Abstract
Previously we reported the cultivation of novel verrucomicrobia, including strain TAV2 (93% 16S rRNA gene identity to its nearest cultivated representative, Opitutus terreae PB90-1) from the gut of the termite Reticulitermes flavipes. To gain better insight into the Verrucomicrobia as a whole and understand the role of verrucomicrobia within the termite gut ecosystem, we analyzed a draft genome and undertook a physiological characterization of TAV2. Strain TAV2 is an autochthonous member of the R. flavipes gut microbiota and groups phylogenetically among diverse Verrucomicrobia from R. flavipes and other termites that are represented by 16S rRNA gene sequences alone. TAV2 is a microaerophile, possessing a high-affinity cbb3-type terminal oxidase-encoding gene and exhibiting an optimum growth rate between 2 and 8% (vol/vol) oxygen. It has the genetic potential to degrade cellulose, an important function within termite guts, but its in vitro substrate utilization spectrum was limited to starch and a few mono- and disaccharides. Growth occurred on nitrogen-free medium, and genomic screening revealed genes for dinitrogenases, heretofore detected in only a few members of the Verrucomicrobia. This represents the first (i) characterization of a verrucomicrobial species from the termite gut, (ii) report of nif and anf genes in a nonacidophilic verrucomicrobial species, and (iii) description of a microaerophilic genotype and phenotype in this phylum of bacteria. The genetic and physiological distinctiveness of TAV2 supports its recognition as the type strain of a new genus and species, for which the name Diplosphaera colitermitum gen. nov., sp. nov., is proposed.
INTRODUCTION
The bacterial phylum Verrucomicrobia (25, 30) together with the Planctomycetes, Chlamydia, and sister phyla Poribacteria and Lentisphaera form a monophyletic, deeply rooted “superphylum” (1, 8, 67). The abundance of verrucomicrobia in many habitats (37) suggests that they have the potential to exert a considerable ecological impact. For example, verrucomicrobial 16S rRNA gene sequences represent a sizeable percentage of sequences in soil clone libraries (23, 56), with total verrucomicrobial cell number estimates ranging from 106 to 108 cells per gram (dry weight) of soil (20), thereby contributing up to 0.2% of the total DNA in soil (45). Moreover, verrucomicrobial rRNA comprised 2.2% to 10% of the total rRNA extracted from soil samples (11), implying that they are metabolically active members of the microbial communities in both agricultural and native soils.
Despite their ubiquity and implied ecological importance, only the heavily piliated Verrucomicrobium spinosum (1), several species of Prosthecobacter (25, 61, 62), and three strains isolated from rice paddy soils (32) were obtained in pure culture prior to the new millennium. However, new strategies for isolation, as well as novel methods for detection of sought-after microorganisms on solid media (64), have yielded more than 40 additional Verrucomicrobia cultivars (13, 14, 16, 31, 34, 50, 54–57, 64, 74–81). Nevertheless, the verrucomicrobia remain defined primarily through the more than 3,900 16S rRNA gene sequences from various terrestrial, aquatic, and host-associated environments which have been deposited in public databases. Hence our knowledge of the in situ physiology, function, and importance of this ubiquitous group of microorganisms remains meager. Perhaps the most surprising recent discovery is that three verrucomicrobial isolates, “Methylacidiphilum” strain V4, “Methylacidiphilum” strain SolV, and “Methylacidiphilum” strain Kam1, have the ability to oxidize methane (a capacity previously known only within the proteobacteria) and can do so under extremely acidic conditions (pH 0.8 to 2.0) (18, 31, 46, 50). Additionally, these isolates contain genes for nitrogen fixation, and nitrogen-fixing activity and diazotrophic growth have been demonstrated for strain SolV and strain Kam1, the first such finding within the phylum Verrucomicrobia (31, 35).
Cultivation-dependent and -independent studies have revealed that some verrucomicrobia live in close association with eukaryotes. Members of the candidate genus Xiphinematobacter populate the intestinal epithelia of juvenile nematode worms (66), ectosymbiotic verrucomicrobia on the marine ciliate Euplotidium act as defensive cells (47), and novel verrucomicrobia have been isolated from the human intestine (16), the gut of the sea cucumber (54), marine sponges (57), and the digestive tract of a marine clamworm (14). Previously, we reported the isolation of several novel verrucomicrobia from the guts of wood-eating termites, including the strain we further investigate here, TAV2, which is a member of the Opitutaceae (64).
For over 100 years, termites and their gut microorganisms have been a model for host-microbe nutritional interactions. In hindguts, bacterial symbionts perform four functions important to termite vitality: (i) they contribute to cellulose and hemicellulose digestion; (ii) they contribute to termite nitrogen nutrition via dinitrogen fixation and the recycling of excretory (uric acid) N, as the lignocellulosic food of termites is typically nitrogen poor; (iii) they produce acetate from the fermentative degradation of lignocellulose, which supports up to 100% of the insect's respiratory requirements; and (iv) peripheral, gut wall-associated symbionts consume inwardly diffusing oxygen, creating anoxia in the lumen that supports fermentative production of acetate by bacteria (and anaerobic protozoa) (10). Here, we sought to infer the possible and probable ecological roles of strain TAV2 in termite guts and in so doing increase our limited understanding of the Verrucomicrobia in general.
MATERIALS AND METHODS
Cultivation, nutrition, and physiological studies.
Strain TAV2, along with three other verrucomicrobia (TAV1, -3, and -4), was previously isolated from hindgut homogenates of the common Eastern subterranean termite, Reticulitermes flavipes (Kollar) (64). For routine maintenance, verrucomicrobial strains were grown on solid R2A medium (Becton-Dickinson Co., Sparks, MD) (53). For liquid cultures, cells were grown with shaking (at 200 rpm in air) in 50-ml sterile Erlenmeyer flasks containing 15 ml R2B medium, which was identical to R2A but lacked agar. Unless indicated otherwise, all incubations were at 25°C.
Substrate utilization by TAV strains was examined by using 18-mm anaerobe tubes (Bellco, Vineland, NJ) containing 5 ml of basal salt solution (BSS) (pH 7.0) (71) amended with 5 mg/ml yeast extract and 0.01% each of a trace element solution and a vitamin solution (71) and a 4 mM final concentration (unless otherwise noted in parentheses) of the substrate to be tested. The test substrates included d-glucose, d-fructose, d-galactose, d-mannose, d-trehalose, sucrose, d-ribose, d-xylose, l-arabinose, d-mannitol, d-sorbitol, d-maltose, d-raffinose, d-cellobiose, starch (0.1%), methylcellulose (0.1%), carboxy methylcellulose (0.1%), microcrystalline cellulose (0.1%), xylan (0.2%, previously extracted with 70% cold ethanol), sodium dl-lactate, sodium pyruvate, sodium fumarate, sodium acetate, allantoin, d-glucuronate, d-galacturonate (0.1%), d-gluconic acid, xanthine, tannic acid (0.1%), resorcinol, vanillic acid, sodium benzoate, and trimethoxybenzoate. Test cultures were inoculated with 1% (vol/vol) of an exponential-phase culture growing in R2B and then were incubated at 25°C with the tubes held nearly horizontal in a rotary cell culture drum operating at 35 rpm. Cell growth was measured by using a Milton-Roy Spectronic 20 colorimeter at a wavelength of 600 nm. An increase of at least 50% in optical density above that of the “no-substrate” control was considered evidence of the ability to utilize the substrate.
To determine whether strain TAV2 produces acetate, lactate, formate, succinate, or butyrate during growth on glucose, TAV2 was cultivated in triplicate in BSS medium amended with 5 mM glucose (described above) in butyl rubber-stoppered 18-mm anaerobe tubes under atmospheres of 21% O2 (balance N2), 2% O2 (balance N2), or 100% N2. Simultaneously with growth measurements, 1.0 ml of culture fluid was removed and centrifuged at 12,000 × g for 10 min, and the supernatant was filtered through a 0.22-μm-pore-size filter and stored at −20°C until used for organic acid analysis. Organic acids were quantified by high-performance liquid chromatography (HPLC) (Waters, Milford, MA) on a 300- by 7.8-mm Aminex HPX-87H column (Bio-Rad, Hercules, CA) at 23°C with 4 mM H2SO4 as the eluent (0.6 ml/ml). Organic acids were detected with a Waters 2487 UV detector at 210 nm and calibrated with homologous standards.
The temperature range for growth was determined by using 250-ml Erlenmeyer flasks containing 50 ml of R2B medium and possessing a laterally attached 18-mm anaerobe tube for measuring optical density. Each flask was equilibrated to the test temperature and then inoculated with 1% (vol/vol) of a mid-exponential-phase culture of TAV2 grown in R2B at a growth-supporting temperature close to the test temperature. Growth, while cultures were shaken, was monitored by measurements of optical density at 600 nm (OD600) (as described above) until stationary phase was reached. The pH tolerance of TAV2 was tested by using 50-ml Erlenmeyer flasks containing 10 ml of R2B medium ranging in pH from 4.0 to 9.0 in increments of 0.5 pH unit. The medium was buffered with 10 mM citric acid (for a pH range of 4.0 to 4.5), 2-(N-morpholino)ethanesulfonic acid (for a pH range of 5.0 to 5.5), 3-(N-morpholino)propanesulfonic acid (MOPS) (for a pH range of 6.0 to 7.5), or Tris-HCl (for a pH range of 8.0 to 9.0). The salt tolerance of TAV2 was determined by using 18-mm tubes containing 5 ml of R2B amended with sodium chloride at concentrations varying from 0 to 4% in increments of 0.5%.
