Abstract
While the Pycnoporus cinnabarinus laccase (PcL) is one of the most promising high-redox-potential enzymes for environmental biocatalysis, its practical use has to date remained limited due to the lack of directed evolution platforms with which to improve its features. Here, we describe the construction of a PcL fusion gene and the optimization of conditions to induce its functional expression in Saccharomyces cerevisiae, facilitating its directed evolution and semirational engineering. The native PcL signal peptide was replaced by the α-factor preproleader, and this construct was subjected to six rounds of evolution coupled to a multiscreening assay based on the oxidation of natural and synthetic redox mediators at more neutral pHs. The laccase total activity was enhanced 8,000-fold: the evolved α-factor preproleader improved secretion levels 40-fold, and several mutations in mature laccase provided a 13.7-fold increase in kcat. While the pH activity profile was shifted to more neutral values, the thermostability and the broad substrate specificity of PcL were retained. Evolved variants were highly secreted by Aspergillus niger (∼23 mg/liter), which addresses the potential use of this combined-expression system for protein engineering. The mapping of mutations onto the PcL crystal structure shed new light on the oxidation of phenolic and nonphenolic substrates. Furthermore, some mutations arising in the evolved preproleader highlighted its potential for heterologous expression of fungal laccases in yeast (S. cerevisiae).
INTRODUCTION
Fungal laccases (EC 1.10.3.2) belong to the widely distributed family of blue-multicopper oxidases, and they participate in multiple processes ranging from fungal morphogenesis to lignification/delignification (44). The potential applications of these enzymes are enormous due to their oxidative versatility and low catalytic requirements: they use oxygen from the air and release water as the sole by-product (66). Indeed, their ability to catalyze both polymerization and degradation processes makes them suitable candidates as green biocatalysts in several sectors of industry, including textile and food industries, bioremediation, and forestry (wood and pulp) (53, 63). Recent findings have also highlighted the potential of fungal laccases to be used for improving the conversion of plant biomass in future integrated lignocellulose biorefineries (16), in organic synthesis (32, 52, 64), and in bioelectrocatalysis (58).
The catalytic mechanism of these generalist enzymes is governed by four copper atoms, one located at the T1 site at which the reducing substrate binds and the remaining three clustered at a trinuclear copper site where molecular oxygen is reduced to water (45). Laccases produced by the basidiomycetes white rot fungi—involved in lignin biodegradation—generally exhibit a higher redox potential at the T1 site (close to +800 mV), which broadens the range of possible substrates to be oxidized by the enzyme (1). Moreover, in the presence of natural or synthetic redox mediators, laccase can transform compounds with higher redox potentials than the laccase itself, as well as complex polymers (lignin, starch, cellulose), circumventing steric hindrance difficulties (8, 30, 40). Among high-redox-potential laccases (HRPLs), the laccase from Pycnoporus cinnabarinus (PcL) shows particularly interesting biochemical characteristics in terms of stability and turnover rates for natural and synthetic substrates (36, 67). Indeed, the last decade has seen exhaustive studies about the potential application of PcL in the delignification of paper pulps, pitch control, food industry, dye decolorization, or degradation of polycyclic aromatic hydrocarbon (14, 15, 24, 25, 29, 33, 57). Optimization of PcL production achieved the highest expression levels yet reported for any HRPL (over 1 g/liter from selected fungal monokaryotic strains and up to 100 mg/liter from heterologous expression in Aspergillus spp.) (4, 26, 37, 51). Besides, the PcL has been crystallized (5). Together these features provide significant opportunities for PcL engineering. However, several bottlenecks are encountered when tailoring basidiomycete laccases by directed evolution, due to poor functional expression in the preferred evolutionary host of eukaryotic proteins (Saccharomyces cerevisiae) and the lack of suitable screening assays based on natural laccase substrates. As such, the development of new tools for HRPL design is crucial for further advances in this field (3, 6, 69).
We recently reported the engineering of an HRPL from the basidiomycete PM1 using molecular evolution and rational approaches (41). Here, we describe a directed-evolution platform for the future guidance of PcL and other HRPLs toward possible different fates (e.g., from laccase chimeragenesis to increasing laccase alkalophilicity or substrate specificity onto certain compounds). The native signal sequence of PcL was replaced by the preproleader of the α-factor mating pheromone from Saccharomyces cerevisiae, and the α-PcL fusion gene was subjected to six rounds of evolution. This process exploited the eukaryotic machinery of S. cerevisiae, both to functionally express laccase and to re-create diversity via in vivo DNA recombination. A multiscreening assay based on the oxidation of natural and artificial substrates was validated so as not to limit the development of the enzyme toward dependence on a specific substrate. This strategy was combined with searches for active mutants at more neutral pHs. The mutations identified were mapped and are discussed in relation to the newly described PcL crystal structure.
MATERIALS AND METHODS
Reagents and enzymes.
The pGEM-T Easy vector containing native PcL cDNA (lac1 from P. cinnabarinus I-937, GenBank accession no. AF170093) was provided by E. Record (INRA, Marseille, France). 2,2′-Azino-bis (3-ethylbenzothiazoline-6-sulfonic acid) (ABTS), 2,6-dimethoxyphenol (DMP), Taq polymerase, and an S. cerevisiae transformation kit were all purchased from Sigma-Aldrich (Madrid, Spain). Escherichia coli XL2-Blue competent cells and Genemorph I and II random mutagenesis kits were obtained from Stratagene (La Jolla, CA). Protease-deficient S. cerevisiae strain BJ5465 was bought from LGCPromochem (Barcelona, Spain). The uracil-independent and ampicillin resistance shuttle vector pJRoC30 was kindly donated by Novozymes, and the pGAPZα vector containing the α-factor preproleader was from Invitrogen. A Zymoprep yeast plasmid miniprep kit, Zymoclean gel DNA recovery kit, and DNA clean and concentrator TM-5 kit were all obtained from Zymo Research (Orange, CA). A NucleoSpin plasmid kit was purchased from Macherey-Nagel (Germany), and the restriction enzymes BamHI and XhoI were from New England BioLabs (Hertfordshire, United Kingdom). All chemicals were of reagent-grade purity.
Culture media.
Minimal medium contained 100 ml 6.7% sterile yeast nitrogen base, 100 ml 19.2 g/liter sterile yeast synthetic dropout medium supplement without uracil, 100 ml sterile 20% raffinose, 700 ml sterile double-distilled H2O (ddH2O), and 1 ml 25 g/liter chloramphenicol. Yeast extract-peptone (YP) medium contained 10 g yeast extract, 20 g peptone, and ddH2O to 650 ml. Expression medium contained 720 ml YP, 67 ml 1 M KH2PO4, pH 6.0, buffer, 111 ml 20% galactose, 2 mM CuSO4, 25 g/liter ethanol, 1 ml 25 g/liter chloramphenicol, and ddH2O to 1,000 ml. The yeast extract-peptone-dextrose (YPD) solution contained 10 g yeast extract, 20 g peptone, 100 ml 20% sterile glucose, 1 ml 25 g/liter chloramphenicol, and ddH2O to 1,000 ml. Synthetic complete (SC) dropout plates contained 100 ml 6.7% sterile yeast nitrogen base, 100 ml 19.2 g/liter sterile yeast synthetic dropout medium supplement without uracil, 20 g Bacto agar, 100 ml 20% sterile glucose, 1 ml 25 g/liter chloramphenicol, and ddH2O to 1,000 ml.
Construction of α-PcL.
The pGEM-T Easy vector containing the PcL cDNA was used as a template to amplify PcL using the primers NEcoRI sense (5′-CGGAATTCGCCATAGGGCCTGTGGCGG-3′) and CNotI antisense (5′-AAGGAAAAAAGCGGCCGCTCAGAGGTCGCTGGGGTCAAGTGC-3′), which included targets for EcoRI and NotI (underlined), respectively, and the optimized stop codon for Pichia pastoris. The PcL fragment generated lacked its natural signal peptide, which was replaced by the signal leader of the α-factor preproleader, resulting in detectable laccase secretion. The pGAPZα vector (Invitrogen) containing the α-factor preproleader was linearized with EcoRI and NotI. The amplified PcL fragment was digested with EcoRI and NotI and cloned into the linearized pGAPZα, giving rise to pGAPZα-PcL. This pGAPZα-PcL was used to amplify the fusion gene α-PcL using the following primers: NpJBglII sense (5′-GAAGATCTATGAGATTTCCTTCAATTTTTACTGC-3′) and CNotI antisense (5′-AAGGAAAAAAGCGGCCGCTCAGAGGTCGCTGGGGTCAAGTGC-3′), which included the BglII site (compatible with BamHI) and the NotI site (underlined), respectively. The resulting fragment was digested with BglII and NotI. The episomal shuttle vector pJRoC30 was digested with BamHI and XhoI, and α-PcL was ligated with the vector to produce the pJRoC30-α-PcL construct. PCRs were performed in a final volume of 50 μl containing 400 nM each primer, 25 ng of template, deoxynucleoside triphosphates (dNTPs; 0.25 mM each), 4 mM MgCl2, 5 μl Taq polymerase buffer, and 2.5 units of Taq polymerase. The PCR cycles followed were 94°C for 5 min, 55°C for 5 min, and 72°C for 5 min (1 cycle); 95°C for 0.35 min, 50°C for 2 min, and 72°C for 4 min (25 cycles); and 72°C for 10 min (1 cycle).
Laboratory evolution.
For each generation, PCR fragments were cleaned and concentrated, loaded onto a low-melting-point preparative agarose gel, and purified using the Zymoclean gel DNA recovery kit (Zymo Research). The PCR products were cloned under the control of the gal1 promoter of the expression shuttle vector pJRoC30, which was linearized with XhoI and BamHI. The linearized vector was concentrated and purified as described above for the PCR fragments.
(i) First generation: mutagenic PCR.
Two libraries (1,000 mutants each) with different mutation rates were generated by mutagenic PCR with Mutazyme II DNA polymerase, using α-PcL as template. The first mutagenic library was constructed with a mutation rate of between 0 and 4.5 mutations per 1,000 bp, and the second was constructed with a rate of between 4.5 and 9 mutations per 1,000 bp. Error-prone PCR was carried out in a gradient thermocycler (Mycycler; Bio-Rad) in a final volume of 50 μl containing 185 nM each primer, 4.65 μg and 2 μg of template for low- and medium-mutation-rate libraries, respectively, dNTPs (0.2 mM each), 3% dimethyl sulfoxide (DMSO), and 2.5 units of Mutazyme II DNA polymerase. PCRs were performed as follows: 95°C for 2 min (1 cycle); 94°C for 0.45 min, 53°C for 0.45 min, and 74°C for 3 min (28 cycles); and 74°C for 10 min (1 cycle). The primers used for amplification were RMLN sense (5′-CCTCTATACTTTAACGTCAAGG-3′ which binds to bp 160 to 180 of pJRoC30-αPcL) and RMLC antisense (5′-GGGAGGGCGTGAATGTAAGC-3′, which binds to bp 2031 to 2050 of pJRoC30-αPcL). To promote in vivo ligation, overhangs of 40 and 66 bp homologous to the linear vector were designed. The PCR products (400 ng) were mixed with the linearized vector (100 ng) and transformed into competent cells using a yeast transformation kit (Sigma). Transformed cells were plated on SC dropout plates and incubated for 3 days at 30°C. Colonies containing the whole autonomously replicating vector were selected and screened. This protocol was applied for each round of evolution.
