Abstract
We report here the first engineering effort for Escherichia coli biocatalysts to assimilate cellobiose through a phosphorolytic mechanism. Cytoplasmic expression of the Saccharophagus cellobiose phosphorylase was shown to enable E. coli to use cellobiose. Subsequent knockout and complementation studies provided solid evidence that the endogenous LacY was responsible for the transport of cellobiose.
TEXT
Cellulose degradation by cellulases generates cellodextrin intermediates dominated by cellobiose (7, 8). Most wild-type Escherichia coli cells are unable to use cellobiose due to the cryptic nature of the cellobiose-utilizing operons such as bgl and cel (11). However, it was reported that a cellobiose-positive phenotype could be easily obtained by prolonged cultivation using cellobiose as the sole carbon source (4, 6, 11). Efforts to engineer E. coli to use cellobiose are of interest, as commercial cellulase cocktails lack sufficient beta-glucosidase activity, which makes beta-glucosidase supplementation necessary (18). Previously, expressing beta-glucosidase or displaying it on the cell surface was attempted, resulting in some reduction of the need for beta-glucosidase (3). Alternatively, Ingram and coworkers engineered E. coli by expressing the cel operon from Klebsiella oxytoca. In this approach, cellobiose is transported through a phosphotransferase system to yield intracellular cellobiose-phosphate, which is subsequently cleaved by another operon-encoded enzyme, phospho-beta-glucosidase, yielding glucose-6-phosphate and glucose (9). Subsequent phosphorylation of glucose is mediated by the native glucose kinase.
The genome of naturally cellulolytic bacteria such as Saccharophagus degradans encodes a cellobiose phosphorylase (EC 2.4.1.20), which potentially provides another mechanism of cellobiose assimilation by cleaving the disaccharide into glucose and a phosphorylated glucose, glucose-1-phosphate (16). Because the phosphorylation uses inorganic phosphate as a donor, the phosphorolytic mechanism is an ATP-saving mechanism more often associated with some cellulolytic bacteria (5, 12, 15, 20). All known cellobiose phosphorylases are cytoplasmic, as they lack signal peptides, and experimental data support this notion (1, 19). Thus, cellobiose assimilation through phosphorolytic mechanism requires a transporter that delivers the unmodified disaccharide to the cytoplasm. Cellobiose permease in E. coli has not been reported, to the best of our knowledge. Lactose permease, LacY, is the best-known disaccharide transporter. However, whether LacY can also transport cellobiose is not clearly understood. In vitro studies indicated that cellobiose was a poor inhibitor for lactose transport via LacY, suggesting a low affinity of cellobiose to LacY (10, 14). In one study, the authors showed that cellobiose inhibited the transport of a lactose analog only in a LacY mutant carrying mutations at the substrate binding site (17). A recent report by Sadie et al. showed that a yeast lactose permease from Kluyveromyces lactis was able to transport cellobiose in Saccharomyces cerevisiae (13). However, no report was found to show that E. coli LacY was able to transport cellobiose.
Expression of a cellobiose phosphorylase enables E. coli to use cellobiose.
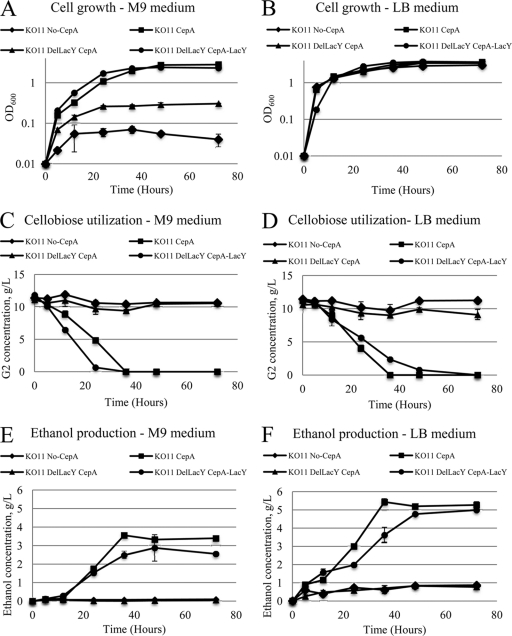
In this study, we cloned and overexpressed a Saccharophagus cellobiose phosphorylase gene in E. coli. To this end, the gene cep94A (ABD80580) (16), was amplified from the genomic DNA using the primers Cep94A-F (5′–CTCGCAGGATCCATGAAATTTGGGCACTTTGACGACAA–3′, BamHI site) and Cep94A-R (5′–CGATGCCTGCAGTTAGCCCAATGTAACTTCTACGTTAC–3′, PstI site). The amplified gene fragment was ligated into BamHI-PstI-linearized pQE80L (a commercial T5-driven expression vector from Qiagen) to obtain pQE80L-cep94A, which was transformed into E. coli KO11 (ATCC 55124), an ethanologen. The resulting KO11 transformant and a control with the same host bearing an empty plasmid were cultured anaerobically in M9 minimal medium in the presence of 1% cellobiose as the carbon source at 250 rpm at 37°C for 