Abstract
Uptake of extracellular oligopeptides in yeast is mediated mainly by specific transporters of the peptide transporter (PTR) and oligopeptide transporter (OPT) families. Here, we investigated the role of potential peptide transporters in the yeast Schizosaccharomyces pombe. Utilization of naturally occurring dipeptides required only Ptr2/SPBC13A2.04c and none of the other 3 OPT proteins (Isp4, Pgt1, and Opt3), whereas only Isp4 was indispensable for tetrapeptide utilization. Both Ptr2 and Isp4 localized to the cell surface, but under rich nutrient conditions Isp4 localized in the Golgi apparatus through the function of the ubiquitin ligase Pub1. Furthermore, the ubiquitin ligase Ubr11 played a significant role in oligopeptide utilization. The mRNA levels of both the ptr2 and isp4 genes were significantly reduced in ubr11Δ cells, and the dipeptide utilization defect in the ubr11Δ mutant was rescued by the forced expression of Ptr2. Consistent with its role in transcriptional regulation of peptide transporter genes, the Ubr11 protein was accumulated in the nucleus. Unlike the situation in Saccharomyces cerevisiae, the oligopeptide utilization defect in the S. pombe ubr11Δ mutant was not rescued by inactivation of the Tup11/12 transcriptional corepressors, suggesting that the requirement for the Ubr ubiquitin ligase in the upregulation of peptide transporter mRNA levels is conserved in both yeasts; however, the actual mechanism underlying the control appears to be different. We also found that the peptidomimetic proteasome inhibitor MG132 was still operative in a strain lacking all known PTR and OPT peptide transporters. Therefore, irrespective of its peptide-like structure, MG132 is carried into cells independently of the representative peptide transporters.
INTRODUCTION
Most organisms utilize oligopeptides, which are hydrolyzed and recycled as amino acids or used as a source of nitrogen or carbon. Extracellular oligopeptides are imported into cells by specific plasma membrane transporters, which are distinct from amino acid transporters. Proteins of the highly conserved peptide transporter (PTR) family or SLC15 family, which are found in eukaryotes and some bacteria, import di- and tripeptides in a proton-coupled manner (49). In the budding yeast Saccharomyces cerevisiae, Ptr2 is a unique member of this PTR family and is responsible for the import of di- and tripeptides (23, 25, 44). Interestingly, PepT1 and PepT2, mammalian representatives of the SLC15 family, import a wide range of substrates from naturally occurring small peptides to various peptidomimetics or even unrelated drugs (7, 14, 46). The fission yeast Schizosaccharomyces pombe has a unique homolog of this family, SPBC13A2.04c/Ptr2, which was investigated in this study. In addition to these widely conserved PTR proteins, fungi and plants express an oligopeptide transporter (OPT) family of proteins (21, 29). In S. pombe, the OPT family protein Isp4 is responsible for the uptake of tetra- and pentapeptides (34). Two other genes encoding OPTs were identified in the S. pombe genome: pgt1/opt1 (glutathione transporter) and the uncharacterized SPCC1840.12 (referred to as opt3 in this study) (16, 52). The human opportunistic pathogen Candida albicans has 8 OPT genes, and some of their encoded proteins can transport longer peptides, up to at least 8 amino acids in length (35, 45). Additionally, the allantoate/ureidosuccinate permease Dal5 can transport several dipeptides in S. cerevisiae (9, 23).
Apart from the biochemical nature of these transporters, the molecular mechanisms regulating their expression have been characterized in detail, especially in S. cerevisiae. Transcription of these transporter genes, and hence utilization of peptides, is repressed in the presence of a preferable rich nitrogen source such as ammonium but is induced by relatively poor nitrogen sources such as allantoin and urea (6, 20). In addition to nitrogen catabolite repression, extracellular dipeptides induce PTR2 expression by accelerating the ubiquitin-dependent degradation of Cup9 protein (8, 53). Cup9 forms a corepressor complex with Tup1-Ssn6 and inhibits the transcription of PTR2 (56). Ubiquitination of Cup9 is mediated by the ubiquitin ligase Ubr1, which is involved in the N-end rule protein degradation pathway (8, 15, 48, 54). The imported dipeptide directly binds and activates Ubr1 by relieving it from autoinhibition through a conformational change and promotes the interaction between Cup9 and Ubr1 (15, 57). Thus, di/tripeptides effectively stimulate their own uptake by increasing the protein levels of the corresponding transporter Ptr2. Furthermore, the presence of amino acids influences peptide utilization via upregulation of peptide transporter expression (5, 24, 44, 55, 56). This amino acid-induced stimulation requires the Ssy1-Ptr3-Ssy5 (SPS) amino acid-sensing system and also depends on the function of Ubr1 (10, 12, 23, 33, 55, 56). Interestingly, amino acids accelerate the Ubr1-mediated degradation of Cup9 but do not stimulate the degradation of the N-end rule substrates themselves.
The ubiquitin ligase Ubr is widely conserved in eukaryotes (48, 54), and a deficiency in the human UBR1 gene leads to the hereditary disease Johanson-Blizzard syndrome (58). We have reported the characterization of the Ubr ubiquitin ligases in S. pombe. Two distinct but homologous Ubr proteins, Ubr1 and Ubr11, exist in this yeast. Both Ubr1 and Ubr11 are dispensable for growth, and strains in which both genes are simultaneously deleted from the genome are viable. The ubr1 strain exhibits pleiotropic phenotypes, including the accumulation of the Mei2 meiosis initiator protein (27), an altered oxidative stress response and drug resistance partly due to the inefficient degradation of the Pap1 transcription factor (19, 28), a localization defect of the nuclear proteasome (51), a defect in invasive growth (13), and a morphology defect and aberrant meiosis (our unpublished results). Inactivation of Ubr11 does not exacerbate the defects in the ubr1 strain. Furthermore, forced expression of Ubr11 from the heterologous nmt promoter fails to rescue the defects, suggesting that Ubr1 and Ubr11 perform unique, nonoverlapping functions. We found that only Ubr11 is responsible for the N-end rule degradation pathway in S. pombe (our unpublished results). Nonetheless, no apparent phenotype has been identified so far in the ubr11 mutant.
In this study, all the known possible peptide transporter proteins in S. pombe were examined for their involvement in oligopeptide utilization. We also investigated the role of Ubr ubiquitin ligase in the regulation of oligopeptide transporters. Our data showed that Ubr11 is important for the expression of Ptr2 and Isp4, which are responsible for transporting di- and tetrapeptides, respectively. To the best of our knowledge, other than for S. cerevisiae, this is the first study to demonstrate the conserved requirement for the Ubr ubiquitin ligase in peptide utilization.
MATERIALS AND METHODS
Yeast strains, media, and culture conditions.
The S. pombe strains used in this study are listed in Table 1. Gene disruption was carried out using the PCR products of the 5′ and 3′ untranslated regions of the corresponding gene with ura4+ or drug resistance genes as selection markers (4, 17, 30, 47). Detailed information on strain construction methods is in the supplemental material. The complete medium YES and minimal medium EMM were used for growth (40). In some experiments, the ammonium chloride (nitrogen source) in EMM was replaced with 0.1 to 0.15% (wt/vol) proline or was completely omitted for nitrogen-free medium. Expression from the nmt promoter was repressed by the addition of 5 μg/ml thiamine (+B1) (38). Oligopeptides were purchased from Bachem AG (Bubendorf, Switzerland), Sigma-Aldrich (St. Louis, MO), Peptide Institute (Osaka, Japan), or Wako Pure Chemical Industries (Osaka, Japan). Most oligopeptides were dissolved in water, sterilized by filtration, and added to the autoclaved medium at 0.2 mM. Z-Ile-Leu (Sigma-Aldrich) was dissolved in dimethyl sulfoxide (DMSO) and added at 0.4 mM. Soy peptides (Hi-Nute AM; Fuji Oil, Osaka, Japan) were dissolved in medium at 0.1% (wt/vol) and autoclaved. S. pombe cells were grown at 28°C in liquid medium or at 31°C on agar solid medium. To inactivate the temperature-sensitive proteasome activity in the mts2 strain, cells were cultured at 36°C for 3.5 h.
Table 1.