(ii) Second generation: mutagenic PCR and in vivo DNA shuffling.
The best mutants from the first generation (1D12, 3D3, 5F7, and 3G10) were subjected to Taq-MnCl2 amplification, and they were recombined by in vivo DNA shuffling (∼2,000 clones). PCR amplification mixtures were prepared in a final volume of 50 μl containing 90 nM each primer (RMLN and RMLC), 4.6 ng of the mutant template, 0.3 mM dNTPs (0.075 mM each), 3% DMSO, 1.5 mM MgCl2, 0.01 mM MnCl2, and 2.5 units of Taq polymerase. The PCRs and the primers used were the same as those used in the previous generation. Mutated PCR products were mixed in equimolar amounts and transformed into S. cerevisiae with the linearized vector (ratio of PCR products/vector = 4:1).
(iii) Third generation: mutagenic PCR and in vivo DNA shuffling.
The best mutants from the 2nd generation (10A7, 19C8, 20C7, 1F10, 1C9, and 2D8) were submitted to Taq-MnCl2 amplification and recombined by in vivo DNA shuffling (2,000 clones), as described for the second generation.
(iv) Fourth generation: mutational exchange by IVOE.
Mutational exchange was carried out using the 7A9 mutant from the 3rd cycle and evolved PM1L template (41) as templates. Mutations were introduced by site-directed mutagenesis/recombination by in vivo overlap extension (IVOE) (3). Mutations R[α2]S, A[α9]D, A240P, and P394H were introduced between both laccase scaffolds by mutational exchange. PCR mixtures were prepared in a final volume of 50 μl containing 0.25 μM each primer, 100 ng mutant template (7A9 or evolved PM1L), 1 mM dNTPs (0.25 mM each), 3% DMSO, and 2.5 units of Pfu Ultra DNA polymerase. The PCRs were performed as follows: 95°C for 2 min (1 cycle); 94°C for 0.45 min, 55°C for 0.45 min, and 74°C for 2 min (28 cycles); and 74°C for 10 min (1 cycle).
(a) P394H mutant.
The primers for PCR 1 were RMLN and 3CP484HREV (5′-GCAAGTGGAAGGGGTGGTGGAAGCCGGGGGCGGCGGAGG-3′, which binds to bp 1639 to 1678 of pJRoC30-αPM1 and where the underlining indicates the targeted codon for mutagenesis). The primers for PCR 2 were 3CP484HFOR (5′-CCTCCGCCGCCCCCGGCTTCCACCACCCCTTCCACTTGC-3′, which binds to bp 1639 to 1678 of pJRoC30-αPM1) and RMLC.
(b) A[α9]D mutant.
The primers for PCR 1 were RMLN-2 (5′-GGTAATTAATCAGCGAAGC-3′, which binds to bp 5 to 24 of pJRoC30-αPM1) and 1C-REVDI (5′-GAGGATGCTGCGAATAAATCATCAGTAAAAATTGAAGG-3′, which binds to bp 219 to 257 of pJRoC30-αPM1). The primers for PCR 2 were 1C-FORDI (5′-CCTTCAATTTTTACTGATGATTTATTCGCAGCATCCTC-3′, which binds to bp 219 to 257 of pJRoC30-αPM1) and RMLC.
(c) Site-directed recombination library R[α2]S, A240P.
the primers for PCR 1 were RMLN and 3SA240P antisense (5′-GCACGAAGGAGTAGCGCTGCGCAGGAAAAATCTGGATTGAATC-3′, which binds to bp 195 to 235 of PjRoC30-αPcL). The primers for PCR 2 were 2SPREAL sense (5′-GGATCCATAAGATCTATGAGTTTTCCTTCAATTTTTACTGC-3′, which binds to bp 1182 to 1224 of PjRoC30-αPcL) and RMLC antisense. Approximately 400 clones were explored.
(v) Fifth generation: mutagenic PCR.
The 7A9 mutant was subjected to mutagenic PCR with Mutazyme II DNA polymerase (∼1,200 clones), using the same conditions described for the construction of the medium-mutation-rate library of the 1st generation.
(vi) Sixth generation: in vivo DNA shuffling and backcrossing recombination and mutagenic PCR.
Two different libraries were prepared in the 6th generation. Library 1 was built by in vivo DNA shuffling of the 7 best mutants from the 5th generation (1H3, 6A10, 12B4, 3B7, 9E2, 7F11, and 8B9) and with the 5D3 mutant from the 3rd generation for backcrossing. The mutants were amplified by PCR in a final volume of 50 μl containing 0.25 mM each primer (RMLN and RMLC), 100 ng of each mutant template, 1 mM dNTPs (0.25 mM each), 3% DMSO, and 2.5 units of Pfu Ultra DNA polymerase. PCRs were performed as follows: 95°C for 2 min (1 cycle); 94°C for 0.45 min, 55°C for 0.45 min, and 74°C for 2 min (28 cycles); and 74°C for 10 min (1 cycle). The amplified products were then cotransformed (100 ng of each of the 8 mutants) with the linearized plasmid (200 ng) using the yeast transformation kit (Sigma). Library 2 was built by error-prone PCR using Taq-MnCl2 and the 1H3 mutant as the parental type. The PCR products (400 ng) were mixed with the linearized vector in a 4:1 ratio and transformed into competent cells using the yeast transformation kit (Sigma).
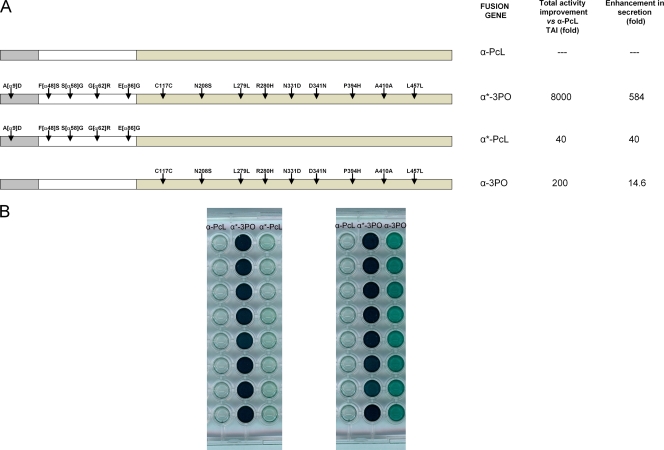
Engineering α*-PcL and α-3PO fusion genes.
Two fusion genes were constructed by using IVOE (3). (i) The α*-PcL fusion gene comprised the evolved α-factor preproleader (harboring mutations A[α9]D, F[α48]S, S[α58]G, G[α62]R, and E[α86]G) plus the native PcL. (ii) The α-3PO fusion gene comprised the native (nonevolved) α-factor preproleader plus the ultimate evolved mature PcL (3PO laccase) harboring C117C, N208S, L279L, R280H, N331D, D341N, P394H, A410A, and L457L.
(i) α*-PcL fusion.
The primers for PCR 1 were RMLN and alpha-rev (5′GCATTGGTAAGGGTCAGGTCC3′, which binds to bp 5′-550 to 521-3′ of pJRoC30-α*-3PO). The primers for PCR 2 were PcL-dir (5′ AATTCGCCATAGGGCCTGTGG 3′, which binds to bp 5′-477 to 499-3′ of pJRoC30-α-PcL) and RMLC. The products from PCR 1 and PCR 2 have overhangs with homologous regions of 44 bp between each other and of 40 bp and 66 bp with the linearized vector for in vivo cloning.
(ii) α-3PO fusion.
The primers for PCR 1 were RMLN and alpha-rev (5′GCATTGGTAAGGGTCAGGTCC3′, which binds to bp 5′-550 to 521-3′ of pJRoC30-α-PcL). The primers for PCR 2 were PcL-dir (5′ AATTCGCCATAGGGCCTGTGG 3′, which binds to bp 5′-501 to 522-3′ of pJRoC30-α*-3PO) and RMLC. The products from PCR 1 and PCR 2 have overhangs with homologous regions of 44 bp between each other and of 40 bp and 66 bp with the linearized vector for in vivo cloning.
For both fusions, the linearized plasmid (100 ng) was mixed with products from PCR 1 and PCR 2 (400 ng each) and transformed into competent S. cerevisiae cells. Individual clones were picked and cultured in 96-well plates (GreinerBio-One, Germany) containing 50 μl of minimal medium per well and subjected to the screening procedure described below. Positive clones were rescreened (see below), the in vivo-repaired plasmid was recovered, and the fusion genes were confirmed by DNA sequencing.
High-throughput (HTP) multiscreening. (i) General aspects.
In the first cycles of evolution, the low secretion levels required performing screenings in liquid format after 5 days of induction. From the 3rd cycle onwards, expression was strong enough to reduce protein induction to 24 h. It is worth noting that in the first rounds of evolution, screening in endpoint mode was required after incubating the supernatants for 24 h in the presence of the substrates. In the last cycles, the improvements were assessed in a few minutes, mainly due to the increases in expression/activity.
(ii) HTP assay.
Individual clones were selected and cultured in 96-well plates (Sero-Wel; Sterilin, Staffordshire, United Kingdom) containing 50 μl minimal medium per well. In each plate, column number 6 was inoculated with the parental type and one well (H1 control) was not inoculated. The plates were sealed to prevent evaporation and incubated at 30°C, 225 rpm, and 80% relative humidity in a humidity shaker (Minitron-INFORS; Biogen, Spain). After 48 h, 160 μl of expression medium was added to each well, and the plates were incubated at 20°C for 5 days in the first two generations and for 24 h in subsequent generations. The plates (master plates) were centrifuged (Eppendorf 5810R centrifuge; Germany) for 5 min at 3,000 × g at 4°C, and 20 μl of the supernatant was transferred from the master plate onto three replica plates with the help of a robot (Liquid Handler Quadra 96-320; Tomtec, Hamden, CT). The first replica plate was filled with 180 μl of 100 mM sodium acetate buffer (pH 5.0) containing 3 mM ABTS. The second replica plate was filled with 180 μl of 100 mM sodium acetate buffer (pH 5.0) containing 3 mM DMP. The third replica plate was filled with 180 μl of 100 mM sodium acetate buffer (pH 5.0) containing 250 μM sinapic acid (SA). The plates were briefly stirred, and the absorption at 418 nm (εABTS·+ = 36,000 M−1 cm−1), 469 nm (εDMP = 27,500 M−1 cm−1), and 512 nm (εSA = 14,065 M−1 cm−1) was recorded in a plate reader (SPECTRAMax Plus 384; Molecular Devices, Sunnyvale, CA). The plates were incubated at room temperature in darkness until the color developed, and the absorption was then remeasured. Relative activities were calculated from the difference in absorption over time, and that of the initial measurement was normalized against the parental type in the corresponding plate. The coefficients of variance (CV) for the screening assays were adjusted throughout the evolution process (resulting in CVs below 11% from the third round of evolution onward).
(iii) First rescreening.