72 h. M9 medium contains (per liter) 12.8 g Na2HPO4 · 7H2O, 3 g KH2PO4, 0.5 g NaCl, and 1 g NH4Cl and 2 mM MgSO4–0.1 mM CaCl2. Ampicillin at a concentration of 100 μg/ml and IPTG at a concentration of 0.2 mM and a small amount of yeast extract (0.005 g/liter) (Fisher Scientific, Pittsburgh, PA) were included in the medium. Cells expressing CepA were able to grow in M9 medium with cellobiose as the sole carbon source (Fig. 1A), and cellobiose was consumed after about 36 h of incubation (Fig. 1C). In contrast, the control, the transformant with an empty plasmid (KO11 No-CepA), did not grow (Fig. 1A), and there was minimal consumption of cellobiose (Fig. 1C).
Fig 1.
Time profiles of cell density, residual cellobiose, and ethanol concentration during anaerobic cultivation in M9 (A, C, and E) and LB (B, D, and F) media.
Extracellular, periplasmic, and intracellular fractions were prepared according to the method described in the pET system manual (EMD Chemicals, San Diego, CA). Briefly, 3 ml culture was harvested and resuspended in 1.5 ml 30 mM Tris-HCl buffer (pH 8.0) containing 20% (wt/vol) sucrose and 1 mM EDTA. The cell suspension was incubated at room temperature for 10 min and pelleted by centrifugation at 10,000 × g at 4°C for 10 min. Cell pellets were resuspended in 0.2 ml ice-cold 5 mM MgSO4 and incubated on ice for 10 min. The cells were pelleted by centrifugation as described above, and the supernatant was collected as a periplasmic fraction. The pellets were resuspended in 0.2 ml PBS solution, sonicated, and the supernatant was saved as a cytoplasmic fraction. Each subcellular sample was analyzed for cellobiose phosphorylase and beta-glucosidase activity. The phosphorylase activity of recombinant CepA was determined by measuring the amount of glucose-1-phosphate generated from cellobiose and phosphate (19). The reaction mixture in a total volume of 200 μl consisted of 50 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] buffer, 8 mM phosphate at pH 7.0, 5 mM MgSO4, 0.5 mM NADP, 4 U of phosphoglucomutase, 2 U of glucose-6-phosphate dehydrogenase (Sigma-Aldrich, St. Louis, MO), 250 μM pure cellobiose, and 50 μl of enzyme solution. The mixture was incubated at 42°C for 10 min and heated to 95°C for 5 min to stop the reaction. The concentration of NADPH formed was determined by measuring the change in absorbance at 340 nm. Glucose-1-phosphate formation was estimated assuming molar equivalence to the NADPH produced (19). The beta-glucosidase activity was determined using pNP-beta-glucoside (Sigma-Aldrich, St. Louis, MO) as the substrate, and released p-nitrophenol was measured spectrophotometrically. The reaction mixture (500 μl) contained 20 mM PIPES at pH 6.5 and 0.15 mM pNP-beta-glucoside and an enzyme fraction prepared as described above. After incubation at 37°C for the indicated time, the reaction was quenched by the addition of 500 μl of 1 M Tris base. The absorbance at 400 nm was used to calculate the amount of product formation. The protein concentration was determined through the Bradford assay. One unit of activity is defined as the amount of enzyme that releases 1 nmol of product per minute of reaction.
An enzyme activity assay on cellobiose phosphorylase and beta-glucosidase showed that there were no beta-glucosidase activities in any subcellular locations, extracellular locations, periplasmic space, or cytoplasm, indicating that cellobiose utilization was not due to the activation of an endogenous beta-glucosidase. Additionally, cellobiose phosphorylase activities were measured for all three subcellular fractions. Cellobiose phosphorylase activities were found in cytoplasm only. For example, at 48 h, the cytoplasmic cellobiose phosphorylase activity was about 420 ± 14.21 U/mg (average of two measurements). No activities were found in periplasmic and extracellular locations, consistent with a gene structure devoid of signal sequence. Taken together with the activity analysis of beta-glucosidase, it appears that the ability to use cellobiose can be attributed to the novel phosphorylase activity introduced via overexpression of CepA.
