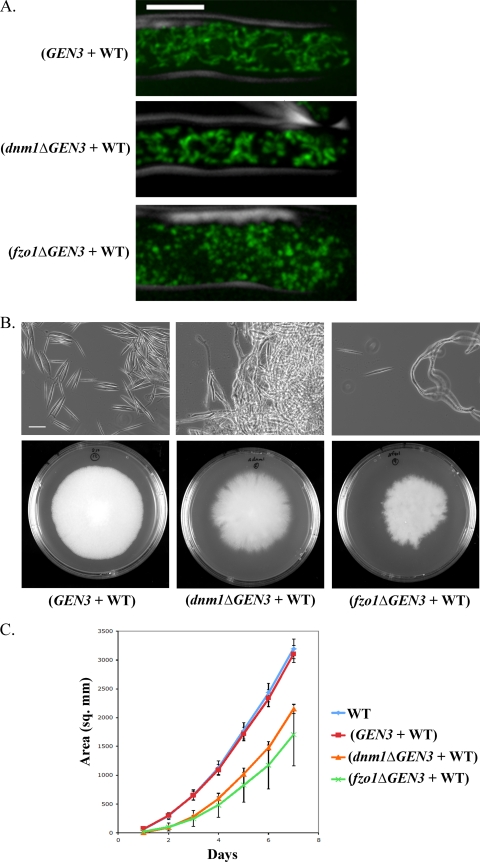
dnm1Δ and fzo1Δ heterokaryon mutants display defects in mitochondrial morphology and colony growth. (A) DNM1 and FZO1 were deleted by genomic integration of a deletion cassette containing the selectable marker GEN3. Deletion strains were heterokaryons and contained both haploid nuclei with deletion alleles and nuclei with WT alleles. Mycelia grown on agar plates of dnm1Δ (AG594) and fzo1Δ (AG595) heterokaryon mutants and a control heterokaryon strain with the same deletion cassette integrated into an intergenic region of chromosome I (AG510) were stained with MitoTracker in liquid AFM. A cell outline from phase-contrast microscopy is shown in gray. The scale bar represents 5 μm. (B) Strains were grown on agar plates with G418 selection for 7 days at 30°C. A mycelium growing on agar plates was spread on a glass slide. Needle-shaped spores are prevalent only in the control heterokaryon. The scale bar represents 25 μm. (C) The colony area was measured daily for strains grown on agar plates for 7 days. Error bars show standard deviations of results from three replicates.
Confocal fluorescent imaging.
Confocal fluorescent images were acquired on a Nikon A1 line-scanning confocal system using Nikon Elements 3.2 software (see Fig. 3 and 4) or a Zeiss LSM 710 microscope using Zeiss ZEN software (see Movie S1 in the supplemental material). On the Nikon A1 system, all images were acquired using a Plan-Apochromat VC 60×/1.4-NA objective. CFP signals were acquired using a 457-nm laser for excitation and a 482/35 filter for emission with a pinhole size of 136 μm (3 arbitrary units [AU]). MitoTracker signals were acquired using a 488-nm laser for excitation and a 525/50 filter for emission with a pinhole size of 33.3 μm. TMRE signals were acquired using a 561-nm laser for excitation and a 595/50 filter for emission with a pinhole size of 33.3 μm. Supplemental Movie S1 was captured on a Zeiss LSM 710 system using a Plan-Apochromat 63×/1.4-NA objective. GFP signal was acquired using a 488-nm laser for excitation and a spectral detector detecting light between 495 and 563 nm for emission, with a pinhole size of 67 μm.
Conversion of ratio values to deviation in ΔΨM.
Because of Nernstian partitioning of cationic dyes across the mitochondrial membrane, differences in mitochondrial membrane potential (ΔΨM) can be calculated between two points within a cell by differences in the ratio of TMRE to MitoTracker Green FM (MitoTracker) fluorescence as previously described (45). Importantly, while TMRE fluorescence depends on both mitochondrial uptake of the dye and the mitochondria's location relative to the focal plane of the microscope, the ratio of TMRE intensity to the intensity of MitoTracker or another ΔΨM-independent signal is insensitive to a mitochondrion's location relative to the focal plane (41, 45). Based on the Nernst equation, the electric potential (E) of an ion of charge z partitioned at equilibrium across a membrane can be described as follows: E = (RT/zF) × ln([ion]in/[ion]out), where R is the universal gas constant, T is the absolute temperature, and F is the Faraday constant. When comparing the differences in membrane potential between two mitochondrial points in a single cell, ΔΨM between two points can be described by a ratio of TMRE to MitoTracker fluorescence intensity as follows: ΔΨM = RT/F × ln(ratio1/ratio2). Ratio1 and ratio2 refer to the ratio of TMRE to MitoTracker signal at two locations within a cell. At room temperature, this can be simplified to ΔΨM = 59.2 mV × log10(ratio1/ratio2). For our analysis, ratio1 was the mean ratio of mitochondrial pixels within the region of interest, and ratio2 was the mean of ratios of mitochondrial pixels within the entire cell (see Fig. 6B and C) or hypha (see Fig. 6A) analyzed. In this case, ΔΨM describes the difference between the mean membrane potential in the region of interest and the mean membrane potential of the entire cell or hypha.