Aliquots (5 μl) of the best clones were removed from the master plates to inoculate 50 μl of minimal medium in new 96-well plates. Columns 1 and 12 (rows A and H) were not used to prevent the appearance of false positives. After 24 h of incubation at 30°C and 225 rpm, 5 μl of the growth medium was transferred to the adjacent well and the plates were incubated for a further 24 h. Finally, 160 μl of expression medium was added and the plates were incubated for 24 h at 20°C. Accordingly, each mutant was grown in 4 wells. Parental types were subjected to the same procedure (lane D, wells 7 to 11). Finally, the plates were assessed using the same screening protocols described above.
(iv) Second rescreening.
An aliquot from the wells with the best clones from the first rescreening was inoculated in 3 ml of YPD and incubated at 30°C for 24 h at 225 rpm. The plasmids from these cultures were extracted (Zymoprep yeast plasmid miniprep kit; Zymo Research), and as the product of the Zymoprep extraction was very impure and the concentration of extracted DNA was very low, the shuttle vectors were transformed into supercompetent E. coli cells (XL2-Blue; Stratagene) and plated onto LB-ampicillin (LB-amp) plates. Single colonies were selected, used to inoculate 5 ml LB-amp medium, and grown overnight at 37°C and 225 rpm, after which the plasmids were extracted (NucleoSpin plasmid kit; Macherey-Nagel, Germany). S. cerevisiae was transformed with plasmids from the best mutants and also with the parental type. Five colonies of each mutant were selected and screened as described above.
Production and purification of PcL variants. (i) Production of laccases in S. cerevisiae.
A single colony from the S. cerevisiae clone containing the parental or mutant laccase genes was selected from an SC dropout plate, inoculated in 3 ml of minimal medium, and incubated for 48 h at 30°C and 225 rpm (Micromagmix shaker; Ovan, Spain). An aliquot of the cells was removed and inoculated in a final volume of 50 ml of minimal medium in a 500-ml flask (optical density at 600 nm [OD600] = 0.25). The cells were incubated for two complete growth phases (6 to 8 h). Thereafter, 250 ml of expression medium was inoculated with the 50-ml preculture in a 1-liter flask (OD600 = 0.1). After incubation for 120 h at 20°C and 225 rpm (laccase activity was maximal; OD600 = 25 to 30), the cells were separated by centrifugation for 20 min at 3,000 × g (4°C) and the supernatant was double filtered (using both a glass membrane and a nitrocellulose membrane of 0.45-μm pore size).
(ii) Laccase purification.
The crude extract was first concentrated and dialyzed in 20 mM Tris-HCl buffer (pH 7.4) by tangential ultrafiltration through a 10-kDa-pore-size membrane (Minisette; Filtron) using a peristaltic pump (Masterflex easy load; Cole-Parmer). The concentrate was then precipitated with ammonium sulfate at 65%, and after centrifugation, the supernatant was dialyzed and concentrated by pressure ultrafiltration through a 10-kDa-pore-size membrane (Amicon; Millipore). The sample was filtered and loaded onto a weak anion-exchange column (HiTrap Q FF; Amersham Bioscience) preequilibrated with Tris-HCl buffer and coupled to an ÄKTA purifier system (GE Healthcare). The proteins were eluted with a linear gradient of from 0 to 1 M NaCl at a flow rate of 1 ml/min in two phases: from 0 to 50% over 75 min and from 50 to 100% over 15 min. Fractions with laccase activity were pooled, concentrated, dialyzed against Tris-HCl buffer, and loaded onto a high-resolution, strong-anion-exchange column (MonoQ HR 5/5; Amersham Bioscience) that was preequilibrated with Tris-HCl buffer. The proteins were eluted with a linear gradient of 0 to 1 M NaCl at a flow rate of 1 ml/min in two phases: from 0 to 25% in 25 min and from 25 to 100% in 1 min. Fractions with laccase activity were again pooled, dialyzed against Tris-HCl buffer, concentrated, and further purified by high-pressure liquid chromatography (HPLC) with a Superose 12 HR 10/30 molecular exclusion column (Amersham Bioscience) preequilibrated with 150 mM NaCl in Tris-HCl buffer at a flow rate of 0.5 ml/min. The fractions with laccase activity were pooled, dialyzed against Tris-HCl buffer, concentrated, and stored at −20°C. Throughout the purification protocol the fractions were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) on 7.5% gels, in which the proteins were stained with Coomassie blue. All protein concentrations were determined using the Bio-Rad protein reagent and bovine serum albumin as a standard.
Overproduction of evolved mutants in Aspergillus niger.
The cDNA corresponding to the 7A9 mutant (3rd generation) was cloned in the expression vector pAN52-4. The sequence encoding the evolved α-factor preproleader of 7A9 for secretion in S. cerevisiae was replaced by the 24-amino-acid glucoamylase preprosequence from Aspergillus niger, under the Emericella nidulans gpd promoter and trpC terminator. Cotransformants were selected on agar plates of selective minimum medium without uridine containing 200 mM ABTS, which gave green colonies when laccase was expressed (51). In order to screen the laccase production in liquid medium, 50 ml of culture medium containing 70 mM NaNO3, 7 mM KCl, 200 mM Na2HPO4, 2 mM MgSO4, 10% (wt/vol) glucose, and trace elements and adjusted to pH 5.0 with a 1 M citric acid solution was inoculated with 1 × 106 spores/ml. The culture was monitored for 11 days at 30°C in a shaker incubator (200 rpm). The pH was adjusted to 5.0 daily with 1 M citric acid. Twenty positive clones were cultured in liquid for each construction. Results for laccase activity ranged from 150 to 2,400 units/liter. The best producer was selected for the purification of laccase. For protein purification, 450-ml cultures were prepared in 1-liter flasks under the same conditions and purified as reported elsewhere (51).
Characterization of evolved laccase variants. (i) Determination of thermostability.
The thermostability of different laccase samples was estimated by assessing their T50 values, defined as the temperature at which the enzyme retains 50% of its activity after 10 min of incubation (18), using 96/384-well gradient thermocyclers. Appropriate laccase dilutions were prepared in such a way that 20-μl aliquots produced a linear response in the kinetic mode. Subsequently, 50-μl samples (three independent incubations for each point in the gradient scale) were subjected to a temperature gradient ranging from 35 to 90°C. The temperature profile was established as follows (in °C): 35.0, 36.7, 39.8, 44.2, 50.2, 54.9, 58.0, 60.0, 61.1, 63.0, 65.6, 69.2, 72.1, 73.9, 75.0, 76.2, 78.0, 80.7, 84.3, 87.1, 89.0, and 90.0. After a 10-min incubation, the samples were chilled on ice for 10 min and incubated at room temperature for a further 5 min. Afterwards, 20-μl samples were added to 180-μl volumes of 100 mM acetate buffer (pH 5) containing 3 mM ABTS, and the activities were measured in triplicate in kinetic mode. The thermostability values were deduced from the ratio between the residual activities for incubations at the different temperature points and the initial activity at room temperature (23).
(ii) Determination of optimum pH.
Appropriate laccase dilutions were prepared in such a way that 10-μl aliquots produced a linear response in the kinetic mode. Plates containing 10 μl of laccase samples and 180 μl of 100 mM Britton and Robinson buffer were prepared at pH values of 2.0, 3.0, 4.0, 5.0, 6.0, 7.0, 8.0, and 9.0. The assay commenced when 10 μl of 60 mM ABTS or DMP was added to each well to give a final substrate concentration of 3 mM. The activities were measured in triplicate in kinetic mode, and the relative activity (in percent) is based on the maximum activity for each variant in the assay.
(iii) Steady-state kinetic constants.
The kinetics parameters were estimated at two different pH values for each substrate. Reactions were carried out in triplicate in a final volume of 250 μl containing the corresponding substrate in 100 mM acetate buffer (pH 5.0) or 100 mM tartrate buffer (pH 3.0). Substrate oxidation was followed by measurement of the absorption at 418 nm for ABTS (ε418 = 36,000 M−1 cm−1), 469 nm for DMP (ε469 = 27,500 M−1 cm−1), and 312 nm for sinapic acid (ε312 = 17,600 M−1 cm−1) using the plate reader. To calculate the values of Km and kcat, the average Vmax was represented versus substrate concentration and fitted to a single rectangular hyperbola function in SigmaPlot (version 10.0) software, where parameter a was equal to kcat and parameter b was equal to Km.
DNA sequencing.
Plasmids containing HRPL variants were sequenced using a BigDye Terminator (version 3.1) cycle sequencing kit. The following primers were designed with Fast-PCR software (University of Helsinki, Helsinki, Finland): RMLN, PcLF1 (5′-CGACGACACTGTCATTACGC-3′), PcLF2 (5′-CGAGGTCGACTTGCATCC-3′), PcLR3 antisense (5′-GCTTCGTAGGAGTAGTC-3′), PcLR4 antisense (5′-ACAAGAACGAGTGGTTTG-3′), and RMLC.
Protein modeling.
A structural model of the P. cinnabarinus laccase crystal structure at a resolution of 1.75 Å (Protein Data Bank [PDB] accession no. 2XYB) was kindly provided by K. Piontek (University of Munich). Mutations selected upon PcL evolution were analyzed by use of a DeepView/Swiss-Pdb viewer (Glaxo SmithKline) and PyMOL viewer (DeLano Scientific LLC). Using the Swiss-Model protein automated modeling server (http://swissmodel.expasy.org/), evolved PM1L was modeled. The crystal structure of Trametes trogii laccase (PDB accession no. 2HRG; 1.58-Å resolution), which shares 97% sequence identity with PM1L (43), served as a template.
RESULTS
Heterologous expression of PcL in S. cerevisiae.
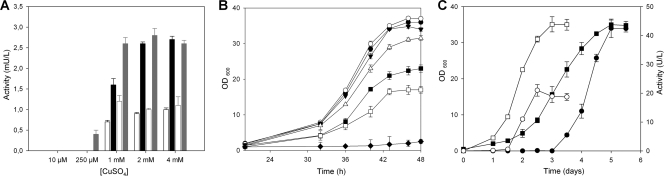
Native PcL, like many HRPLs, is not functionally expressed in S. cerevisiae at levels sufficient for in vitro evolution (32, 41, 42). To circumvent this problem, we constructed a fusion protein where the PcL signal leader peptide (21 amino acids) was exchanged for the preproleader (83 amino acids) of the S. cerevisiae α-mating factor (12) (see Fig. S1 in the supplemental material). This construction (α-PcL) yielded detectable levels of secreted laccase, but they were still low and it was necessary to enhance the laccase production by optimizing certain microfermentation parameters (medium composition, copper uptake, temperature, shaking) to the levels required for the high-throughput format (Fig. 1). When the expression medium was supplemented with ethanol (25 g/liter), both laccase production and S. cerevisiae yeast growth were enhanced (Fig. 1A). We also found that CuSO4 concentrations below 200 μM failed to produce a detectable response, whereas the addition of millimolar concentrations of CuSO4 enhanced laccase activity (Fig. 1A) (28). Beyond 4 mM, copper was toxic for the cell and decreased laccase activity (Fig. 1B). On the other hand, a significant increase in laccase levels was detected by lowering the incubation temperature from 30°C to 20°C (Fig. 1C). By optimization of the fermentation conditions, α-PcL activity levels increased to 0.03 U/liter, which, while still low, fall within the limits of sensitivity for the screening assays used for directed evolution.