To evaluate the growth of strain TAV2 in a liquid medium under defined headspace concentrations of O2, butyl rubber-stoppered 18-mm anaerobe tubes containing 5 ml of R2B medium modified to contain 10 mM total glucose (an increase above the normal 2.7 mM glucose concentration of R2B medium to ensure that growth in all tubes was oxygen, not substrate, limited) were prepared in air (21% O2) or under a headspace of 100% N2. To the tubes containing a 100% N2 headspace, air was injected to attain a final headspace percentage of 2, 4, or 8% O2 (after the overpressure was released). To attain a headspace percentage of 12, 16, or 20% O2, pure oxygen was injected instead of air. The tubes were sterilized by autoclaving, inoculated with a 5% (vol/vol) inoculum of an exponential-phase TAV2 culture growing in a homologous medium in air, and then incubated in a rotary cell culture drum at 35 rpm at 22 to 23°C. Growth was monitored spectrophotometrically as described above.
To evaluate the growth of strain TAV2 in nitrogen-free medium and hence infer dinitrogen fixation capability, TAV2 was grown in a glucose-salts medium (all values are in g/liter: glucose, 10.0; dipotassium phosphate, 1.0, magnesium sulfate, 0.2; calcium carbonate, 1.0; sodium chloride, 0.2; sodium molybdate, 0.005; and ferrous sulfate, 0.1) that contained 0.2% gellan gum. Inoculated tubes were kept for 2 weeks in a 2% O2 environment and inspected daily for growth. The ability to grow was evidenced by comparison with negative (noninoculated) and positive controls after 2 weeks of incubation. Herbaspirillum seropidicae strain ATCC 35892 was used as positive control.
Enzyme assays.
TAV2 cells were grown in 100 ml R2B medium, in air, in 500-ml glass bottles (Bellco, Vineland, NJ) to which an 18-mm anaerobe tube was laterally attached (70) and shaken at 250 rpm. At mid-log phase, cells from the entire culture were harvested by centrifugation at 10,000 × g for 10 min at 4°C, washed with 20 ml sonication buffer (10 mM EDTA, 50 mM Tris-HCl, pH 7.0), recentrifuged, and then resuspended in 10 ml of the same buffer. While held in an ice bath, cells were disrupted by sonication with 3 pulses of 30 s each using a Branson model 450 Sonifier (power setting of 5, 50% duty cycle) equipped with a 1/2-inch threaded-body step horn with a flat tip, followed by centrifugation at 12,000 × g for 60 min at 4°C. The resulting supernatant was used for enzyme assays.
Catalase was assayed by measuring the rate of decrease in A240 of H2O2 as described by Beers and Sizer (4). Superoxide dismutase activity was measured by the xanthine/xanthine oxidase-cytochrome c reduction method (21). NAD(P)H oxidase and peroxidase activities were assayed as described previously (63).
PCR-related procedures.
The autochthony (i.e., indigenousness) of TAV2 for R. flavipes was evaluated as described previously (70). Purified DNA was obtained from approximately 100 resected guts of R. flavipes worker larvae, from termite “nest soil,” and from adjacent but not termite-associated “bulk forest soil” from Dansville, MI (the origin of R. flavipes specimens). The DNA preparations were then PCR amplified with the forward Verrucomicrobia-specific primer Ver53f (5′-TGG CGG CGT GGW TAA GA-3′) (64) combined with the eubacterial reverse primer 1492r (5′-GGT TAC CTT GTT ACG ACT T-3′) (68). Amplifications were performed in a 25-μl reaction mixture containing 0.2 μM each primer, 200 μM deoxynucleoside triphosphates (dNTPs), 1× Taq buffer, 1.5 mM MgCl2, 0.625 U of Taq DNA polymerase (Invitrogen Co., Carlsbad, CA), and 50 ng of DNA template. The PCR was initiated with a 3-min denaturation step at 95°C, followed by 30 cycles of denaturation at 95°C for 30 s, primer annealing at 61°C for 30 s, extension at 72°C for 45 s, and a final extension for 10 min. PCR-amplified DNA was cloned into TOP10 Escherichia coli using the plasmid vector pCR2.1 (TA clone kit; Invitrogen, Carlsbad, CA). Approximately 50 clones from each treatment were randomly selected and the plasmid insert size checked via PCR using the same primers and PCR conditions described above. The PCR products from clones with the appropriate plasmid insert size (approximately 1,439 bp) were chosen for direct sequence analysis. Prior to sequencing, unreacted dNTPs and primers in the PCR mixtures were digested and dephosphorylated using ExoSap-IT (USB, Cleveland, OH).
Partial 16S rRNA gene sequences for each clone were determined by using Applied Biosystems cycle sequencing technology (Applied Biosystems, Foster City, CA), with the primer Ver53f. Sequence chromatograms were checked for quality, and only sequences longer than 400 nucleotides were used for subsequent analyses. The partial 16S rRNA gene sequence of each clone was imported and aligned using the ARB software package (http://www.arb-home.de/) (40). Ambiguities in sequence alignments were corrected manually, where possible, and only unambiguously aligned positions were used for the phylogenetic analysis. Phylogenetic tree construction and bootstrap analyses were done in ARB using TREE-PUZZLE. The statistical similarity of clone libraries was assessed with the program ∫-LIBSHUFF. A nonparametric estimate of library similarity was done according to the method of Yue and Clayton as implemented by SONS (58, 59).
The total number of Verrucomicrobia cells in R. flavipes and their primary anatomical location were estimated by a dilution-to-extinction PCR approach (70). Briefly, DNA was extracted from 50 to 100 freshly collected intact termites, termite hindguts (from which the midguts were removed), and degutted termite bodies by using a MoBio Ultraclean soil DNA extraction kit (MoBio Laboratories, Carlsbad, CA). Purified DNA was normalized on a per-termite-equivalent basis and was serially diluted in buffer (10 mM Tris-HCl [pH 8.0] containing 50 ng/μl calf thymus DNA as a carrier). As a control, 100 ng purified DNA from strain TAV2 was also serially diluted in buffer. Each dilution was used as the template in a PCR with forward primer Ver53f, 63f (5′-CAT GTC GAC GTY TTA AGC ATG CAA GT-3′), or 8f (5′-AGA GTT TGA TCC TGG CTC AG-3′) combined with reverse primer 1492r. These primer pairs target regions of the 16S rRNA gene common to most Verrucomicrobia, Spirochaetes, or Bacteria, respectively. The PCR mixture was identical to that described above, and a total of 30 PCR cycles were used. Five-microliter samples of each PCR mixture were analyzed by electrophoresis on a 1.0% agarose-0.5× Tris-borate-EDTA gel stained with 1× Gelstar nucleic acid stain (Cambrex, East Rutherford, NJ). An estimate of the in situ abundance of the organism was made by comparison of the extinction point (i.e., the smallest amount of template DNA yielding a visible amplimer) of purified TAV2 DNA to that of homologous target DNA in serially diluted termite samples, as well as the determined genome size (see Fig. S1 in the supplemental material) and 16S rRNA gene copy number (see Fig. S2 in the supplemental material) of TAV2 (5.2 Mb and one copy, respectively).
PCR amplification of the N subunit of the cbb3-type cytochrome oxidase from TAV1 was done by using primers CcoNGenF (5′-CTC CAA GCG CAC GGG NCC NGA YYT-3′) and CcoNGenR (5′-CGT GCA CGT GGC CGA YNR TCC ART C-3′) (69). PCR conditions were as described above except that an annealing temperature of 53°C was used. PCR amplification of nifH or anfH was done as described in reference 38 and references therein.
Genomic properties.
The genome size of TAV2 was estimated by pulsed-field gel electrophoresis of PmeI, BlnI, or NotI restriction endonuclease digestions of genomic DNA from an exponential-phase culture (OD600 = 0.2 to 0.3; approximately 8 × 108 cells/ml) according to previously published protocols (6, 22). Electrophoresis was carried out using a CHEF-DRII system (Bio-Rad Laboratories, Richmond, CA). Estimation of 16S rRNA gene copy number for TAV2 was done by methods previously described (36) using the restriction enzymes ApaI, SmaII, and HaeII (New England BioLabs, Ipswich, MA). G+C content was determined by HPLC analysis of P1 nuclease- and alkaline phosphatase-digested samples as described by Mesbah et al. (42), except that DNA was purified by using a Qiagen genomic DNA kit (Qiagen, Valencia, CA) and an Altima C18 column (250 by 4.6 mm; particle size, 5 μm) (Alltech Associates, Deerfield, IL) was used for HPLC.
Preparation of genomic DNA for sequencing.
