Abstract
To identify Escherichia coli proteins involved in adaptation to intestinal inflammation, mice were monoassociated with the colitogenic E. coli strain UNC or with the probiotic E. coli strain Nissle. Intestinal inflammation was induced by treating the mice with 3.5% dextran sodium sulfate (DSS). Differentially expressed proteins in E. coli strains collected from cecal contents were identified by 2-dimensional difference gel electrophoresis. In both strains, acute inflammation led to the downregulation of pathways involved in carbohydrate breakdown and energy generation. Accordingly, DSS-treated mice had lower concentrations of bacterial fermentation products in their cecal contents than control mice. Differentially expressed proteins also included the Fe-S cluster repair protein NfuA, the tryptophanase TnaA, and the uncharacterized protein YggE. NfuA expression was 3-fold higher in E. coli strains from DSS-treated than from control mice. Reporter experiments confirmed the induction of nfuA in response to iron deprivation, mimicking Fe-S cluster destruction by inflammation. YggE expression, which has been reported to reduce the intracellular level of reactive oxygen species, was 4- to 8-fold higher in E. coli Nissle than in E. coli UNC. This was confirmed by in vitro reporter gene assays indicating that Nissle is better equipped to cope with oxidative stress than UNC. Nissle isolated from DSS-treated and control mice had TnaA levels 4- to 7-fold-higher than those of UNC. Levels of indole resulting from the TnaA reaction were higher in control animals associated with E. coli Nissle. Because of its anti-inflammatory effect, indole is hypothesized to be involved in the extension of the remission phase in ulcerative colitis described for E. coli Nissle.
INTRODUCTION
Inflammatory bowel disease (IBD) comprises two forms of intestinal inflammation: ulcerative colitis (UC) and Crohn's disease (CD). The pathogenesis of IBD is not completely understood but is considered to result from an aberrant immune response to the intestinal microbiota in genetically predisposed subjects (39). In both IBD patients and animal models of gut inflammation, intestinal Escherichia coli was reported to become a dominant species of the gut microbiota (5, 10, 18, 24, 43, 44, 49).
For example, interleukin 10-deficient (IL-10−/−) mice, which develop inflammation in the cecum and colon, displayed reduced microbial diversity and elevated E. coli numbers. E. coli was represented by one predominant strain with an O7:H7:K1 serotype, which outcompeted other E. coli strains in diassociation experiments in gnotobiotic mice (49). In IL-2−/− mice, which also develop colitis, E. coli represents as much as 10% of the mucosa-associated microbiota (42). Some E. coli strains are capable of inducing intestinal inflammation in genetically susceptible mice. UNC, a murine strain of E. coli randomly isolated from wild-type mice raised under specific-pathogen-free conditions, induces mild cecal inflammation in IL-10−/− mice after 3 weeks of monoassociation (22). IL-2−/− mice monoassociated with E. coli mpk develop colitis accompanied by upregulation of gamma interferon, tumor necrosis factor alpha (TNF-α), cluster of differentiation 14 (CD14), and IL-10, while IL-2−/− mice monoassociated with either Bacteroides vulgatus mpk or the probiotic E. coli strain Nissle are not affected (47). In addition, E. coli Nissle is as effective as the standard medication at keeping chronic ulcerative colitis patients in remission (37). Nissle modulates several elements of the immune response (8, 9, 16, 34, 45, 50), but the bacterial components mediating these immunomodulatory effects have not yet been identified. There are some hints that the flagellin of E. coli Nissle contributes to its probiotic function (40).
To better understand how different E. coli strains adapt to the inflammatory conditions in the intestinal tract and to find out whether this adaptation possibly leads to an exacerbation of inflammation, the proteomes of E. coli strains Nissle and UNC isolated from the ceca of healthy or inflamed mice were compared, and bacterial proteins expressed in response to inflammation were tested for their possible roles in adaptation to this condition. The results suggest that E. coli develops an energy deficiency in the state of acute inflammation and that the Fe-S biogenesis protein NfuA and the uncharacterized protein YggE play roles in the adaptive response of E. coli to environmental stress caused by inflammation.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The colitogenic E. coli strain UNC was a kind gift from Dirk Haller (Technical University Munich, Munich, Germany). The commercial product Mutaflor (Ardeypharm, Germany) served as the source for E. coli Nissle. E. coli K-12 MG1655 (CGSC 6300) was kindly provided by K. Schnetz (University of Cologne, Cologne, Germany). For association of germ-free mice with E. coli, strain Nissle or UNC was grown in Luria-Bertani (LB) medium (10 g/liter tryptone, 5 g/liter yeast extract, 5 g/liter NaCl) at 37°C to an optical density at 600 nm (OD600) of 0.5 to 0.7 (SmartSpec Plus; Bio-Rad). Bacteria were collected by centrifugation (at 10,000 × g and 4°C for 5 min), and the pellet was resuspended in sterile phosphate-buffered saline (PBS; 37 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.47 mM KH2PO4 [pH 7.4]) and was administered by oral gavage.
Gnotobiotic animal model.
The germ-free status of the animals was confirmed before each experiment by microscopic inspection of Gram-stained feces. Furthermore, fresh fecal material was plated on Columbia sheep blood agar (bioMérieux, Germany) and was incubated under aerobic and anaerobic conditions at 37°C. 129/SvEv mice (8 to 9/group; 6 to 10 weeks of age) were kept in individually ventilated cages (IVC) for the duration of the experiment. Each mouse was orogastrically inoculated with 107 E. coli Nissle or UNC cells. The mice had free access to sterile food and autoclaved water. Mice of the control groups were housed under these conditions for 21 days. The animals treated with dextran sodium sulfate (DSS) received sterile drinking water for the first 7 days after association; then they received drinking water supplemented with 3.5% (wt/vol) DSS (MP Biomedicals) for another 7 days, followed by 2 days of sterile drinking water. Mice were subsequently killed by cervical dislocation. The colon length was determined, and tissue samples from the cecum and the colon were taken for histopathological analyses or, alternatively, for the preparation of RNA. Cecal and colonic contents were collected, weighed, and diluted 1:10 (wt/vol) with PBS containing a protease inhibitor mixture (GE Healthcare, Sweden) at a 100-fold dilution. After homogenization of intestinal contents by agitation with a Uniprep 24 gyrator (speed 2; UniEquip, Germany) in the presence of glass beads (diameter, 2.85 to 3.33 mm), the samples were centrifuged (at 300 × g and 4°C for 5 min) to remove coarse particles originating from the feed. Cell counts in the supernatants were determined by plating on LB-Lennox agar (31).
Isolation of bacteria from cecal contents.
The supernatants mentioned above were centrifuged (at 10,000 × g and 4°C for 5 min), and the pelleted cells were resuspended in washing buffer (10 mM Tris [pH 8], 5 mM magnesium acetate, 30 mg/liter chloramphenicol, protease inhibitor mixture diluted 1:100). The resulting supernatants were collected and were stored at −20°C for the determination of concentrations of bacterial fermentation products and indole. The cells were subsequently isolated by Nycodenz (Axis-Shield, Norway) gradient centrifugation as described by Vogel-Scheel et al. (46). Washed cells were stored at −80°C.