TMRE/MitoTracker ratio processing and quantification.
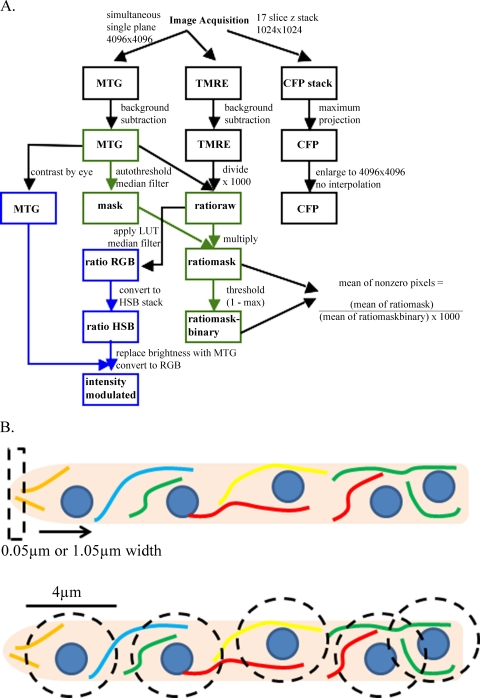
Images were processed entirely in Fiji (version 1.44; NIH [http://pacific.mpi-cbg.de/]), a package of ImageJ with additional plug-ins, according to the schematic shown (see Fig. 5A). Colored pathways in Fig. 5A indicate that Fiji macros were developed to automate this process, while processes linked by black lines were performed manually in Fiji. These processes used background-subtracted images of the MitoTracker Green (MTG) and TMRE channels to yield a ratio image, which was either intensity modulated by the MTG signal for display or converted into a mask for quantification that eliminated nonmitochondrial pixels from analysis. The stack of CFP signals was converted to a maximum projection and enlarged to 4,096 by 4,096 pixels without interpolation. For images used for quantification, but not display, cells other than the cell of interest and autofluorescent extracellular material were removed from the MTG image to ensure accuracy in further processing. The background signals of MTG and TMRE channels were determined by averaging the means of 4 intracellular regions and manually subtracting this value from the image. The TMRE channel was then divided by the MTG channel and multiplied by 1,000 to yield a 16-bit image called “ratioraw.” The MTG channel was autothresholded, and a 1-pixel median filter was applied to generate a binary mask, which was multiplied by the ratioraw image to yield an image called “ratiomask.” A copy of ratiomask was thresholded from 1 to the maximum value to generate a binary image called “ratiomask-binary,” which indicated the position of nonzero pixels in ratiomask. These nonzero pixels are referred to as mitochondrial pixels. Ratiomask and ratiomask-binary were used to quantify mean ratios of nonzero pixels in regions of interest. The intensity-modulated images were generated by applying a rainbow look-up table to the ratioraw image with appropriate contrast settings, applying a median filter with a 1-pixel radius, and converting the image to RGB color. This image, called ratio RGB, was converted to a hue, saturation, brightness (HSB) stack, and the brightness channel was replaced with a linearly contrast-enhanced image of the MTG channel. Converting this new HSB stack to an RGB image yielded an intensity-modulated ratio image used for display.
Fig 5.
Quantification and processing of TMRE/MitoTracker ratio images. (A) TMRE and MitoTracker signals were processed according to the protocol outlined here to generate ratio images for visualization (intensity modulated) or quantification of ΔΨMs (ratiomask and ratiomask-binary). Colored boxes indicate steps that were automated using Fiji (Image J) macros. LUT, look-up table. (B) Models demonstrate the methods of one-dimensional (top) and two-dimensional (bottom) quantification of ΔΨMs shown in Fig. 6.
Ratio images were quantified using a one-dimensional trace method (see Fig. 6A) or a two-dimensional perinuclear region analysis method (see Fig. 6B and C; the results of analysis methods are depicted in Fig. 5B). To quantify line traces along a hypha, the “plot profile” command in Fiji was used to calculate mean pixel values for linear columns of pixels in a rectangular region of interest in both ratiomask and ratiomask-binary, and these values were imported into Microsoft Excel. Because the mean of ratiomask values was dependent on the amount of mitochondrial pixels in the column, these values were divided by the mean of ratiomask-binary values to yield an average of mitochondrial pixels in ratiomask by the relationships x̅mitochondrial pixels = x̅all pixels/proportion of mitochondrial pixels and x̅mitochondrial pixels = x̅ratiomask/x̅ratiomask-binary. The average of mitochondrial pixels in ratiomask was plotted versus distance from the hyphal tip for each column of pixels in the image. To smooth the curve, a moving average that calculated the mean of all mitochondrial pixels within 10 pixels (0.5 μm) of the column of interest was used. Because the height of the column was fixed, this could be done by weighting the contribution of each column's mean by the proportion of mitochondrial pixels within the column. Ratios were then converted to deviation from mean ΔΨM using the mean ratio of mitochondrial pixels in the entire region analyzed. The position of nuclei was determined by manually aligning the CFP maximum projections on top of the linear plot in Microsoft PowerPoint based on fitting the start and endpoint of the intracellular CFP background of the region analyzed to the start and endpoint of the graphical line trace. Nuclei were manually drawn onto the chart.