TAV1 and TAV2 cells were grown to mid-exponential phase, and genomic DNA was isolated via two phenol-chloroform extractions followed by precipitation with isopropanol (0.6, vol/vol) and salt removal with cold 80% ethanol. DNA was resuspended in Tris-EDTA (TE) (pH 8.0) containing RNase A (1 mg/ml), and the final concentration was measured with the DyNA QUANT2000 fluorometer (Amersham Pharmacia Biotech UK Ltd., Buchinghamshire, United Kingdom). The DNA was quality checked on a 1.0% agarose-0.5× Tris-borate-EDTA gel stained with 1× Gelstar nucleic acid stain. After repeated attempts, only DNA from strain TAV2 passed quality standards for size and purity required by the Joint Genome Institute for full genome sequencing. Therefore, TAV2 DNA was submitted to the Joint Genome Institute Microbial Sequencing program for sequencing and annotation (http://genome.jgi-psf.org/opiba/opiba.info.html). TAV1 DNA was usable for PCR procedures (see above).
Microscopy.
Phase-contrast micrographs were prepared from wet mounts on agar-coated slides (48). Negative stains of TAV2 were prepared according to established protocols using 7% nigrosin (5), and images were captured on a Zeiss Axioskop microscope (Carl Zeiss, Inc., Thornwood, NY) equipped with a SPOT charge-coupled-device digital camera (Diagnostic Instruments, Inc., Sterling Heights, MI). Transmission electron microscopy and scanning electron microscopy were done at the MSU center for advanced microscopy as described in reference 70.
Nucleotide sequence accession and strain deposition numbers.
The partial 16S rRNA gene sequences obtained from PCR amplification and cloning of DNA from termite guts, termite nest soil, and non-termite-associated soil obtained in this study have been deposited in the GenBank database under accession numbers HQ830036 through HQ830155. Partial ccoN and anfH gene sequences obtained from TAV1 DNA have been deposited under accession numbers JQ064561 and JQ064562, respectively. The genome of TAV2 can be accessed through GenBank under accession number ABEA00000000. Type strain TAV2 has been deposited in the ATCC (accession no. BAA-2264) and USDA Agriculture Research Service (accession no. NRRL B-59605) culture collections.
RESULTS AND DISCUSSION
Abundance, diversity, and autochthony of Verrucomicrobia in R. flavipes termite guts.
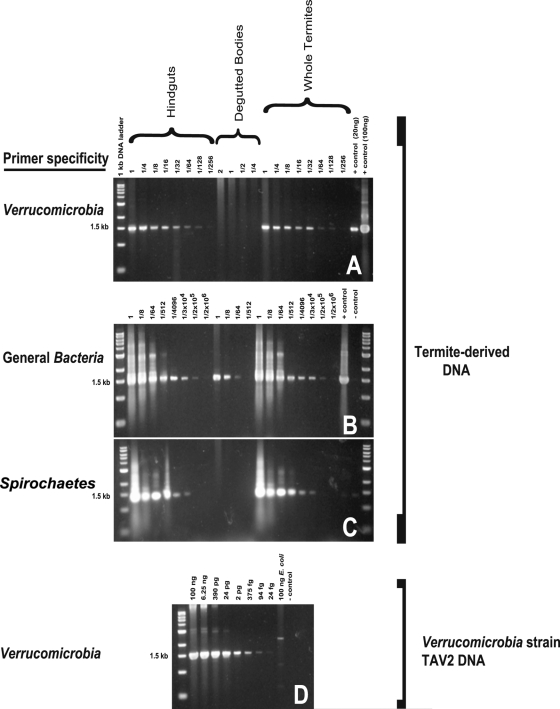
Estimates of the location and abundance of verrucomicrobia associated with R. flavipes, based on a dilution-to-extinction PCR method (Fig. 1), revealed that, like spirochetes, they are located exclusively within the termite hindgut. However, assuming a 5.2-Mb genome with one 16S RNA gene copy per cell (as determined for TAV2 [see Fig. S1 and S2 in the supplemental material]), verrucomicrobia are but a small portion of the total gut microbiota, contributing only about 1.2 × 103 cells out of an estimated total of 6 × 106 bacterial cells per gut (Fig. 1). Although they represent only a small portion of this microbial community, preliminary physiological analysis of strain TAV1 coupled with genomic and physiological analyses of strain TAV2, described below, suggest that verrucomicrobia may nevertheless fill an important ecological niche in the termite hindgut.
Fig 1.
(A to C) Dilution-to-extinction PCR of DNA from termite hindguts, degutted termite bodies, and whole termites amplified with Verrucomicrobia (A)-, general Bacteria (B)-, and Spirochaetes (C)-specific primers. Numbers above each lane represent the amount of DNA added on a “per-termite-equivalent” basis. (D) Purified DNA from Verrucomicrobia strain TAV2 was serially diluted and amplified as a control. Assuming that the average nucleotide has a molecular mass of 324 g/mol, one 5.2-Mb TAV2 genome with one 16S rRNA gene copy is slightly over five femtograms. A PCR product was detected in termite gut DNA diluted to the equivalent of 1/256 of a gut (A). A PCR product was detected when control TAV2 DNA was diluted to 24 fg, approximating 4.8 cells (D). Thus, we can estimate that there are 4.8 verrucomicrobia cells per 1/256 of a gut, yielding a total of 1.2 × 103 verrucomicrobial cells per termite gut.
To determine whether termite-associated verrucomicrobia were indigenous to the guts of R. flavipes and to assess their phylogenetic diversity, 16S rRNA gene clone libraries were prepared from termite guts, termite nest soil, and bulk forest soil by using PCR with primers targeting the Verrucomicrobia phylum. Analysis of the clone libraries revealed that sequences obtained from R. flavipes guts were phylogenetically distinct from verrucomicrobial sequences amplified from termite nest soil or bulk forest soil (Fig. 2). Sequences from R. flavipes guts shared 94.4% ± 6.2% 16S rRNA gene identity to each other. In contrast, the nucleotide identities of the hindgut clones were only 76.9% ± 4.2 and 76.2% ± 4.9% with clones obtained from nest soil and forest soil, respectively. Likewise, ∫-LIBSHUFF analysis also indicated that the clone library from termite guts was significantly different (P < 0.001) than the libraries obtained from both nest and forest soils. Taken together, the above results substantiate the autochthonous nature of the R. flavipes gut verrucomicrobia.
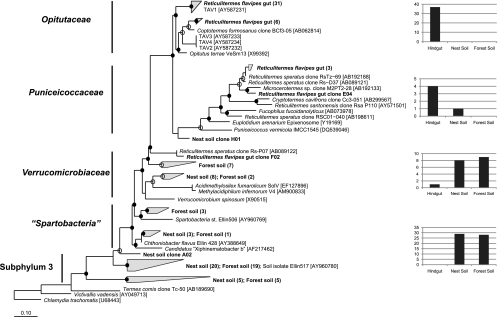
Fig 2.
Maximum-likelihood-based phylogeny of PCR-amplified 16S rRNA gene clones obtained with a Verrucomicrobia phylum-targeting primer set in this study (boldface). Clones were obtained from R. flavipes hindguts (42 clones; GenBank accession numbers HQ830113 to HQ830155), nest soil (38 clones; GenBank accession numbers HQ830074 to HQ830112), and adjacent but non-termite-associated bulk forest soil (37 clones; GenBank accession numbers HQ830036 to HQ830073). The numbers of clones obtained from gut, nest soil, or forest soil DNA for each of the five subphyla within the Verrucomicrobia are given in the bar graphs (subphyla 3 and “Spartobacteria” were combined). The phylogeny is based on 426 aligned nucleotides. Groups of closely related clones are condensed and indicated by gray blades, with the number of clones indicated in parentheses. Branch points with >75% support are indicated by filled circles. Branch points with 50 to 74% support are indicated by open circles. Accession numbers of reference species are shown in brackets. Scale bar, 0.10 change per nucleotide.
Further phylogenetic analysis clearly indicated the presence of at least three distinct groupings of verrucomicrobia 16S rRNA gene clones within the guts of R. flavipes (Fig. 2). Most clones clustered within the family Opitutaceae and were most closely related to strain TAV1 (98.7% ± 0.8% 16S rRNA gene identity), an isolate obtained concomitantly with TAV2 (64). Six clones, also within the family Opitutataceae, grouped most closely with a clone (BCf3-05) from Coptotermes formosanus termites (93.9% ± 0.8% 16S rRNA gene identity) (60). Additionally, four clones grouped within the family Puniceicoccaceae and shared 76.4% ± 1.0% identity with TAV2. Together, these R. flavipes gut clones shared an average 87.5% ± 2.9% 16S rRNA gene identity with the isolate TAV2, indicating that R. flavipes organisms collected in Dansville, MI, harbor a phylogenetically diverse assemblage of Verrucomicrobia within their guts.
Interestingly, R. flavipes gut-derived clones within the family Puniceicoccaceae grouped with several other termite-derived 16S rRNA gene clones obtained from the guts of Reticulitermes santonensis (suggested to be synonymous with R. flavipes [2]) collected in France, Reticulitermes speratus collected in Japan (27, 28, 43), Cryptotermes cavifrons collected in the United States (26), and Microcerotermes sp. collected in Thailand (27) (Fig. 2). These clones shared 83.5% ± 4.2% 16S rRNA gene identity with the isolate TAV2. A single R. flavipes-derived clone outside the families Opitutaceae and Puniceicoccaceae grouped with clone Rs-P07 obtained from R. speratus (28). While more detailed studies are needed to indicate whether cospeciation has occurred between termites and their gut verrucomicrobia, the similarity of verrucomicrobial clones from R. flavipes to those of other termites and the implied autochthony of these bacteria suggest that the niches occupied by these symbionts are important for the vitality of their host and/or gut microbial community stability. Accordingly, we sought to further examine termite gut verrucomicrobia through physiological characterization and draft genome sequence analysis.