S. pombe strains used in this study
| Strain | Genotype | Source and/or reference |
|---|---|---|
| KSP2 | h− | Our stock |
| KSP634 | h+leu1-32 | Our stock |
| KSP635 | h−leu1-32 | Our stock |
| KSP1058 | h−ura4-D18 leu1-32 | Our stock |
| KSP1923 | h−ubr1::ura4+ura4-D18 leu1-32 | 27 |
| KSP2222 | h−ubr11::ura4+ura4-D18 leu1-32 | 27 |
| KSP2263 | h+ubr11::hphMX6 leu1-32 | This study |
| KSP2321 | h−ubr11::ura4+ura4-D18 | 27 |
| KSP2335a | h+ubr11::hphMX6 leu1+-[pDUAL-HFG41]inthis2 | This study |
| KSP2335b | h+ubr11::hphMX6 leu1+-[Pnmt41-HFG-ptr2+]inthis2 | this study |
| KSP2335c | h+ubr11::hphMX6 leu1+-[Pnmt41-6HisUb-Flag-halo7-ubr1+]inthis2 | This study; 28 |
| KSP2422 | h+ptr2::ura4+ura4-C190T | This study |
| KSP2472 | h−ubr11::hphMX6 | This study |
| KSP2698 | h+ptr2::ura4+ura4-D18 his7-366 leu1+-[pDUAL-HFG41]int | This study |
| KSP2699 | h+ptr2::ura4+ ura4-D18 his7-366 leu1+-[Pnmt41-HFG-ptr2+]int | This study |
| KSP2817 | h−ubr11::hphMX6 ura4-D18 leu1-32[pDUAL(Pnmt41-6HisUb-Flag-halo7-ubr11+)-ura4+]multicopy | This study; 28 |
| KSP2863 | h+leu1+-[Pnmt41-UbG76V-GFP]intmei2+-3HA-kanMX6 ura4-D18 | This study |
| KSP2874 | h+ubr11::hphMX6 leu1+-[Pnmt41-6HisUb-Flag-halo7-ubr11+]inthis2 | This study; 28 |
| KSP2882 | h−isp4::hphMX6 ura4-D18 leu1-32 | This study |
| KSP2884 | h−ptr2::ura4+ura4-D18 leu1-32 | this study |
| KSP2886 | h−tup11::ura4+tup12::ura4+ura4-C190T leu1-32 | This study; 37 |
| KSP2887 | h−tup11::ura4+tup12::ura4+ura4-C190T leu1-32 | This study; 37 |
| KSP2888 | h−ubr11::hphMX6 tup11::ura4+tup12::ura4+ura4-C190T leu1-32 | This study |
| KSP2889 | h−ubr11::hphMX6 tup11::ura4+tup12::ura4+ura4-C190T leu1-32 | This study |
| KSP2920 | h+ptr2::ura4+isp4::hphMX6 pgt1::natMX6 opt3::pSVEM-bsd leu1+-[Pnmt41-UbG76V-GFP]intmei2+-3HA-kanMX6 ura4-D18 | This study |
| KSP2924 | h−opt3/SPCC1840.12::kanMX6 ura4-D18 leu1-32 | This study |
| KSP2925 | h−pgt1::natMX6 ura4-D18 leu1-32 | This study |
| KSP2934 | h−isp4::hphMX6 pgt1::natMX6 opt3::kanMX6 ura4-D18 leu1-32 | This study |
| KSP2971 | h−isp4::hphMX6 ura4-D18 leu1+-[Pnmt41-HFG-isp4+]int | This study |
| KSP2972 | h−isp4::hphMX6 ura4-D18 leu1+-[Pnmt41-HFG-isp4+]int p(gms1-RFP; ura4+) | This study |
| KSP2996 | h−pub1::ura4+isp4::hphMX6 leu1+-[Pnmt41-HFG-isp4+]intura4-D18 | This study; YGRCa |
| KSP2999 | h−isp4::hphMX6 | This study |
| 591-1C | h+isp4::hphMX6 lys1-131 ura4-D18 leu1-32 | This study |
| 597-1C | h+isp4::hphMX6 leu1-32 | This study |
| 598-1A | h+ptr2::ura4+dal5h1::hphMX6 dal5h2::natMX6 leu1-32 | This study |
| 598-1C | h−isp4::kanMX6 leu1-32 | This study |
| 598-13CL | h−ptr2::ura4+isp4::kanMX6 dal5h1::hphMX6 dal5h2::natMX6 | This study |
| 2885L | h−isp4::hphMX6 ptr2::ura4+ura4-D18 | This study |
| 2928L | h−ptr2::ura4+isp4::hphMX6 pgt1::natMX6 opt3::kanMX6 ura4-D18 | This study |
YGRC, Yeast Genetic Resource Center of Japan (YGRC/NBRP) (http://yeast.lab.nig.ac.jp/nig/).
Plasmid construction.
All pDUAL vector-derived plasmids were digested with NotI and integrated into the leu1-32 loci of the host strains or used as an episomal multicopy plasmid (ura4+ selection) if indicated (36). These are distinguished by the suffix int or multicopy in the genotypes in Table 1. To express the N-terminally green fluorescent protein (GFP)-tagged Ptr2 protein, the coding region of ptr2 was amplified by PCR using oligonucleotides ptr2N and ptr2C (see Table S2 in the supplemental material for the nucleotide sequence) and inserted into the BglII site of the pDUAL-HFG41 vector using the “in fusion” reaction (Clontech, CA). The GFP-Isp4-expressing plasmid was made in similar way using oligonucleotides isp4N and isp4C. For plasmids expressing Dal5-like genes, coding regions of each gene were amplified with dal5h1N and dal5h1C for dal5h1/SPCC417.10 and with dal5h2N and dal5h2C for dal5h2/SPBC1773.15 and then inserted between the NdeI and BglII sites in the pDUAL-HFG41 vector.
To construct a plasmid encoding a proteasome activity reporter substrate, the ubiquitinG76V-GFP fusion gene (including the stop codon) was excised from the Ub-G67V-GFP plasmid (plasmid 11941, obtained from Addgene) (11, 36) by digestion with NheI and XbaI and inserted into the NheI site of the pDUAL-HFF41 vector to yield pDUAL(nmt41-UbG76V-GFP). Due to the mutation of the last (76th) amino acid from glycine to valine in the ubiquitin moiety, the peptide bond between ubiquitinG76V and GFP is resistant to cleavage by deubiquitinase. As a result, UbG76V-GFP is expressed as a fusion protein, ubiquitinated by the ubiquitin ligase Ufd4, and degraded by the proteasome (UFD degradation pathway) (22, 26).
Plasmids expressing Gms1-red fluorescent protein (RFP) and Halo7-tagged Ubr proteins have been described previously (28, 42). Plasmids harboring the genomic ubr11, ptr2, or isp4 gene in the episomal multicopy vector pAL-SK were obtained from the Yeast Genetic Resource Center of Japan (YGRC/NBRP) (http://yeast.lab.nig.ac.jp/nig/).
RT-PCR.
To monitor the mRNA levels of the ubr11 gene or peptide transporter genes ptr2 and isp4, total RNA was extracted and analyzed by reverse transcription-PCR (RT-PCR), as described previously (28). The cdc2 and nda3 (β-tubulin) genes were also analyzed as a control for RNA input levels.
UbG76V-GFP fusion protein expression, flow cytometry, and Western blotting.
Cells were treated with the proteasomal inhibitor MG132 (50 μM; Peptide Institute, Osaka, Japan) or its solvent DMSO (1%, vol/vol) for 3.5 h where indicated. To monitor the intensity of UbG76V-GFP fluorescence, live cells were directly analyzed with a FACSCalibur (Becton Dickinson, NJ). UbG76V-GFP protein levels were also examined by Western blotting using anti-GFP antibody GF200 (Nakalai Tesque, Kyoto, Japan) and anti-Cdc2 antibody sc-53 (Santa Cruz Biotechnology, Santa Cruz, CA), as described previously (28).
Fluorescence microscopy.