Histopathology scoring.
Material for histopathology scoring was obtained from the ceca and colons (Swiss rolls) of only three randomly chosen animals, because the tissue was scarce. Intestinal tissue material was fixed in 10% neutral buffered formalin, embedded in paraffin, sectioned at a thickness of 4 μm, and stained with hematoxylin and eosin (H&E). To quantify intraepithelial lymphocytes, the tissue sections were stained with an anti-CD3 antibody (Dako, Germany). The severity of inflammation was assessed on blinded samples according to the method of Burich et al. (7) using a scale of zero (no inflammation) to 28 (severe inflammation). Each section was scored for the following categories: cellular infiltration, crypt hyperplasia, goblet cell depletion, edema, architectural distortion, and the area involved. The scoring was performed by Susanne Mauel (Freie Universität Berlin, Institut für Tierpathologie).
Determination of bacterial fermentation product concentrations.
Concentrations of the bacterial fermentation products formate, lactate, and succinate in cecal supernatants were determined by enzymatic assays (Boehringer Mannheim/R-Biopharm, Germany) according to the manufacturers' instructions.
Preparation of whole-bacterial-cell protein extracts.
Bacterial protein extracts were prepared according to the method of Vogel-Scheel et al. (46). Briefly, frozen cells were thawed, resuspended in 0.8 ml lysis buffer {8 M urea, 30 mM Tris, 4% [wt/vol] 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate [CHAPS] [pH 8.5]}, and mechanically disrupted with zirconia-silica beads (diameter, 0.1 mm; Roth, Germany) in an FP120 FastPrep cell disruptor (Thermo Scientific, Waltham, MA). Unbroken cells were removed by centrifugation (at 14,000 × g and 4°C for 20 min). Components interfering with proteomic analysis were removed by selective precipitation of proteins (2-D cleanup kit; GE Healthcare). The concentration of resuspended proteins was determined by the Bradford assay (Bio-Rad, Spain). The pH of the protein solution was adjusted to 8.5 with 50 mM NaOH for optimal reactions with CyDyes in the following step.
Two-dimensional DIGE.
The protein extracts from 5 randomly chosen samples per group were labeled according to the manufacturer's instructions with CyDyes (GE Healthcare, Sweden) for difference gel electrophoresis (DIGE) and were applied to immobilized pH gradient strips (pH range, 4 to 7; length, 24 cm) in an Ettan IPGphor 3 isoelectric focusing unit (GE Healthcare, Sweden). Active rehydration (30 V for 10 h) was followed by isoelectric focusing of the samples for a total of 61.05 kV · h at 20°C. The second dimension was run on 12.5% sodium dodecyl sulfate (SDS) gels in an Ettan-Dalt II apparatus at 1 W per gel for 45 min, followed by 17 W per gel for 3.5 h. The proteins were visualized with a Typhoon Trio laser scanner, and image analysis was performed with DeCyder software, version 6.5 (both from GE Healthcare, Sweden).
In-gel protein digestion.
In-gel protein digestion was performed according to the method of Vogel-Scheel et al. (46). Briefly, preparative gels were stained with ruthenium II tris(bathophenanthroline disulfonate) (36) and were matched to the DIGE gels. Proteins of interest were excised automatically using an Ettan Dalt spot picker (GE Healthcare, Sweden) and were subjected to tryptic digestion in 96-well plates. The tryptically digested peptides were dried by vacuum centrifugation.
Identification of proteins.
Proteins were identified by nano-liquid chromatography-electrospray ionization-tandem mass spectrometry (MS-MS) as described previously (46), except that ProteinLynx Global Server software, version 2.3 (Waters Corporation), and Swiss-Prot, version 57.2 (http://www.expasy.org/sprot/), were used for processing of the MS-MS data and subsequent data bank searching. To eliminate false-positive results, a second search was performed using the randomized protein database.
Generation of luciferase reporter constructs.
Luciferase reporter constructs were generated in pKEST06, which carries the luxAB genes from Photorhabdus luminescens (see Fig. S2 in the supplemental material). The bglB-bglG homology region was excised from the vector by digestion with the restriction enzymes EcoRI and XbaI (both from Fermentas, Germany). The nfuA and yggE promoter regions were amplified by PCR from E. coli MG1655, Nissle, and UNC by using primers n1/n2 for nfuA or y1/y2 for yggE (Table 1). After digestion with restriction enzymes, PCR products were ligated into pKEST06 at the EcoRI and XbaI sites. The final constructs were transformed into the respective E. coli strain. All the constructs obtained were verified by sequencing (Eurofins MWG Operon, Germany) of the cloned DNA fragments.
Table 1.
Oligonucleotides used in this study
| Primer name | Primer sequence (5′ → 3′)a |
|---|---|
| n1 | CGGAATTCACGTACACGGGCCACTGCGA |
| n2 | GCTCTAGAGCGTGCCAGGGTTAATCACA |
| y1 | CGGAATTCCAGCGCATTATCAATCTGGT |
| y2 | GCTCTAGATGACAATATGCGGTCCATCC |
| d1 | ATCGGGCAATCTACAAAAGAGGGGATAACTTAGTAGTAGGAGTGTTCGCCtgtaggctggagctgcttcg |
| d2 | TGGGCGTATTATAACCAACTAAAATAGTCAACTATTAGGCCATTACTATGattccggggatccgtcgacc |
| nfuA1 | CGAAGCTTACGTACACGGGCCACTGCGA |
| nfuA2 | GCGGATCCTGCGGCACCACGACTGATAC |
Boldface letters indicate restriction sites, and lowercase letters indicate priming sites P1 (d1) and P2 (d2) for the generation of deletion mutants. The nomenclature for extensions and priming sites is based on the Keio database (http://www.shigen.nig.ac.jp/ecoli/strain/nbrp/explanation/keioCollection.jsp).
Luciferase reporter gene assay.
All three E. coli strains containing the respective pKEST06 constructs were grown aerobically for 1 h in LB medium containing 50 μg/ml carbenicillin. Subsequently, 8-ml aliquots of the culture were transferred to 6-well plates, incubated at 37°C, shaken at 215 rpm, and stimulated for 1 h with 250 μM 2,2′-dipyridyl (Sigma-Aldrich, Germany) or 300 μM H2O2 (Roth, Germany). Bacteria were harvested by centrifugation (at 10,000 × g and 4°C for 3 min), and the pellet was resuspended in 200 μl PBS containing 30 mg/liter chloramphenicol. For luciferase reporter gene assays, 5 × 109 cells were used. Measurements were carried out in triplicate following the addition of 100 μl 2% (vol/vol) decanal (Sigma-Aldrich, Germany) using a Luminoscan Ascent plate luminometer (Labsystems, Finland). Luminescence was measured immediately after the addition of the substrate, and the results are expressed as relative light units (RLU). To calculate the relative luminescence, the luminescence signal of the stimulated bacterial cells was normalized to the luminescence of the controls.