Fig 1.
Study of the fermentation conditions for the directed evolution of α-PcL in S. cerevisiae. (A) Effect of ethanol and copper on laccase secretion. The expression medium was tested with 25 g/liter ethanol (filled bars) or without ethanol (empty bars) and assayed at various concentrations of copper. Activity on ABTS (black bars) and DMP (gray bars) is represented as the average values from five independent experiments (the 95% confidence limits are shown). (B) Effect of copper concentration on yeast growth (black circle, 1 mM CuSO4; white circle, 2 mM CuSO4; black inverted triangle, 3 mM CuSO4; white triangle, 4 mM CuSO4; black square, 6 mM CuSO4; white square, 8 mM CuSO4; black diamond, 13 mM CuSO4). Average values are from three independent experiments with 95% confidence limits. (C) Effect of incubation temperature on yeast growth (squares) and laccase secretion (circles) by S. cerevisiae. Solid symbols, 20°C; open symbols, 30°C. Average values from three independent experiments with 95% confidence limits are shown.
Directed evolution of α-PcL in S. cerevisiae.
Six rounds of directed evolution were performed in order to improve the functional expression, kinetics, and pH profile of PcL. The evolution of P. cinnabarinus laccase was carefully planned to better adjust the exogenous enzyme to the yeast secretory route. Accordingly, the whole α-PcL fusion gene (α-factor preproleader plus PcL cDNA) was targeted for random mutagenesis and/or DNA recombination in conjunction with semirational approaches. Briefly, mutations that accumulated after each round of evolution were initially evaluated in terms of the total activity improvement (TAI) in S. cerevisiae microcultures, i.e., the product of secretion levels and kcat values compared to the parental type. These two parameters were assessed after purifying the best mutants (see below), which were selected using a multiscreening colorimetric assay for the oxidation of phenolic and nonphenolic compounds of natural or artificial origin. Thus, a fine-tuning assay based on the natural phenolic mediator sinapic acid, the artificial nonphenolic mediator ABTS, and the phenolic substrate DMP was adjusted and validated (with coefficients of variance of less than 10%; see Fig. S2 in the supplemental material). The use of the three substrates with different chemical characteristics to screen laccase activity guaranteed that the broad laccase substrate specificity was retained. Sinapic acid (a p-hydroxycinnamic acid) was chosen for the assay because (i) it is a natural redox mediator of biotechnological relevance, and (ii) its prompt oxidation by the enzyme generates a pool of phenoxyl radicals whose strong tendency for β-β′ coupling produces the formation of pinkish dilactones detectable in the visible spectrum (16). HRPLs typically exhibit optimal activity at acidic pH, which differs considerably from the pH values used in many of their potential applications (32). In order to alter the PcL pH activity profile (in which the optimum pH versus ABTS is 2.0), the three colorimetric methods used in the screening assay were buffered at pH 5.0, where native PcL retained ∼30% of its activity.
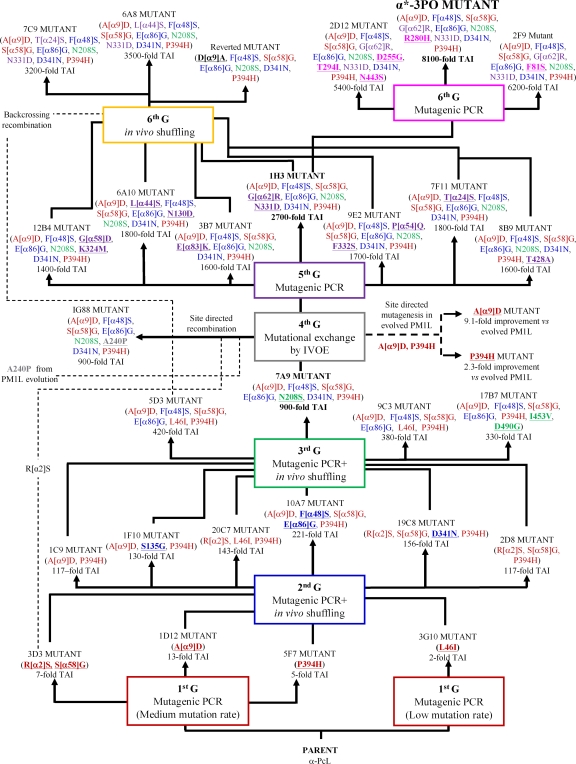
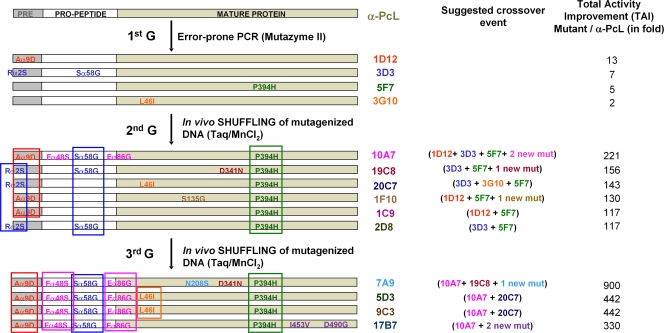
Diversity was generated by classical random mutagenesis methods (i.e., Mutazyme and Taq-MnCl2 polymerases adjusted to different mutational rates) in combination with in vivo approaches for DNA recombination. As we have previously demonstrated, the high frequency of homologous DNA recombination in S. cerevisiae can be extremely useful to promote beneficial crossover events for directed-evolution experiments (2, 3, 41, 69). In vivo DNA shuffling, backcrossing recombination, and in vivo overlap extension (IVOE) for site-directed mutagenesis/recombination were all employed in the present study (Fig. 2; see Table S1 in the supplemental material). In particular, the shuffling of mutagenic libraries from different parental types (in the 2nd and 3rd cycles) proved a suitable approach to recombine best mutations and discover new mutations in the same round of evolution (Fig. 3). In the 4th round, we performed mutational exchange with another highly related basidiomycete laccase recently evolved in our laboratory (PM1L; sequence identity, 77% [41]) in order to check the compatibility of beneficial mutations between both laccase scaffolds, including that of the α-factor preproleader. The mutations R[α2]S, A[α9]D, A240P, and P394H were all analyzed using this new strategy (Fig. 2). During the 1st cycle of α-PcL evolution, the A[α9]D mutation was introduced into the α-factor preleader, producing a 13-fold improvement in secretion, and it was maintained from round 3 onwards in all selected mutants (Fig. 3). The P394H mutation was also discovered in the 1st cycle and induced a 5-fold improvement in specific activity, and it was conserved from round 2 onwards (Fig. 3). Sequence alignment of PcL with other fungal laccases (including PM1L) indicated that P394 belongs to a highly conserved region in HRPLs. Both the P394H and A[α9]D mutations were introduced and analyzed in the evolved PM1L, provoking improvements similar to those seen in PcL (with TAI values of 2.3-fold and 9.1-fold for P394H and A[α9]D, respectively). The A240P mutation (A239P according to PM1L numbering) was discovered in the 2nd round of PM1L evolution, and it resulted in a 12-fold improvement in specific activity (41). This position is also highly conserved in both PcL and PM1L sequences. The A240P mutation was introduced by site-directed recombination/IVOE in the 7A9 mutant (4th generation), along with R[α2]S, which improved secretion. The latter was selected in the 1st round of evolution of α-PcL (3D3 mutant) and accumulated in those selected mutants of the 2nd generation lacking the A[α9]D mutation. Finally, R[α2]S was lost during DNA shuffling in the 3rd round due to its proximity to the selected beneficial mutation A[α9]D, which compromised the crossover event (Fig. 3). After constructing and exploring the site-directed recombination library, no further improvements in the activity of the 7A9 mutant were detected. The A240P mutation showed no beneficial effects on the activity of mutants with that mutation, while the R[α2]S mutation could not be found (either alone or in combination with A240P) in any of the clones sequenced. Hence, R[α2]S and A[α9]D would seem to be incompatible for the secretion of active laccase. The following rounds of evolution combined mutagenic PCR with in vivo DNA shuffling and backcrossing recombination, which gave rise to the ultimate variant, the α*-3PO mutant, with an 8,000-fold increase in the total activity over the original parental-type α-PcL (Fig. 2; see Table S1 in the supplemental material).
Fig 2.
Family tree for the directed evolution of α-PcL. Nonrational approaches (random mutagenesis with different mutational rates and in vivo DNA recombination) were combined with a rational approach (mutational exchange with evolved PM1L through site-directed mutagenesis and site-directed recombination/IVOE). For each round of evolution, new mutations are underlined and presented in bold. TAI indicates the overall improvement in laccase activity detected in S. cerevisiae microcultures for each mutant compared to the parental-type α-PcL. G, generation.
Fig 3.
Schematic illustration of the possible crossover events in selected mutants from the 1st to 3rd evolution cycles. Silent mutations are not included. mut, mutation.
Biochemical characterization.
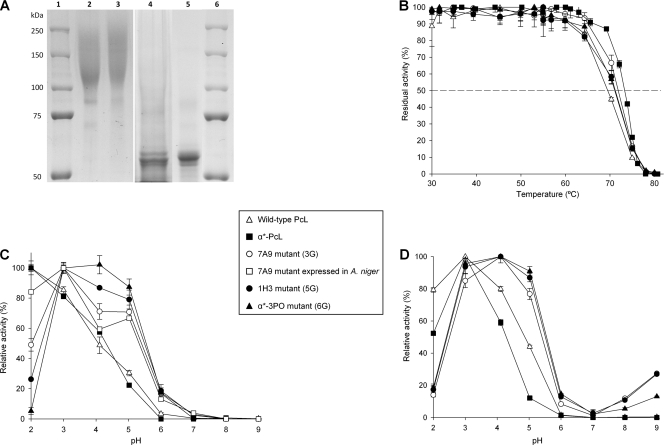
Laccases from the evolution process (7A9 mutant, 3rd generation; α*-3PO mutant, 6th generation) were purified and biochemically characterized. The final mutant, α*-3PO, showed a total activity value of 300 U/liter and secretion levels of ∼2 mg/liter. The molecular mass of the evolved laccases was ∼120,000 Da, with a degree of glycosylation of about 50% being evident in deglycosylation gels (Fig. 4A). Unlike evolved PcL expressed in yeast (α-PcL), the laccase from the fungus (wild-type PcL) resulted in a much lower molecular mass (∼70,000 Da), with glycosylation contributing about 10% (51). The main consequence of the hyperglycosylation was an improvement in the thermal stability of α-PcL expressed by yeast, which displayed a T50 value 3.9°C higher than that of the same enzyme expressed in the original host (wild-type PcL; Fig. 4B). However, the evolved α*-3PO mutant decreased its thermal stability, showing a T50 of 71.2°C (i.e., similar to that of the wild-type PcL from the fungus). The accumulation of beneficial mutations in this particular evolutionary pathway had destabilizing effects, as often occurs when enzymes are evolved in the laboratory (10, 55). Unfortunately, the very weak expression of the parental-type α-PcL in S. cerevisiae (0.003 ABTS U/liter) hampered its purification to homogeneity and biochemical characterization. To circumvent this shortcoming and estimate the contribution of the evolved α-factor preproleader to the laccase secretion levels in yeast, two new fusion genes were engineered by in vivo overlap extension (IVOE) (2): (i) α*-PcL, which comprised the evolved α-factor preproleader plus the native PcL, and (ii) α-3PO, which comprised the native (nonevolved) α-factor preproleader plus the ultimate evolved mature PcL (3PO laccase). The TAIs for α*-PcL and α-3PO were about 40- and 200-fold versus the parental-type α-PcL, respectively (Fig. 5). The evolved α-factor preproleader (α*) enhanced the secretion as much as 40-fold, whereas some mutations in the 3PO mature protein (see Discussion) improved the secretion levels about 14-fold. Altogether the functional expression of the evolved α*-3PO increased 584-fold with respect to that of the parental-type α-PcL expressed by yeast (Table 1). Taking advantage of the notable enhancement conferred by the evolved preproleader, we used α*-PcL transformants to produce and purify the native PcL expressed in yeast and compare it with the PcL of the evolved variants. The activity (kcat) of native PcL was similar regardless of whether the laccase was expressed in S. cerevisiae or P. cinnabarinus (Table 1). Thus, the final α*-3PO mutant showed a 13.7-fold better kcat for ABTS than the native laccase expressed by yeast or P. cinnabarinus. In particular, α*-3PO kinetics improved with all the substrates tested compared to the wild-type PcL from the fungus: the catalytic efficiencies for ABTS, sinapic acid, and DMP increased 20-, 5.4-, and 1.6-fold, respectively. This indicates that the overall activity of the evolved PcL was conserved after evolution. These considerable activity improvements with all the substrates tested laid the ground for further experimentation to drive evolution toward specific destinies.