Expression of GFP-Ptr2, GFP-Isp4, and Flag-Halo7-Ubr11 proteins from the nmt41 promoter was induced in the absence of thiamine (−B1). The Flag-Halo7-Ubr11 protein (Halo-Ubr11) was labeled with the red fluorescent TMR ligand (Promega, Madison, WI), as described previously (28). The nuclear chromatin region was stained with Hoechst 33342 at 1 μg/ml. Cells were observed under a fluorescence microscope (model TE2U; Nikon, Tokyo, Japan), and fluorescent signals were captured by a charge-coupled-device (CCD) camera (ORCA-ER) with AQUA-Lite software (Hamamatsu Photonics, Shizuoka, Japan).
RESULTS
Roles of the PTR and OPT family transporters in oligopeptide utilization in S. pombe.
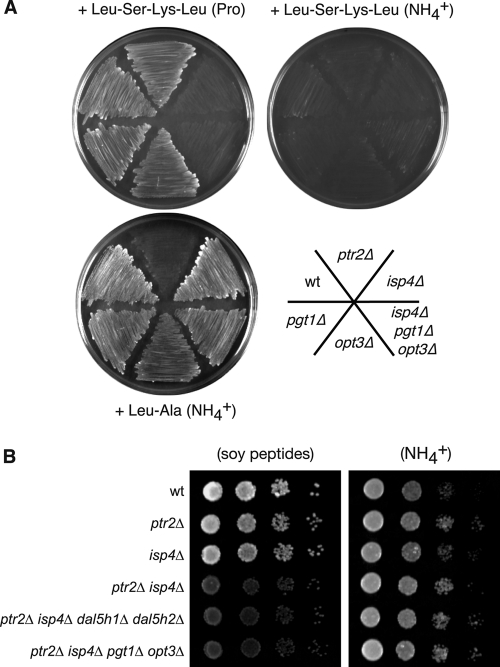
First, we investigated the roles of PTR and OPT proteins in oligopeptide utilization. S. pombe has a single PTR-type transporter gene, SPBC13A2.04c (referred to as ptr2 based on its homology to PTR2 in S. cerevisiae), that has not yet been characterized. Strains carrying individual deletions of the entire ptr2 gene or one of the three OPT family genes (isp4, pgt1, and opt3/SPCC1840.12) were prepared. To monitor oligopeptide utilization in the above-described transporter mutants, di- and tetrapeptides containing leucine were used to determine if they could support the growth of peptide transporter mutants harboring the leucine-auxotrophic leu1 mutation (Fig. 1A). As previously reported (34), strains lacking the isp4 gene failed to grow in minimal medium containing tetrapeptide (Leu-Ser-Lys-Leu) as a source of leucine. Other strains grew well under the same condition. For dipeptide utilization, only the ptr2Δ strain failed to grow in the dipeptide (Leu-Ala) minimal medium. Therefore, Ptr2 has an essential role in dipeptide- but not tetrapeptide-dependent growth. None of the 3 OPT transporters has a role in dipeptide (Leu-Ala) utilization, because even the triple OPT mutant isp4Δ pgt1Δ opt3Δ strain grew as efficiently as the control wild-type strain. Neither the pgt1Δ nor the opt3Δ mutation affected growth in di- or tetrapeptide medium. Given that peptide utilization is under the control of nitrogen catabolite repression, we examined the effects of different nitrogen sources. Although dipeptide utilization was observed even in nitrogen-rich (ammonium [NH4+]) medium, tetrapeptides was imported only when a poor nitrogen source (proline) was used (Fig. 1A; see Fig. 3B). We also examined the roles of these transporters by monitoring the utilization of soy peptides, a mixture of various oligopeptides derived from soybeans. When soy peptides were used as a sole nitrogen source, cells lacking either ptr2 or isp4 could grow well, but simultaneous inactivation of both ptr2 and isp4 significantly slowed the growth under this condition (Fig. 1B). By Blast search using the S. cerevisiae Dal5 protein as a query, we found two Dal5-like proteins from the S. pombe genome database (referred to as Dal5h1/SPCC417.10 and Dal5h2/SPBC1773.15). The retarded growth in the ptr2Δ isp4Δ strain in soy peptide medium was not further exacerbated by additionally deleting both DAL5-like genes (dal5h1 and dal5h2) or by deleting the remaining two OPT genes (pgt1 and opt3) (Fig. 1B), indicating that Ptr2 and Isp4 are major peptide transporters in S. pombe within the peptide substrates tested in this study.
Fig 1.
Ptr2 and Isp4 are major transporters for oligopeptide utilization. (A) Leucine-auxotrophic leu1 strains lacking the possible peptide transporter(s) were grown in minimal medium containing the indicated di- or tetrapeptides as a source of leucine. The nitrogen source in each medium is shown in parentheses: Pro, proline; NH4+, ammonium chloride. Strains: wild type (wt), KSP1058; ptr2Δ, KSP2884; isp4Δ, KSP2882; pgt1Δ, KSP2925; opt3Δ, KSP2924; isp4Δ pgt1Δ opt3Δ, KSP2934. (B) Utilization of soy peptides by transporter mutants. Prototrophic strains lacking the indicated gene(s) were serially diluted and spotted to medium containing soy peptides or NH4+ as a sole nitrogen source. Strains: wt, KSP2; ptr2Δ, KSP2422; isp4Δ, KSP2999; ptr2Δ isp4Δ, 2885L; ptr2Δ isp4Δ pgt1Δ opt3Δ, 2928L; ptr2Δ isp4Δ dal5h1Δ dal5h2Δ, 598-13CL.
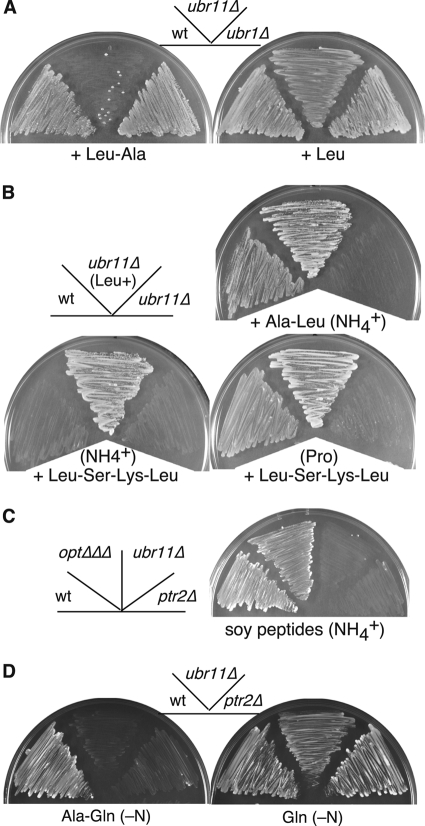
Fig 3.
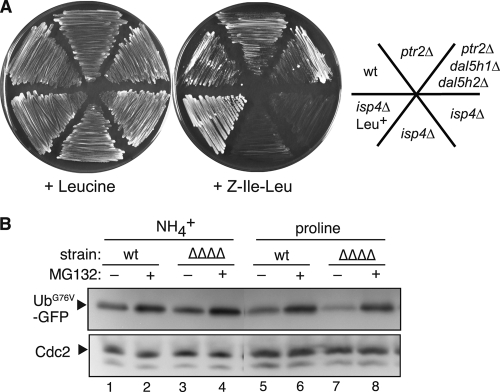
Ubr11 is essential for oligopeptide utilization. (A) Wild-type (KSP634), ubr11Δ (KSP2222), and ubr1Δ (KSP1923) strains, which are auxotrophic for leucine, were grown in minimal medium containing Leu-Ala dipeptide (left) or leucine (right). Colonies of the ubr11Δ strain in the Leu-Ala dipeptide medium are spontaneous suppressor mutants. (B) The same wild-type or ubr11Δ strains used for panel A were grown in the presence of Ala-Leu dipeptide or Leu-Ser-Lys-Leu tetrapeptide. The nitrogen source used in each plate is indicated in parentheses. To obtain prototrophic (Leu+) cells, the same ubr11Δ strain (KSP2222) was transformed with a vector that rescued the auxotrophy for leucine. (C) The indicated strains were grown in minimal medium containing soy peptides as a source of leucine. The optΔΔΔ strain lacks all the genes of the OPT family (isp4Δ pgt1Δ opt3Δ). Strains: wt (KSP1058), ptr2Δ (KSP2884), ubr11Δ (KSP2263), optΔΔΔ (KSP2934). Only Ptr2, and none of the OPTs, is responsible for the uptake of soy peptides containing leucine under this condition. (D) The indicated prototrophic strains were grown in a medium containing the Ala-Gln dipeptide (left) or monomeric glutamine (right) as the sole nitrogen source. Strains; wt, KSP2; ubr11Δ, KSP2472; ptr2Δ, KSP2422. The ubr11Δ and ptr2Δ strains failed to grow in the Ala-Gln dipeptide medium, but their growth was not affected if glutamine was used instead of the dipeptide.
