Abstract
A cellulosome-microbe complex was assembled ex vivo on the surface of Bacillus subtilis displaying a miniscaffoldin that can bind with three dockerin-containing cellulase components: the endoglucanase Cel5, the processive endoglucanase Cel9, and the cellobiohydrolase Cel48. The hydrolysis performances of the synthetic cellulosome bound to living cells, the synthetic cellulosome, a noncomplexed cellulase mixture with the same catalytic components, and a commercial fungal enzyme mixture were investigated on low-accessibility recalcitrant Avicel and high-accessibility regenerated amorphous cellulose (RAC). The cell-bound cellulosome exhibited 4.5- and 2.3-fold-higher hydrolysis ability than cell-free cellulosome on Avicel and RAC, respectively. The cellulosome-microbe synergy was not completely explained by the removal of hydrolysis products from the bulk fermentation broth by free-living cells and appeared to be due to substrate channeling of long-chain hydrolysis products assimilated by the adjacent cells located in the boundary layer. Our results implied that long-chain hydrolysis products in the boundary layer may inhibit cellulosome activity to a greater extent than the short-chain products in bulk phase. The findings that cell-bound cellulosome expedited the microbial cellulose utilization rate by 2.3- to 4.5-fold would help in the development of better consolidated bioprocessing microorganisms (e.g., B. subtilis) that can hydrolyze recalcitrant cellulose rapidly at low secretory cellulase levels.
INTRODUCTION
Biofuels and commodity chemicals produced from cellulosic biomass are of interest as sustainable substitutes for functionally similar molecules based on petroleum. The primary obstacle to biological production of such products is cost-effective technology to overcome the recalcitrance of cellulosic biomass (19, 22, 37).
Consolidated bioprocessing (CBP), in which saccharolytic enzyme production, plant cell wall solubilization, and fermentation occur in a single step, is widely seen as a promising low-cost processing route (18, 22, 24, 37). CBP microorganisms can be developed according to three strategies: (i) engineering naturally occurring cellulolytic microorganisms for improved product formation-related properties, such as Clostridium thermocellum (6), Clostridium cellulovorans (29), and Clostridium phytofermentans ISDg (30); (ii) engineering natural high-yield-product-forming microorganisms by expressing recombinant cellulases, such as Saccharomyces cerevisiae (16, 31, 34); and (iii) engineering one host with both recombinant-product-forming and cellulose-utilizing abilities, such as Escherichia coli (15) and Bacillus subtilis (2, 26, 37).
Nature has evolved two distinctive cellulase systems for degrading cellulosic material: noncomplexed cellulase mixtures and complexed cellulases, called cellulosomes. Aerobic fungi (e.g., Trichoderma reesei) usually secrete high levels (e.g., >1 to 10 g protein/liter) of several different functionally distinct cellulase components. In contrast, some anaerobic bacteria, such as C. thermocellum and C. cellulovorans, produce low levels of cellulosomes (i.e., ∼0.1 g/liter), in which many glycoside hydrolases are linked together by nonhydrolytic scaffoldins through the high-affinity interaction between cohesins in scaffoldins and enzyme-borne dockerins (3, 7–10). C. thermocellum exhibits among the highest growth rates on cellulose among described microbes (24), although it produces less cellulase per cell mass than aerobic microorganisms. This observation raises an interesting question: how anaerobic cellulolytic microorganisms can hydrolyze cellulose rapidly and effectively without the production of ample secretory cellulase, where the biosynthesis of cellulase means a large bioenergetic burden for anaerobic cellulolytic bacteria. Recently, in vitro evidence pertaining to designer cellulosomes suggests that designer cellulosomes exhibit a higher hydrolysis rate than their noncomplexed counterparts due to an enzyme proximity synergy (25, 32, 41). Zverlov et al. (43) reported that a C. thermocellum mutant featuring a completely defective scaffoldin protein exhibited a 15-fold reduction in specific cellulase activity on crystalline cellulose. Furthermore, Lu et al. (21) found that C. thermocellum along with cell-bound cellulosome exhibited ca. 2.8- to 4.7-fold-enhanced cellulose hydrolysis rates on Avicel compared to purified cellulosome in the presence of another soluble sugar-utilizing microorganism (21). Several recent studies have expressed minicellulosomes on the surfaces of microorganisms, such as B. subtilis (1, 5) and S. cerevisiae (31, 34), but did not quantitatively evaluate the enzyme-microbe synergy.