Growth data in LB medium supplemented with 1% (wt/vol) cellobiose showed that cells expressing CepA and the control without expressing CepA both grew, but with the former achieving about 20% higher cell density (Fig. 1B). Apparently, the control used a carbon source present in LB medium other than cellobiose for growth whereas cells expressing CepA could additionally use cellobiose for growth, reaching a higher cell density. The measurement of cellobiose during growth on LB medium supports this claim. The concentrations of cellobiose and ethanol were measured by a high-performance liquid chromatography (HPLC) method on Agilent 1100 HPLC equipped with a refractive index detector (Agilent, Palo Alto, CA). An Aminex HPX-87H column (Bio-Rad, Hercules, CA) was used for separation, and 5 mM H2SO4 at a flow rate of 0.4 ml/min was used as the mobile phase. The consumption of cellobiose by the control was minimal, whereas the cells expressing CepA consumed all the sugar within about 36 h (Fig. 1D). The ability to use cellobiose in cells expressing CepA and the control does not seem to be influenced whether minimal or rich medium was used (Fig. 1C and D), as almost exactly the same sugar profiles were observed for both types of cells in either minimal or rich medium.
Taken together, these data suggest that expressing cellobiose phosphorylase is sufficient to enable E. coli cells to grow on cellobiose through the phosphorolytic mechanism. But how did the cellobiose get in the cells?
LacY is the disaccharide permease responsible for cellobiose transport.
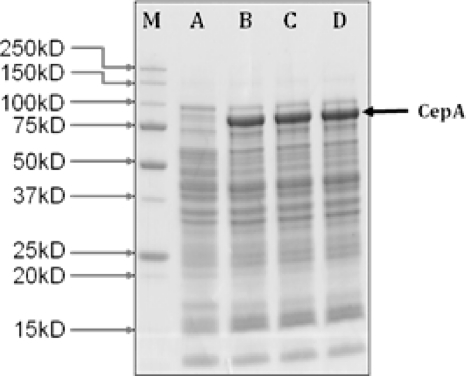
We hypothesized that LacY may play an important role in the transport. Using the PCR knockout method developed by Datsenko and Wanner (2), a lacY deletion mutant was generated, KO11 DelLacY. Briefly, the FLP recombination target (FRT)-flanked kanamycin resistance gene was PCR amplified using primers with 40-nucleotide extensions homologous to the adjacent regions of lacY. Homologous recombination was mediated by the plasmid-borne (pKD46) phage λ Red recombinase. The kanamycin gene was then excised by helper plasmid pCP20 containing the FLP recombinase. Finally, both plasmids were cured by growth at 42°C, as they contain temperature-sensitive replicons. Subsequently, the knockout strain expressing CepA, or KO11 DelLacY-CepA, was tested for cell growth in both M9 and LB as described above. No growth was observed in M9 medium. Also, there was no consumption of cellobiose for the knockout in either M9 or LB medium (Fig. 1D). The full gene lactose permease (lacY, ACT30314) was amplified from the genome of E. coli KO11 via PCR and ligated into PstI-NcoI-linearized pQE80L-cepA to form the plasmid pQE80L-cepA-lacY, which was later transformed into E. coli KO11 DelLacY. Complementation by coexpressing LacY with CepA restored the growth (Fig. 1A). The final cell density reached for cells with plasmid-borne LacY was about 2.4, compared to 2.8 for cells having an intact chromosomal LacY (Fig. 1A). On LB medium, all four strains grew with only small differences in final cell density. This is expected, as in LB medium cell growth was not solely dependent on the ability to use cellobiose. The cellobiose consumption profiles, as shown in Fig. 1C and Fig. 1D for M9 and LB medium, respectively, are evidence that complementation of knockout by plasmid-borne LacY restored the ability of the cells to use cellobiose. These results indicate that LacY is the permease responsible for cellobiose transport. Interestingly, expression from the chromosome was sufficient for effective transport. LacY is part of an inducible lactose operon. Presumably, the induction of transcription was triggered by the addition of IPTG. As IPTG is an analog of lactose, the authentic substrate for LacY, the response of the expression of recombinant cellobiose phosphorylase to IPTG induction in lacY deletion mutants may be retarded. This was investigated using SDS-PAGE. Cells of all four strains, KO11 with an empty plasmid, KO11 CepA, KO11 DelLacY CepA, and KO11 DelLacY CepA-LacY, were harvested after 24 h of cultivation in LB medium containing 0.2 mM IPTG as described above, and cell pellets were then suspended in Laemmli buffer (Bio-Rad). A 20-μg sample of proteins was loaded to the protein gel. The arrow (Fig. 2) indicates a prominent band corresponding to the expected molecular weight of CepA, which was absent from lane A for the control. As shown in Fig. 2, the band intensities for CepA from the other three strains were similar, regardless whether lacY was deleted or not (lanes B, C, and D). Therefore, we could conclude that IPTG induction was not affected by lacY deletion.