The Opt2 protein in S. cerevisiae is essential for the detoxification of certain drugs (2). In the presence of ZnSO4 (0.75 to 6 mM) or spermine (0.25 to 2 mM), which are reported to retard the growth of the S. cerevisiae opt2 strain, growth-inhibitory profiles of the two drugs were indistinguishable in the wild type and each of the opt mutant strains (data not shown). Therefore, it is unclear whether the OPT family proteins have a similar role in drug resistance in S. pombe.
Cellular localization of Ptr2 and Isp4.
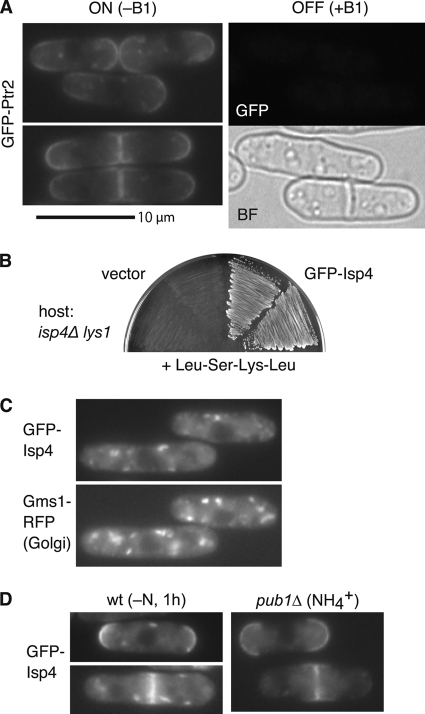
Cellular localization of Ptr2 and Isp4 proteins was monitored by fusing GFP to the N terminus of the corresponding transporters. Functionality of these GFP-fused transporters was confirmed by their rescue of the corresponding mutants (Fig. 2B; see Fig. 4B). GFP-Ptr2 was localized at the plasma membrane, particularly at both ends of the cell and also at the center in dividing cells (Fig. 2A). In contrast, GFP-Isp4 signals were observed as cytoplasmic punctate dots which largely overlapped with those of Gms1, a UDP-galactose transporter protein localized in the Golgi apparatus (42), in growing cells when NH4+ was used as a nitrogen source (Fig. 2C; see Fig. S1 in the supplemental material). This localization is consistent with the fact that utilization of tetrapeptide via Isp4 was not operative in the NH4+ medium (Fig. 1A). Upon nitrogen starvation, however, GFP-Isp4 moved to the plasma membrane (Fig. 2D, left panel). Interestingly, GFP-Isp4 localized at the plasma membrane even under a rich nitrogen condition in the pub1Δ ubiquitin ligase mutant (Fig. 2D, right panel). These localization behaviors of Isp4 are quite similar to those of permeases Aat1 and Cat1 (3, 43). Localization of both Ptr2 and Isp4 on the cell surface is consistent with their role in the uptake of extracellular oligopeptides.
Fig 2.
Cell surface localization of Ptr2 and Isp4. (A) GFP-Ptr2 localizes to the cell surface at both ends and also to the central region of dividing cells. Left panels, expression of GFP-Ptr2 was induced in the absence of thiamine (ON, −B1), and the GFP signals were observed. Right panels, GFP and bright-field (BF) images of the same cells in the uninduced condition (OFF, +B1). Strain, KSP2699. For the functionality of the GFP-Ptr2, see Fig. 4B. (B) The N-terminally GFP-tagged Isp4 is functional. A lysine-auxotrophic isp4Δ strain (591-1C) was transformed with a control GFP vector or the GFP-Isp4-expressing plasmid. Utilization of the Leu-Ser-Lys-Leu tetrapeptide as lysine via the GFP-Isp4 was monitored. (C) Isp4 localizes in the Golgi apparatus under rich nitrogen (NH4+) conditions. Localization of the functional GFP-Isp4 overlapped with that of the Golgi-localized Gms1-RFP protein. Strain, KSP2972. (D) Membrane localization of GFP-Isp4 is regulated by nutrients through the function of the ubiquitin ligase Pub1. (Left panel) GFP-Isp4 was localized at the plasma membrane after 1 h in nitrogen-free medium (−N). Strain, KSP2971. (Right panel) In the pub1Δ strain (KSP2996), GFP-Isp4 membrane localization was detected even in the presence of the rich nitrogen source NH4+.
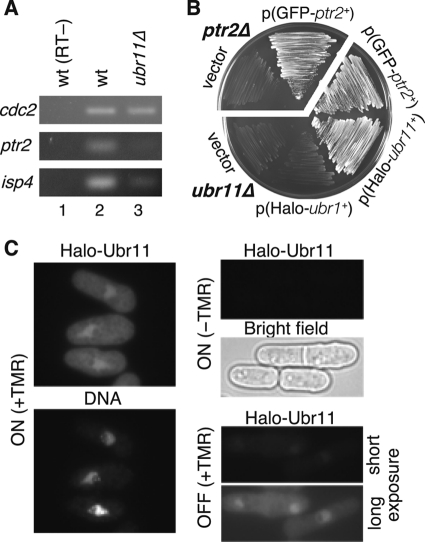
Fig 4.
Ubr11 is required for the expression of ptr2 and isp4. (A) The mRNA levels of the peptide transporter genes in each strain were compared by RT-PCR. Cells were cultured in EMM-N (proline) medium. The cdc2 gene was used as an internal control for the input RNA levels. Lane 1, the same total RNA used in lane 2 was similarly processed but without reverse transcription, as a control. Lane 2, wild-type cells (KSP2); lane 3, ubr11Δ cells (KSP2321). (B) Ectopic expression of the dipeptide transporter Ptr2 rescues the peptide utilization defect in the ubr11Δ strain. The ptr2Δ and ubr11Δ strains, both auxotrophic for histidine, were transformed with the indicated plasmids or a control vector. Growth of each strain in minimal medium containing the His-Leu dipeptide as a source of histidine was examined. The GFP-tagged Ptr2 was functional because it could rescue the peptide uptake defect in a ptr2Δ strain (KSP2698 or KSP2699). Only the ubr11Δ mutant expressing ectopic Halo-tagged Ubr11 (KSP2874) or GFP-Ptr2 (KSP2335b), but not control GFP only (KSP2335a) or Halo-Ubr1 (KSP2335c), grew under this condition. (C) Ubr11 accumulates in the nuclear chromatin region. The Halo-Ubr11 protein was induced in a ubr11Δ strain (KSP2817) and labeled with a fluorescent TMR ligand for the halo tag (ON, +TMR). Fluorescence specificity was verified by observing induced cells without labeling (ON, −TMR) or uninduced cells (OFF, +TMR). Residual weak expression from the nmt promoter occurred even under the uninduced condition; therefore, the Halo-Ubr11 signal was detected after a longer exposure time.
Ubr11 is essential for oligopeptide utilization.
Next we examined whether the Ubr ubiquitin ligases had a role in peptide utilization in S. pombe, as reported for S. cerevisiae (1, 8). Leucine-auxotrophic leu1 strains were tested for their ability to grow in medium containing the dipeptide Leu-Ala as a source of leucine. Only the ubr11Δ mutant, and not the wild-type and ubr1Δ strains, failed to grow in the Leu-Ala dipeptide medium (Fig. 3A). The same ubr11Δ strain grew well in the presence of monomeric leucine. We also tested the other leucine-containing dipeptides, Ala-Leu, His-Leu, Lys-Leu, and Tyr-Leu, and obtained similar results (Fig. 3B and data not shown). Furthermore, both the Ala-Leu and Leu-Ala dipeptides did not inhibit the growth of the prototrophic ubr11Δ strain (Fig. 3B and data not shown), indicating that the ubr11Δ strain was unable to utilize leucine-containing dipeptides. Similarly, the ubr11Δ leu1 cells also failed to grow in the Leu-Ser-Lys-Leu tetrapeptide medium (Fig. 3B). We also tested the effect of soy peptides and found that soy peptides supported the growth of leucine-auxotrophic (but otherwise wild-type) strains. In contrast, the ubr11Δ leu1 mutant failed to grow in the same medium (Fig. 3C), indicating that the soy peptides did not contain enough monomeric leucine as amino acid to support the growth of the ubr11Δ leu1 mutant and, more importantly, that Ubr11 was essential to utilize soy peptides in which various different kinds of leucine-containing oligopeptides should exist. Similar to that of the ubr11Δ strain, growth of the ptr2Δ leu1 mutant, but not that of the isp4Δ pgt1Δ opt3Δ leu1 mutant, was severely retarded in the soy peptide medium (Fig. 3C), indicating that under this condition Ptr2, but not the OPT proteins, plays a major role.