In this study, mini-CipA was displayed on the cell surface of B. subtilis through a cell wall-binding module (CBM) of a B. subtilis cell wall hydrolase, LytE. A trifunctional minicellulosome was assembled ex vivo on the cell surface of B. subtilis. The hydrolysis performances of a three-enzyme mixture, a cell-free minicellulosome, a cell-bound minicellulosome, and a commercial fungal cellulase mixture were compared on low-accessibility Avicel and high-accessibility regenerated amorphous cellulose (RAC).
MATERIALS AND METHODS
Chemicals.
All chemicals were reagent grade or higher and were purchased from Sigma (St. Louis, MO) or Fisher Scientific (Pittsburgh, PA), unless otherwise noted. Microcrystalline cellulose (Avicel PH105; 20 μm) was purchased from FMC (Philadelphia, PA). RAC was prepared from Avicel as previously described (19, 38). The oligonucleotides were synthesized by Integrated DNA Technologies (Coraville, IA). The PCR enzyme was high-fidelity Phusion DNA polymerase from New England BioLabs (Ipswich, MA). A commercial Trichoderma cellulase mixture (50013) was a gift from Novozymes North America (Franklinton, NC). The purified fungal enzymes of cellobiohydrolase I (CBH I) (Cel7A) and endoglucanase II (EG II) (Cel5) from Trichoderma spp. were purchased from Megazyme (Wicklow, Ireland).
Strains and media.
The strains and plasmids used in this study are listed in Table 1. E. coli JM109 was used as a host cell for DNA manipulation. E. coli BL21 Star (DE3) (Invitrogen, Carlsbad, CA) and B. subtilis WB600 (35) were used as the hosts for recombinant protein expression. B. subtilis was transformed through a new simple and fast transformation technology as described elsewhere (38). Luria-Bertani (LB) medium was used for E. coli cell culture and recombinant protein expression, and 2× Mal medium was used for B. subtilis recombinant protein expression (38). The final concentrations of antibiotics for E. coli were 100 mg/liter ampicillin and 25 mg/liter chloramphenicol. The chloramphenicol concentration for B. subtilis was 5 mg/liter.
Table 1.
Strains and plasmids in this study
| Strain or plasmid | Characteristics | Reference or source |
|---|---|---|
| E. coli strains | ||
| JM109 | recA1 supE44 endA1 hsdR17 (rK− mK+) gyrA96 relA1 thi Δ(lac-proAB) F′(traD36 proAB+lacIqlacZΔM15) | |
| BL21 Star (DE3) | F−ompT hsdSB (rB− mB−) gal dcm rne-131 (DE3) | Invitrogen, Carlsbad, CA |
| B. subtilis WB600 | nprE aprA epr bpf mpr nprB | 35 |
| Plasmids | ||
| pNWP43N | Cmr, pNW33N derivate | 38 |
| pNWP43N-LysM | Cmr, with LysM expression cassette cloned | This work |
| pNWP43N-LysM-mini-CipA | Cmr, with LysM-mini-CipA expression cassette cloned | This work |
| pET20b | Ampr, overexpression vector containing T7-dependent promoter | Novagen, Madison, WI |
| pET20b-mini-CipA | Ampr, with mini-CipA expression cassette cloned | This work |
| pET20b-Bscel5′ | Ampr, with Bscel5′ expression cassette cloned | This work |
| pET20b-Ctcel9 | Ampr, with Ctcel9 expression cassette cloned | This work |
| pET20b-Cpcel48 | Ampr, with Cpcel48 expression cassette cloned | This work |
Construction of plasmids.