Generation of nfuA deletion mutants.
Chromosomal sequences comprising the nfuA gene were replaced by a kanamycin resistance cassette according to the technique of Datsenko and Wanner (11) by using pKD13 as a template for the antibiotic resistance gene. The primers (d1/d2) for the construction of the deletion mutants are listed in Table 1. Mutant candidates were tested for the loss of the target gene by PCR with kanamycin (k1/k2)- and locus-specific primers. In addition, the genotype of each mutant was confirmed by sequencing and characterization of the growth phenotype.
Characterization of nfuA deletion mutants.
Cells were precultured aerobically overnight in LB medium and were inoculated freshly into LB medium alone or containing 300 μM paraquat (Sigma-Aldrich, Germany). The cultures were incubated aerobically at 37°C for 8 h, and growth was monitored by measuring the OD600. The effect of 250 μM 2,2′-dipyridyl was tested in the same way, except that 350 μM FeSO4 (Merck, Germany) was added after 16 h, and growth was monitored for 12 h.
Complementation of the nfuA deletion.
nfuA was amplified by PCR from MG1655 with primers nfuA1 and nfuA2 (Table 1), which contain restriction sites for HindIII and BamHI, respectively. A 1,127-nucleotide (nt) HindIII-BamHI fragment was cloned into the low-copy-number plasmid pSU19 (4). The insert including both promoter regions of nfuA was verified by sequencing. In addition, a 1,070-nt HindIII-KpnI fragment of the nfuA region was recovered from the pSU19 construct, inserted into pBRINTs-Cat2, and recombined into the chromosomal lacZ gene of the ΔnfuA strain as described by Le Borgne et al. (30). For complementation experiments, MG1655 carrying pSU19, MG1655 ΔnfuA carrying either pSU19 or pSU19-nfuA, or MG1655 ΔnfuA lacZ::(nfuA cat) was grown to the exponential-growth phase in LB medium (with chloramphenicol at 20 μg/ml), collected by centrifugation, and resuspended in fresh medium. Cell suspensions of equal OD600s were serially diluted, and 5 μl of each dilution was spotted onto LB plates containing 20 μg/ml chloramphenicol or onto plates containing, in addition, either 2,2′-dipyridyl (375 μM) or paraquat (300 μM).
Growth experiments for the determination of strain differences in tolerance to reactive oxygen species (ROS).
Wild-type E. coli strains MG1655, Nissle, and UNC were precultured aerobically overnight in LB medium and were inoculated into LB medium alone or containing 300 μM paraquat. The cultures were grown aerobically at 37°C for 8 h, and growth was monitored by measuring the OD600.
Determination of indole concentrations in cecal supernatants.
The procedure for the determination of indole concentrations in cecal supernatants is based on the method of Mattivi et al. (32), with major modifications. Cecal supernatants were centrifuged (at 20,000 × g and 4°C for 5 min). For high-performance liquid chromatography (HPLC) analysis, 20 μl of the liquid phase was used. Indole was separated on a LiChrospher 100 RP-18 column (length, 250 mm; inner diameter, 4 mm; particle size, 5 μm; Merck, Germany). Solvent A was 1% acetic acid (vol/vol), and solvent B was methanol (MeOH). The following gradient was used for elution: 45 to 72% solvent B in 10 min, 72 to 76% solvent B in 10 min, 76 to 80% solvent B in 3 min, and 80 to 100% solvent B in 5 min at 20°C and a flow rate of 0.5 ml/min. Fluorescence was detected at an excitation wavelength of 225 nm and an emission wavelength of 365 nm (821-FP fluorescence detector; Jasco). The indole concentration was calculated by using a calibration curve produced with indole (Sigma-Aldrich, Germany) standards.
Statistical analysis.
Results are expressed as medians. The significance of differences was analyzed by the Mann-Whitney U test (GraphPad Prism, version 5; GraphPad, La Jolla, CA).
RESULTS
Characterization of acute colitis.
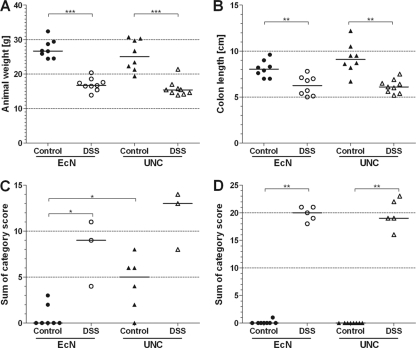
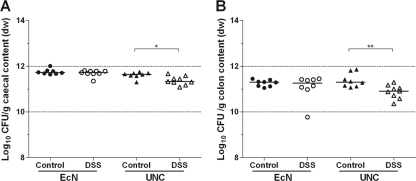
The disease severity of DSS-treated and control mice was determined by measuring body weight and colon length and by histopathological scoring of cecal and colonic tissues. The body weights and colon lengths of the DSS-treated mice were lower than those of the control mice, whether they were monoassociated with E. coli Nissle or with E. coli UNC (Fig. 1A and B). There was no weight difference between mice associated with strain Nissle and mice associated with strain UNC in either the DSS group or the control group (Fig. 1A). However, the DSS-induced weight loss tended to be greater in animals associated with UNC than in those associated with Nissle (P = 0.06) (see Fig. S3 in the supplemental material). Histopathological scoring of cecal and colonic tissues revealed that DSS caused inflammation preferentially in the colon and to a lesser extent in the cecum. There were no differences in the histopathological scores for the cecum or colon between the DSS-treated mice, whether they were monoassociated with E. coli Nissle or with E. coli UNC (Fig. 1C and D). However, the ceca of the control animals associated with UNC were mildly inflamed, whereas the ceca of Nissle-associated animals did not show elevated inflammation scores (Fig. 1C). Different concentrations of strains Nissle and UNC cannot account for the higher histological score, since the cecal and colonic concentrations of Nissle and UNC in the control animals were equal. Only the intestinal UNC concentrations in the DSS-treated animals were approximately 0.25 log (cecum) and 0.5 log (colon) lower than those in the control mice (Fig. 2). Although these differences are statistically significant, it is disputable whether they are biologically relevant.
Fig 1.
Characterization of acute colitis. Germ-free mice (129/SvEv) were monoassociated with E. coli strain Nissle (EcN) or UNC. Mice of the DSS group received sterile drinking water with 3.5% DSS; control mice received sterile drinking water without DSS. (A) Body weight; (B) colon length; (C) histological score in the cecum; (D) histological score in the colon. Data are expressed as medians. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Fig 2.
Analysis of intestinal bacterial cell numbers. Germ-free mice (129/SvEv) were monoassociated with E. coli strain Nissle or UNC. Mice of the DSS group received sterile drinking water with 3.5% DSS; control mice received sterile drinking water without DSS. Bacterial cell numbers (CFU) in cecal (A) and colonic (B) contents were determined by plating on LB-Lennox agar. dw, dry weight. Data are expressed as medians. *, P ≤ 0.05; **, P ≤ 0.01.