Fig 4.
Biochemical characterization of evolved laccases. (A) SDS-PAGE. Lanes: 1 and 6, protein markers; 2, purified 7A9 variant (3rd generation); 3, purified α*-3PO variant (6th generation); 4, 7A9 deglycosylated with peptide:N-glycosidase F (PNGase F); 5, α*-3PO deglycosylated with PNGase F. Samples were resolved on a 7.5% SDS-polyacrylamide gel and stained with Coomassie brilliant blue. (B) T50 of evolved laccases from the 3rd, 5th, and 6th generations. Each value and the standard deviation are derived from three independent experiments. (C and D) pH activity profiles of evolved laccases. Activities were measured in 100 mM Britton and Robinson buffer at different pHs with 3 mM ABTS (C) or DMP (D) as the substrate. Laccase activity was normalized to the optimum activity value. Each value, including the standard deviation, is from three independent experiments. Wild-type PcL, original laccase homologously expressed in P. cinnabarinus containing the native secretion leader; α*-PcL, original PcL fused to the evolved α-factor preproleader for secretion in S. cerevisiae; α*-3PO, ultimate variant of the whole evolution process (containing the evolved α-factor preproleader plus the evolved PcL).
Fig 5.
(A) Different fusion genes engineered for the directed evolution of PcL. (B) Aspect of the screening assay for the different constructs and differences in total activity levels between α-PcL, α*-3PO α*-PcL, and α-3PO fusions. S. cerevisiae cells were transformed with pJRoC30 harboring the corresponding fusion gene and plated on SC dropout plates. Individual colonies were picked and inoculated in a 96-well plate. Eight individual clones were evaluated for each fusion gene using the ABTS screening assay (see Materials and Methods for details). The breakdown for secretion was calculated using the kcat improvement value for ABTS (Table 1).
Table 1.
Kinetic constants at pH 5.0 of the purified PcL variants obtained by directed evolution and TAI breakdown
| Laccase | Substrate | Km (mM) | kcat (s−1) | kcat/Km (mM−1 s−1) | TAIa (fold) vs α-PcL | Improvement vs α-PcLb |
|
|---|---|---|---|---|---|---|---|
| kcat | Expression | ||||||
| Wild-type PcL | ABTS | 0.035 ± 0.002 | 38.0 ± 0.4 | 1,085 | |||
| Sinapic acid | 0.013 ± 0.000 | 21.6 ± 0.2 | 1,662 | ||||
| DMP | 0.027 ± 0.002 | 16.3 ± 0.3 | 581 | ||||
| α*-PcL | ABTS | 0.020 ± 0.001 | 35.1 ± 0.9 | 1,755 | 40 | 40 | |
| Sinapic acid | 0.020 ± 0.002 | 19.9 ± 0.5 | 948 | ||||
| DMP | 0.012 ± 0.002 | 10.0 ± 0.3 | 833 | ||||
| 7A9 (3rd generation) | ABTS | 0.013 ± 0.001 | 205.1 ± 4.5 | 15,777 | 900 | 5.8 | 155 |
| Sinapic acid | 0.020 ± 0.003 | 124.4 ± 5.6 | 6,193 | 6.1 | |||
| DMP | 0.106 ± 0.004 | 90.6 ± 0.8 | 858 | 9 | |||
| α*-3PO (6th generation) | ABTS | 0.024 ± 0.002 | 482.6 ± 10.2 | 19,944 | 8,000 | 13.7 | 584 |
| Sinapic acid | 0.023 ± 0.002 | 197.7 ± 4.7 | 8,457 | 10.4 | |||
| DMP | 0.213 ± 0.013 | 196.9 ± 3.1 | 923 | 19.6 | |||
The total activity improvement (TAI) was measured using 3 mM ABTS as the substrate in supernatants of cultures grown in 96-well plates. Values represent the average of five measurements.
The improvement in expression is defined as the ratio of the total increase in activity and kcat with ABTS as the substrate. α-PcL, parent type used in the directed evolution in S. cerevisiae (formed by the native α-factor preproleader and the native PcL); wild-type PcL, original laccase homologously expressed in P. cinnabarinus containing the native secretion leader; α*-PcL, native PcL fused to the evolved α-factor preproleader for secretion in S. cerevisiae; α*-3PO, ultimate variant of the whole evolution process (containing the evolved α-factor preproleader plus the evolved PcL).
Regardless of the substrate, the pH activity profile shifted noticeably toward more neutral values, a consequence of the selective pressure applied in the evolution (since screening was carried out at pH 5.0; Fig. 4C and D). The activity of the original PcL (either α*-PcL secreted by yeast or the wild-type PcL secreted by P. cinnabarinus) was optimal at pH 2.0 for ABTS (pH 3.0 for DMP), and it retained only ∼30% of its relative activity at pH 5.0. The α*-3PO variant exhibited a switch in optimum pH to 4.0, retaining over 90% of its relative activity between pH 3 and 5. At pH 6.0, α*-3PO displayed over 10% residual activity for both DMP and ABTS. This constitutes a promising starting point to engineer alkalophilic laccases with high redox potential.
Although the screening assay was performed at pH 5.0 and the pH profile was broader after evolution, the net improvement in kinetics was not dependent on pH (Tables 1 and 2). Thus, catalytic efficiencies of evolved PcL variants were enhanced not only at the pH used in the screening assay (pH 5.0) but also at the optimum pH of wild-type PcL (pH 3.0; Table 2). It is noteworthy that the turnover rates of the evolved variants for ABTS oxidation at pH 3.0 increased throughout evolution and they were again significantly higher than that of the wild-type PcL, even considering the 5-fold increase in catalytic efficiency of wild-type PcL to oxidize ABTS at pH 3.0 versus pH 5.0. The improved oxidation of ABTS at an acidic pH (close to the optimum pH value of the wild-type PcL for this substrate) was responsible for this difference (65). Interestingly, the evolved variants also displayed better turnovers for the oxidation of phenolic substrates at pH 3.0. Thus, the kcats of the α*-3PO variant were 6.6- and 15.8-fold greater than the kcat of the wild-type PcL when oxidizing DMP and sinapic acid, respectively. However, the affinity for phenolic substrates decreased (being notably lower at this pH than at pH 5.0), with Km values being up to 4.2- and 4.7-fold higher for DMP and sinapic acid, respectively. At pH 5.0, deprotonation of a highly conserved aspartate residue (Asp206; side chain pKa, 3.9) that interacts with the reducing substrate at the T1 Cu site (9) may have contributed to this effect (19).
Table 2.
Kinetic constants of purified PcL variants at pH 3.0
| Laccase | Substrate | Km (mM) | kcat (s−1) | kcat/Km (mM−1 s−1) |
|---|---|---|---|---|
| Wild-type PcL | ABTS | 0.028 ± 0.001 | 144.6 ± 1.1 | 5,164 |
| Sinapic acid | 0.085 ± 0.004 | 52.8 ± 0.1 | 621 | |
| DMP | 0.324 ± 0.015 | 26.4 ± 0.4 | 81 | |
| α*-PcL | ABTS | 0.076 ± 0.001 | 108.6 ± 1.4 | 1,429 |
| Sinapic acid | 0.174 ± 0.011 | 92.9 ± 3.2 | 533 | |
| DMP | 0.449 ± 0.002 | 36.5 ± 0.4 | 81 | |
| 7A9 (3rd generation) | ABTS | 0.021 ± 0.001 | 287.7 ± 3.1 | 13,766 |
| Sinapic acid | 0.186 ± 0.011 | 222.2 ± 7.9 | 1,196 | |
| DMP | 1.002 ± 0.026 | 86.5 ± 0.9 | 86 | |
| α*-3PO (6th generation) | ABTS | 0.030 ± 0.002 | 707.3 ± 15.2 | 23,977 |
| Sinapic acid | 0.401 ± 0.045 | 778.5 ± 61.5 | 1,943 | |
| DMP | 1.357 ± 0.034 | 159.2 ± 1.7 | 117 |
Taken together, these findings indicate that the enhanced catalytic efficiency of P. cinnabarinus laccase through in vitro evolution was not pH dependent. However, a remarkable increase of laccase activity at more neutral pH values was observed. A shift in the optimum pH of T. versicolor laccase following directed mutagenesis of the conserved Asp206 residue (D206A) has been reported (39). The shift in the optimum pH (ΔpH = 1.4) that results from this single mutation applied only to DMP as a substrate and not ABTS, while the mutation decreased the catalytic efficiency toward DMP due to the notable increase in Km. In contrast, we considerably enhanced the catalytic efficiency of the P. cinnabarinus laccase and simultaneously shifted its optimal pH toward more neutral values both for phenolic substrates and for ABTS.
An intermediate variant of the evolutionary route (mutant 7A9, 3rd generation) was used to test its overexpression in Aspergillus niger. Accordingly, the evolved α-factor preproleader of 7A9 mutant was replaced by the glucoamylase preprosequence from A. niger to facilitate the transit and secretion in the new host. This fusion was under the glyceraldehyde-3-phosphate dehydrogenase (gpdA) gene promoter of A. nidulans. Functional expression levels of 23 mg/liter were achieved, without optimizing the fermentation conditions. The mutant expressed by A. niger displayed similar biochemical features as the variant expressed by S. cerevisiae, including the remarkable shift in the pH profile (Fig. 4C).