The primers used in this study are listed in Table 2. For constructing pNWP43N-LysM, the DNA sequence encoding the B. subtilis cell wall hydrolase (LysM, GenBank accession number U38819, amino acids 25 to 230) was amplified from the genomic DNA of B. subtilis 168 by a primer pair of LysM_For and LysM_Rev_Flag; the DNA sequence encoding a vector pNWP43N was amplified from pNWP43N-BsCel5 (38) by a primer pair of pNWP43N_For and pNWP43N_Rev. The two PCR products were both digested with NheI/XhoI and then ligated, yielding pNWP43N-LysM. For constructing pNWP43N-LysM-mini-CipA (pNWP43N-LMC), the DNA sequence encoding LysM was amplified by using a primer pair of LysM_For and LysM_Rev based on the B. subtilis genomic DNA by PCR, followed by double digestion by XhoI/EcoRV. The DNA sequence encoding truncated mini-CipA (GenBank accession number L08665, amino acids 26 to 723) was amplified from the genomic DNA of C. thermocellum by a primer pair of MC_For and MC_Rev_Flag, followed by double digestion by EcoRV/NheI. The two resultant fragments were ligated into the XhoI/NheI-digested vector pNWP43N to produce pNWP43N-LMC. The DNA sequence encoding truncated mini-CipA was amplified from the genomic DNA of C. thermocellum ATCC 27405 by a primer pair of mini-CipA_For and mini-CipA_Rev. The PCR product was digested with NdeI/XhoI and then ligated into the NdeI/XhoI-digested vector pET20b (Novagen, Madison, WI), yielding pET20b-mini-CipA.
Table 2.
Primers used to amplify gene fragments
| Gene | Template | Primer name | Sequencea | Restriction enzyme site |
|---|---|---|---|---|
| LysM | Genomic DNA of B. subtilis 168 (ATCC 23857) | LysM_For | GAGCAGCTCGAGGCACAAAGCATTAAGGTGAAAAAAGG | XhoI |
| LysM_Rev_Flag | GCTGCTGCTAGCTTATTATTTGTCATCGTCATCTTTATAATCGACTAACGCTTTTGCATCAGAAACCAGCTTG | NheI | ||
| LysM-mini-CipA | Genomic DNA of B. subtilis 168 (ATCC 23857) | LysM_For | GAGCAGCTCGAGGCACAAAGCATTAAGGTGAAAAAAGG | XhoI |
| LysM_Rev | GCTGCTGATATCGACTAACGCTTTTGCATCAGAAACC | EcoRV | ||
| Genomic DNA of C. thermocellum (ATCC 27405) | MC_For | GTAAGTAGATATCGTATCGGCGGCCACAATGACAGTCG | EcoRV | |
| MC_Rev_Flag | GCAGTAGCTAGCTTATTATTTGTCATCGTCATCTTTATAATCATTCGAATCATCTGTCGGTGTTGTTACAGG | NheI | ||
| pNWP43N | pNWP43N-BsCel5 | pNWP43N_For | GCCGACGCTAGCTTAAGCTTTTTTTTGGCGGACATCAGTAAC | NheI |
| PNWP43N_Rev | GACTATCTCGAGACCTGCAGCTGAGGCATGTGTTACAAAAAC | XhoI | ||
| Mini-CipA | Genome of Clostridium thermocellum | Mini-CipA_For | GTAGTACATATGGTATCGGCGGCCACAATGACAG | NdeI |
| Mini-CipA_Rev | GCAGTACTCGAGATTCGAATCATCTGTCGGTGTTG | XhoI | ||
| BsCel5′ | Genome of Bacillus subtilis | BsCel5_For | CCTCAGCATATGGCAGGGACAAAAACGCC | NdeI |
| BsCel5′_Rev | CTCCGGTTCTTCTGGGTCTACTCCTCCAGAAATACCATTT TCCTGTGTGGGTTTATC | |||
| Genome of Clostridium thermocellum | DocK_For | GGAGGAGTAGACCCAGAAGAACCGGAGGTTATTTATG | ||
| DocK_Rev | GCCGCCCTCGAGTTTATGTGGCAATACATCTATC | XhoI | ||
| CtCel9 | Genome of Clostridium thermocellum | CtCelF_For | GCTTCACATATGGCGGATTTCAACTATGGTGAGGCAC | NdeI |
| CtCelF_Rev | GGACCATCTCGAGCTGTTCAGCCGGGAATTTTTCAATAAG | XhoI | ||
| CpCel48 | pP43N-Cpcel48 | Cpcel48_For | CCTCTGCATATGGGTGAAACTGAGCAAGC | NdeI |
| Cpcel48_Rev | GTAGAGGACCCACCTCCTCCAGATCCTGGTTCGATACCCCAATTAAGTTTTCC | |||
| Genome of Clostridium thermocellum | DocS_For | GGATCTGGAGGAGGTGGGTCCTCTACTAAATTATACGGCGACGTC | ||
| DocS_Rev | GCATTACTCGAGGTTCTTGTACGGCAATGTATC | XhoI |
Restriction enzyme sites included in primer sequences for cloning purposes are indicated in bold, the Flag tag sequences are indicated by underlining, and the overlapping sequences are indicated by italics.