Animals that were not subjected to histological scoring were analyzed for mRNA levels of various proinflammatory cytokines, including TNF-α, in cecal and colonic tissues (see Fig. S4 in the supplemental material). The results indicate concordantly that the intestines of the DSS-treated animals were inflamed.
Adaptation of E. coli to intestinal inflammation.
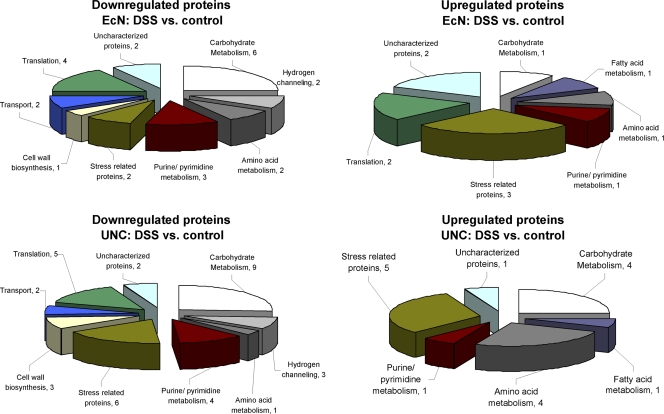
To identify bacterial proteins involved in the adaptation of E. coli to intestinal inflammation, the expression of E. coli proteins in the ceca of DSS-treated mice was compared to that in the ceca of control mice. In E. coli Nissle, 35 proteins were differentially expressed (≥3-fold; P ≤ 0.05), of which 24 were downregulated while 11 were upregulated (Fig. 3; see also Table S1 in the supplemental material). Besides stress-related proteins, proteins involved in pyrimidine conversion (Dcd), amino acid metabolism (GlnA), fatty acid metabolism (FabA), carbohydrate scavenging (Agp), and translation (RL15, RS6) were upregulated during intestinal inflammation. Proteins downregulated included enzymes involved in carbohydrate catabolism (GapA, TpiA, PckA, TktA, PflB, MelA), translation (LysU, TufA, RL6, RL9), and purine/pyrimidine synthesis (PurC) and nucleotide salvage (DeoD, Upp). Furthermore, proteins involved in ATP synthesis (AtpA) and transport processes (HybC, MglB, RbsB) were repressed.
Fig 3.
Classification of proteins regulated by intestinal inflammation in E. coli strains Nissle and UNC. The proteins considered were differentially expressed by a factor of ≥3 in cecal samples isolated from DSS-treated animals versus control animals (n = 5; P ≤ 0.05).
In the colitogenic E. coli strain UNC, 35 proteins were downregulated while 16 were upregulated. Most of the latter are stress-related proteins. Other upregulated proteins play a role in amino acid (TdcE, GlnA, AspA, TnaA) or carbohydrate (TktA, LacZ, SdhA, PflB) metabolism. As with E. coli Nissle, the repressed proteins are mainly involved in carbohydrate metabolism (Eno, Pgk, GapA, TpiA, FbaA, GalT, GalK, RbsK, RbsD) and to a lesser extent also in translational processes (LysU, ArgS, TufA, RS6, RL11) and purine/pyrimidine metabolism (CpdB, PurE, PurC, Upp) (Fig. 3; see also Table S1 in the supplemental material).
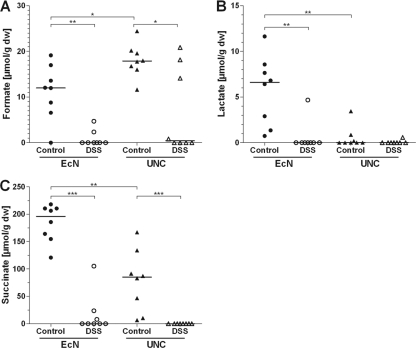
To check if the downregulation of enzymes related to central energy-generating pathways, as observed in both bacterial strains in response to DSS treatment, results in changes in carbon catabolism, the concentrations of bacterial fermentation products in intestinal contents were measured. In agreement with the proteomic data, the concentrations of formate, lactate, and succinate in the cecal water of DSS-treated animals were lower than in that of control mice (Fig. 4). In contrast, the cecal carbohydrate (glucose and galactose) levels were not affected by intestinal inflammation (see Fig. S5 in the supplemental material).
Fig 4.
Concentrations of bacterial fermentation products. Germ-free mice (129/SvEv) were monoassociated with E. coli strain Nissle or UNC. Mice of the DSS group received sterile drinking water containing 3.5% DSS; control mice received sterile drinking water without DSS. Concentrations of the bacterial fermentation products formate (A), lactate (B), and succinate (C) were measured in cecal water. Data are expressed as medians. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Role of NfuA in bacterial adaptation to intestinal inflammation.
Among the E. coli proteins upregulated under inflammatory conditions were several stress-related proteins, such as GroL, RecA, and NfuA in strain Nissle, as well as Tpx, AhpF, NfuA, ClpB, and NusA in strain UNC (Fig. 3; see also Table S1 in the supplemental material). The Fe-S biogenesis protein NfuA was upregulated by a factor of 3 in both E. coli strains (see Table S1). Previous publications showed that NfuA plays a critical role in the adaptation of E. coli MG1655 to oxidative stress and iron starvation under in vitro conditions, since it binds iron-sulfur clusters and transfers them to the corresponding apoprotein (1). Therefore, the role of NfuA in adaptation to inflammatory conditions was investigated in more detail.
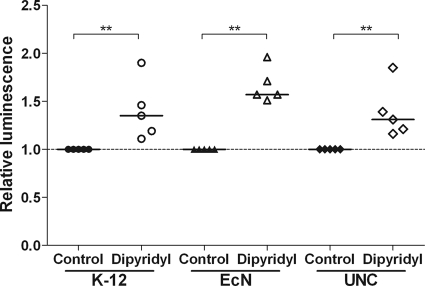
Luciferase reporter gene constructs were generated in E. coli strains Nissle, UNC, and MG1655 (used as a control strain), and nfuA promoter activation by 2,2′-dipyridyl treatment was determined. 2,2′-Dipyridyl chelates iron (Fe2+) and thereby causes the destruction of Fe-S clusters. Treatment with 2,2′-dipyridyl resulted in the activation of the nfuA promoter in all three strains (Fig. 5), suggesting a role for NfuA in the repair of Fe-S clusters destroyed either by 2,2′-dipyridyl (in vitro) or by inflammatory conditions (in vivo).
Fig 5.
Activation of the nfuA promoters in the presence of 2,2′-dipyridyl. Luciferase reporter gene constructs of the nfuA promoters were generated for E. coli K-12 MG1655, E. coli Nissle, and E. coli UNC. Cells were grown aerobically in LB medium to early-exponential phase. Luciferase activity was measured 60 min after stimulation with 250 μM 2,2′-dipyridyl. Data are expressed as medians. *, P ≤ 0.05; **, P ≤ 0.01.