DISCUSSION
The differences between the basidiomycete machinery of P. cinnabarinus and that of the ascomycete S. cerevisiae (one of the preferred hosts for the directed evolution of eukaryotic proteins) seem to hamper the successful PcL exportation by the yeast. We overcame this hurdle by constructing the α-PcL fusion gene, composed of the α-factor preproleader from S. cerevisiae and the mature laccase, and by optimizing the fermentation conditions to promote laccase production. Moreover, α-PcL was subjected to directed evolution in order to further enhance its functional expression and catalytic efficiency and to shift its pH profile. The presence of ethanol in the expression medium was necessary to detect laccase activity in microcultures. Although inhibition of growth in response to increased ethanol concentrations due to changes in yeast physiology and medium redox balance has been reported (21), we found that the use of ethanol as an extra carbon source was beneficial for the heterologous production of laccase and yeast growth (62). The improved secretion of laccase in the presence of ethanol was in accordance with previous reports of laccase production in other organisms (35, 37). In addition to increasing cytoplasmic membrane permeability (38, 48), ethanol may generate a stress response in the cell, inducing the expression of the chaperones that are involved in the protein folding/secretion process (62). Copper uptake is essential for laccase expression (28), establishing a compromise between protein secretion and copper toxicity when choosing the final copper concentration (in our case, 2 mM CuSO4). Unlike native producers in which copper is involved in the transcriptional regulation of these enzymes (20, 47), in heterologous hosts, high copper concentrations appear to be necessary for the correct folding and assembly of the laccase during posttranscriptional phases. Under conditions of copper deficiency, misfolded laccase apoprotein may be produced and ultimately degraded (28). Furthermore, the apoprotein heterologously produced under low-copper conditions can be reconstituted by exogenous added copper (34). Temperature is a key factor for optimizing heterologous expression of proteins in yeast, with low fermentation temperatures positively regulating laccase production (17). Our results support the positive effect of low temperatures (20°C) on laccase production. Low temperatures can benefit protein folding, as they reduce cell growth rates, allowing non-rate-limiting protein folding (27). Although the S. cerevisiae strain used in this work is protease deficient, reduced activity of residual proteases at low temperature cannot be ruled out.
Filamentous fungi, including Aspergillus (A. awamori, A. niger, or A. oryzae) and Trichoderma reesei, are widely used in the biotechnology sector because of their superior ability to secrete large quantities of heterologous proteins. When the 7A9 mutant was expressed in A. niger, high secretion levels were achieved (up to 23 mg/liter under nonoptimized conditions). This productivity could be further enhanced by molecular and bioprocess engineering (i.e., in a bioreactor) (49). Alternatively, homologous overproduction could be carried out. In fact, the native PcL was already successfully produced by homologous transformation (4). The expression system consisted of the Schizophyllum commune gpdA or the PcL gene promoters. Production of 1.2 g · liter−1 of laccase was achieved, yielding a laccase with the same biochemical and physical characteristics as the native protein. Therefore, it seems plausible that a similar approach can be designed for the overproduction of PcL mutants evolved in S. cerevisiae.
Mutational effect on laccase secretion.
After exploring over 10,500 clones during six generations of laboratory evolution, the final α*-3PO mutant selected harbored 14 mutations: 5 mutations in the α-factor preproleader and the remaining 9 in the mature protein gene, 4 of which were synonymous. All synonymous mutations favored codon usage, which can potentially enhance secretion levels (see Table S1 in the supplemental material) (22, 54). Mutations in the α-factor preproleader probably help expression by adjusting the foreign protein with the secretory leader to the subtleties of the heterologous host. The α-factor preproleader encodes an 83-amino-acid polypeptide, of which the first 19 residues constitute the preleader and the remaining 64 the proleader containing three sites for Asn-linked glycosylation. The preleader inserts the nascent polypeptide into the endoplasmic reticulum (ER), where it is cleaved by an endopeptidase. The protein is then transported to the Golgi compartment, where the proleader is removed by the action of the proteases, KEX2, STE13, and KEX1 (7, 54, 59). Up to 13 positions were mutated in the α-factor preproleader during the entire α-PcL evolution, of which 5 were ultimately conserved in the α*-3PO mutant: A[α9]D, F[α48]S, S[α58]G, G[α62]R, and E[α86]G. Four of these mutations were first introduced at the beginning of evolution (in the 1st and 2nd generations), further recombined, and thus maintained in subsequent generations. Of the preproleader mutations, the A[α9]D mutation produced the greatest improvement that could be attributed to a single mutational change over the entire evolutionary route (up to 13-fold). When this mutation was reverted in the 6th generation, secretion dropped dramatically. A[α9]D is located at the hydrophobic core of the canonical preleader, which is involved in the orientation and insertion of the nascent polypeptide during translocation to the ER. In the directed evolution of α-PM1L (41), a similar beneficial mutation (V[α10]D) was introduced in the same region, suggesting that reduced hydrophobicity in this area may be beneficial for secretion. Moreover, when the A[α9]D mutation was subjected to mutational exchange with α-PM1L, the same improvement was achieved independently of the laccase attached to the signal sequence. These results suggest that individual changes in the hydrophobic domain of the α-factor preleader involving charged carboxylic residues enhance the interaction between the preleader and the signal recognition particle, thereby improving the translocation of the polypeptide chain into the ER (11, 46). The F[α48]S mutation was located in the proleader, and interestingly, the same mutation (same change and position) was also reported in a study of the directed evolution of the α-factor preproleader that sought to enhance heterologous secretion levels of a variety of proteins (50). The S[α58]G mutation was located at the second of the three N-glycosylation sites in the proleader (Asn-Ser-Thr). This site was not removed upon mutation, although the replacement of Ser by Gly may have altered the affinity for sugar anchoring. Results from the parallel in vitro evolution process with α-PM1L revealed that removal of the first glycosylation site of the proleader (by N[α23]K mutation) improved the secretion of active laccase (41). While glycosylation of the proleader may facilitate transit from the ER to the Golgi compartment, our results do not support this hypothesis. Finally, the E[α86]G mutation was located at the processing site of STE13, a dipeptidyl aminopeptidase that removes the spacer peptide (Glu-Ala-Glu-Ala) and that lies between the α-factor proleader and the mature protein. The singular conformational state adopted at the spacer residue upon mutation may alter the action of KEX2 at its cleavage target, Lys-Arg. Notably, during the directed evolution of α-PM1L, a beneficial mutation (A[α87]T) was also selected at the STE13 processing site (41). In view of the similarities between the mutational profiles and regions identified by in vitro evolution of the α-factor preproleader attached to different HRPLs or even other unrelated proteins (50), it seems plausible to evolve this secretory leader as a universal signal peptide for the heterologous expression of HRPLs in yeast.
Mutations in mature evolved PcL.
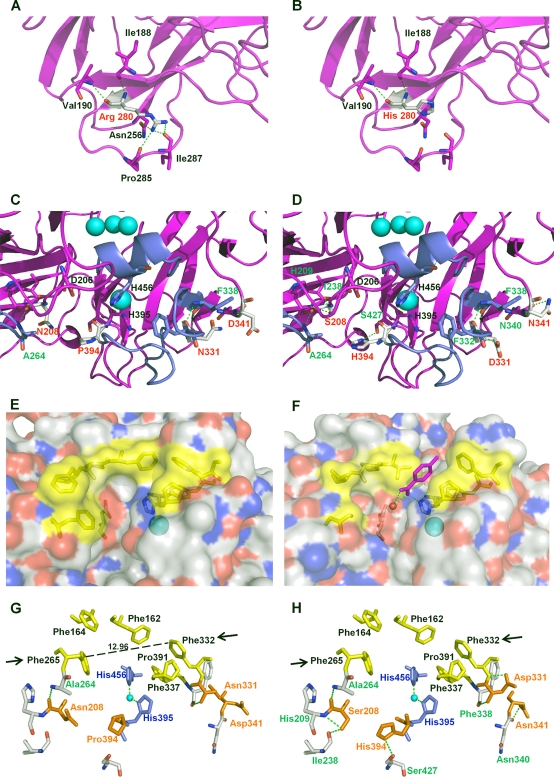
Laccases are organized into three cupredoxin-like domains (D1, D2, and D3). The trinuclear Cu cluster T2/T3 is embedded between D1 and D3, with both domains providing residues for the coordination of the copper ions. D3 contains the T1 mononuclear site, while D3 and D2 contain residues that participate in substrate binding. The mature 3PO laccase harbored five amino acid substitutions, all of which were located in D2 and D3 (N208S, R280H, N331D, D341N, and P394H). Interestingly, taking into account the sequence similarities between both laccase scaffolds (75%) and their parallel artificial evolutionary pathways, the positions mutated in the evolved PM1L were also exclusively located in D2 and D3 (see Fig. S3 in the supplemental material). The relative positions of the mutations in the 3PO variant and their possible interactions with nearby residues were evaluated in light of the PcL crystal structure recently determined at a resolution of 1.75 Å (PDB accession no. 2XYB; K. Piontek, personal communication).
The R280H and D341N mutations are located at the protein surface, with the former being very far from catalytic coppers, whereas the remaining three 3PO mutations are placed in the vicinity of the reducing substrate binding pocket (Table 3; Fig. 6). Arg280 is located at the end of a distal β sheet, where it forms up to 5 H bonds with neighboring residues in native PcL. Analysis of the three-dimensional (3D) protein structure suggests that the R280H mutation may interrupt 3 of the 5 H bonds, thereby enhancing the flexibility of this region (Fig. 6A and B). This effect could facilitate folding during the posttranslational stages. The D341N mutation also maps to an external coil (Fig. 6C and D). Apart from a new H bond formed with a nearby residue, the most notable implication of this mutation is the generation of a new N-glycosylation site, which may aid protein stability or augment the secretion of heterologous proteins otherwise prone to aggregation in ER-derived vesicles (56). Interestingly, this position was initially mutated in the second generation, when activity levels were too low to be assessed in kinetic mode, and the stability of the mutant hits was compromised during screening.
Table 3.
Mutations in mature 3PO variant
| Mutation | Domain | Secondary structure motif | Relative position | Distance (Å) to: |
H bonding with surrounding residuesa |
||
|---|---|---|---|---|---|---|---|
| T1 site | T3 siteb | Before mutation | After mutation | ||||
| N208S | D2 | Beta sheet | Near D206 (responsible for binding phenolic substrates at the T1 site) | 10.13 | 15.81, T3 (2) | A264 | H209, I238, A264 |
| R280H | D2 | End of distal beta sheet | Surface | 34.15 | 30.56, T3 (2) | I188, V190, N256, P285, I287 | I188, V190 |
| N331D | D3 | Beta sheet (substrate binding loop) | Contiguous to F332 (key residue of the binding pocket) | 9.20 | 17.77, T3 (1) | F338 | F332, F338 |
| D341N | D3 | Coil | Surface | 15.16 | 19.64, T3 (1) | N340 | |
| P394H | D3 | Coil (substrate binding loop) | Contiguous to H395 (T1 Cu ligand) | 5.53 | 14.96, T3 (1) | S427 | |
Underlining, interrupted bonds after mutation; bold, bonds newly formed after mutation.
T3(1), first copper atom; T3(2), second copper atom.
Fig 6.