CFP signal and mean ratios of perinuclear mitochondria were quantified in Volocity 5 (Improvision-PerkinElmer). Ratiomask, ratiomask-binary, and CFP maximum projections were imported as separate channels in a single image sequence in Volocity. A single, manually determined registration correction based on the intracellular background intensity in the CFP channel and the localization of mitochondria was used to align the CFP signal with the mitochondrial signals for all images. For all nuclei in an image, a circular region of interest (ROI) with a diameter of 1.5 μm was placed at the center of each nucleus to measure the mean CFP intensity of the nucleus. Subsequently, a circular ROI with a diameter of 4 μm was placed at the center of each nucleus to measure the mean TMRE/MitoTracker ratio of perinuclear mitochondrial pixels by dividing the mean of ratiomask values for the region by the mean of ratiomask-binary values. To convert mean ratios of regions to deviations from mean ΔΨM, the mean ratio of mitochondrial pixels in ratiomask for the entire cell was calculated. Because mitochondrial signal was acquired from an image of a single plane and CFP was acquired as a Z stack through an entire cell, mitochondrial signal was not captured for nuclei outside the mitochondrial focal plane. For 58 nuclei of the 526 measured, fewer than 5% of pixels in the 4-μm ROI were mitochondrial elements, so these nuclei were excluded from all analyses due to an increased contribution of noise.
Statistical analysis.
All statistical analyses and generation of graphical figures were performed in JMP (version 8; SAS) and Microsoft Excel (version 11.6).
RESULTS
Heterogeneity in mitochondrial morphology is not associated with nuclear division state.
To visualize mitochondria in A. gossypii, we expressed green fluorescent protein (GFP) with a mitochondrial targeting sequence (mitoGFP) from a replicating plasmid. Mitochondria in A. gossypii formed a dynamic, branched, tubular network (Fig. 1 and see Movie S1 in the supplemental material). Within single hyphae, mitochondrial morphology appeared heterogeneous, varying in density of clumps or nets of tubules (Fig. 2A). To test whether mitochondrial morphology and nuclear division state were associated, we scored over 250 nuclei for both division state and the morphology of surrounding mitochondria. We visualized the nuclear division cycle state by tagging gamma-tubulin (Tub4), a component of the spindle pole body (SPB), with mCherry, as has been done previously (28). Because the spindle pole body duplicates and segregates with dividing nuclei, it serves as a marker of nuclear division state in A. gossypii (12, 28). We scored nuclei with a single, round SPB as G1, a single elongated SPB as S, two resolvable SPBs as G2, and two SPBs segregated across the nucleus as M (Fig. 2B). We categorized mitochondrial morphology by the appearance of mitochondria in the immediate vicinity (∼2 to 3 μm from the periphery) of a nucleus (Fig. 2B). Nuclei within a dense clump or net of mitochondria were termed “dense,” nuclei surrounded by mitochondrial tubules were termed “intermediate,” and nuclei in regions with a low density of mitochondrial tubules or fragments were termed “sparse.” All of our scoring was performed using maximum-projection images of multiple Z slices through the depth of the cell. There was no apparent association between category of mitochondrial morphology and nuclear division state, such that nuclei of all division states existed in each category of mitochondrial morphology (Fig. 2C). Although the fraction of S-phase nuclei surrounded by dense mitochondria was greater than that of mitotic nuclei (47.0% versus 28.6%), the fraction of S-phase nuclei surrounded by sparse mitochondria was also greater than for mitotic nuclei (18.0% versus 14.3%). Therefore, mitochondria do not appear to undergo specific morphological changes associated with the division state of nearby nuclei.
Heterogeneity in ΔΨM is not associated with nuclear division state.
Because mitochondrial morphology appeared independent of nuclear division state, we investigated whether mitochondrial function, as measured by mitochondrial membrane potential (ΔΨM), was associated with nuclear division state. To observe ΔΨM, we utilized the organic dyes MitoTracker Green FM (MitoTracker) and tetramethylrhodamine ethyl ester (TMRE). TMRE is a cationic fluorescent dye which accumulates in active mitochondria based on ΔΨM, and its accumulation can be described by the Nernst equation (see Materials and Methods). Conversely, MitoTracker accumulates in mitochondria based on mass, and its accumulation is relatively independent of changes in ΔΨM (6, 31, 45). To confirm the response of MitoTracker and TMRE to changes in ΔΨM in A. gossypii, we stained cells simultaneously with both dyes before treating them with oligomycin or carbonyl cyanide m-chlorophenyl hydrazone (CCCP). Oligomycin increases ΔΨM by inhibition of ATP synthase, and CCCP is a protonophore which reduces ΔΨM by allowing proton diffusion across the inner mitochondrial membrane. Cells treated with oligomycin displayed increased TMRE fluorescence, especially at their hyphal tips, and CCCP treatment essentially abolished TMRE fluorescence (Fig. 3A). However, levels of MitoTracker staining were similar among CCCP-treated, oligomycin-treated, and untreated cells, indicating its insensitivity to changes in ΔΨM in this system.