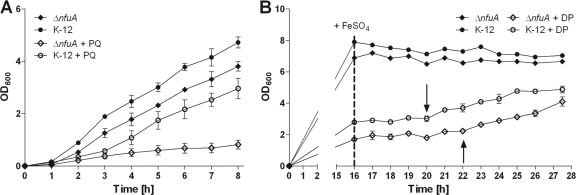
Since all E. coli strains behaved similarly, nfuA knockout mutants were generated only for E. coli MG1655. The deletion of nfuA resulted in growth retardation when the strain was grown in the presence of the superoxide generator paraquat or the iron chelator 2,2′-dipyridyl. In contrast, the growth of the deletion mutant under control conditions (LB medium) was not affected (Fig. 6). In complementation experiments, the pSU19-nfuA plasmid was able to restore the growth of the nfuA deletion strain in the presence of either paraquat or 2,2′-dipyridyl, indicating that the growth defect was due to the deletion of nfuA (see Fig. S1 in the supplemental material).
Fig 6.
Growth retardation of a ΔnfuA mutant by paraquat (PQ) or 2,2′-dipyridyl (DP). nfuA deletion mutants (ΔnfuA) were generated for E. coli K-12 MG1655. (A) Cells were grown aerobically in LB medium without (filled symbols) or with (open symbols) 300 μM paraquat for 8 h. (B) For the experiments with 2,2′-dipyridyl, cells were grown aerobically in LB medium without (filled symbols) or with (open symbols) 250 μM 2,2′-dipyridyl for 16 h; then 350 μM FeSO4 was added, and growth was monitored for 12 h. Arrows indicate the starting points of growth for the different genotypes after the addition of FeSO4. Experiments were run in triplicate. Data are expressed as means ± standard deviations.
Strain-specific expression of E. coli Nissle and UNC proteins.
The proteomic data were also analyzed for differences in protein expression between the two E. coli strains under both control and inflammatory conditions. In the control group, 26 proteins were upregulated and 39 proteins were downregulated in UNC versus Nissle, while under DSS treatment, 21 proteins were upregulated and 32 proteins were downregulated in UNC versus Nissle (see Table S2 in the supplemental material). The differentially expressed proteins are mostly related to the central metabolism (carbohydrate, fatty acid, and amino acid metabolism). Tryptophanase (TnaA) was 7-fold (control mice) and 4-fold (DSS mice) upregulated in E. coli Nissle versus E. coli UNC. TnaA catalyzes the cleavage of l-tryptophan to pyruvate and indole (48). Recently, indole was reported to exert anti-inflammatory effects (2, 3). Therefore, the cecal indole concentrations of control and DSS-treated mice monoassociated with either Nissle or UNC were determined. Control animals monoassociated with Nissle showed cecal indole concentrations 40% higher than those of mice associated with UNC. All DSS-treated mice had lower cecal indole concentrations than control mice, whether the mice were associated with strain Nissle or strain UNC (Fig. 7).
Fig 7.
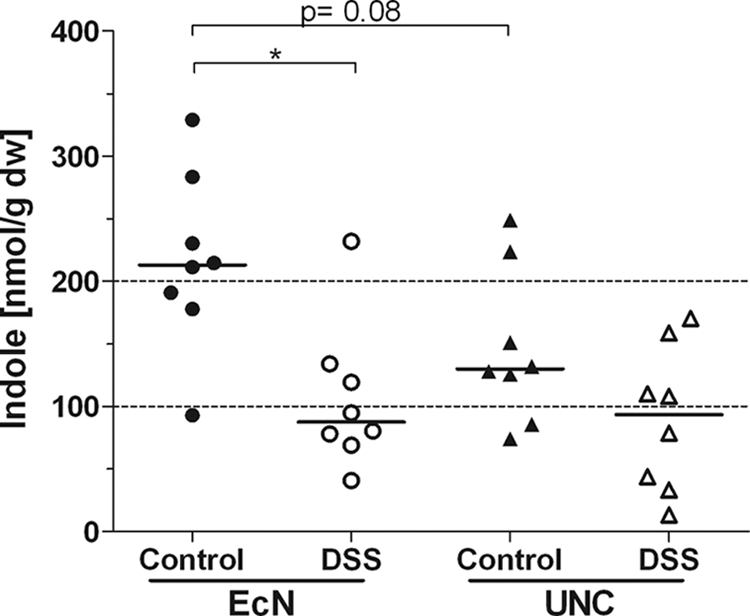
Indole concentrations in the cecal water. Germ-free mice (129/SvEv) were monoassociated with E. coli strain Nissle or UNC and were either left untreated or treated with 3.5% DSS in drinking water. Indole concentrations in the cecal water were measured by HPLC. Data are expressed as medians. *, P ≤ 0.05.
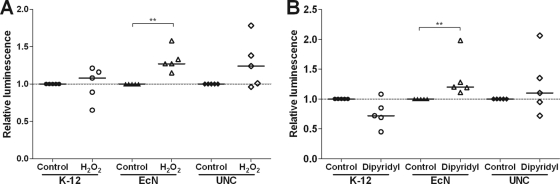
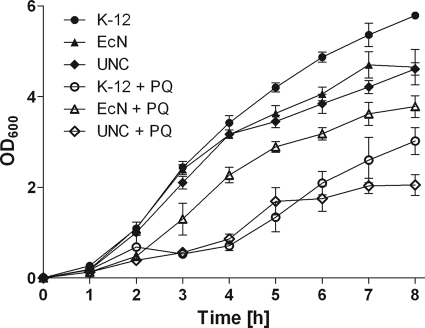
YggE was 8.1-fold upregulated in E. coli Nissle versus UNC under DSS treatment and was 4.4-fold upregulated under control conditions. YggE has been predicted to function as an auxiliary defense system against oxidative stress (23). We therefore hypothesized that YggE is a fitness factor for E. coli Nissle. To test this hypothesis, luciferase reporter gene assays were conducted with the three E. coli strains Nissle, UNC, and MG1655. Significant activation of the yggE promoter was observed only for Nissle after stimulation with 300 μM H2O2 (1.3-fold induction; P ≤ 0.01) or 250 μM 2,2′-dipyridyl (1.2-fold induction; P ≤ 0.01), not for UNC or MG1655 (Fig. 8). Growth experiments with wild-type E. coli Nissle, UNC, and MG1655 demonstrated that paraquat affected the growth of Nissle to a lesser extent than that of the other strains, indicating that these E. coli strains differ in their tolerance of superoxide (Fig. 9).
Fig 8.
Activation of the yggE promoter under different stress conditions. Luciferase reporter gene constructs of the strain-specific yggE promoters were generated for E. coli K-12 MG1655, Nissle, and UNC. Cells were grown aerobically in LB medium to early-exponential phase. Luciferase activity was measured 60 min after stimulation with 300 μM hydrogen peroxide (A) or 250 μM 2,2′-dipyridyl (B). Data are expressed as medians. **, P ≤ 0.01.
Fig 9.
Growth of different E. coli strains under superoxide stress. E. coli K-12 MG1655, Nissle, and UNC were grown aerobically in LB medium without (filled symbols) or with (open symbols) 300 μM paraquat (PQ). Growth was monitored by measuring the optical density at 600 nm (OD600) over 8 h. Experiments were run in triplicate. Data are expressed as means ± standard deviations.