Isolate TAV1 appears to best represent the majority of verrucomicrobia within the R. flavipes hindgut as inferred by 16S rRNA gene sequencing (Fig. 2). Unfortunately, it produces copious amounts of exopolysaccharides (64), which stymied most physiological analyses and resulted in low DNA quality (low purity and small fragment sizes) insufficient for complete genome sequencing. Therefore, we chose TAV2 (which is the same species as TAV3 and TAV4, sharing 99.5% 16S rRNA gene identity [64]) for full-genome sequencing and physiological analyses. Preliminary comparisons of TAV1 and TAV2 demonstrated they were highly similar in substrate utilization spectra and in presence of a cbb3-type terminal cytochrome oxidase and nitrogen fixation genes (see below). This indicates that TAV2 is an appropriate model for making ecological inferences regarding verrucomicrobia in the termite gut.
Substrate utilization and acetate production.
Of the Verrucomicrobia isolates examined to date, most utilize components of plant biomass as energy sources, including a variety of oligo- and polysaccharides (33, 55). Owing to the importance of microbial symbionts within the termite gut to the conversion of plant polysaccharides to acetate (the major oxidizable energy source of termites), we examined the genome of TAV2 for genes relevant to polysaccharide hydrolysis. We found that the TAV2 genome contains a gene predicted to encode a catalytic cellulase protein (EC 3.2.1.4; glycohydrolase family 5) that shared 74% amino acid identity with a cellulase from the closely related organism Opitutus terrae PB90-1 (GenBank accession number YP_001819625), a β-1,4-xylanase (EC 3.2.1.8; glycohydrolase family 16) that shared 34% identity with a xylanase from O. terrae PB90-1 (YP_001820089), and a starch phosphorylase (EC 2.4.1.1; glycosyltransferase family 35) that shared 72% amino acid identity with a starch phosphorylase from O. terrae PB90-1 (YP_001820917).
Additionally, analysis of the TAV2 genome revealed nearly all genes necessary for glycolysis, with the exception of the gene for glucose-6-phosphate isomerase (responsible for conversion of glucose-6-phosphate to fructose-6-phosphate). It may be that the seeming absence of this gene is due to the fact the genome has yet to be closed. However, an extensive BLAST search of the TAV2 genome with homologs from O. terrae and other bacteria failed to reveal even a partial match for this gene. It is possible that TAV2 expresses an as-yet-unidentified enzyme that can catalyze this reaction or that it forms fructose-6-phopsphate via the pentose phosphate pathway, for which it has all of the necessary enzyme-encoding genes. TAV2 also contains a gene encoding a PPi-dependent 6-phosphofructokinase (EC 2.7.1.90), an alternative to the “traditional” ATP-dependent enzyme (12, 72). TAV2 also contains all the genes necessary for the tricarboxylic acid (TCA) cycle and thus has the potential to completely oxidize glucose to CO2.
These genomic data, including the discovery of genes for cellulose, xylan, and starch degradation, prompted in vitro experiments to test the ability of TAV1 and TAV2 to grow on 33 substrates representing different classes of chemical compounds. Of these, glucose, galactose, maltose, cellobiose, and starch supported both TAV1 and TAV2 growth (Table 1). Xylose and arabinose were growth supporting for TAV1 but not TAV2. This, along with the genomic data from TAV2 (see above), suggests that TAV2 may have recently lost the ability to oxidize monomeric pentoses, a capability that may be retained in the majority of verrucomicrobia in the hindgut. In total, hindgut verrucomicrobia appear to have become adapted to further oxidize pentose and hexose sugars that may be released during polysaccharide hydrolysis by other bacteria and/or protozoa.
Table 1.
Phenotypic characteristics useful for distinguishing between Diplosphaera colitermitum strain TAV2T and other cultivated representatives of Verrucomicrobiaa
| Characteristic | F. fucoidanolyticus | P. vermicola | A. agarolyticus | O. terrae | TAV1 | TAV2 |
|---|---|---|---|---|---|---|
| Subphylum | 4 | 4 | 4 | 4 | 4 | 4 |
| Environment | Sea cucumber gut | Clamworm gut | Hot spring | Paddy soil | Termite gut | Termite gut |
| Cell shape | Coccus | Coccus | Coccus | Coccus | Diplococcus | Diplococcus |
| Cell size (μm) | 1.2–1.6 | 0.6–1.0 | 0.8–0.9 | 0.4–0.6 | 0.5–0.6 | 0.5–0.6 |
| Color | Yellow or beige | Pale red | White | Colorless | Colorless | Colorless |
| Anaerobic growth | − | + | + | + | − | − |
| Oxidase | − | − | + | − | − | − |
| Catalase | + | − | + | − | − | − |
| Motility | − | − | + | + | − | − |
| Temp range (°C) | 10–40 | 8–37 | 40–55 | 10–37 | ND | 15–35 |
| pH range | 5.0–12.0 | 5.5–9.5 | ND | 5.5–9.0 | ND | 5.5–7.5 |
| Growth in NaCl (%) | 10 | 10 | 2.0 | 3.0 | ND | 1.5 |
| Carbon sources | ||||||
| Glucose | ND | + | + | + | + | + |
| Galactose | ND | + | + | + | + | + |
| Maltose | ND | ND | + | + | + | + |
| Cellobiose | ND | + | + | − | + | + |
| Xylose | ND | − | + | − | + | − |
| Arabinose | ND | + | ND | + | + | − |
| Starch | ND | − | ND | + | + | + |
| GC content (mol %) | 52 | 52.1 | 65.8 | 73.7 | ND | 60.5 |
In R. flavipes guts, the major end product of polysaccharide dissimilation is acetate, with other identifiable end products being lactate, formate, succinate, and butyrate (65). Thus, we investigated whether TAV2 could produce acetate or other organic acid end products from glucose when grown under a headspace of 21% O2 (balance N2), 2% O2 (balance N2), or 100% N2. We were unable to detect any acetate or other identifiable soluble end products during growth on glucose under aerobic (21% O2) or microaerobic (2% O2) conditions. TAV2 (and TAV1) did not grow anaerobically (100% N2) even though a variety of alternative electron acceptors were added to the medium (see below). Thus, TAV2 appears to completely oxidize glucose to CO2 during aerobic growth in vitro and probably does so in situ.
Relationship of strain TAV2 to oxygen.
Oxygen concentrations at the termite gut periphery are estimated to be approximately 4% (vol/vol), decreasing to anoxia at 100 to 200 μm inwardly from the gut wall (9). Though TAV2 can grow on agar media under atmospheric (21%, vol/vol) concentrations of oxygen, its autochthony in the termite gut and apparent lack of catalase, peroxidase, and superoxide dismutase activities, as well as an O2-protective capsule (evidenced by nigrosin negative staining [Fig. 3]), suggested that it may be better adapted to lower-than-atmospheric concentrations of oxygen. Thus, we investigated the growth of TAV2 on glucose under conditions favorable for anaerobic (fermentation and anaerobic respiration) and aerobic growth. The results (Table 2) revealed that under complete anoxia, TAV2 failed to grow, even if nitrite, nitrate, sulfate, or sulfite was added to the medium to support anaerobic respiration. Under 2% (vol/vol) oxygen (initial concentration), the generation time during exponential phase was 7.7 ± 0.7 h, and the final cell yield was 2.5 × 109 ± 7.4 ×107 cells/ml. At initial O2 concentrations of 4 and 8%, the generation times and cell yields were similar. However, if the initial O2 concentration was increased to 12% and 20%, the generation times during exponential phase decreased to 9.2 ± 0.6 h and 11.7 ± 1.4 h, respectively. Thus, under the conditions tested, TAV2 appears to reach an optimal generation time at initial O2 concentrations of between 2 and 8% (mean, 7.8 ± 0.6 h), decreasing with O2 concentrations of between 12 and 20% O2 (mean, 10.0 ± 1.6 h) (P < 0.0001 by Student's t test; n = 6 for each O2 concentration). When cells were grown in the same tubes left unsealed (and thus continually exposed to an atmospheric concentration of oxygen), the generation time increased significantly to 14 ± 0.9 h (P = 0.038; n = 3 for tubes incubated in air and n = 6 for sealed tubes incubated under 20% initial O2). The inability to grow under anoxia, with optimum growth at between 2 and 8% initial O2 indicates that TAV2 can be considered a microaerophile and that, in situ, cells probably reside on or near the hindgut wall (9). The microaerophilic growth of TAV2 may be partially explained by the apparent lack of catalase, superoxide dismutase, and peroxidase activities (Table 1). However, TAV2 is not an obligate microaerophile, owing to its ability to grow at atmospheric concentrations of O2, which suggests that it must possess other mechanisms for the removal of toxic oxygen species.