We confirmed the defects in a different way by examining whether cells were competent to utilize dipeptides as a nitrogen source. For this purpose, growth of prototrophic strains was monitored in minimal medium containing dipeptides (Ala-Gln) as the sole nitrogen source. A wild-type strain was able to grow in this dipeptide medium (Fig. 3D). In marked contrast, the ptr2Δ peptide transporter and, importantly, the ubr11Δ mutant failed to grow in the same medium. All the strains grew well when the amino acid glutamine, one of the constituents of the Ala-Gln dipeptide, was used as a nitrogen source. Collectively, Ubr11 is indispensable for the utilization of both di- and tetrapeptides.
Ubr11 regulates the mRNA levels of ptr2 and isp4.
As described above, peptide utilization depends on specific transporters, Ptr2 and Isp4. It is likely that the expression or function of these peptide transporters is affected in the ubr11Δ strain. Indeed, RT-PCR analyses clearly revealed that the mRNA levels of ptr2 and isp4 were significantly reduced in ubr11Δ cells (Fig. 4A). In contrast to the case for these two transporters, expression of the pgt1 and opt3 genes was not affected by the lack of the ubr11 gene (data not shown).
We found that forced expression of the ptr2 gene from the heterologous nmt promoter suppressed the dipeptide utilization defect in ubr11Δ cells (Fig. 4B), supporting the notion that the ubr11Δ mutant was defective for the expression of peptide transporters mostly at the transcriptional level. This indicates that insufficient expression of these peptide transporters leads to the peptide utilization defect in the ubr11Δ mutant.
Considering the observation that Ubr11 regulates the transcription of ptr2 and isp4, we hypothesized that Ubr11 may be a nuclear protein. We monitored the cellular localization of Ubr11 that was functionally marked by a Halo7 tag at the N terminus (28). Labeling of the Halo-Ubr11 fusion protein by the fluorescent ligand confirmed that Ubr11 was enriched in the nucleus, especially in the Hoechst-stained chromatin region (Fig. 4C).
Tup11/12 corepressors are required for efficient peptide utilization.
In S. cerevisiae, the Tup1-Ssn6 corepressor system complexed with Cup9 inhibits PTR2 gene expression, and the peptide uptake defect in the ubr1 strain is rescued by inactivating Tup1 or Ssn6 (56). S. pombe has two TUP1-like genes, tup11 and tup12 (41). We examined whether the phenotype of the ubr11Δ cells was also suppressed by inactivation of both the tup genes, as in S. cerevisiae. For reasons that are unclear, inactivation of both tup11 and tup12 resulted in an inability of the cells to use the Leu-Ala dipeptide (Fig. 5A). This growth defect was not observed when soy peptides were used as a source of leucine (Fig. 5B). In contrast to the parental strain, the ubr11Δ tup11Δ tup12Δ triple mutant failed to grow in soy peptide medium containing ammonium (Fig. 5B) or proline (data not shown) as a nitrogen source, suggesting that inactivation of the Tup11/12 complex does not suppress the defects in the ubr11Δ strain. Because of the inviability of ssn6Δ cells of S. pombe (18), we did not test the effect of Ssn6 inactivation on the suppression of defects in the ubr11Δ strain.
Fig 5.
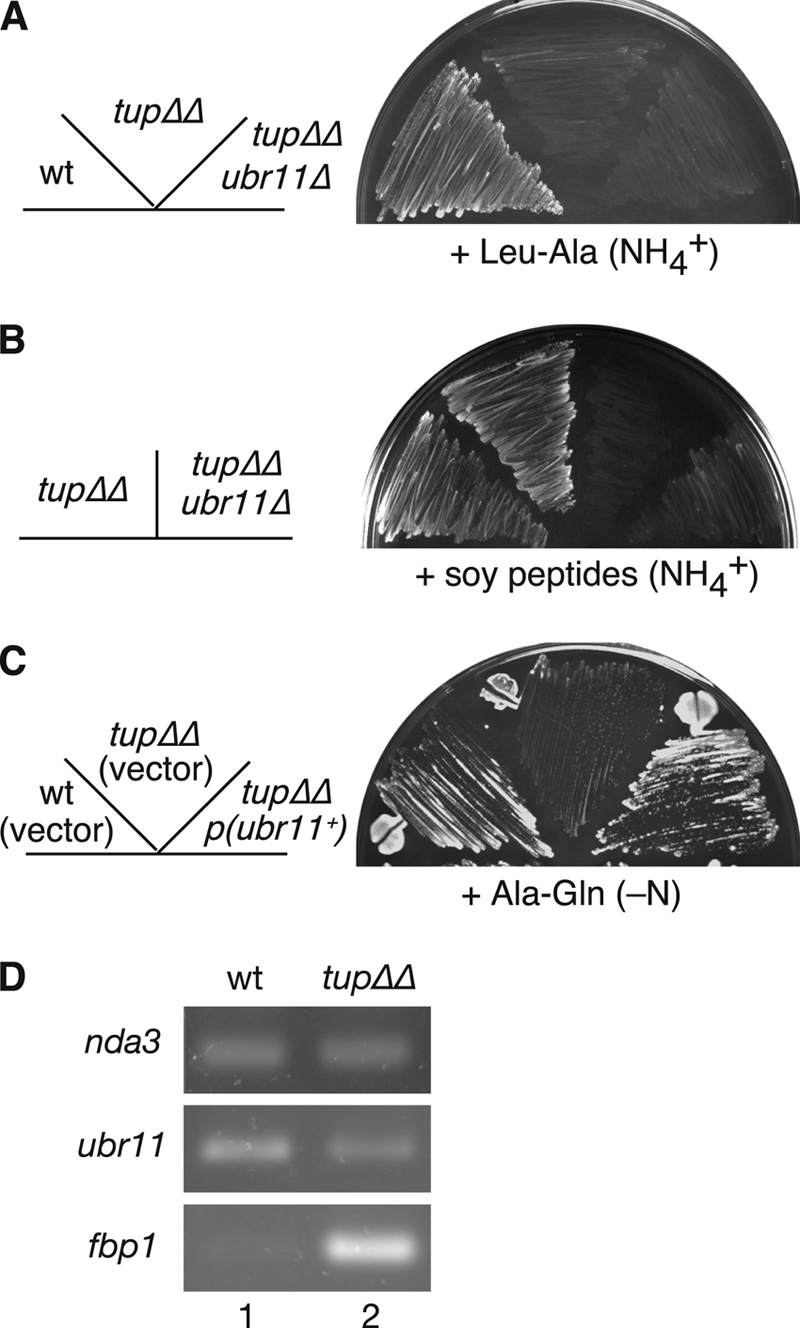
The Tup corepressor proteins are required for efficient peptide utilization. (A and B) The indicated leucine-auxotrophic strains were grown in minimal medium containing Leu-Ala dipeptide (A) or soy peptides (B) as a source of leucine. tupΔΔ, tup11Δ tup12Δ double disruptant. Strains: wt, KSP635; tupΔΔ, KSP2886 and -2887; tupΔΔ ubr11Δ, KSP2888 and -2889. (C) Bypass of Tup protein function by increasing Ubr11 protein expression. The same strains used for panels A and B (KSP635 and KSP2886) were transformed with a plasmid harboring the genomic ubr11+ gene or a control vector, and grown in medium containing Ala-Gln dipeptide as the sole source of nitrogen. (D) ubr11 mRNA levels in the wild-type (KSP635, lane 1) and the tup11Δ tup12Δ (KSP2886, lane 2) strains. The nda3 (β-tubulin) gene was used as an internal control. Expression levels of the fbp1 gene, whose transcription is derepressed in the tup11Δ tup12Δ strain, are also provided.