Because TMRE, but not MitoTracker, is sensitive to changes in ΔΨM, the ratio of TMRE to MitoTracker signal allows for comparison of differences in ΔΨM of mitochondria within a single cell. To determine how ΔΨM varied within a single A. gossypii cell, we stained wild-type cells simultaneously with TMRE and MitoTracker and analyzed the ratio of TMRE to MitoTracker signal intensity (Fig. 3B). We then analyzed ΔΨM in an A. gossypii strain with a CFP-tagged histone, H4, to visualize nuclei alongside ΔΨM. From these data, it is clear that nuclei exist in different environments of ΔΨM within a single cell (Fig. 4).
In order to quantify local differences in ΔΨM, we developed a method to classify pixels in an image as mitochondrial on the basis of MitoTracker signal intensity, TMRE signal intensity, and connectivity to other mitochondrial pixels (Fig. 5A and see Materials and Methods). By applying a binary mask of mitochondrial elements to a ratio image of TMRE/MitoTracker signal, we quantified spatial heterogeneity in membrane potential independently of mitochondrial density both one dimensionally along a hypha and two dimensionally within entire cells (Fig. 5B). Furthermore, because TMRE's mitochondrial uptake can be described by the Nernst equation, we were able to convert differences in unitless ratio values to real differences in potential measured in millivolts (see Materials and Methods). By applying this analysis to individual hyphae, we plotted spatial variations from the mean ΔΨM of a hypha (Fig. 6A). This demonstrates that although neighboring nuclei can experience similar ΔΨMs, differences in ΔΨMs on the order of 5 mV are possible for neighbors.
To test whether this intracellular variability in ΔΨM was associated with the division state of nuclei, we quantified the mean ratio intensity of mitochondrial pixels within a 2-μm radius of 468 nuclei in 6 independent cells. We then calculated the difference in potential between each perinuclear region and the mean of the cell in which it resided. This normalization eliminates the effect of differential dye uptake between independent cells. Across the population, we found that the deviation of mean ΔΨMs of perinuclear mitochondria from mean ΔΨMs of the cell was distributed with a standard deviation of 3.7 mV and encompassed a range of −14.2 mV to 11.9 mV (Fig. 6B). Because the intensity of H4-CFP increases as nuclei progress through the division cycle, we also quantified the mean CFP intensity of each nucleus in the maximum projection image as a marker of division state (12). There was no modulation of ΔΨM with changes in CFP intensity, nor was there a linear association between the CFP intensity of a nucleus and a ΔΨM deviation of the surrounding mitochondria (linear-regression two-tailed t test, P = 0.75) (Fig. 6C). Thus, while ΔΨM varies substantially within single A. gossypii cells and neighboring nuclei inhabit regions of different ΔΨMs, this variability is not linked to the nearby nuclear division cycle.
G2/M arrest does not fragment mitochondria.
After finding no relationship between mitochondrial morphology or ΔΨM and local nuclear division state, we tested whether we can induce changes in mitochondrial morphology by manipulating the cell cycle. Nocodazole induces G2/M arrest by inhibiting microtubule polymerization. In A. gossypii, nocodazole has been shown to depolymerize microtubules within 1 h, and treatment for 4 h results in >70% of nuclei being in G2/M arrest (12, 21). Cells treated with nocodazole for 180 min displayed widespread G2/M arrest and increased distances between nuclei, indicative of cell growth without nuclear division (Fig. 7). Mitochondria in these cells were not fragmented, but morphology was disrupted, and regions of clumped mitochondria were intermixed with regions lacking mitochondria. Because microtubules are known to be important for mitochondrial transport in other fungi, we also imaged cells after 30 min in nocodazole in order to depolymerize cytoplasmic microtubules without inducing widespread cell cycle arrest (44). Mitochondria of cells treated with nocodazole for 30 min appeared similar to those of cells treated for 180 min, suggesting that the altered morphology is a result of microtubule depolymerization rather than arrest of nuclear division. These data support a role for microtubules, but not for the division state of nuclei, in modulating mitochondrial morphology.
Mitochondrial fusion and fission dynamics are important for cell growth and sporulation.