pET20b-Bscel5′ was obtained by using overlap extension PCR. The DNA sequence encoding mature BsCel5 (GenBank accession number CAA82317) was amplified from genomic DNA of B. subtilis 168 by a primer pair of BsCel5_For/BsCel5′_Rev. The DNA fragment encoding a dockerin module (DocK, amino acids 821 to 895) of C. thermocellum CelK (NCBI reference sequence YP_001036843) was amplified from the genomic DNA of C. thermocellum by a primer pair of DocK_For/DocK_Rev. The two resultant fragments were assembled by using a primer pair of BsCel5_For/DocS_Rev through overlap extension PCR. These resultant fragments were cloned into NdeI/XhoI-digested pET20b, generating pET20b-Bscel5′. pET20b-Ctcel9 was obtained by using PCR amplification and overlap extension PCR. The DNA encoding the mature C. thermocellum Cel9 (GenBank accession number CAA43035) was amplified from the genomic DNA of C. thermocellum by a primer pair of CtCelF_For/CtCelF_Rev. The PCR product was digested with NdeI/XhoI and ligated into the corresponding sites of the vector pET20b, yielding pET20b-Ctcel9. The DNA sequence encoding a mature C. phytofermentans Cel48 (GenBank accession number ABX43721) was amplified from pP43N-Cpcel48 (39) by a primer pair of CpCel48_For/CpCel48_Rev. Plasmid pET20b-Cpcel48 was constructed in the same way as pET20b-Bscel5′. The dockerin of Cpcel48 was DocS of the C. thermocellum Cel48S (GenBank accession number L06942, amino acids 673 to 741). All plasmid sequences were verified by DNA sequencing. The resulting plasmids are listed in Table 1.
Production of dockerin-containing cellulases in E. coli.
The strain E. coli BL21 Star (DE3) containing the protein expression plasmid was cultivated in LB medium supplemented with 1.2% glycerol at 37°C. Protein expression and purification were conducted routinely as published elsewhere (19, 37, 38).
Removal of the B. subtilis surface proteins by LiCl.
B. subtilis cells harboring pNWP43N-LMC were precultured in LB medium at 37°C until the A600 reached about 1.2, which remained at a logarithmic growth phase. Two hundred microliters of the cell culture was inoculated into 50 ml of 2× Mal medium and then grown at 30°C until the A600 reached 3. Two milliliters of culture of the B. subtilis cells was washed two times in buffer A (50 mM HEPES buffer [pH 7.5] containing 50 mM NaCl and 10 mM CaCl2). The cell pellets were resuspended in 80 μl of buffer B (50 mM HEPES buffer [pH 7.5] containing 5 M LiCl, 50 mM NaCl, and 10 mM CaCl2). After incubation for 20 min on ice followed by centrifugation at 8,000 × g at 4°C for 10 min, a fraction of the supernatant after 10% trichloroacetic acid precipitation was loaded for SDS-PAGE. The other fraction of the supernatant was diluted 5-fold in buffer A and then mixed with 50 μg RAC. After centrifugation, the LMC adsorbed by RAC was examined by SDS-PAGE, as described elsewhere (37, 38). For validation of the formation of an ex vivo minicellulosome, the resuspended cells with bound LMC were mixed with 0.05 mg purified dockerin-tagged cellulases (BsCel5′ or cellulase mixture with equimolar BsCel5′, CtCel9, and CpCel48) at 4°C for 1 h. The cells were washed in buffer A two times. The cell-bound minicellulosome was eluted by LiCl, adsorbed by RAC, and examined by SDS-PAGE, as described above.
Confocal immunofluorescence microscopy.
B. subtilis cells (200 μl of cell culture at an A600 of 3.0) having surface-displayed LMC or cell-bound minicellulosome were washed in ice-cold phosphate-buffered saline (PBS) (8 g/liter NaCl, 0.2 g/liter KCl, 1.44 g/liter Na2HPO4, and 0.24 g/liter KH2PO4) two times and then mixed with 4% paraformaldehyde at 4°C for 30 min. After being washed in 1 ml of PBS two times, the cells were resuspended in 250 μl of PBS containing 1 mg/ml of bovine serum albumin (BSA) and 0.5 μg of monoclonal anti-Flag M2 (Sigma F1804) or monoclonal anti-His (Sigma H1029) antibody with occasional mixing for 2 h. The cells were washed in 1 ml of PBS two times, followed by resuspension in 250 μl of PBS containing 1 mg/ml BSA and 0.5 μg anti-mouse IgG conjugated with fluorescein isothiocyanate (FITC) (Sigma F9137). After incubation for 2 h, cells were washed with 1 ml of PBS two times and then resuspended in PBS to obtain a cell solution with an A600 of 1. The cells were examined with a Zeiss LSM 510 confocal laser microscope (Zeiss, Thornwood, NY).