DISCUSSION
High concentrations of intestinal E. coli play an important role in the onset and perpetuation of chronic intestinal inflammation (38). In the present study, a DSS-based gnotobiotic mouse model of acute intestinal inflammation was used to identify proteins that help E. coli to cope with this form of environmental stress. Two strains of E. coli, the probiotic strain Nissle and the colitogenic strain UNC, were used to evaluate strain-specific differences. Our experiments show that disease severity did not differ between animals associated with Nissle or UNC (Fig. 1A, B, C, and D). The inability of the probiotic E. coli strain Nissle to ameliorate acute inflammation is in agreement with its preferential use in keeping ulcerative colitis patients in remission (26, 27, 37). Even though administration of E. coli Nissle reduces proinflammatory cytokine secretion in conventionally raised mice with DSS-induced colitis, the histological markers are not affected (41). In contrast, Grabig et al. (14) observed an amelioration of DSS-induced colitis and a decrease in proinflammatory cytokine levels. Ukena et al. (45) reported a protective effect of E. coli Nissle against acute DSS-mediated leakiness of the gut.
Differences in histological scores were observed only for the ceca of control mice. While UNC induced mild inflammation in the ceca of these mice, Nissle did not (Fig. 1C). UNC was reported to induce cecal inflammation in IL-10−/− mice after 3 weeks of monoassociation (22). Our experiments suggest that UNC induces cecal inflammation not only in genetically susceptible IL-10−/− mice but also in healthy wild-type mice. Since the concentrations of E. coli Nissle and UNC cells in the ceca of control animals were equal (Fig. 2), differences in cell concentrations cannot account for the higher histological score of the ceca of control mice associated with UNC.
During severe intestinal inflammation, as induced by DSS, interstitial macrophages and phagocytic leukocytes (monocytes, eosinophils, polymorphonuclear neutrophils) produce reactive oxygen and nitrogen species (ROS and RNS), as well as lactoferrin (6, 35); epithelial cells and Paneth cells produce antimicrobial proteins and peptides such as peroxidases, lysozyme, and defensins to prevent the translocation of bacteria into the tissue (13). E. coli Nissle is endowed with a large number of fitness factors ensuring its survival under adverse conditions (15). Our experiments identified the uncharacterized protein YggE as a factor that enables E. coli Nissle to cope with the hostile environment in the inflamed intestine (Fig. 8; see also Table S2 in the supplemental material). YggE is a putative periplasmic protein (17), which is upregulated in E. coli cells exposed to UVA irradiation and thermal elevation (33). In E. coli cells expressing monoamine oxidase (MAO), overexpression of yggE alleviates MAO-derived oxidative stress (33). Moreover, spontaneously derived superoxide dismutase (SOD)-deficient E. coli cells induce yggE (23). Therefore, yggE is proposed to act as an auxiliary defense system against oxidative stress (23). In accordance with this notion, intracellular ROS levels were lower in SOD-deficient E. coli strains that overexpressed yggE than in E. coli strains that did not do so (25). All these observations confirm the ROS-scavenging function of YggE. Interestingly, we observed reductions in the levels of superoxide dismutase [Fe] (SodB) in E. coli from DSS-treated mice (see Table S1 in the supplemental material); this reduction was strain independent. In contrast, YggE expression was elevated only in strain Nissle and not in strain UNC (see Table S2 in the supplemental material). This supports our hypothesis that YggE enables Nissle to reach higher cell concentrations than the other E. coli strains in the inflamed gut and also in vitro in the presence of paraquat (Fig. 2, Fig. 9).
TnaA is another protein that becomes upregulated in E. coli Nissle versus UNC under both control and inflammatory conditions (see Table S2 in the supplemental material). The conversion of tryptophan by TnaA produces indole, ammonia, and pyruvate. Tryptophan supplementation in a porcine model of DSS-induced colitis improves histological markers and lowers intestinal permeability and levels of proinflammatory cytokines (21). Recently, indole was reported to reduce the chemotaxis, motility, and attachment of pathogenic E. coli to epithelial cells (3). Owing to its ability to modulate gene expression, indole is also involved in the reinforcement of the mucosal barrier and the stimulation of mucin production (2). In intestinal epithelial cells, indole attenuates the TNF-α-mediated activation of NF-κB and the expression of the proinflammatory chemokine IL-8, whereas it increases the expression of the anti-inflammatory cytokine IL-10 (2). Because of the enhanced expression of TnaA in E. coli Nissle isolated from the ceca of control and DSS-treated mice, we hypothesized that increased indole production by Nissle contributes to its beneficial effect. Control mice monoassociated with Nissle had cecal indole concentrations 40% higher than those of mice associated with UNC (Fig. 7). Since E. coli Nissle exerts its beneficial effect mainly in the remission phase of ulcerative colitis (26, 27, 37), it is conceivable that the observed elevated basal TnaA level, which, in turn, leads to increased indole production, contributes to the extension of this phase. All DSS-treated mice displayed lower cecal indole concentrations than control mice, whether they were associated with Nissle or with UNC. A possible reason for the finding that TnaA upregulation in UNC did not result in higher indole concentrations is the loss of indole due to increased permeability of the epithelial layer in the inflamed animals. We also cannot exclude the possibility that factors other than TnaA influence intestinal indole concentrations.
The proteomic analysis of E. coli also revealed a number of proteins that are repressed in the state of acute intestinal inflammation. In particular, enzymes of the central energy metabolism were repressed (Fig. 3). Glycolytic enzymes such as triose-phosphate isomerase and glyceraldehyde-3-phosphate dehydrogenase A were downregulated in both E. coli strains (see Table S1 in the supplemental material). In agreement with these findings, the concentrations of the bacterial fermentation products formate, lactate, and succinate in the ceca of DSS-treated mice were reduced with both E. coli strains (Fig. 4). Interestingly, lactate formation was observed only in E. coli Nissle-associated control mice, not in the corresponding DSS-treated mice (Fig. 4B).
In some instances, proteins belonging to the same metabolic categories (such as carbohydrate metabolism or nucleotide synthesis) were regulated oppositely. We do not have a definite explanation for these observations. However, inflammation may lead to the release from damaged epithelial cells of metabolites, which can in part be used by E. coli. This may, for example, require the upregulation of salvage enzymes and the downregulation of de novo synthesis of enzymes within the same category.