Location of different residues in the parental PcL and the mutations introduced by molecular evolution. (A) Residue R280 in PcL (PDB accession no. 2XYB, from K. Piontek). (B) Mutation R280H in α*-3PO variant. (C) Residues D341, N208, and P394 in PcL. (D) Mutations D341N, P394H, and N208S in the α*-3PO variant. (E and F) Detail of the electrostatic surface of PcL crystal structure (E) compared with the electrostatic surface of PM1L (F) modeled on the basis of the crystal structure of Trametes troggi laccase (PDB accession no. 2HRG), showing the differences in the amino acid residues that delimit the binding-site cavities (in yellow). A p-toluate molecule is modeled in the T1 Cu site of PM1L according to 2HRG. (G and H) Detail of the binding pocket in PcL (G) and the α*-3PO variant (H). Residues defining the binding-site cavity are depicted in yellow, and the mutated positions are depicted in orange. Arrows indicate Phe332 and Phe265, marking the entrance path of the substrate. Coordinating His residues are depicted in blue. Blue spheres represent the copper atoms.
The P394H mutation is located at the T1 Cu site, where the reducing substrate is oxidized (Fig. 6C and D). Pro394 is a highly conserved residue in fungal laccases and is located contiguous to His395, one of the ligands of T1 Cu (61). The P394H mutation might modify the coordinating sphere of Cu T1 by the formation of a new H bond with Ser427, which may pull down His395 and provoke an elongation of the T1 Cu Nδ H395 bond. These changes may affect the catalysis and/or the redox potential at the T1 copper site. Indeed, it has previously been reported that the increased distance between the T1 Cu and the coordinating His456 contributes to the enhanced redox potential of the T1 Cu site in PcL and other HRPLs (49). Nonetheless, other structural characteristics, such as the nature of the second-sphere residues that influence solvent accessibility and H bonding around the T1 site, may also affect the redox potential of laccases. The impact of the P394H mutation on laccase activity was further supported by a mutational exchange carried out during the evolution of PM1L (41). P394H was introduced into the evolved PM1L by site-directed mutagenesis, improving the kinetic constants of the enzyme.
Nδ H395 bond. These changes may affect the catalysis and/or the redox potential at the T1 copper site. Indeed, it has previously been reported that the increased distance between the T1 Cu and the coordinating His456 contributes to the enhanced redox potential of the T1 Cu site in PcL and other HRPLs (49). Nonetheless, other structural characteristics, such as the nature of the second-sphere residues that influence solvent accessibility and H bonding around the T1 site, may also affect the redox potential of laccases. The impact of the P394H mutation on laccase activity was further supported by a mutational exchange carried out during the evolution of PM1L (41). P394H was introduced into the evolved PM1L by site-directed mutagenesis, improving the kinetic constants of the enzyme.
The N208S mutation is located in a β sheet next to the reducing substrate binding pocket (Fig. 6C and D). The binding pocket of laccases is relatively wide, surrounding the T1 Cu ion that is not exposed to solvent. Inside the cavity, the His456 (PcL numbering) coordinating the T1 Cu and the acidic residue at position 206 (Asp/Glu fully conserved in fungal laccases) are in close proximity, and they are both involved in substrate binding. His456 mediates the initiation of the catalytic cycle by retiring one electron from the reducing substrate, while Asp206 is responsible for removing a proton from the  OH or
OH or  NH2 group (31, 43). In the 3PO mutant, the N208S mutation appears to form two new H bonds with His209 and Ile238, which may affect the relative position of Asp206 at the bottom of the binding pocket. The dramatic increase in Km values for DMP observed over the course of evolution (Table 2) may be due to a weaker interaction of Asp206 with the phenolic substrate (60), making the transfer of the proton more difficult. Unlike DMP, it seems like the interaction of Asp206 with bulky phenolic substrates such as sinapic acid was not hindered, which explains the similar affinity for sinapic acid through evolution. ABTS affinity also was not affected since its oxidation occurs via a mechanism not involving proton transfer (31).
NH2 group (31, 43). In the 3PO mutant, the N208S mutation appears to form two new H bonds with His209 and Ile238, which may affect the relative position of Asp206 at the bottom of the binding pocket. The dramatic increase in Km values for DMP observed over the course of evolution (Table 2) may be due to a weaker interaction of Asp206 with the phenolic substrate (60), making the transfer of the proton more difficult. Unlike DMP, it seems like the interaction of Asp206 with bulky phenolic substrates such as sinapic acid was not hindered, which explains the similar affinity for sinapic acid through evolution. ABTS affinity also was not affected since its oxidation occurs via a mechanism not involving proton transfer (31).
Residues from the enzymatic pocket are also of catalytic significance due to their key role in substrate binding, even though they do not directly contribute for electron abstraction. When comparing the binding pockets of PcL and PM1L, the most noticeable differences were the wider size and less hydrophobic nature of the cavity in PM1L (Fig. 6E and F). The steric threshold in laccases is reportedly dependent upon the distance between residues 332 and 265 (PcL numbering), which regulates the entrance of the reducing substrate to the binding pocket (60). The distance between both positions in PcL is 12.96 Å, almost half the distance in PM1L (21.86 Å) (Fig. 6G). These features could facilitate the traffic and oxidation by PM1L of bulky substrates with polar groups, such as ABTS. Indeed, we recently produced and characterized the PM1L wild type (unpublished material), which exhibited better kinetics for ABTS (kcat/Km = 33,580 s−1 mM−1, Km = 8.1 μM) than wild-type PcL (Tables 1 and 2). The 3PO PcL variant from the current study dramatically enhanced ABTS oxidation, with a catalytic efficiency of ≥20,000 s−1 mM−1 at both pH 5.0 and pH 3.0. The N331D mutation is located on one of the binding substrate loops (Fig. 6C, D, G, and H), and it appears to form a new H bond with Phe332. The movement of Phe332 may broaden the entrance to the 3PO binding pocket, improving the fitting of ABTS (and other bulky substrates, such as sinapic acid) (Fig. 6G and H).
Conclusions.
Ten years ago, the directed evolution of HRPLs was considered by many to be too littered with obstacles for success. The lack of functional expression in S. cerevisiae, the absence of reliable and specific screening assays, and limited knowledge of the mutational robustness of laccases discouraged researchers from adopting this approach. This changed when the first low-redox-potential laccase was evolved in the laboratory (13). Since then, we have learned how to exploit the eukaryotic machinery of S. cerevisiae, which supports a high frequency of DNA recombination, both to produce laccase mutants and to generate diversity (42, 68). PcL is a highly active and thermostable HRPL that can be easily produced in large amounts in heterologous hosts. In addition, its crystal structure enables rational/semirational design. The laboratory evolution platform presented in this study permits PcL to be evolved toward specific destinies. To name a few, the design of more robust and efficient HRPLs will enable the engineering of 3D nanobiodevices for biomedical application and the use of evolved laccases for improving the use of plant biomass (15, 37, 41).
Supplementary Material
ACKNOWLEDGMENTS
We thank Klaus Piontek (University of Freiburg) for providing the PcL crystal structure.
This research was funded by EU projects (NMP2-CT-2006-026456, NMP4-SL-2009-229255, COST Action CM0701, and KBBE-2010-4-26537) and national projects (CTQ2005-08925-C02-02, CCG08-CSIC/PPQ-3706, PIE 200920I207, and BIO2010-19697). A.I.C. was supported by a national grant from the Ministerio de Ciencia e Innovación.
Footnotes
Published ahead of print 30 December 2011
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Alcalde M. 2007. Laccases: biological functions, molecular structure and industrial applications, p 459–474 In Polaina J, MacCabe AP. (ed), Industrial enzymes: structure, function and applications. Springer, Berlin, Germany [Google Scholar]
- 2. Alcalde M. 2010. Mutagenesis protocols in Saccharomyces cerevisiae by in vivo overlap extension, p 3–15 In Bramman J. (ed), In vitro mutagenesis protocols. Humana Press, Totowa, NJ: [DOI] [PubMed] [Google Scholar]
- 3. Alcalde M, Zumárraga M, Polaina J, Ballesteros A, Plou FJ. 2006. Combinatorial saturation mutagenesis by in vivo overlap extension for the engineering of fungal laccases. Comb. Chem. High Throughput Scr. 9:719–727 [DOI] [PubMed] [Google Scholar]
- 4. Alves AM, et al. 2004. Highly efficient production of laccase by the basidiomycete Pycnoporus cinnabarinus. Appl. Environ. Microbiol. 70:6379–6384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Antorini M, et al. 2002. Purification, crystallisation and X-ray diffraction study of fully functional laccases from two ligninolytic fungi. BBA Protein Struct. Mol. Enzym. 1594:109–114 [DOI] [PubMed] [Google Scholar]
- 6. Arnold FH, Georgiou G. 2003. Directed enzyme evolution. Screening and selection methods. Humana Press, Totowa, NJ [Google Scholar]
- 7. Barr PJ, Brake AJ, Shuster JR. 1987. Heterologous gene expression in the yeast Saccharomyces cerevisiae. Abstr. Pap. Am. Chem. Soc. 194:116-MBTD [Google Scholar]
- 8. Barreca AM, Fabbrini M, Galli C, Gentili P, Ljunggren S. 2003. Laccase/mediated oxidation of a lignin model for improved delignification procedures. J. Mol. Catal. B Enzym. 26:105–110 [Google Scholar]
- 9. Bertrand T, et al. 2002. Crystal structure of a four-copper laccase complexed with an arylamine: insights into substrate recognition and correlation with kinetics. Biochemistry 41:7325–7333 [DOI] [PubMed] [Google Scholar]
- 10. Bloom JD, Arnold FH. 2009. In the light of directed evolution: pathways of adaptive protein evolution. Proc. Natl. Acad. Sci. U. S. A. 106:9995–10000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Boyd D, Beckwith J. 1990. The role of charged amino-acids in the localization of secreted and membrane-proteins. Cell 62:1031–1033 [DOI] [PubMed] [Google Scholar]
- 12. Brake AJ. 1990. Alpha factor leader directed secretion of heterologous proteins from yeast. Methods Enzymol. 185:408–421 [DOI] [PubMed] [Google Scholar]
- 13. Bulter T, et al. 2003. Functional expression of a fungal laccase in Saccharomyces cerevisiae by directed evolution. Appl. Environ. Microbiol. 69:987–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Camarero S, et al. 2004. Efficient bleaching of non-wood high-quality paper pulp using laccase-mediator system. Enzyme Microb. Technol. 35:113–120 [Google Scholar]