We next generated mutants with perturbed mitochondrial morphology to assess the effects of a hyperfused or a hyperfragmented mitochondrial network on A. gossypii cells. Dnm1p and Fzo1p are highly conserved GTPases required for the fission and fusion, respectively, of mitochondria (8). Therefore, inhibition of Dnm1p activity results in hyperfused mitochondria, while inhibition of Fzo1p activity results in fragmented mitochondria. To alter mitochondrial morphology, we generated dnm1Δ and fzo1Δ heterokaryon mutants in which the gene was disrupted by a cassette containing the mitoGFP and the GEN3 gene, which confers resistance to the compound G418, in a subset of nuclei. To control for the effects of the deletion cassette, we generated another heterokaryon strain (AG510) in the same background as the mutants with the same deletion cassette inserted into an intergenic region in chromosome I. Because A. gossypii cells are multinucleated, initial genetic transformants are heterokaryons, which contain a mixture of haploid nuclei that are either wild type or mutant. Within a heterokaryon mycelium, genetic manipulation, even deletion of essential genes, is generally tolerated due to some expression of the wild-type allele (43). Surprisingly, fzo1Δ and dnm1Δ heterokaryon mutants exhibited strong phenotypes despite the presence of wild-type nuclei in a shared cytoplasm.
As was expected because of gene disruption, fzo1Δ heterokaryon cells had fragmented mitochondria, and dnm1Δ heterokaryon cells had clumped mitochondria (Fig. 8A). fzo1Δ heterokaryon mutants sporulated poorly compared to control cells, and dnm1Δ heterokaryon mutants failed to sporulate at all (Fig. 8B). Because of this sporulation defect, these strains were imaged from a colony growing on solid medium rather than from young cells grown from spores in liquid culture, as in all previous experiments. The difference in growth conditions makes it difficult to make direct comparisons of these mutant strains to wild-type cells grown in liquid media. Furthermore, both mutants displayed severe colony morphogenesis defects, suggesting a deficiency in hyphal branching or polarized growth (Fig. 8B). Mutant strains also grew more slowly than either WT or control strains (Fig. 8C). The poor growth and lack of sporulation of these heterokaryon mutants precluded a more thorough investigation of nuclear and mitochondrial dynamics in these strains. Additionally, the lack of spores produced made the construction of homokaryons impossible. However, such dominant phenotypes in heterokaryon mutants indicate a possible need for local expression of Fzo1p and Dnm1p from each nucleus in a heterokaryon.
DISCUSSION
In this study, we tested the hypothesis that mitochondrial morphology and membrane potential is associated with the division state of a nearby nucleus. Using a quantitative analysis of mitochondrial morphology and a ratiometric technique to visualize changes in membrane potential, we demonstrated that mitochondria are spatially heterogeneous within single A. gossypii cells. Notably, this heterogeneity is not associated with the division state of nearby nuclei. These results suggest that the nuclear cycle does not direct changes in mitochondrial morphology and that the nuclear cycle is apparently insensitive to spatially localized fluctuations in aerobic respiration. We also characterized heterokaryon mutants with deletions of the mitochondrial fusion gene FZO1 and the mitochondrial fission gene DNM1. These mutants showed defects in colony growth and morphogenesis as well as sporulation, which suggest the importance of dynamic mitochondrial morphology in a wide range of essential cellular processes in this system.
Our finding that mitochondrial membrane potential varies by approximately ±10 mV within a single A. gossypii cell is comparable with previous measurements of variability within single mammalian cells and between genetically identical cells in culture (19, 45). Given estimates of mitochondrial membrane potential of 150 to 180 mV, our work suggests that potential varies spatially by more than ±5% within single A. gossypii cells (31). Theoretically, mitochondrial ATP output is highly sensitive to fluctuation in the proton motive force (PMF). At constant [Pi], a 14-mV decrease in proton motive force (PMF = ΔΨM − 59.2ΔpHM) corresponds to an approximately 10-fold decrease in the maximal ATP/ADP ratio that could be maintained by mitochondria (29). The specific effects of modulations in the proton motive force are not well understood, but the spatial heterogeneity in mitochondrial membrane potential that we observe in A. gossypii is likely of a physiologically relevant magnitude.
On the molecular scale, little is known about the regulation of mitochondrial heterogeneity or its implication in cellular physiology, but such heterogeneity may be relevant to cell polarity. Modulation of mitochondrial morphology has been shown to affect the localization of mitochondria in neural synapses and to affect migration in leukocytes (5, 42). In A. gossypii cells, which are constitutively polarized after germination, intracellular mitochondrial heterogeneity may contribute to the maintenance of polarized growth at hyphal tips and establishment of new sites at branches, causing the colony morphogenesis defects that we observed in Δfzo1 and Δdnm1 heterokaryon mutants. In Neurospora, mitochondria are highly enriched at growing tips, where their role is more likely as a calcium store rather than a site of ATP synthesis, and there they may promote polarized growth (3, 22, 35).
The lack of relationship between mitosis and mitochondria in A. gossypii may be explained by the lack of p53 in fungi, a protein required for integrating metabolic signals at the G1/S checkpoint in Drosophila and mammalian cells (23, 26). Despite the lack of p53, the growth of a filamentous fungus in nature allows different regions of a single cell to explore areas of variable nutrient concentration, and it seems likely that A. gossypii is able to translate local metabolic information to the nuclear division cycle. Because our analysis of mitochondrial heterogeneity is limited to a single moment in time, it is still possible that nuclei may be influenced by the amount of time that they spend near mitochondria of certain morphology or metabolic output even though nuclear division state at a given moment is independent of surrounding mitochondria.