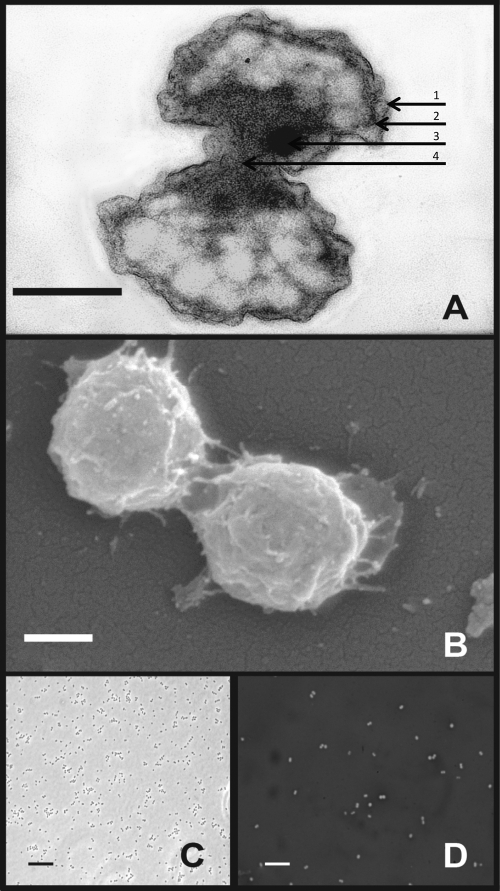
Fig 3.
Transmission electron (A), scanning electron (B), phase-contrast (C), and phase-contrast nigrosin stain (D) micrographs of TAV2. Arrows indicate the outer membrane (1), inner membrane (2), and an unidentified intracellular inclusion (3). Note that both the inner and outer membranes encircle each cell (4). Scale bars, 0.25 μm (A and B) and 10 μm (C and D).
Table 2.
Effect of headspace oxygen concentration on generation time, final cell yield, and final medium pH of Diplosphaera colitermitum strain TAV2a
| Headspace O2, % (no. of replicates) | Mean ± SD |
||
|---|---|---|---|
| Generation time, h | Final cell yield, cells/ml | Final pH | |
| 0 (3) | NGb | ||
| 2 (6) | 7.7 ± 0.7 | 2.5 × 109 ± 7.4 × 107 | 6.0 ± 0.1 |
| 4 (6) | 8.3 ± 0.4 | 3.6 × 109 ± 2.7 × 108 | 6.3 ± 0.1 |
| 8 (6) | 7.7 ± 0.3 | 3.3 × 109 ± 2.3 × 108 | 6.3 ± 0.1 |
| 12 (6) | 9.2 ± 0.6 | 3.7 × 109 ± 5.3 × 108 | 6.2 ± 0.1 |
| 16 (6) | 8.9 ± 0.8 | 3.2 × 109 ± 2.3 × 108 | 6.2 ± 0.1 |
| 20 (6) | 11.7 ± 1.4 | 2.6 × 109 ± 1.2 × 108 | 6.2 ± 0.1 |
| Air (3) | 14.0 ± 0 | 3.7 × 109 ± 6.0 × 107 | 6.6 ± 0.1 |
Growth at O2 concentrations of 0 to 20% was measured in enclosed anaerobe tubes without replacement of the atmosphere. Growth in air was measured in anaerobe tubes with caps that allowed continual influx of atmosphere.
NG, no growth.
Analysis of the TAV2 genome for genes relevant to respiratory activity further supports the microaerophilic nature of TAV2. Many microaerophiles that spend at least part of their life in hypoxic environments, such as the N2-fixing root nodule symbiont Bradyrhizobium japonicum, the gastric mucosa-colonizing pathogen Helicobacter pylori, and the recently characterized obligate microaerophile (from termite guts) Stenoxybacter acetivorans, utilize a high-affinity cbb3-type terminal cytochrome oxidase (41, 49, 51, 69). Similarly, the genome sequence of TAV2 reveals it to have all of the necessary genes to express a cbb3-type terminal oxidase, and the catalytic (N) subunit of a cbb3 oxidase was also identified in TAV1 (95% amino acid sequence identity to TAV2) by PCR amplification and sequencing (Fig. 4). This is one of the few instances where organisms outside the proteobacteria have been shown to possess a cbb3-type terminal oxidase (49), and it is the first report of such genes within the phylum Verrucomicrobia. In contrast to the well-studied cytochrome aa3 oxidase, which has an estimated Km for oxygen of 0.1 to 1 μM, the cbb3 oxidases have estimated Kms in the nanomolar range (e.g., 7 nM for B. japonicum) (51). A bd-type cytochrome has an even lower Km, at or near 3 nM O2 (17). However, the TAV2 genome does not appear to contain either of the genes (CydA and CydB) necessary for expression of a bd-type terminal oxidase. The ability of organisms utilizing these high-affinity enzymes to consume oxygen to very low concentrations presumably minimizes the production of reactive oxygen species (to which microaerophiles may be particularly sensitive) and helps also to reconcile the absence of genes encoding catalase, peroxidase, and superoxide dismutase in TAV2, as well as the absence of such activities in TAV2 cell extracts. The typically greater energy yield of aerobic versus anaerobic metabolic processes may give a competitive edge to organisms with a cbb3-type terminal oxidase in low-O2 environments. It may also allow O2-labile processes such as N2 fixation to occur (see below) (52).
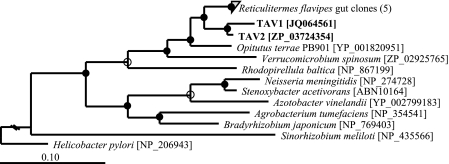
Fig 4.
Maximum-likelihood-based phylogenetic analysis of the deduced amino acid sequence (122 positions) of the catalytic subunit of the cbb3 terminal cytochrome oxidase (ccoN) from the draft genome sequence of TAV2 and targeted PCR amplification of TAV1 DNA (boldface). The TAV sequences cluster with five others obtained from the guts of Reticulitermes flavipes as described in reference 69. The GenBank accession numbers for these five clones are ABN10029, ABN10043, ABN10046, ABN10058, and ABN10088. Accession numbers for TAV and reference species are given in brackets. Branch points with >75% support are indicated by filled circles. Branch points with 50 to 74% support are indicated by open circles. Scale bar, 0.1 change per amino acid.
The TAV2 genome contains genes encoding subunits of cytochrome aa3 oxidase, but it lacks several genes encoding enzymes that catalyze the conversion of protoheme to the heme A prosthetic group required for the aa3 oxidase activity as well as genes encoding aa3 assembly proteins (e.g., COX10, COX11, and COX15). Therefore, the genomic data suggest that the cbb3 terminal oxidase may be the sole oxidase used by TAV2. Interestingly, TAV2 does not appear to contain a bc1-type cytochrome complex or a cytochrome c reductase, which are normally responsible for the transfer of electrons from a ubiquininone or menaquinone pool onto the terminal cytochrome oxidase. How TAV2 transfers electrons from NADH dehydrogenase and succinate dehydrogenase/fumarate reductase down the electron transport chain onto the cbb3 terminal oxidase remains an interesting aspect of its physiology to be studied.
Genomic evidence for dinitrogen fixation potential in TAV2.
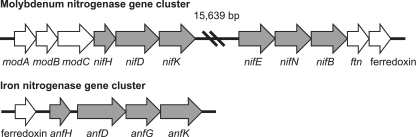
The paradigmatic enzyme for fixation of atmospheric dinitrogen into ammonia is a molybdenum-containing nitrogenase enzyme complex (Mo nitrogenase) encoded by the nifHDK operon. However, some diazotrophs also contain “alternative,” molybdenum-independent nitrogenases, such as iron-only or vanadium-dependent nitrogenases encoded by the anfHDGK and vnfHDGK operons, respectively (7, 44). Analysis of the TAV2 genome revealed two regions containing genes initially identified as “nitrogenase.” To further explore this, Pfam (3) and TIGRFAM (24) model comparisons were done, as well as COG and KEGG database searches. Together, these data strongly suggest that TAV2 contains both a nifHDK operon and an anfHDGK operon (Fig. 5). Furthermore, genes in close proximity to the nifHDK operon are predicted to encode molybdenum ABC transport and permease proteins as well as to synthesize ferredoxin, a common electron donor for nitrogenase enzymes (19). It was also confirmed that TAV2 contains nifA and anfA genes, the transcriptional activators at nif and anf promoters (39). TAV2 does not seem to contain a gene for nifL, a transcriptional repressor of nifA under unfavorable growth conditions, but does contain glnB, a nifA activator in some diazotrophic bacteria (39).
Fig 5.
Organization of open reading frames predicted to encode molybdenum and iron nitrogenases, as well as associated genes in TAV2. Coding regions: mod, molybdenum transport; nif, molybdenum nitrogenase; ftn, ferretin; anf, iron nitrogenase.