- 15. Cañas A, et al. 2007. Transformation of polycyclic aromatic hydrocarbons by laccase is strongly enhanced by phenolic compounds present in soil. Environ. Sci. Technol. 41:2964–2971 [DOI] [PubMed] [Google Scholar]
- 16. Cañas AI, Camarero S. 2010. Laccases and their natural mediators: biotechnological tools for sustainable eco-friendly processes. Biotechnol. Adv. 28:694–705 [DOI] [PubMed] [Google Scholar]
- 17. Cassland P, Jönsson LJ. 1999. Characterization of a gene encoding Trametes versicolor laccase A and improved heterologous expression in Saccharomyces cerevisiae by decreased cultivation temperature. Appl. Microbiol. Biotechnol. 52:393–400 [DOI] [PubMed] [Google Scholar]
- 18. Cirino PC, Georgescu R. 2011. Screening for thermostability, p 117–125 In Arnold FH, Georgiou G. (ed), Directed enzyme evolution. Screening and selection methods, vol 230 Humana Press, Totowa, NJ [Google Scholar]
- 19. Colao MC, Caporale C, Silvestri F, Ruzzi M, Buonocore V. 2009. Modeling the 3-D structure of a recombinant laccase from Trametes trogii active at a pH close to neutrality. Protein J. 28:375–383 [DOI] [PubMed] [Google Scholar]
- 20. Collins PJ, Dobson ADW. 1997. Regulation of laccase gene transcription in Trametes versicolor. Appl. Environ. Microbiol. 63:3444–3450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cortassa S, Aon JC, Aon MA. 1995. Fluxes of carbon, phosphorylation, and redox intermediates during growth of Saccharomyces cerevisiae on different carbon-sources. Biotechnol. Bioeng. 47:193–208 [DOI] [PubMed] [Google Scholar]
- 22. Dix DB, Thompson RC. 1989. Codon choice and gene-expression—synonymous codons differ in translational accuracy. Proc. Natl. Acad. Sci. U. S. A. 86:6888–6892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. García-Ruiz E, Maté D, Ballesteros A, Martínez AT, Alcalde M. 2010. Evolving thermostability in mutant libraries of ligninolytic oxidoreductases expressed in yeast. Microb. Cell Fact. 9:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Georis J, et al. 2003. Pycnoporus cinnabarinus laccases: an interesting tool for food or non-food applications. Commun Agric. Appl. Biol. Sci. 68:263–266 [PubMed] [Google Scholar]
- 25. Gutiérrez A, et al. 2006. Enzymatic removal of free and conjugated sterols forming pitch deposits in environmentally sound bleaching of eucalypt paper pulp. Environ. Sci. Technol. 40:3416–3422 [DOI] [PubMed] [Google Scholar]
- 26. Herpoël I, Moukha S, Lesage-Meessen L, Sigoillot JC, Asther M. 2000. Selection of Pycnoporus cinnabarinus strains for laccase production. FEMS Microbiol. Lett. 183:301–306 [DOI] [PubMed] [Google Scholar]
- 27. Hong F, Meinander NQ, Jönsson LJ. 2002. Fermentation strategies for improved heterologous expression of laccase in Pichia pastoris. Biotechnol. Bioeng. 79:438–449 [DOI] [PubMed] [Google Scholar]
- 28. Hoshida H, Fujita T, Murata K, Kubo K, Akada R. 2005. Copper-dependent production of a Pycnoporus coccineus extracellular laccase in Aspergillus oryzae and Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 69:1090–1097 [DOI] [PubMed] [Google Scholar]
- 29. Ibarra D, Camarero S, Romero J, Martínez MJ, Martínez AT. 2006. Integrating laccase-mediator treatment into an industrial-type sequence for totally chlorine free bleaching eucalypt kraft pulp. J. Chem. Technol. Biotechnol. 81:1159–1165 [Google Scholar]
- 30. Ibarra D, et al. 2007. Structural modification of eucalypt pulp lignin in a totally chlorine free bleaching sequence including a laccase-mediator stage. Holzforschung 61:634–646 [Google Scholar]
- 31. Kallio JP, et al. 2009. Structure-function studies of a Melanocarpus albomyces laccase suggest a pathway for oxidation of phenolic compounds. J. Mol. Biol. 392:895–909 [DOI] [PubMed] [Google Scholar]
- 32. Kunamneni A, et al. 2008. Engineering and applications of fungal laccases for organic synthesis. Microb. Cell Fact. 7:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Labat E, Morel MH, Rouau X. 2000. Effects of laccase and ferulic acid on wheat flour doughs. Cereal Chem. 77:823–828 [Google Scholar]
- 34. Larrondo LF, Avila M, Salas L, Cullen D, Vicuña R. 2003. Heterologous expression of laccase cDNA from Ceriporiopsis subvermispora yields copper-activated apoprotein and complex isoform patterns. Microbiology 149:1177–1182 [DOI] [PubMed] [Google Scholar]
- 35. Lee IY, Jung KH, Lee CH, Park YH. 1999. Enhanced production of laccase in Trametes versicolor by the addition of ethanol. Biotechnol. Lett. 21:965–968 [Google Scholar]
- 36. Li KC, Xu F, Eriksson KEL. 1999. Comparison of fungal laccases and redox mediators in oxidation of a nonphenolic lignin model compound. Appl. Environ. Microbiol. 65:2654–2660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lomascolo A, et al. 2003. Overproduction of laccase by a monokaryotic strain of Pycnoporus cinnabarinus using ethanol as inducer. J. Appl. Microbiol. 94:618–624 [DOI] [PubMed] [Google Scholar]
- 38. Madeira A, et al. 2010. Effect of ethanol on fluxes of water and protons across the plasma membrane of Saccharomyces cerevisiae. FEMS Yeast Res. 10:252–258 [DOI] [PubMed] [Google Scholar]
- 39. Madzak C, et al. 2006. Shifting the optimal pH of activity for a laccase from the fungus Trametes versicolor by structure-based mutagenesis. Protein Eng. Des. Sel. 19:77–84 [DOI] [PubMed] [Google Scholar]
- 40. Marzorati M, Danieli B, Haltrich D, Riva S. 2005. Selective laccase-mediated oxidation of sugars derivatives. Green Chem. 7:310–315 [Google Scholar]
- 41. Maté D, et al. 2010. Laboratory evolution of high redox potential laccases. Chem. Biol. 17:1030–1041 [DOI] [PubMed] [Google Scholar]
- 42. Maté D, García-Ruiz E, Camarero S, Alcalde M. 2011. Directed evolution of fungal laccases. Curr. Genomics 12:113–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Matera I, et al. 2008. Crystal structure of the blue multicopper oxidase from the white-rot fungus Trametes trogii complexed with p-toluate. Inorg. Chim. Acta 361:4129–4137 [Google Scholar]
- 44. Mayer AM, Staples RC. 2002. Laccase: new functions for an old enzyme. Phytochemistry 60:551–565 [DOI] [PubMed] [Google Scholar]
- 45. Morozova V, Shumakovich GP, Gorbacheva MA, Shleev SV, Yaropolov AI. 2007. “Blue” laccases. Biochemistry (Mosc.) 72:1136–1150 [DOI] [PubMed] [Google Scholar]
- 46. Nothwehr SF, Gordon JI. 1990. Targeting of proteins into the eukaryotic secretory pathway—signal peptide structure-function-relationships. Bioessays 12:479–484 [DOI] [PubMed] [Google Scholar]
- 47. Palmieri G, Giardina P, Bianco C, Fontanella B, Sannia G. 2000. Copper induction of laccase isoenzymes in the ligninolytic fungus Pleurotus ostreatus. Appl. Environ. Microbiol. 66:920–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Petrov VV, Okorokov LA. 1990. Increase of the anion and proton permeability of Saccharomyces carlsbergensis plasmalemma by normal alcohols as a possible cause of its deenergization. Yeast 6:311–318 [DOI] [PubMed] [Google Scholar]
- 49. Piontek K, Antorini M, Choinowski T. 2002. Crystal structure of a laccase from the fungus Trametes versicolor at 1.90-Å resolution containing a full complement of coppers. J. Biol. Chem. 277:37663–37669 [DOI] [PubMed] [Google Scholar]
- 50. Rakestraw JA, Sazinsky SL, Piatesi A, Antipov E, Wittrup KD. 2009. Directed evolution of a secretory leader for the improved expression of heterologous proteins and full length antibodies in Saccharomyces cerevisiae. Biotechnol. Bioeng. 103:1192–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Record E, et al. 2002. Expression of the Pycnoporus cinnabarinus laccase gene in Aspergillus niger and characterization of the recombinant enzyme. Eur. J. Biochem. 269:602–609 [DOI] [PubMed] [Google Scholar]
- 52. Riva S. 2006. Laccases: blue enzymes for green chemistry. Trends Biotechnol. 24:219–226 [DOI] [PubMed] [Google Scholar]
- 53. Rodríguez-Couto S, Toca JL. 2006. Industrial and biotechnological applications of laccases: a review. Biotechnol. Adv. 24:500–513 [DOI] [PubMed] [Google Scholar]
- 54. Romanos MA, Scorer CA, Clare JJ. 1992. Foreign gene expression in yeast—a review. Yeast 8:423–488 [DOI] [PubMed] [Google Scholar]
- 55. Romero PA, Arnold FH. 2009. Exploring protein fitness landscapes by directed evolution. Nat. Rev. Mol. Cell Biol. 10:866–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sagt C, et al. 2000. Introduction of an N glycosylation site increases secretion of heterologous proteins in yeasts. Appl. Environ. Microbiol. 66:4940–4944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Schultz A, Jonas U, Hammer E, Schauer F. 2001. Dehalogenation of chlorinated hydroxybiphenyls by fungal laccase. Appl. Environ. Microbiol. 67:4377–4381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shleev S, Ruzgas T. 2008. Transistor-like behavior of a fungal laccase. Angew. Chem. 47:7270–7274 [DOI] [PubMed] [Google Scholar]
- 59. Shuster JR. 1991. Gene expression in yeast: protein secretion. Curr. Opin. Biotechnol. 2:685–690 [DOI] [PubMed] [Google Scholar]
- 60. Tadesse MA, d'Annibale A, Galli C, Gentili P, Sergi F. 2008. An assessment of the relative contributions of redox and steric issues to laccase specificity towards putative substrates. Org. Biomol. Chem. 6:868–878 [DOI] [PubMed] [Google Scholar]
- 61. Valderrama B, Oliver P, Medrano-Soto A, Vázquez-Duhalt R. 2003. Evolutionary and structural diversity of fungal laccases. Antonie Van Leeuwenhoek 84:289–299 [DOI] [PubMed] [Google Scholar]
- 62. van de Laar T, et al. 2007. Increased heterologous protein production by Saccharomyces cerevisiae growing on ethanol as sole carbon source. Biotechnol. Bioeng. 96:483–494 [DOI] [PubMed] [Google Scholar]
- 63. Widsten P, Kandelbauer A. 2008. Laccase applications in the forest products industry: a review. Enzyme Microb. Technol. 42:293–307 [Google Scholar]
- 64. Witayakran S, Ragauskas AJ. 2009. Synthetic applications of laccase in green chemistry. Adv. Synth. Catal. 351:1187–1209 [Google Scholar]
- 65. Xu F. 1997. Effects of redox potential and hydroxide inhibition on the pH activity profile of fungal laccases. J. Biol. Chem. 272:924–928 [DOI] [PubMed] [Google Scholar]
- 66. Xu F. 2005. Applications of oxidoreductases: recent progress. Ind. Biotechnol. 1:38–50 [Google Scholar]
- 67. Xu F, et al. 2000. Redox chemistry in laccase-catalyzed oxidation of N-hydroxy compounds. Appl. Environ. Microbiol. 66:2052–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zumárraga M, et al. 2007. In vitro evolution of a fungal laccase in high concentrations of organic cosolvents. Chem. Biol. 14:1052–1064 [DOI] [PubMed] [Google Scholar]
- 69. Zumárraga M, et al. 2008. Altering the laccase functionality by in vivo assembly of mutant libraries with different mutational spectra. Proteins 71:250–260 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.