Transmission of metabolic information to the nuclear division cycle may also occur independently of mitochondria. Previous work in A. gossypii suggests that starvation inhibits the G2/M transition via activation of the Wee1 homolog Swe1p, and it is possible that this pathway is independent of mitochondrial morphology or membrane potential (16). It is also possible that metabolic factors are able to regulate nuclear division indirectly through pathways affecting cell growth, allowing hyphae in nutrient-rich regions to grow more quickly than hyphae in nutrient-poor regions. If nuclear division were coupled to cell size or growth rate, nuclei would preferentially divide in regions of increased cell growth without receiving direct signals about regional metabolic or nutrient status.
Our data also suggest that the nuclear cycle does not control local mitochondrial function. Most simply, this could be an indication that key regulators of the nuclear division pathway are spatially restricted to nuclei and exist only briefly in the cytosol. In this case, nuclear cycle regulators would not encounter mitochondria in the cytosol and would, therefore, be unable to contribute to mitochondrial dynamics. It is also possible that the independence that we observe between nuclei and mitochondria is due to the absence of any molecular connection between mitochondria and nuclear cycle progression. For example, Drp1, the mammalian homolog of Dnm1p, is activated by phosphorylation during mitosis on a particular serine residue that is not conserved in budding yeast or A. gossypii (40). If this is the case, the division cycle may still act in islands of cytoplasm around nuclei and be large enough to contain a functional shell of organelles. However, mitochondria on these functional shells would fail to undergo cell cycle-dependent changes in morphology or membrane potential.
Although seemingly independent of the nuclear division cycle, modulation of mitochondrial morphology appears essential for proper cell growth and proliferation in A. gossypii. Because A. gossypii is haploid and multinucleate, gene deletion by homologous recombination results in integration of the deletion cassette into the genome of some, but not all, nuclei in a cell. Therefore, initial transformants contain nuclei with the mutated allele and the drug resistance gene used for selection and also nuclei containing the wild-type allele and no drug resistance (43). Our discovery of strong mutant phenotypes in Δfzo1 and Δdnm1 heterokaryon mutant strains is striking given that these strains contain nuclei with both wild-type and mutant alleles, and dominant phenotypes in a heterokaryon deletion mutant have never been reported in A. gossypii. These data suggest that either the regulation of mitochondrial morphology by Fzo1p and Dnm1p is dosage dependent in A. gossypii or is dependent on local expression from each nucleus, such that WT gene expression from a nucleus in the same cytoplasm cannot complement a defect. Null or strong loss-of-function mutants of homologues of Fzo1p and Dnm1p are viable in budding yeast, Caenorhabditis elegans, and Drosophila, although there are growth and development defects (4, 15, 17, 30, 42). Deletion of Dnm1 is also tolerated in the filamentous fungus Podospora anserina (37). Therefore, it seems unlikely that Dnm1p and Fzo1p are required with dosage dependency in A. gossypii. The alternative possibility, a dependence on locally transcribed Fzo1p and Dnm1p around each nucleus, would support a model by which nuclei inhabit regions of cytoplasm with distinct properties. Supersuppressor (ssu) alleles in Neurospora crassa have been shown genetically to act in a nuclearly autonomous manner in heterokaryon cells, suggesting the possible existence of spatially restricted gene products in a syncytium (14).
The sporulation defect that we discovered in Δfzo1 and Δdnm1 heterokaryon mutants is intriguing given the spermatogenesis defects seen in a Drosophila mutant of the FZO1 homolog and sporulation defects in mitochondrial morphology mutants in budding yeast and the filamentous fungus Neurospora crassa (13, 15, 32). These data also are consistent with specific changes visualized in mitochondrial morphology during sporulation in budding yeast (27). Clearly, the importance of mitochondrial dynamics in meiotic division is highly conserved, yet it is interesting that such a defect exists in A. gossypii cells, which produce only asexual ascospores, presumably without reduction of chromosome number.
We demonstrated that mitochondria in A. gossypii are heterogeneous in both their morphology and their membrane potential, but this heterogeneity is independent of nuclear progression through the division cycle. We also observed dominant phenotypes in Δdnm1 and Δfzo1 heterokaryon mutants, suggesting the possible importance of local modulation of mitochondrial morphology around each nucleus in a syncytial organism.
This study is the first quantitative description of mitochondrial dynamics and function in the model multinucleate fungal system A. gossypii. The imaging methods presented here can be readily applied to diverse filamentous fungi, and the large mycelial networks of filamentous fungi are ideal for understanding the functional consequences of heterogeneous mitochondrial form and potential.
Supplementary Material
ACKNOWLEDGMENTS
We thank Cori D'Ausilio for critically reading the manuscript, members of the Gladfelter lab for helpful discussions, and Ann Lavanway for imaging support.