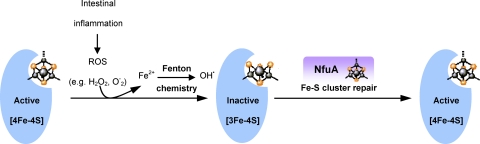
Intestinal inflammation also resulted in the upregulation of several bacterial stress response proteins in both E. coli strains (Fig. 3). Among these is the Fe-S biogenesis protein NfuA (see Table S1 in the supplemental material). As pointed out above, intestinal inflammation results in the destruction of Fe-S clusters (12, 19, 20, 28). Iron-sulfur proteins fulfill many important functions in metabolism, so that their inactivation has various adverse consequences for the bacterial cell. NfuA binds iron-sulfur clusters and transfers them to the corresponding apoprotein (1). We therefore propose that the upregulation of NfuA in response to intestinal inflammation reflects the cell's effort to repair damaged Fe-S proteins (Fig. 10). This view is supported by the demonstration of the activation of nfuA promoters following the destruction of Fe-S proteins by the iron chelator 2,2′-dipyridyl (Fig. 5). Moreover, the growth of nfuA deletion mutants was inhibited under conditions of superoxide stress (paraquat) and iron starvation (2,2′-dipyridyl) (Fig. 6). NfuA (previously YhgI) is also upregulated in Klebsiella pneumoniae during infection of mice (29). As we do here, the authors of this study suggested that on entering the host, bacterial pathogens have to cope with iron deprivation and ROS produced by the immune cells and that the induction of genes such as nfuA might help the bacteria to deal with these stress factors (29). Our experimental results indicate that NfuA induction is a general mechanism to protect E. coli against the adverse effects of intestinal inflammation. This response might contribute to the elevated numbers of E. coli cells found in the intestines of IBD patients and in animal models of gut inflammation (5, 10, 18, 24, 43, 44, 49).
Fig 10.
Proposed role of NfuA. Intestinal inflammation leads to the production of ROS and hydroxyl radicals, which, in turn, cause damage to Fe-S clusters. NfuA would serve as a scaffolding protein, which can repair Fe-S clusters by reinserting either complete clusters or Fe2+.
In conclusion, acute inflammation disturbs energy production in cecal E. coli strains and leads to the production of ROS and subsequently to the induction of NfuA, enabling E. coli to cope with the consequences of acute inflammation. Expression of the uncharacterized protein YggE by E. coli Nissle in the inflamed gut probably reduces intracellular ROS levels and thereby contributes to better survival of strain Nissle. Increased indole concentrations in the intestines of mice monoassociated with E. coli Nissle may indicate that indole contributes to the ability of Nissle to extend the remission phase in colitis patients. This work demonstrates the multifaceted and partly strain specific response of E. coli to inflammatory conditions in the mouse intestine.
Supplementary Material
ACKNOWLEDGMENTS
We thank K. Schnetz (University of Cologne, Cologne, Germany) for providing E. coli MG1655 and pKEST06, G. Gosset (UNAM, Mexico) for providing pBRINTs-Cat2, I. Grüner and U. Lehmann for taking care of the animals, and B. Gruhl, S. Schaan, and P. Albrecht for skillful technical assistance.
The project was supported by grant BL 257/7-1 from the Deutsche Forschungsgemeinschaft.
Footnotes
Published ahead of print 30 December 2011
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Angelini S, et al. 2008. NfuA, a new factor required for maturing Fe/S proteins in Escherichia coli under oxidative stress and iron starvation conditions. J. Biol. Chem. 283:14084–14091 [DOI] [PubMed] [Google Scholar]
- 2. Bansal T, Alaniz RC, Wood TK, Jayaraman A. 2010. The bacterial signal indole increases epithelial-cell tight-junction resistance and attenuates indicators of inflammation. Proc. Natl. Acad. Sci. U. S. A. 107:228–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bansal T, et al. 2007. Differential effects of epinephrine, norepinephrine, and indole on Escherichia coli O157:H7 chemotaxis, colonization, and gene expression. Infect. Immun. 75:4597–4607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bartolomé B, Jubete Y, Martinez é, de la Cruz F. 1991. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene 102:75–78 [DOI] [PubMed] [Google Scholar]
- 5. Baumgart M, et al. 2007. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn's disease involving the ileum. ISME J. 1:403–418 [DOI] [PubMed] [Google Scholar]
- 6. Birgens HS. 1985. Lactoferrin in plasma measured by an ELISA technique: evidence that plasma lactoferrin is an indicator of neutrophil turnover and bone marrow activity in acute leukaemia. Scand. J. Haematol. 34:326–331 [DOI] [PubMed] [Google Scholar]
- 7. Burich A, et al. 2001. Helicobacter-induced inflammatory bowel disease in IL-10- and T cell-deficient mice. Am. J. Physiol. Gastrointest. Liver Physiol. 281:G764–G778 [DOI] [PubMed] [Google Scholar]
- 8. Cross ML, Ganner A, Teilab D, Fray LM. 2004. Patterns of cytokine induction by gram-positive and gram-negative probiotic bacteria. FEMS Immunol. Med. Microbiol. 42:173–180 [DOI] [PubMed] [Google Scholar]
- 9. Cukrowska B, et al. 2002. Specific proliferative and antibody responses of premature infants to intestinal colonization with nonpathogenic probiotic E. coli strain Nissle 1917. Scand. J. Immunol. 55:204–209 [DOI] [PubMed] [Google Scholar]
- 10. Darfeuille-Michaud A, et al. 1998. Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn's disease. Gastroenterology 115:1405–1413 [DOI] [PubMed] [Google Scholar]
- 11. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Flint DH, Tuminello JF, Emptage MH. 1993. The inactivation of Fe-S cluster containing hydro-lyases by superoxide. J. Biol. Chem. 268:22369–22376 [PubMed] [Google Scholar]
- 13. Ganz T, Weiss J. 1997. Antimicrobial peptides of phagocytes and epithelia. Semin. Hematol. 34:343–354 [PubMed] [Google Scholar]
- 14. Grabig A, et al. 2006. Escherichia coli strain Nissle 1917 ameliorates experimental colitis via toll-like receptor 2- and toll-like receptor 4-dependent pathways. Infect. Immun. 74:4075–4082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Grozdanov L, et al. 2004. Analysis of the genome structure of the nonpathogenic probiotic Escherichia coli strain Nissle 1917. J. Bacteriol. 186:5432–5441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guzy C, et al. 2008. The probiotic Escherichia coli strain Nissle 1917 induces γδ T cell apoptosis via caspase- and FasL-dependent pathways. Int. Immunol. 20:829–840 [DOI] [PubMed] [Google Scholar]