A prototrophic tup11Δ tup12Δ strain grew very weakly in medium containing the dipeptide Ala-Gln as the sole nitrogen source. Interestingly, this slow growth was significantly improved by increasing the expression of Ubr11 from a multicopy plasmid (Fig. 5C). Although the endogenous ubr11 mRNA levels were slightly decreased in the tup11Δ tup12Δ strain (0.6- to 0.7-fold compared to those in the wild-type strain), sufficient levels of mRNA could be still detected (Fig. 5D).
MG132 is imported independently of PTR and OPT proteins.
Since the human peptide transporters have broad substrate specificity, we examined the effect of nonnative oligopeptides in S. pombe and found that extracellular Ile-Leu dipeptide that is benzyloxycarbonylated at the N terminus (Z-Ile-Leu) could support the growth of leucine-auxotrophic strain only if the Isp4 was functional (Fig. 6A). The peptide aldehyde MG132 [(benzyloxycarbonyl)leucyl-leucyl-leucine aldehyde], which is widely used as a proteasome inhibitor (31), has the same chemical modification with Z-Ile-Leu at its N terminus. Given their structural similarities and the role of Isp4 in the utilization of Z-Ile-Leu, we examined whether the peptidomimetic MG132 was also carried into cells via peptide transporters. UbG76V-GFP was used as a reporter substrate for monitoring proteasomal activity (11). We confirmed that UbG76V-GFP was stabilized in the mts2 proteasome mutant (see Fig. S2A in the supplemental material). The fluorescence intensity of UbG76V-GFP determined by flow cytometry correlated well with its protein levels detected by Western blotting (see Fig. S2C in the supplemental material). As expected, instability of this monitoring substrate depended on the ubiquitin ligase Ufd4, one of the central players in the UFD degradation pathway (see Fig. S2B in the supplemental material) (22, 26).
Fig 6.
MG132 is imported independently of the PTR and OPT proteins. (A) Isp4-dependent utilization of the N-terminally modified nonnative dipeptide Z-Ile-Leu [(benzyloxycarbonyl)isoleucyl-leucine]. Z-Ile-Leu could support the growth of the leucine-auxotrophic strain and thus was used as a source of leucine only if the Isp4 was functional. Consistent with the use of Isp4, Z-Ile-Leu could support growth under the poor (proline) but not rich (NH4+) nitrogen condition (data not shown). The prototrophic (Leu+) isp4Δ strain could grow under this condition, suggesting that Z-Ile-Leu did not inhibit the growth of the isp4Δ mutant. Strains: wt, KSP635; ptr2Δ, KSP2884; ptr2Δ dal5h1Δ dal5h2Δ, 598-1A), isp4Δ Leu+, KSP2999; isp4Δ, 597-1C and 598-1C. (B) MG132 is operative in the peptide transporter-deficient strain. UbG76V-GFP was induced in the wild-type strain (KSP2863) and a quadruple transporter mutant (ΔΔΔΔ, ptr2Δ isp4Δ pgt1Δ opt3Δ) (KSP2920) under rich (NH4+) or poor (proline) nitrogen conditions. Cells were treated with MG132 (lanes 2, 4, 6, and 8) or DMSO (lanes 1, 3, 5, and 7) during the last 3.5 h before the extracts were prepared. UbG76V-GFP protein levels were analyzed by immunoblotting with the anti-GFP antibody.
In the wild-type strain, MG132 prevented the degradation of UbG76V-GFP (Fig. 6B; see Fig. S2A and C in the supplemental material). We tested the effect of MG132 in a strain lacking all the known PTR and OPT proteins (ΔΔΔΔ [ptr2Δ isp4Δ pgt1Δ opt3Δ]). Interestingly, a similar degree of inhibition by MG132 was also observed in this quadruple transporter mutant strain (Fig. 6B). The effect of MG132 was observed under both rich (NH4+) and poor (proline) nitrogen conditions, suggesting that PTR and OPTs are not required for the import of MG132. We also examined the effect of MG132 in a quadruple mutant simultaneously lacking Ptr2, Isp4, and two Dal5-like proteins (Dal5h1 and Dal5h2). However, MG132 still effectively inhibited the degradation of UbG76V-GFP in this mutant (see Fig. S2D in the supplemental material).
DISCUSSION
The fission yeast S. pombe has 1 PTR and 3 OPT peptide transporter family genes. In this study, we investigated the necessity of these transporters and confirmed that Ptr2 (PTR family) and Isp4 (OPT family) are essential for dipeptide and tetrapeptide (Leu-Ser-Lys-Leu) utilization, respectively (Fig. 1A). For Dal5-related transporters (Dal5h1 and Dal5h2), we have no evidence that either protein has a role in peptide utilization in S. pombe. First, inactivation of Ptr2 completely abrogated, but simultaneous inactivation of both Dal5h1 and Dal5h2 did not affect, the utilization of three dipeptides (see Fig S3A and B in the supplemental material). Second, forced expression of Dal5h1 or Dal5h2 in the ptr2Δ strain did not suppress the utilization defect of Ala-Gln dipeptide, a preferred substrate of Dal5 in S. cerevisiae (see Fig. S3C in the supplemental material). Although only a limited number of defined oligopeptides were used in this study, we also examined the utilization of soy peptides, a mixture of various peptides (see Fig. S4 in the supplemental material). When soy peptides were used as a sole nitrogen source, growth of the ptr2Δ isp4Δ double mutant was significantly retarded, though each parental single mutant grew normally (Fig. 1B). Besides, slow growth in the ptr2Δ isp4Δ strain was not further aggravated by additional inactivation of other possible transporters. Collectively, these data indicate that Ptr2 and Isp4 play a major role in general peptide transport in S. pombe, at least with the peptide substrates used in this study. Other transporters may be specialized for specific peptides such as glutathione (Pgt1) (16, 52) or may have a role in different environmental situations not examined in this study. Since the ptr2Δ isp4Δ strain still grows in the soy peptide medium (Fig. 1B), one attractive idea is that an uncharacterized transporter is involved in the peptide uptake under this condition. Alternatively, this residual growth may rely on the utilization of amino acids that could be included in the soy peptides used in this study. However, the amount of free amino acids in the soy peptides seems to be small, since they are insufficient to support the growth of ptr2Δ and ubr11Δ strains that are auxotrophic for several different amino acids (Fig. 2A; see Fig. S4 in the supplemental material).
The most important finding in this study is that the ubiquitin ligase Ubr11 is required for the expression of Ptr2 and Isp4 in S. pombe (Fig. 4A). The defect in peptide utilization is the first apparent phenotype of the ubr11Δ mutant. Involvement of the Ubr protein in peptide utilization was first described for S. cerevisiae, and our finding in S. pombe is the second example of such a role. We found that Ubr11, but not Ubr1, is the ubiquitin ligase (N-recognin) in the N-end rule pathway in S. pombe (our unpublished data), strongly suggesting that peptide utilization is an N-recognin-specific function. Regardless of the conservation of Ubr proteins in eukaryotes, studies on their physiological roles are still limited. The ubr mutants exhibit various phenotypes in both S. cerevisiae and S. pombe, but curiously, only the peptide utilization defect is common to both yeasts. It will be interesting to see if Ubr proteins also play a role in peptide utilization in other organisms. Prior to utilization, oligopeptides are digested to amino acid monomers by an intracellular peptidase(s) (6, 39); this process may be defective in the ubr11Δ mutant. However, the mRNA levels of the ptr2 and isp4 transporter genes are apparently reduced in the ubr11Δ mutant (Fig. 4A), and peptide utilization was rescued by the ectopic expression of Ptr2 from the heterologous nmt promoter (Fig. 4B). Although we have not directly measured peptide uptake in the ubr11Δ strain, it is likely that the peptide utilization defect in the ubr11Δ mutant is caused by the deficiency of peptide import due to inefficient expression of the transporters and that the action of peptidases is irrelevant.