RAC and Avicel hydrolysis.
All cellulose hydrolysis experiments were conducted in 50-ml serum bottles with a rotary shaking rate of 250 rpm at 37°C. Equimolar amounts of BsCel5′, CtCel9, and CpCel48 were premixed together, where the molecular weights of BsCel5′, CtCel9, and CepCel48 were 44,918, 80,108, and 107,184, respectively. An equimolar amount of mini-CipA was mixed with the three-cellulase mixture for the formation of a trifunctional minicellulosome. Similarly, the number of LMC molecules on the surface of B. subtilis was determined as described elsewhere (4). The cell culture containing an equimolar amount of LMC was mixed with the three-cellulase mixture for the formation of a cell-bound cellulosome. For RAC hydrolysis, the LMC-displaying B. subtilis cells collected from 33.3 ml of the cell culture (A600 = 3.0) were mixed with 0.1 mg (total) three-cellulase mixture in 10 ml of ice-cold buffer A containing 0.4% RAC, followed by hydrolysis at 37°C. The LysM-displaying B. subtilis cells collected from 33.3 ml of the cell culture (A600 = 3.0) were mixed with trifunctional minicellulosome containing 0.1 mg (total) three-cellulase mixture in 10 ml of ice-cold buffer A containing 0.4% RAC, followed by hydrolysis at 37°C. For Avicel hydrolysis, the LMC-displaying B. subtilis cells collected from 133.3 ml of the B. subtilis cell culture (A600 = 3.0) were mixed with 0.4 mg of the three-cellulase mixture in 10 ml of ice-cold buffer A containing 0.4% Avicel, followed by hydrolysis at 37°C. The LysM-displaying B. subtilis cells collected from 133.3 ml of the cell culture (A600 = 3.0) were mixed with trifunctional minicellulosome containing 0.4 mg (total) three-cellulase mixture in 10 ml of the ice-cold buffer A containing 0.4% Avicel, followed by hydrolysis at 37°C. Cellulose hydrolysis by the same amount of the LMC-displaying B. subtilis cells or LysM-displaying B. subtilis cells without heterologously added cellulase was performed as a negative control. The same cellulase (mass) concentrations of minicellulosome and the three-cellulase mixture were also used to hydrolyze RAC and Avicel in 10 ml buffer A in the presence of 60 units of β-glucosidase (Bgl) (Novozymes 188; Sigma) per gram of cellulose, respectively. A commercial Novozymes cellulase and a two-enzyme Trichoderma fungal cocktail containing EG II and CBH I at the same mass concentration as the bacterial three-cellulase mixture were used to hydrolyze RAC and Avicel at 37°C in 50 mM citrate buffer (pH 5.0) containing 50 mM NaCl and 10 mM CaCl2 in the presence of 60 units of β-glucosidase per gram of cellulose, respectively. In the two-enzyme cocktail, the mass amount of EG II was the same as the sum of BsCel5′ and CtCel9 and the mass amount of CBH I was the same as CpCel48. Cellulose hydrolysis with the same amount of β-glucosidase was performed as a negative control. One milliliter of the reaction sample was withdrawn at the indicated time intervals. The concentration of soluble sugars in the supernatant was measured by using the phenol-sulfuric acid method with glucose as the standard, while the residual cellulose was determined by quantitative saccharification with glucose as the standard (42). All hydrolysis experiments were performed in triplicate.
Other assays.
Protein mass concentration was measured by the Bio-Rad Bradford protein dye reagent method with bovine serum albumin as a reference. The protein masses, based on the Bradford method, were calibrated by UV absorbance at 280 nm in 6 M guanidine hydrochloride (38). The purity of protein samples was examined by SDS-PAGE followed by Coomassie blue staining. The activities of individual cellulases were measured as described elsewhere (19).