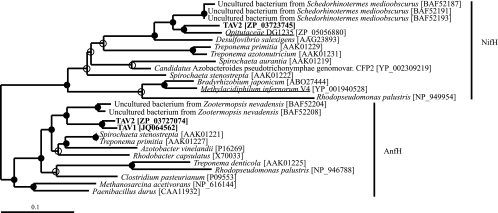
Sequence comparisons were made and a phylogenetic tree was constructed for the putative nifH and anfH genes of TAV2 by using a database of 12,344 nifH and anfH sequences collected from GenBank (Fig. 6). These results support the finding that TAV2 has both nifH and anfH genes, as each clustered strongly with other known nifH and anfH sequences. The validity of these clusters is supported by empirical evidence, particularly with regard to mutational and functional analyses of nifH and anfH in Rhodopseudomonas palustris (44). Interestingly, preliminary data show TAV1 to have an anfH gene that shares 97% amino acid identity with the anfH from TAV2 (Fig. 6). More targeted amplification and/or genome sequencing will be needed to determine if TAV1 also has a nifH gene. Surprisingly, the nifH gene from TAV2 shared 83% identity over 274 amino acids to the nifH gene from Verrucomicrobium strain DG1235 (unpublished; GenBank accession number ABSI00000000), collected from the marine dinoflagellate Scrippsiella trochidae (Fig. 5). Based on 16S rRNA phylogeny, TAV1, TAV2, and DG1235 are members of the Opitutaceae family of the Verrucomicrobia, suggesting common ancestry of the nifH gene within this family and the possibility that many other members may have homologous genes, though it does not seem to be present in the free-living Opitutus terrae PB90-1 (GenBank accession number CP001032). Three nifH clones sequenced from DNA obtained from the hindguts of the termite Zootermopsis navadensis clustered with TAV2 nifH, and two anfH clones from the hindguts of the termite Schedorhinotermes medioobscurus (73) clustered with TAV2 anfH, indicating they may also be derived from termite gut verrucomicrobia. The deduced amino acid sequence of the nitrogenase iron-protein NifH and AnfH of TAV2 revealed motifs such as the Walker-A motif, as well as the Fe4S4 binding sites that are present in all nitrogenase iron-proteins (15). This, together with positive growth of TAV2 on nitrogen-free medium (data not shown), indicates that one or both of these enzymes are fully functional. Nitrogen fixation is a particularly important functional attribute within the gut ecosystem, as the termite diet has a low N content. Although spirochetes, being numerically abundant, appear to play an important role in providing nitrogen to the termite host (38), verrucomicrobia may contribute the N pool within the gut ecosystem as well. Studies investigating the expression of TAV2 nifH and anfH in vivo and in vitro, particularly with regard to oxygen concentration, are ongoing.
Fig 6.
Maximum-likelihood-based phylogenetic analysis of the deduced amino acid sequence (74 positions) of nifH and anfH from draft genome sequence analysis of TAV2 and targeted PCR amplification of TAV1 DNA (boldface). nifH sequences from two other verrucomicrobia are underlined (the nifH from “Methylacidiphilum” strain SolV [GenBank accession number GU299762] shares 98% amino acid sequence homology with the nifH from “Methylacidiphilum” strain V4 [YP_001940528], and hence we represent only strain SolV here). Despite exhaustive searching, the dinitrogenase reductase from “Methylacidiphilum” strain Kam1 was not available through NCBI GenBank, though it is assumed to cluster with those of strains SolV and V4 based on a previous publication (34). Branch points with >75% support are indicated by filled circles. Branch points with 50 to 74% support are indicated by open circles. Accession numbers of reference species are shown in brackets. Scale bar, 0.1 change per amino acid.
TAV2 is the fourth published member of the phylum Verrucomicrobia found to possess nifHDK genes (46); the three others belong to the genus “Methylacidiphilum,” whose members characteristically have C1 metabolism and extreme acid tolerance. Two of the isolates can fix N2, as inferred directly through acetylene reduction assays (for “Methylacidiphilum” strain SolV [35]) or through growth in nitrogen-free medium (for “Methylacidiphilum” strain Kam1 [31]). No physiological evidence for N2 fixation in “Methylacidiphilum” strain V4 has been published (18, 29). Hence, TAV2 is the first reported nonacidiphilic, obligately heterotrophic, host-associated Verrucomicrobia strain that has nifHDK genes and expresses a functional nitrogenase. Furthermore, it is the first Verrucomicrobia strain found to have anfHDGK genes. Taking these findings together with genomic data from the as-yet-unpublished Verrucomicrobium strain DG1235, it is intriguing to consider if verrucomicrobia may exert a greater impact with regard to nitrogen availability in certain ecosystems (host associated, acidic, and oligotrophic) than previously realized. With the advent of new low-cost, high-throughput sequencing technologies, an assessment of the distribution of nitrogenase genes within members of the phylum Verrucomicrobia (and the related Planctomyces, etc.) along with increased cultivation efforts and nitrogenase activity assays would be very revealing with regard to the importance of verrucomicrobia to nitrogen fixation in other ecosystems.
Proposal for a new taxon, Diplosphaera colitermitum gen. nov., sp. nov.
The genomic and physiological properties of Verrucomicrobia strain TAV2 are sufficiently distinct from those of any other known bacterium to warrant its classification as a new genus and species within the phylum Verrucomicrobia. The relatively distant 16S rRNA gene similarity (93%) to the closest named relative, Opitutus terrae PB90-1, coupled with the biochemical characteristics outlined in Table 2 and its microaerophilic phenotype, narrow substrate utilization spectrum, and autochthony within the termite gut further support this. Accordingly, we propose the name Diplosphaera colitermitum for this bacterium (see below).
Description of Diplosphaera gen. nov.
Diplosphaera gen. nov. (Di.plo.spha.e′ra. Gr. adj. diploos, double; L. fem. n. sphaera, globe, sphere; N.L. fem. n. diplosphaera, double sphere or diplococcus). The genus description is, at present, the same as for the type species, Diplosphaera colitermitum.
Description of Diplosphaera colitermitum sp. nov.
Diplosphaera colitermitum sp. nov. (co.li.term.i′tum. L. n. colon -i, colon, part of the large intestine; L. n. termes -itis, wood-eating worm, termite; N. L. gen. pl. n. colitermitum, of the gut of termites). Cells are coccoid (0.25 μm to 0.5 μm in diameter) and occur almost exclusively in pairs, with a Gram-negative cell wall morphology that includes an outer membrane (Fig. 3). Cells are nonmotile, obligate aerobes and are microaerophilic. The shortest generation times occur in liquid medium under an atmosphere of 2 to 8% O2 (balance N2). On solid R2A medium, colonies are 2 to 4 mm in diameter, have an entire margin and a low convex, mucoid morphology, and are cream colored. Cells do not possess catalase, superoxide dismutase, or NADH/NADPH peroxidase activity. Nitrogenase activity is inferred through growth on nitrogen-free medium. Growth occurs in liquid media between 15 and 35°C (optimum, 30°C); there is no growth at 37°C or 4°C. Growth occurs at a pH range of 5.5 to 7.5 (optimum, 7.0); there is no growth at a pH of ≤5 or ≥8. Substrates utilized as energy sources include starch, d-cellobiose, d-maltose, d-glucose, d-galactose, and one or more components present in yeast extract. Microcrystalline cellulose, methylcellulose, carboxy methylcellulose, xylan, d-fructose, d-mannose, d-trehalose, sucrose, d-ribose, d-xylose, l-arabinose, d-mannitol, d-sorbitol, d-raffinose, dl-lactate, sodium pyruvate, sodium fumarate, sodium acetate, allantoin, d-glucuronate, d-galacturonate, d-gluconic acid, xanthine, tannic acid, resourcinol, vanillic acid, sodium benzoate, and trimethylbenzoate are not utilized. The genome of type strain TAV2 is 5.2 Mb in size, contains 60.5 mol% G+C, and possesses one 16S rRNA gene copy (see Fig. S2 in the supplemental material). The type strain, isolated from guts of Reticulitermes flavipes (Kollar) collected in Dansville, MI, is TAV2.
Supplementary Material
ACKNOWLEDGMENTS
We sincerely thank the Joint Genome Institute Microbial Sequencing program for full genome sequencing, annotation, and partial finishing of TAV2. We thank David Wunder and Scott Prentice for help with organic acid analyses. We thank Kwi Kim for help with determination of rRNA copy number and genome size and fosmid library construction for TAV2. We are also grateful to C. Flegler and A. Pastor-Lecha for scanning and transmission electron microscopy and to Reuben Bell for advice on nomenclature, etymology, and construction of Latinized names.
This research was supported by grants from the National Science Foundation (MCB-0731913 to T.M.S. and IBN-0114505 to J.A.B.), by an MSU College of Natural Science Recruiting Fellowship and Calvin Research Fellowships (to J.T.W.), and by a Calvin College Star Fellowship (to E.K.).