Consistent with the fact that Ubr11 regulates peptide transporter gene expression, the Ubr11 protein is enriched in the nucleus, especially in the chromatin region (Fig. 4C). In S. cerevisiae, Ubr1 facilitates peptide uptake by ubiquitination, thereby promoting the proteasome-dependent degradation of Cup9, a transcriptional repressor of the PTR2 gene (8, 53). The Tup1-Ssn6 corepressor complex, together with Cup9, represses transcription of the PTR2 gene (56). In S. pombe, the peptide uptake defect is not rescued by the inactivation of the Tup11/12 complex (Fig. 5B), unlike in S. cerevisiae. Furthermore, there is no apparent Cup9 homolog in the S. pombe genome. Therefore, although the requirement for the Ubr ubiquitin ligase in peptide utilization is conserved between the two evolutionarily distant yeast species, the target substrate for the Ubr protein and the actual mechanism of transporter gene derepression are different. We identified a suppressor mutation that completely restored ptr2 and isp4 mRNA levels, and consequently peptide utilization, in the S. pombe ubr11Δ mutant (our unpublished results). Our attempts to clone the suppressor gene have been unsuccessful so far. Although the identity of this suppressor gene remains unknown, it is expected to play a very important role in peptide transporter gene expression that is antagonistic to that of Ubr11. Unexpectedly, the tup mutant of S. pombe was inefficient in peptide utilization. Interestingly, increasing the expression of Ubr11 substantially cured the peptide utilization defect in the tup11Δ tup12Δ strain. The fact that the requirement for the Tup11/12 corepressor complex in peptide utilization can be bypassed by a high dosage of Ubr11 reinforces the importance of Ubr11's role in peptide utilization.
Finally, we addressed the question of whether import of the peptide-like proteasome inhibitor MG132 required peptide transporters. Despite the utilization of (benzyloxycarbonyl)isoleucyl-leucine (Z-Ile-Leu) via Isp4 and its structural resemblance to MG132, none of the possible peptide transporters tested in this study was involved; MG132 inhibited proteasome activity even in the strain that lacked all the PTR and OPT transporters (Fig. 6B) and also in a quadruple mutant lacking two dal5-like genes together with being ptr2Δ and isp4Δ (see Fig. S2D in the supplemental material). Since MG132 is a modified trileucine, its amino acid length or aldehyde modification of the third leucine may interfere with its recognition by PTR or OPT transporters. MG132 is generally ineffective in wild-type cells of S. cerevisiae, but it is active if a rich nitrogen source, such as ammonium, in the growth medium is replaced by proline together with the addition of a small amount of sodium dodecyl sulfate (32). Given that the nature of the nitrogen source greatly affects the expression profiles of many genes, this presumably implies that MG132 is actively imported by a specific carrier that is expressed only under the proline condition and not by simple passive diffusion. Most of the proteasomal subunit genes are essential for growth. Unlike the proteasome mutations, MG132 treatment does not cause cell cycle arrest in S. pombe (50). Unraveling the MG132 transport system is an important challenge to more effectively block proteasome activity without using special membrane ergosterol synthesis or efflux pump mutants, which may greatly affect the cellular physiology.
ACKNOWLEDGMENTS
We thank S. Fukutome for technical support, S. Kitagawa for the kind gift of soy peptides, and A. Erler, A. Matsuyama, M. Yoshida, and the Yeast Genetic Resource Center of Japan (YGRC/NBRP) for plasmids and strains. We also thank I. Yamashita and N. Tanaka for discussion and support.
This study was supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, and Culture of Japan.
Footnotes
Published ahead of print 6 January 2012
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Alagramam K, Naider F, Becker JM. 1995. A recognition component of the ubiquitin system is required for peptide transport in Saccharomyces cerevisiae. Mol. Microbiol. 15:225–234 [DOI] [PubMed] [Google Scholar]
- 2.Aouida M, Khodami-Pour A, Ramotar D. 2009. Novel role for the Saccharomyces cerevisiae oligopeptide transporter Opt2 in drug detoxification. Biochem. Cell Biol. 87:653–661 [DOI] [PubMed] [Google Scholar]
- 3.Aspuria PJ, Tamanoi F. 2008. The Tsc/Rheb signaling pathway controls basic amino acid uptake via the Cat1 permease in fission yeast. Mol. Genet. Genomics 279:441–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bähler J, et al. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14:943–951 [DOI] [PubMed] [Google Scholar]
- 5.Basrai MA, Zhang H-L, Miller D, Naider F, Becker JM. 1992. Toxicity of oxalysine and oxalysine-containing peptides against Candida albicans: regulation of peptide transport by amino acids. J. Gen. Microbiol. 138:2353–2362 [DOI] [PubMed] [Google Scholar]
- 6.Becker JM, Naider F. 1977. Peptide transport in yeast: uptake of radioactive trimethionine in Saccharomyces cerevisiae. Arch. Biochem. Biophys. 178:245–255 [DOI] [PubMed] [Google Scholar]
- 7.Brandsch M. 2009. Transport of drugs by proton-coupled peptide transporters: pearls and pitfalls. Expert Opin. Drug Metab. Toxicol. 5:887–905 [DOI] [PubMed] [Google Scholar]
- 8.Byrd C, Turner GC, Varshavsky A. 1998. The N-end rule pathway controls the import of peptides through degradation of a transcriptional repressor. EMBO J. 17:269–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai H, Hauser M, Naider F, Becker JM. 2007. Differential regulation and substrate preferences in two peptide transporters of Saccharomyces cerevisiae. Eukaryot. Cell 6:1805–1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai H, Kauffman S, Naider F, Becker JM. 2006. Genomewide screen reveals a wide regulatory network for di/tripeptide utilization in Saccharomyces cerevisiae. Genetics 172:1459–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dantuma NP, Lindsten K, Glas R, Jellne M, Masucci MG. 2000. Short-lived green fluorescent proteins for quantifying ubiquitin/proteasome-dependent proteolysis in living cells. Nat. Biotechnol. 18:538–543 [DOI] [PubMed] [Google Scholar]
- 12.Didion T, Regenberg B, Jørgensen MU, Kielland-Brandt MC, Andersen HA. 1998. The permease homologue Ssy1p controls the expression of amino acid and peptide transporter genes in Saccharomyces cerevisiae. Mol. Microbiol. 27:643–650 [DOI] [PubMed] [Google Scholar]
- 13.Dodgson J, et al. 2009. Functional genomics of adhesion, invasion, and mycelial formation in Schizosaccharomyces pombe. Eukaryot. Cell 8:1298–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Döring F, et al. 1998. Delta-aminolevulinic acid transport by intestinal and renal peptide transporters and its physiological and clinical implications. J. Clin. Invest. 101:2761–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Du F, Navarro-Garcia F, Xia Z, Tasaki T, Varshavsky A. 2002. Pairs of dipeptides synergistically activate the binding of substrate by ubiquitin ligase through dissociation of its autoinhibitory domain. Proc. Natl. Acad. Sci. U. S. A. 99:14110–14115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dworeck T, Wolf K, Zimmermann M. 2009. SpOPT1, a member of the oligopeptide family (OPT) of the fission yeast Schizosaccharomyces pombe, is involved in the transport of glutathione through the outer membrane of the cell. Yeast 26:67–73 [DOI] [PubMed] [Google Scholar]
- 17.Erler A, Maresca M, Fu J, Stewart AF. 2006. Recombineering reagents for improved inducible expression and selection marker re-use in Schizosaccharomyces pombe. Yeast 23:813–823 [DOI] [PubMed] [Google Scholar]