This work was supported by the National Institutes of Health under grant R01GM081506 (to A.S.G.) and the National Science Foundation under grant MCB-0719126 (to A.S.G.), supplemented by a Research Experience for Undergraduates grant (to J.P.G.) and by the Laura and Arthur Colwin and the Lucy B. Lemann Fellowships at the Marine Biological Laboratory (to A.S.G.).
Footnotes
Published ahead of print 20 January 2012
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1. Alberti-Segui C, Dietrich F, Altmann-Jöhl R, Hoepfner D, Philippsen P. 2001. Cytoplasmic dynein is required to oppose the force that moves nuclei towards the hyphal tip in the filamentous ascomycete Ashbya gossypii. J. Cell Sci. 114:975–986 [DOI] [PubMed] [Google Scholar]
- 2. Altmann-Jöhl R, Philippsen P. 1996. AgTHR4, a new selection marker for transformation of the filamentous fungus Ashbya gossypii, maps in a four-gene cluster that is conserved between A. gossypii and Saccharomyces cerevisiae. Mol. Gen. Genet. 250:69–80 [DOI] [PubMed] [Google Scholar]
- 3. Bowman BJ, Draskovic M, Freitag M, Bowman EJ. 2009. Structure and distribution of organelles and cellular location of calcium transporters in Neurospora crassa. Eukaryot. Cell 8:1845–1855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Breckenridge DG, et al. 2008. Caenorhabditis elegans drp-1 and fis-2 regulate distinct cell-death execution pathways downstream of ced-3 and independent of ced-9. Mol. Cell 31:586–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Campello S, et al. 2006. Orchestration of lymphocyte chemotaxis by mitochondrial dynamics. J. Exp. Med. 203:2879–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Collins TJ, Berridge MJ, Lipp P, Bootman MD. 2002. Mitochondria are morphologically and functionally heterogeneous within cells. EMBO J. 21:1616–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Demeter J, Lee SE, Haber JE, Stearns T. 2000. The DNA damage checkpoint signal in budding yeast is nuclear limited. Mol. Cell 6:487–492 [DOI] [PubMed] [Google Scholar]
- 8. Detmer SA, Chan DC. 2007. Functions and dysfunctions of mitochondrial dynamics. Nat. Rev. Mol. Cell Biol. 8:870–879 [DOI] [PubMed] [Google Scholar]
- 9. Di Talia S, Skotheim JM, Bean JM, Siggia ED, Cross FR. 2007. The effects of molecular noise and size control on variability in the budding yeast cell cycle. Nature 448:947–951 [DOI] [PubMed] [Google Scholar]
- 10. Frederick RL, McCaffery JM, Cunningham KW, Okamoto K, Shaw JM. 2004. Yeast Miro GTPase, Gem1p, regulates mitochondrial morphology via a novel pathway. J. Cell Biol. 167:87–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Frescas D, Mavrakis M, Lorenz H, Delotto R, Lippincott-Schwartz J. 2006. The secretory membrane system in the Drosophila syncytial blastoderm embryo exists as functionally compartmentalized units around individual nuclei. J. Cell Biol. 173:219–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gladfelter AS, Hungerbuehler AK, Philippsen P. 2006. Asynchronous nuclear division cycles in multinucleated cells. J. Cell Biol. 172:347–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gorsich SW, Shaw JM. 2004. Importance of mitochondrial dynamics during meiosis and sporulation. Mol. Biol. Cell 15:4369–4381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Griffiths AJ. 1976. Evidence for nuclear restriction of supersuppressor gene products in Neurospora heterokaryons. Can. J. Genet. Cytol. 18:35–38 [DOI] [PubMed] [Google Scholar]
- 15. Hales KG, Fuller MT. 1997. Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell 90:121–129 [DOI] [PubMed] [Google Scholar]
- 16. Helfer H, Gladfelter AS. 2006. AgSwe1p regulates mitosis in response to morphogenesis and nutrients in multinucleated Ashbya gossypii cells. Mol. Biol. Cell 17:4494–4512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hermann GJ, et al. 1998. Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J. Cell Biol. 143:359–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hinchcliffe E, Thompson E, Miller F, Yang J, Sluder G. 1999. Nucleo-cytoplasmic interactions that control nuclear envelope breakdown and entry into mitosis in the sea urchin zygote. J. Cell Sci. 112:1139–1148 [DOI] [PubMed] [Google Scholar]
- 19. Huang H-M, Fowler C, Zhang H, Gibson GE. 2004. Mitochondrial heterogeneity within and between different cell types. Neurochem. Res. 29:651–658 [DOI] [PubMed] [Google Scholar]
- 20. Kaufmann A, Philippsen P. 2009. Of bars and rings: Hof1-dependent cytokinesis in multiseptated hyphae of Ashbya gossypii. Mol. Cell. Biol. 29:771–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lang C, et al. 2010. Mobility, microtubule nucleation and structure of microtubule-organizing centers in multinucleated hyphae of Ashbya gossypii. Mol. Biol. Cell 21:18–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Levina NN, Lew RR. 2006. The role of tip-localized mitochondria in hyphal growth. Fungal Genet. Biol. 43:65–74 [DOI] [PubMed] [Google Scholar]