Fig 2.
SDS-PAGE for expression of recombinant cellobiose phosphorylase from E. coli strains with and without LacY mutation. Cell pellets were harvested after 24 h of cultivation, and protein samples were prepared according to the manufacturer's manual (Bio-Rad). Each lane was loaded with 20 μg proteins. Lanes: M, Mw marker; A, KO11 No-CepA (KO11/pQE80L); B, KO11 CepA; C, KO11 DelLacY CepA; D, KO11 DelLacY CepA-LacY.
Phosphorolytic mechanism provides an efficient route of cellobiose conversion to ethanol.
We demonstrated in the experiments described above that a heterologous cellobiose phosphorylase and the endogenous LacY effectively form in E. coli a new pathway for cellobiose assimilation. To demonstrate a potential utility in biofuel production, ethanol production from both M9 and LB medium were quantified. As shown in Fig. 1E and F and Table 1, a quantitative conversion of cellobiose to ethanol was achieved in LB medium for KO11 expressing CepA and the complement strains carrying both LacY and CepA on a plasmid. The theoretical yield of 0.537 g ethanol/g cellobiose was used in all calculations. In M9 medium, however, the yield of ethanol was considerably lower, reaching about 63.22% ± 0.18% for KO11 expressing CepA and 47.48% ± 1.84% for the knockout/complement strain, despite the complete consumption of cellobiose in both cases. Apparently, when cells grew on M9, a significant amount of cellobiose was used to support cell growth, whereas when they grew on LB medium, cell growth was supported by other components available in LB medium and cellobiose was quantitatively converted to ethanol.
Table 1.
Ethanol concentration yielda
| Medium | Strain | Ethanol concn (g/liter) | Ethanol yield (%) |
|---|---|---|---|
| M9 | KO11 No-CepA | 0.1 ± 0.04 | 1.76 ± 0.93 |
| KO11 CepA | 3.4 ± 0.01 | 63.22 ± 0.18 | |
| KO11 DelLacY CepA | 0.05 ± 0.01 | 0.93 ± 0.37 | |
| KO11 DelLacY CepA-LacY | 2.55 ± 0.07 | 47.48 ± 1.84 | |
| LB | KO11 No-CepA | 0.87 ± 0.05 | 16.2 ± 1.31 |
| KO11 CepA | 5.27 ± 0.05 | 98.13 ± 1.31 | |
| KO11 DelLacY CepA | 0.77 ± 0.06 | 14.33 ± 1.48 | |
| KO11 DelLacY CepA-LacY | 4.99 ± 0.04 | 92.92 ± 1.11 |
Data shown are average results of two experiments.
Overall, the data demonstrate that assimilation of cellobiose through a phosphorylase-mediated mechanism is efficient, and the novel pathway could be used to convert cellobiose to useful products such as bioethanol.
To our best knowledge, this is the first engineering effort for an E. coli biocatalyst that uses a phosphorolytic mechanism to assimilate cellobiose. This study also contributes to the understanding of sugar transport. Specifically, we provided here solid experimental evidence that cellobiose is transported through endogenous LacY. Further, we demonstrated that the engineered E. coli biocatalysts could be used to convert cellobiose to biofuel through the novel mechanism. The catalysts could be useful in cellulosic ethanol production because they require no beta-glucosidase supplement, which should result in a savings of costs associated with enzymes. Furthermore, the principle of cellobiose assimilation through a phosphorolytic mechanism could be extended to other biofuel products and biorefinery products.