- 18.Fagerström-Billai F, Durand-Dubief M, Ekwall K, Wright AP. 2007. Individual subunits of the Ssn6-Tup11/12 corepressor are selectively required for repression of different target genes. Mol. Cell. Biol. 27:1069–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gatti L, et al. 2011. Ubiquitin-proteasome genes as targets for modulation of cisplatin sensitivity in fission yeast. BMC Genomics 12:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Godard P, et al. 2007. Effect of 21 different nitrogen sources on global gene expression in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 27:3065–3086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hauser M, Narita V, Donhardt AM, Naider F, Becker JM. 2001. Multiplicity and regulation of genes encoding peptide transporters in Saccharomyces cerevisiae. Mol. Membr. Biol. 18:105–112 [PubMed] [Google Scholar]
- 22.Heessen S, Dantuma N, Tessarz P, Jellne M, Masucci MG. 2003. Inhibition of ubiquitin/proteasome-dependent proteolysis in Saccharomyces cerevisiae by a Gly-Ala repeat. FEBS Lett. 555:397–404 [DOI] [PubMed] [Google Scholar]
- 23.Homann OR, Cai H, Becker JM, Lindquist SL. 2005. Harnessing natural diversity to probe metabolic pathways. PLoS Genet. 1:715–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Island MD, Naider F, Becker JM. 1987. Regulation of dipeptide transport in Saccharomyces cerevisiae by micromolar amino acid concentrations. J. Bacteriol. 169:2132–2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Island MD, Perry JR, Naider F, Becker JM. 1991. Isolation and characterization of S. cerevisiae mutants deficient in amino acid-inducible peptide transport. Curr. Genet. 20:457–463 [DOI] [PubMed] [Google Scholar]
- 26.Johnson ES, Ma PC, Ota IM, Varshavsky A. 1995. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J. Biol. Chem. 270:17442–17456 [DOI] [PubMed] [Google Scholar]
- 27.Kitamura K, et al. 2001. Phosphorylation of Mei2 and Ste11 by Pat1 kinase inhibits sexual differentiation via ubiquitin proteolysis and 14-3-3 protein in fission yeast. Dev. Cell 1:389–399 [DOI] [PubMed] [Google Scholar]
- 28.Kitamura K, Taki M, Tanaka N, Yamashita I. 2011. Fission yeast Ubr1 ubiquitin ligase influences the oxidative stress response via degradation of active Pap1 bZIP transcription factor in the nucleus. Mol. Microbiol. 80:739–755 [DOI] [PubMed] [Google Scholar]
- 29.Koh S, et al. 2002. An oligopeptide transporter gene family in Arabidopsis. Plant Physiol. 128:21–29 [PMC free article] [PubMed] [Google Scholar]
- 30.Krawchuk MD, Wahls WP. 1999. High-efficiency gene targeting in Schizosaccharomyces pombe using a modular, PCR-based approach with long tracts of flanking homology. Yeast. 15:1419–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee DH, Goldberg AL. 1998. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 8:397–403 [DOI] [PubMed] [Google Scholar]
- 32.Liu C, Apodaca J, Davis LE, Rao H. 2007. Proteasome inhibition in wild-type yeast Saccharomyces cerevisiae cells. Biotechniques 42:158–162 [DOI] [PubMed] [Google Scholar]
- 33.Ljungdahl PO. 2009. Amino-acid-inducing signalling via the SPS-sensing pathway in yeast. Biochem. Soc. Trans. 37:242–247 [DOI] [PubMed] [Google Scholar]
- 34.Lubkowitz MA, et al. 1998. Schizosaccharomyces pombe isp4 gene encodes a transporter representing a novel family of oligopeptide transporters. Mol. Microbiol. 28:729–741 [DOI] [PubMed] [Google Scholar]
- 35.Lubkowitz MA, Hauser L, Breslav M, Naider F, Becker JM. 1997. An oligopeptide transport gene from Candida albicans. Microbiology 143:387–396 [DOI] [PubMed] [Google Scholar]
- 36.Matsuyama A, et al. 2004. pDUAL, a multipurpose, multicopy vector capable of chromosomal integration in fission yeast. Yeast 21:1289–1305 [DOI] [PubMed] [Google Scholar]
- 37.Matsuzawa T, et al. 2010. The gld1+ gene encoding glycerol dehydrogenese is required for glycerol metabolism in Schizosaccharomyces pombe. Appl. Microbiol. Biotechnol. 87:715–727 [DOI] [PubMed] [Google Scholar]
- 38.Maundrell K. 1993. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene 123:127–130 [DOI] [PubMed] [Google Scholar]
- 39.Moneton P, Sarthou P, Le Goffic F. 1986. Transport and hydrolysis of peptides in Saccharomyces cerevisiae. J. Gen. Microbiol. 132:2147–2153 [DOI] [PubMed] [Google Scholar]
- 40.Moreno S, Klar A, Nurse P. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795–823 [DOI] [PubMed] [Google Scholar]
- 41.Mukai Y, Matsuo E, Roth SY, Harashima S. 1999. Conservation of histone binding and transcriptional repressor functions in a Schizosaccharomyces pombe Tup1p homolog. Mol. Cell. Biol. 19:8461–8468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakase M, et al. 2010. Mannosylinositol phosphorylceamide is a major sphingolipid component and is required for proper localization of plasmamembrane proteins in Schizosaccharomyces pombe. J. Cell Sci. 123:1578–1587. [DOI] [PubMed] [Google Scholar]
- 43.Nakase M, et al. Intracellular trafficking and ubiquitination of the Schizosaccharomyces pombe amino acid permease Aat1p. Microbiology, in press [DOI] [PubMed] [Google Scholar]
- 44.Perry JR, Basrai MA, Steiner HY, Naider F, Becker JM. 1994. Isolation and characterization of a Saccharomyces peptide transport gene. Mol. Cell. Biol. 14:104–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reuss O, Morschhäuser J. 2006. A family of oligopeptide transporters in required for growth of Candida albicans on proteins. Mol. Microbiol. 60:795–812 [DOI] [PubMed] [Google Scholar]
- 46.Rubio-Allaga I, Daniel H. 2008. Peptide transporters and their roles in physiological processes and drug disposition. Xonobiotica 38:1022–1042 [DOI] [PubMed] [Google Scholar]
- 47.Sato M, Dhut S, Toda T. 2005. New drug-resistant cassettes for gene disruption and epitope tagging in Schizosaccharomyces pombe. Yeast 22:583–591 [DOI] [PubMed] [Google Scholar]
- 48.Sriram SM, Kim BY, Kwon YT. 2011. The N-end rule pathway: emerging functions and molecular principles of substrate recognition. Nat. Rev. Mol. Cell Biol. 21:735–747 [DOI] [PubMed] [Google Scholar]
- 49.Steiner H-Y, Naider F, Becker JM. 1995. The PTR family: a new group of peptide transporter. Mol. Microbiol. 16:825–834 [DOI] [PubMed] [Google Scholar]
- 50.Takeda K, Mori A, Yanagida M. 2011. Identification of genes affecting the toxicity of anti-cancer drug Bortezomib by genome-wide screening in S. pombe. PLoS One 6:e22021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takeda K, Yanagida M. 2005. Regulation of nuclear proteasome by Rhp6/Ubc2 through ubiquitination and destruction of the sensor and anchor Cut8. Cell 122:393–405 [DOI] [PubMed] [Google Scholar]
- 52.Thakur A, Kaur J, Bachhawat AK. 2008. Pgt1, a glutathione transporter from the fission yeast Schizosaccharomyces pombe. FEMS Yeast Res. 8:916–929 [DOI] [PubMed] [Google Scholar]
- 53.Turner GC, Du F, Varshavsky A. 2000. Peptides accelerate their uptake by activating a ubiquitin-dependent proteolytic pathway. Nature 405:579–583 [DOI] [PubMed] [Google Scholar]
- 54.Varshavsky A. 2011. The N-end rule pathway and regulation by proteolysis. Protein Sci. 20:1298–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wiles AM, Cai H, Naider F, Becker JM. 2006. Nutrient regulation of oligopeptide transport in Saccharomyces cerevisiae. Microbiology 152:3133–3145 [DOI] [PubMed] [Google Scholar]
- 56.Xia Z, Turner GC, Hwang CS, Varshavsky A. 2008. Amino acids induce peptide uptake via accelerated degradation of CUP9, the transcriptional repressor of the PTR2 peptide transporter. J. Biol. Chem. 283:28958–28968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xia Z, et al. 2008. Substrate-binding sites of UBR1, the ubiquitin ligase of the N-end rule pathway. J. Biol. Chem. 283:24011–24028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zenker M, et al. 2005. Deficiency of UBR1, a ubiquitin ligase of the N-end rule pathway, causes pancreatic dysfunction, malformations and mental retardation (Johanson-Blizzard syndrome). Nat. Genet. 37:1345–1350 [DOI] [PubMed] [Google Scholar]