Footnotes
Published ahead of print 22 December 2011
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Albrecht W, Fischer A, Smida J, Stackebrandt E. 1987. Verrucomicrobium spinosum, an eubacterium representing an ancient line of descent. Syst. Appl. Microbiol. 10:57–62 [Google Scholar]
- 2. Austin JW, et al. 2005. Genetic evidence for the synonymy of two Reticulitermes species: Reticulitermes flavipes and Reticulitermes santonensis. Ann. Entomol. Soc. Am. 98:395–401 [Google Scholar]
- 3. Bateman A, et al. 2002. The Pfam protein families database. Nucleic Acids Res. 30:276–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beers RF, Sizer IW. 1952. A spectrophotometric method for measuring the breakdown of hydrogen peroxide by catalase. J. Biol. Chem. 195:133–140 [PubMed] [Google Scholar]
- 5. Benson HJ. 1990. Negative staining, p 44–45 In Jaffe EG. (ed), Microbiological applications, 5th ed William C. Brown Publishers, Dubuqe, IA [Google Scholar]
- 6. Birren B, Lai E. 1993. Pulsed field gel electrophoresis: a practical guide. Acedemic Press, Inc., San Diego, CA [Google Scholar]
- 7. Bishop PE, Premakumar R. 1992. Alternative nitrogen fixation systems, p 736–762 In Stacey G, Burris RH, Evans HJ. (ed), Biological nitrogen fixation. Chapman and Hall, New York, NY [Google Scholar]
- 8. Brochier C, Philippe H. 2002. Phylogeny: a non-hyperthermophilic ancestor for bacteria. Nature 417:244. [DOI] [PubMed] [Google Scholar]
- 9. Brune A, Emerson D, Breznak J. 1995. The termite gut microflora as an oxygen sink: microelectrode determination of oxygen and pH gradients in guts of lower and higher termites. Appl. Environ. Microbiol. 61:2681–2687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brune A, Ohkuma M. 2011. Role of the termite gut microbiota in symbiotic digestion, p 439–476 In Bignell DE., Roisin Y, Lo N. (ed), Biology of termites: a modern synthesis. Springer, New York, NY [Google Scholar]
- 11. Buckley DH, Schmidt TM. 2001. Environmental factors influencing the distribution of rRNA from Verrucomicrobia in soil. FEMS Microbiol. Ecol. 35:105–112 [DOI] [PubMed] [Google Scholar]
- 12. Chi A. 2000. The primordial high energy compound: ATP or inorganic pyrophosphate? J. Biol. Chem. 275:35677–35679 [DOI] [PubMed] [Google Scholar]
- 13. Chin K-J, Liesack W, Janssen PH. 2001. Opitutus terrae gen. nov., sp. nov., to accommodate novel strains of the division Verrucomicrobia isolated from rice paddy soil. Int. J. Syst. Bacteriol. 51:1965–1968 [DOI] [PubMed] [Google Scholar]
- 14. Choo YJ, Lee K, Song J, Cho JC. 2007. Puniceicoccus vermicola gen. nov., sp. nov., a novel marine bacterium, and description of Puniceicoccaceae fam nov., Puniceicoccales ord. nov., Opitutaceae fam. nov., Opitutales ord. nov., and Opitutae classis nov. in the phylum Verrucomicrobia. Int. J. Syst. Bacteriol. 57:532–537 [DOI] [PubMed] [Google Scholar]
- 15. Dean DR, Jacobson MR. 1992. Biological nitrogen fixation. Chapman & Hall, New York, NY [Google Scholar]
- 16. Derrien M, Vaughan EE, Plugge CM, de Vos WM. 2004. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int. J. Syst. Evol. Microbiol. 54:1469–1476 [DOI] [PubMed] [Google Scholar]
- 17. D'Mello R, Hill S, Poole R. 1995. The oxygen affinity of cytochrome bo′ in Escherichia coli determined by the deoxygenation of oxyleghemoglobin and oxymyoglobin: Km values for oxygen are in the submicromolar range. J. Bacteriol. 177:867–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dunfield PF, et al. 2007. Methane oxidation by an extremely acidophilic bacterium of the phylum Verrucomicrobia. Nature 450:879–882 [DOI] [PubMed] [Google Scholar]
- 19. Edgren T, Nordlund S. 2006. Electron transport to nitrogenase in Rhodospirillum rubrum: the major pathway is dependent on the fix gene products. FEMS Microbiol. Lett. 260:30–35 [DOI] [PubMed] [Google Scholar]
- 20. Felske A, Akkermans ADL. 1998. Prominant occurance of ribosomes from an uncultured bacterium of the Verrucomicrobiales cluster in grassland soils. Lett. Appl. Microbiol. 26:219–223 [DOI] [PubMed] [Google Scholar]
- 21. Flohe L, Becker R, Brigelius R, Lengfelder E, Otting F. 1988. Convenient assays for superoxide dismutase, p 287–293 In Miqeul J. (ed), CRC handbook of free radicals and antioxidants in biomedicine, vol 3 CRC Press, Boca Raton, FL [Google Scholar]
- 22. Graber JR, Leadbetter JR, Breznak JA. 2004. Description of Treponema azotonutricium sp. nov. and Treponema primitia sp. nov., the first spirochetes isolated from termite guts. Appl. Environ. Microbiol. 70:1315–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hackl E, Zechmeister-Boltenstern S, Bodrossy L, Sessitsch A. 2004. Comparison of diversities and compositions of bacterial populations inhabiting natural forest soils. Appl. Environ. Microbiol. 70:5057–5065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Haft D, Selengut J, White O. 2003. The TIGRFAMs database of protein families. Nucleic Acids Res. 31:371–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hedlund BP, Gosink JJ, Staley JT. 1997. Verrucomicrobia div. nov., a new division of the Bacteria containing three new species of Prosthecobacter. Antonie Van Leeuwenhoek 72:29–38 [DOI] [PubMed] [Google Scholar]
- 26. Hongoh Y, et al. 2007. The motility symbiont of the termite gut flagellate Caduceia versatilis is a member of the Synergistes group. Appl. Environ. Microbiol. 73:6270–6276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hongoh Y, et al. 2005. Intra- and interspecific comparisons of bacterial diversity and community structure support coevolution of gut microbiota and termite host. Appl. Environ. Microbiol. 71:6590–6599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hongoh Y, Ohkuma M, Kudo T. 2003. Molecular analysis of bacterial microbiota in the gut of the termite Reticulitermes speratus (Isoptera; Rhinotermitidae). FEMS Microbiol. Ecol. 44:231–242 [DOI] [PubMed] [Google Scholar]
- 29. Hou SB, et al. 2008. Complete genome sequence of the extremely acidophilic methanotroph isolate V4, Methylacidiphilum infernorum, a representative of the bacterial phylum Verrucomicrobia. Biol. Direct. 3:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hugenholtz P, Goebel BM, Pace NR. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765–4774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Islam T, Jensen S, Reigstad LJ, Larsen O, Birkeland N-K. 2008. Methane oxidation at 55° C and pH 2 by a thermoacidophilic bacterium belonging to the Verrucomicrobia phylum. Proc. Natl. Acad. Sci. U. S. A. 105:300–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Janssen PH, Yates PS, Grinton BE, Taylor PM, Sait M. 2002. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl. Environ. Microbiol. 68:2391–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Janssen P, Schuhmann A, Morschel E, Rainey F. 1997. Novel anaerobic ultramicrobacteria belonging to the Verrucomicrobiales lineage of bacterial descent isolated by dilution culture from anoxic rice paddy soil. Appl. Environ. Microbiol. 63:1382–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kasai H, et al. 2007. Rubritalea squalenifaciens sp. nov., a squalene-producing marine bacterium belonging to subdivision 1 of the phylum “Verrucomicrobia.” Int. J. Syst. Evol. Microbiol. 57:1630–1634 [DOI] [PubMed] [Google Scholar]
- 35. Khadem AF, Pol A, Jetten MSM, Op den Camp HJM. 2010. Nitrogen fixation by the verrucomicrobial methanotroph Methylacidiphilum fumariolicum SolV. Microbiology 156:1052–1059 [DOI] [PubMed] [Google Scholar]
- 36. Klappenbach JA, Dunbar JM, Schmidt TM. 2000. rRNA operon copy number reflects ecological strategies of bacteria. Appl. Environ. Microbiol. 66:1328–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lee SY, Bollinger J, Bezdicek D, Ogram A. 1996. Estimation of the abundance of an uncultured soil bacterial strain by a competitive quantitative PCR method. Appl. Environ. Microbiol. 62:3787–3793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lilburn TG, et al. 2001. Nitrogen fixation by symbiotic and free-living spirochetes. Science 292:2495–2498 [DOI] [PubMed] [Google Scholar]
- 39. Little R, Martinez-Argudo I, Shearer N, Johnson P, Dixon R. 2005. Genetic regulation of nitrogen fixation: integration of multiple signals, p 53–55 In Wang Y-P, et al. (ed), Biological nitrogen fixation, sustainable agriculture and the environment. Springer, Dordrecht, Netherlands [Google Scholar]
- 40. Ludwig W, et al. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Marcelli SW, et al. 1996. The respiratory chain of Helicobacter pylori: identification of cytochromes and the effects of oxygen on cytochrome and menaquinone levels. FEMS Microbiol. Lett. 138:59–64 [DOI] [PubMed] [Google Scholar]
- 42. Mesbah M, Premachandran U, Whitman WB. 1989. Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatography. Int. J. Syst. Bacteriol. 39:159–167 [Google Scholar]
- 43. Nakajima H, Hongoh Y, Usami R, Kudo T, Ohkuma M. 2005. Spatial distribution of bacterial phylotypes in the gut of the termite Reticulitermes speratus and the bacterial community colonizing the gut epithelium. FEMS Microbiol. Ecol. 54:247–255 [DOI] [PubMed] [Google Scholar]