ACKNOWLEDGMENT
R.S. acknowledges a graduate student fellowship from the Institute of Paper Science and Technology at Georgia Tech.
Footnotes
Published ahead of print 22 December 2011
REFERENCES
- 1. Alexander J. 1961. Characteristics of cellobiose phosphorylase. J. Bacteriol. 81:903–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ha SJ, et al. 2011. Engineered Saccharomyces cerevisiae capable of simultaneous cellobiose and xylose fermentation. Proc. Natl. Acad. Sci. U. S. A. 108:504–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hall BG, Betts PW. 1987. Cryptic genes for cellobiose utilization in natural isolates of Escherichia coli. Genetics 115:431–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kawaguchi T, et al. 1998. Cloning, nucleotide sequence, and expression of the Clostridium thermocellum cellodextrin phosphorylase gene and its application to synthesis of cellulase inhibitors. J. Ferment. Bioeng. 85:144–149 [Google Scholar]
- 6. Keyhani NO, Roseman S. 1997. Wild-type Escherichia coli grows on the chitin disaccharide, N,N′-diacetylchitobiose, by expressing the cel operon. Proc. Natl. Acad. Sci. U. S. A. 94:14367–14371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lynd LR, Van Zyl WH, McBride JE, Laser M. 2005. Consolidated bioprocessing of cellulosic biomass: an update. Curr. Opin. Biotechnol. 16:577–583 [DOI] [PubMed] [Google Scholar]
- 8. Lynd LR, Weimer PJ, Van Zyl WH, Pretorius IS. 2002. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 66:506–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Moniruzzaman M, Lai XK, York SW, Ingram LO. 1997. Isolation and molecular characterization of high-performance cellobiose-fermenting spontaneous mutants of ethanologenic Escherichia coli KO11 containing the Klebsiella oxytoca casAB operon. Appl. Environ. Microbiol. 63:4633–4637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Olsen SG, Brooker RJ. 1989. Analysis of the structural specificity of the lactose permease toward sugars. J. Biol. Chem. 264:15982–15987 [PubMed] [Google Scholar]
- 11. Parker LL, Hall BG. 1990. Mechanisms of activation of the cryptic cel operon of Escherichia coli K12. Genetics 124:473–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Reichenbecher M, Lottspeich F, Bronnenmeier K. 1997. Purification and properties of a cellobiose phosphorylase (Cep94A) and cellodextrin phosphorylase (Cep94B) from the cellulolytic thermophile Clostridium stercorarium. Eur. J. Biochem. 247:262–267 [DOI] [PubMed] [Google Scholar]
- 13. Sadie CJ, Rose SH, den Haan R, van Zyl WH. 2011. Co-expression of a cellobiose phosphorylase and lactose permease enables intracellular cellobiose utilisation by Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 90:1373–1380 [DOI] [PubMed] [Google Scholar]
- 14. Sandermann H. 1977. Beta-D-galactoside transport in Escherichia coli: substrate recognition. Eur. J. Biochem. 80:507–515 [DOI] [PubMed] [Google Scholar]
- 15. Tanaka K, Kawaguchi T, Imada Y, Ooi T, Arai M. 1995. Purification and properties of cellobiose phosphorylase from Clostridium thermocellum. J. Ferment. Bioeng. 79:212–216 [Google Scholar]
- 16. Taylor LE, et al. 2006. Complete cellulase system in the marine bacterium Saccharophagus degradans strain 2-40T. J. Bacteriol. 188:3849–3861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Varela MF, Wilson TH. 1996. Molecular biology of the lactose carrier of Escherichia coli. Biochim. Biophys. Acta 1276:21–34 [DOI] [PubMed] [Google Scholar]
- 18. Xin Z, Yinbo Q, Peiji G. 1993. Acceleration of ethanol-production from paper-mill waste fiber by supplementation with beta-glucosidase. Enzyme Microb. Technol. 15:62–65 [Google Scholar]
- 19. Zhang YH, et al. 2011. Hydrolytic and phosphorolytic metabolism of cellobiose by the marine aerobic bacterium Saccharophagus degradans 2-40T. J. Ind. Microbiol. Biotechnol. 38:1117–1125 [DOI] [PubMed] [Google Scholar]
- 20. Zhang YH, Lynd LR. 2004. Kinetics and relative importance of phosphorylatic and hydrolytic cleavage of cellodextrins and cellobiose in cell extracts of Clostridium thermocellum. Appl. Microbiol. Biotechnol. 70:1563–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]