- 23. Mandal S, Freije WA, Guptan P, Banerjee U. 2010. Metabolic control of G1-S transition: cyclin E degradation by p53-induced activation of the ubiquitin-proteasome system. J. Cell Biol. 188:473–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mandal S, Guptan P, Owusu-Ansah E, Banerjee U. 2005. Mitochondrial regulation of cell cycle progression during development as revealed by the tenured mutation in Drosophila. Dev. Cell 9:843–854 [DOI] [PubMed] [Google Scholar]
- 25. Mavrakis M, Rikhy R, Lippincott-Schwartz J. 2009. Plasma membrane polarity and compartmentalization are established before cellularization in the fly embryo. Dev. Cell 16:93–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mitra K, Wunder C, Roysam B, Lin G, Lippincott-Schwartz J. 2009. A hyperfused mitochondrial state achieved at G1-S regulates cyclin E buildup and entry into S phase. Proc. Natl. Acad. Sci. U. S. A. 106:11960–11965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Miyakawa I, Aoi H, Sando N, Kuroiwa T. 1984. Fluorescence microscopic studies of mitochondrial nucleoids during meiosis and sporulation in the yeast, Saccharomyces cerevisiae. J. Cell Sci. 66:21–38 [DOI] [PubMed] [Google Scholar]
- 28. Nair DR, D'Ausilio CA, Occhipinti P, Borsuk ME, Gladfelter AS. 2010. A conserved G1 regulatory circuit promotes asynchronous behavior of nuclei sharing a common cytoplasm. Cell Cycle 9:3771–3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nicholls DG. 2004. Mitochondrial membrane potential and aging. Aging Cell 3:35–40 [DOI] [PubMed] [Google Scholar]
- 30. Otsuga D, et al. 1998. The dynamin-related GTPase, Dnm1p, controls mitochondrial morphology in yeast. J. Cell Biol. 143:333–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Perry SW, Norman JP, Barbieri J, Brown EB, Gelbard HA. 2011. Mitochondrial membrane potential probes and the proton gradient: a practical usage guide. Biotechniques 50:98–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Prokisch H, Neupert W, Westermann B. 2000. Role of MMM1 in maintaining mitochondrial morphology in Neurospora crassa. Mol. Biol. Cell 11:2961–2971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rao PN, Johnson RT. 1970. Mammalian cell fusion: studies on the regulation of DNA synthesis and mitosis. Nature 225:159–164 [DOI] [PubMed] [Google Scholar]
- 34. Rieder CL, et al. 1997. Mitosis in vertebrate somatic cells with two spindles: implications for the metaphase/anaphase transition checkpoint and cleavage. Proc. Natl. Acad. Sci. U. S. A. 94:5107–5112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Riquelme M, et al. 2011. Architecture and development of the Neurospora crassa hypha—a model cell for polarized growth. Fungal Biol. 115:446–474 [DOI] [PubMed] [Google Scholar]
- 36. Sambrook J, Russell D. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 37. Scheckhuber CQ, et al. 2007. Reducing mitochondrial fission results in increased life span and fitness of two fungal ageing models. Nat. Cell Biol. 9:99–105 [DOI] [PubMed] [Google Scholar]
- 38. Schieke SM, McCoy JP, Finkel T. 2008. Coordination of mitochondrial bioenergetics with G1 phase cell cycle progression. Cell Cycle 7:1782–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sikorski RS, Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Taguchi N, Ishihara N, Jofuku A, Oka T, Mihara K. 2007. Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J. Biochem. 282:11521–11529 [DOI] [PubMed] [Google Scholar]
- 41. Twig G, et al. 2006. Tagging and tracking individual networks within a complex mitochondrial web with photoactivatable GFP. Am. J. Physiol. Cell Physiol. 291:C176–C184 [DOI] [PubMed] [Google Scholar]
- 42. Verstreken P, et al. 2005. Synaptic mitochondria are critical for mobilization of reserve pool vesicles at Drosophila neuromuscular junctions. Neuron 47:365–378 [DOI] [PubMed] [Google Scholar]
- 43. Wendland J, Ayad-Durieux Y, Knechtle P, Rebischung C, Philippsen P. 2000. PCR-based gene targeting in the filamentous fungus Ashbya gossypii. Gene 242:381–391 [DOI] [PubMed] [Google Scholar]
- 44. Westermann B, Prokisch H. 2002. Mitochondrial dynamics in filamentous fungi. Fungal Genet. Biol. 36:91–97 [DOI] [PubMed] [Google Scholar]
- 45. Wikstrom JD, et al. 2007. Beta-cell mitochondria exhibit membrane potential heterogeneity that can be altered by stimulatory or toxic fuel levels. Diabetes 56:2569–2578 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.