- 17. Hagiwara D, et al. 2003. Genome-wide analyses revealing a signaling network of the RcsC-YojN-RcsB phosphorelay system in Escherichia coli. J. Bacteriol. 185:5735–5746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Heimesaat MM, et al. 2007. Shift towards pro-inflammatory intestinal bacteria aggravates acute murine colitis via Toll-like receptors 2 and 4. PLoS One 2:e662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jang S, Imlay JA. 2007. Micromolar intracellular hydrogen peroxide disrupts metabolism by damaging iron-sulfur enzymes. J. Biol. Chem. 282:929–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Keyer K, Imlay JA. 1997. Inactivation of dehydratase [4Fe-4S] clusters and disruption of iron homeostasis upon cell exposure to peroxynitrite. J. Biol. Chem. 272:27652–27659 [DOI] [PubMed] [Google Scholar]
- 21. Kim CJ, et al. 2010. l-Tryptophan exhibits therapeutic function in a porcine model of dextran sodium sulfate (DSS)-induced colitis. J. Nutr. Biochem. 21:468–475 [DOI] [PubMed] [Google Scholar]
- 22. Kim SC, et al. 2005. Variable phenotypes of enterocolitis in interleukin 10-deficient mice monoassociated with two different commensal bacteria. Gastroenterology 128:891–906 [DOI] [PubMed] [Google Scholar]
- 23. Kim SY, et al. 2005. The gene yggE functions in restoring physiological defects of Escherichia coli cultivated under oxidative stress conditions. Appl. Environ. Microbiol. 71:2762–2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kotlowski R, Bernstein CN, Sepehri S, Krause DO. 2007. High prevalence of Escherichia coli belonging to the B2+D phylogenetic group in inflammatory bowel disease. Gut 56:669–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Krishnaiah D, et al. 2007. Physiological responses of Escherichia coli cells cultivated under a sublethal oxidative stress condition. Malaysian J. Microbiol. 3:14–18 [Google Scholar]
- 26. Kruis W, et al. 2004. Maintaining remission of ulcerative colitis with the probiotic Escherichia coli Nissle 1917 is as effective as with standard mesalazine. Gut 53:1617–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kruis W, et al. 1997. Double-blind comparison of an oral Escherichia coli preparation and mesalazine in maintaining remission of ulcerative colitis. Aliment. Pharmacol. Ther. 11:853–858 [DOI] [PubMed] [Google Scholar]
- 28. Kuo CF, Mashino T, Fridovich I. 1987. α,β-Dihydroxyisovalerate dehydratase. A superoxide-sensitive enzyme. J. Biol. Chem. 262:4724–4727 [PubMed] [Google Scholar]
- 29. Lai YC, Peng HL, Chang HY. 2001. Identification of genes induced in vivo during Klebsiella pneumoniae CG43 infection. Infect. Immun. 69:7140–7145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Le Borgne S, Palmeros B, Bolivar F, Gosset G. 2001. Improvement of the pBRINT-Ts plasmid family to obtain marker-free chromosomal insertion of cloned DNA in E. coli. Biotechniques 30:252–254, 256 [DOI] [PubMed] [Google Scholar]
- 31. Lennox ES. 1955. Transduction of linked genetic characters of the host by bacteriophage P1. Virology 1:190–206 [DOI] [PubMed] [Google Scholar]
- 32. Mattivi F, Vrhovsek U, Versini G. 1999. Determination of indole-3-acetic acid, tryptophan and other indoles in must and wine by high-performance liquid chromatography with fluorescence detection. J. Chromatogr. A 855:227–235 [DOI] [PubMed] [Google Scholar]
- 33. Ojima Y, Kawase D, Nishioka M, Taya M. 2009. Functionally undefined gene, yggE, alleviates oxidative stress generated by monoamine oxidase in recombinant Escherichia coli. Biotechnol. Lett. 31:139–145 [DOI] [PubMed] [Google Scholar]
- 34. Patzer SI, Baquero MR, Bravo D, Moreno F, Hantke K. 2003. The colicin G, H and X determinants encode microcins M and H47, which might utilize the catecholate siderophore receptors FepA, Cir, Fiu and IroN. Microbiology 149:2557–2570 [DOI] [PubMed] [Google Scholar]
- 35. Pavlick KP, et al. 2002. Role of reactive metabolites of oxygen and nitrogen in inflammatory bowel disease. Free Radic. Biol. Med. 33:311–322 [DOI] [PubMed] [Google Scholar]
- 36. Rabilloud T, Strub JM, Luche S, van Dorsselaer A, Lunardi J. 2001. A comparison between Sypro Ruby and ruthenium II tris (bathophenanthroline disulfonate) as fluorescent stains for protein detection in gels. Proteomics 1:699–704 [DOI] [PubMed] [Google Scholar]
- 37. Rembacken BJ, Snelling AM, Hawkey PM, Chalmers DM, Axon AT. 1999. Non-pathogenic Escherichia coli versus mesalazine for the treatment of ulcerative colitis: a randomised trial. Lancet 354:635–639 [DOI] [PubMed] [Google Scholar]
- 38. Rolhion N, Darfeuille-Michaud A. 2007. Adherent-invasive Escherichia coli in inflammatory bowel disease. Inflamm. Bowel Dis. 13:1277–1283 [DOI] [PubMed] [Google Scholar]
- 39. Sartor RB. 2006. Mechanisms of disease: pathogenesis of Crohn's disease and ulcerative colitis. Nat. Clin. Pract. Gastroenterol. Hepatol. 3:390–407 [DOI] [PubMed] [Google Scholar]
- 40. Schlee M, et al. 2007. Induction of human β-defensin 2 by the probiotic Escherichia coli Nissle 1917 is mediated through flagellin. Infect. Immun. 75:2399–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schultz M, et al. 2004. Preventive effects of Escherichia coli strain Nissle 1917 on acute and chronic intestinal inflammation in two different murine models of colitis. Clin. Diagn. Lab. Immunol. 11:372–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schuppler M, Lotzsch K, Waidmann M, Autenrieth IB. 2004. An abundance of Escherichia coli is harbored by the mucosa-associated bacterial flora of interleukin-2-deficient mice. Infect. Immun. 72:1983–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sepehri S, Kotlowski R, Bernstein CN, Krause DO. 2007. Microbial diversity of inflamed and noninflamed gut biopsy tissues in inflammatory bowel disease. Inflamm. Bowel Dis. 13:675–683 [DOI] [PubMed] [Google Scholar]
- 44. Swidsinski A, et al. 2002. Mucosal flora in inflammatory bowel disease. Gastroenterology 122:44–54 [DOI] [PubMed] [Google Scholar]
- 45. Ukena SN, et al. 2007. Probiotic Escherichia coli Nissle 1917 inhibits leaky gut by enhancing mucosal integrity. PLoS One 2:e1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vogel-Scheel J, Alpert C, Engst W, Loh G, Blaut M. 2010. Escherichia coli must synthesize purines and pyrimidines to establish in the mouse intestine. Appl. Environ. Microbiol. 76:5181–5187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Waidmann M, et al. 2003. Bacteroides vulgatus protects against Escherichia coli-induced colitis in gnotobiotic interleukin-2-deficient mice. Gastroenterology 125:162–177 [DOI] [PubMed] [Google Scholar]
- 48. Watanabe T, Snell EE. 1972. Reversibility of the tryptophanase reaction: synthesis of tryptophan from indole, pyruvate, and ammonia. Proc. Natl. Acad. Sci. U. S. A. 69:1086–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wohlgemuth S, Haller D, Blaut M, Loh G. 2009. Reduced microbial diversity and high numbers of one single Escherichia coli strain in the intestine of colitic mice. Environ. Microbiol. 11:1562–1571 [DOI] [PubMed] [Google Scholar]
- 50. Zyrek AA, et al. 2007. Molecular mechanisms underlying the probiotic effects of Escherichia coli Nissle 1917 involve ZO-2 and PKCζ redistribution resulting in tight junction and epithelial barrier repair. Cell. Microbiol. 9:804–816 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.