- 44. Oda Y, et al. 2005. Functional genomic analysis of three nitrogenase isozymes in the photosynthetic bacterium Rhodopseudomonas palustris. J. Bacteriol. 187:7784–7794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. O'Farrell KA, Janssen PH. 1999. Detection of verrucomicrobia in a pasture soil by PCR-mediated amplification of 16S rRNA genes. Appl. Environ. Microbiol. 65:4280–4284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Op den Camp HJM, et al. 2009. Environmental, genomic and taxonomic perspectives on methanotrophic Verrucomicrobia. Environ. Microbiol. Rep. 1:293–306 [DOI] [PubMed] [Google Scholar]
- 47. Petroni G, Spring S, Schleifer K-H, Verni F, Rosati G. 2000. Defensive extrusive ectosymbionts of Euplotidium (Ciliophora) that contain microtubule-like structures are bacteria related to Verrucomicrobia. Proc. Natl. Acad. Sci. U. S. A. 97:1813–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pfenning N, Wagener S. 1986. An improved method of preparing wet mounts for photomicrographs of microorganisms. J. Microbiol. Methods 4:303–306 [Google Scholar]
- 49. Pitcher RS, Watmough NJ. 2004. The bacterial cytochrome cbb3 oxidases. Biochim. Biophys. Acta Bioenergetics 1655:388–399 [DOI] [PubMed] [Google Scholar]
- 50. Pol A, et al. 2007. Methanotrophy below pH 1 by a new Verrucomicrobia species. Nature 450:874–878 [DOI] [PubMed] [Google Scholar]
- 51. Preisig O, Zufferey R, Thony-Meyer L, Appleby C, Hennecke H. 1996. A high-affinity cbb3-type cytochrome oxidase terminates the symbiosis-specific respiratory chain of Bradyrhizobium japonicum. J. Bacteriol. 178:1532–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Preisig O, Anthamatten D, Hennecke H. 1993. Genes for a microaerobically induced oxidase complex in Bradyrhizobium japonicum are essential for a nitrogen-fixing endosymbiosis. Proc. Natl. Acad. Sci. U. S. A. 90:3309–3313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Reasoner DJ, Geldreich EE. 1985. A new medium for the enumeration and subculture of bacteria from potable water. Appl. Environ. Microbiol. 49:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sakai T, Ishizuka K, Kato I. 2003. Isolation and characterization of a fucoidan-degrading marine bacterium. Mar. Biotechnol. 5:409–416 [DOI] [PubMed] [Google Scholar]
- 55. Sangwan P, Chen X, Hugenholtz P, Janssen PH. 2004. Chthoniobacter flavus gen. nov., sp. nov., the first pure-culture representative of subdivision two, Spartobacteria classis nov., of the phylum Verrucomicrobia. Appl. Environ. Microbiol. 70:5875–5881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sangwan P, Kovac S, Davis KER, Sait M, Janssen PH. 2005. Detection and cultivation of soil verrucomicrobia. Appl. Environ. Microbiol. 71:8402–8410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Scheuermayer M, Gulder TAM, Bringmann G, Hentschel U. 2006. Rubritalea marina gen. nov., sp. nov., a marine representative of the phylum Verrucomicrobia, isolated from a sponge (Porifera). Int. J. Syst. Bacteriol. 56:2119–2124 [DOI] [PubMed] [Google Scholar]
- 58. Schloss PD, Handelsman J. 2006. Introducing SONS, a tool for operational taxonomic unit-based comparisons of microbial community memberships and structures. Appl. Environ. Microbiol. 72:6773–6779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Schloss PD, Larget BR, Handelsman J. 2004. Integration of microbial ecology and statistics: a test to compare gene libraries. Appl. Environ. Microbiol. 70:5485–5492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Shinzato N, Muramatsu M, Matsui T, Watanabe Y. 2005. Molecular phylogenetic diversity of the bacterial community in the gut of the termite Coptotermes formosanus. Biosci. Biotechnol. Biochem. 69:1145–1155 [DOI] [PubMed] [Google Scholar]
- 61. Staley JT, Bont JA, Jonge K. 1976. Prosthecobacter fusiformis nov. gen. et sp., the fusiform caulobacter. Antonie Van Leeuwenhoek 42:333–342 [DOI] [PubMed] [Google Scholar]
- 62. Staley JT, Mandel M. 1973. Deoxyribonucleic acid base composition of Prosthecomicrobium and Ancalomicrobium strains. Int. J. Syst. Bacteriol. 23:271–273 [Google Scholar]
- 63. Stanton TB. 1989. Glucose metabolism and NADH recycling by Treponema hyodysenteriae, the agent of swine dysentery. Appl. Environ. Microbiol. 55:2365–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Stevenson BS, Eichorst SA, Wertz JT, Schmidt TM, Breznak JA. 2004. New strategies for cultivation and detection of previously uncultured microbes. Appl. Environ. Microbiol. 70:4748–4755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tholen A, Brune A. 2000. Impact of oxygen on metabolic fluxes and in situ rates of reductive acetogenesis in the hindgut of the wood-feeding termite Reticulitermes flavipes. Environ. Microbiol. 2:436–449 [DOI] [PubMed] [Google Scholar]
- 66. Vandekerckhove TTM, Coomans A, Cornelis K, Baert P, Gillis M. 2002. Use of the verrucomicrobia-specific probe EUB338-III and fluorescent in situ hybridization for detection of “Candidatus Xiphinematobacter” cells in nematode hosts. Appl. Environ. Microbiol. 68:3121–3125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wagner M, Horn M. 2006. The Planctomycetes, Verrucomicrobia, Chlamydiae and sister phyla comprise a superphylum with biotechnological and medical relevance. Cur. Opin. Biotechnol. 17:241–249 [DOI] [PubMed] [Google Scholar]
- 68. Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wertz JT, Breznak JA. 2007. Physiological ecology of Stenoxybacter acetivorans, an obligate microaerophile in termite guts. Appl. Environ. Microbiol. 73:6829–6841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wertz JT, Breznak JA. 2007. Stenoxybacter acetivorans gen. nov., sp. nov., an acetate oxidizing obligate microaerophile among diverse O2-consuming bacteria from termite guts. Appl. Environ. Microbiol. 73:6819–6828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Widdel F, Bak F. 1991. Gram-negative mesophilic sulfate-reducing bacteria, p 3352–3378 Balows A, Truper HG, Dworkin M, Harder W, Scheifer K-H. (ed), The prokaryotes, 2nd ed Springer, New York, NY [Google Scholar]
- 72. Wood H. 1985. Inorganic pyrophosphate and polyphosphates as sources of energy. Curr. Top. Cell. Regul. 26:355–369 [DOI] [PubMed] [Google Scholar]
- 73. Yamada A, Inoue T, Noda S, Hongoh Y, Ohkuma M. 2007. Evolutionary trend of phylogenetic diversity of nitrogen fixation genes in the gut community of wood-feeding termites. Mol. Ecol. 16:3768–3777 [DOI] [PubMed] [Google Scholar]
- 74. Yoon J, et al. 2008. Description of Persicirhabdus sediminis gen. nov., sp. nov., Roseibacillus ishigakijimensis gen. nov., sp. nov., Roseibacillus ponti sp. nov., Roseibacillus persicicus sp. nov., Luetolibacter pohnpeiensis gen. nov., sp. nov., and Luteolibacter algae sp. nov., six marine members of the phylum Verrucomicrobia and emended descriptions of the class Verrucomicrobiae, the order Verrucomicrobiales and the family Verrucomicrobiaceae. Int. J. Syst. Bacteriol. 58:998–1007 [DOI] [PubMed] [Google Scholar]
- 75. Yoon J, et al. 2008. Haloferula rosea gen. nov., sp. nov., Haloferula harenae, sp. nov., Haloferula phyci sp. nov., Haloferula helveola sp. nov. and Haloferula sargassicola sp. nov., five marine representatives of the family Verrucomicobiaceae within the phylum Verrucomicrobia. Int. J. Syst. Bacteriol. 58:2491–2500 [DOI] [PubMed] [Google Scholar]
- 76. Yoon J, et al. 2007. Rubritalea spongiae sp. nov., and Rubritalea tangerina sp. nov., two carotenoid- and squalene-producing marine bacteria of the family Verrucomicrobiaceae within the phylum “Verrucomicrobia,” isolated from marine animals. Int. J. Syst. Evol. Microbiol. 57:2337–2343 [DOI] [PubMed] [Google Scholar]
- 77. Yoon J, et al. 2007. Cerasicoccus arenae gen. nov., sp. nov., a carotenoid-producing marine representative of the family Puniceicoccaceae within the phylum “Verrucomicrobia,” isolated from marine sand. Int. J. Syst. Evol. Microbiol. 57:2067–2072 [DOI] [PubMed] [Google Scholar]
- 78. Yoon J, et al. 2008. Rubritalea sabuli sp. nov., a carotenoid- and squalene-producing member of the family Verrucomicrobiaceae, isolated from marine sediment. Int. J. Syst. Bacteriol. 58:992–997 [DOI] [PubMed] [Google Scholar]
- 79. Yoon J, Oku N, Matsuda S, Kasai H, Yokota A. 2007. Pelagiciccus croceus sp. nov., a novel marine member of the family Puniceicoccaceae within the phylum Verrucomicrobia isolated from seagrass. Int. J. Syst. Bacteriol. 57:2874–2880 [DOI] [PubMed] [Google Scholar]
- 80. Yoon J, et al. 2007. Coraliomargarita akajimensis gen. nov., sp. nov., a novel member of the phylum “Verrucomicrobia” isolated from seawater in Japan. Int. J. Syst. Evol. Microbiol. 57:959–963 [DOI] [PubMed] [Google Scholar]
- 81. Yoon J, et al. 2007. Pelagicoccus mobilis gen. nov., sp. nov., Pelagicoccus albus sp. nov., and Pelagicoccus litoralis sp. nov., three novel members of subdivision 4 within the phylum “Verrucomicrobia” isolated from seawater by in situ cultivation. Int. J. Syst. Evol. Microbiol. 57:1377–1385 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.