RESULTS
Functional display of mini-CipA on the B. subtilis cell surface.
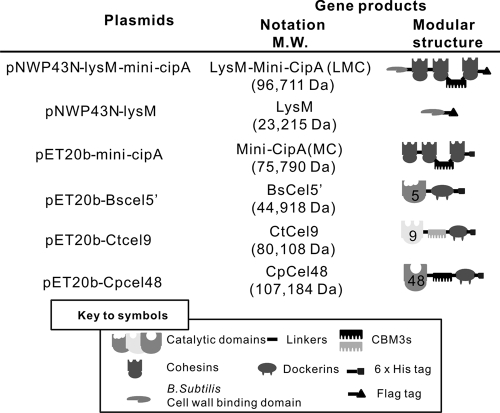
Mini-CipA, a fragment of C. thermocellum CipA containing three cohesins and one family 3b cellulose binding module (CBM3b) (10, 13), was expressed in B. subtilis using the B. subtilis-E. coli shuttle vector pNWP43N-LMC. This vector had an expression cassette containing an NprB signal peptide-encoding sequence, a B. subtilis cell wall-binding module (LysM) from the Bacillus subtilis cell wall hydrolase LytE (4, 36), a mini-CipA, and a C-terminal Flag tag, called LMC, under the control of a strong constitutive P43 promoter (Fig. 1). Because the cell wall hydrolase LytE is located at cell separation sites and poles of B. subtilis through its cell wall-binding module (LysM) (4, 36), LMC can be displayed on the cell wall of B. subtilis. Controls included plasmid pNWP43N-LysM, which expressed a surface-displayed LysM with a C-terminal Flag tag, and plasmid pNWP43N, which did not produce any related surface-displayed protein.
Fig 1.
Schematic representation of the recombinant proteins used in this study.
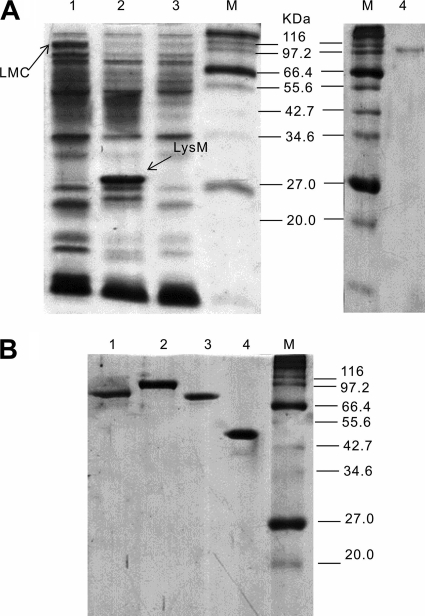
After cell cultivation, B. subtilis cells harboring pNWP43N-LMC and pNWP43N-LysM produced cell surface-bound LMC and LysM, respectively. Through LiCl elution, the cell wall protein solutions containing cell surface-displayed LMC and LysM were examined by SDS-PAGE (Fig. 2A, lanes 1 and 2). By the addition of RAC, which binds with high specificity to CBM3b-containing LMC, the LMC (Fig. 2A, lane 4) was easily separated from other cell wall proteins. The apparent molecular weights for LMC (∼105,000) and LysM (∼30,000) determined by SDS-PAGE were a little higher than their calculated values (96,711 and 23,215, respectively) based on their deduced amino acid sequences, perhaps due to the serine-rich linker sequence in LysM (20). The LMC concentration was estimated to be 1.2 mg/liter of cell culture (A600 = 3.0) based on the band intensity in the SDS-PAGE, as described elsewhere (4). Approximately 20,000 molecules of LMC were estimated to be displayed on the surface of each B. subtilis cell.
Fig 2.
SDS-PAGE of cell wall proteins eluted from the cell surfaces of B. subtilis strains (A) and the purified recombinant cellulases and mini-CipA produced by E. coli (B). (A) Lane 1, LiCl-eluted supernatant from B. subtilis(pNWP43N-LMC) cells; lane 2, LiCl-eluted supernatant from B. subtilis(pNWP43N-LysM) cells; lane 3, LiCl-eluted supernatant from B. subtilis(pNWP43N) cells; lane 4, adsorbed LMC eluted supernatant from B. subtilis(pNWP43N-LMC) cells by using RAC. (B) Lane 1, mini-CipA; lane 2, CpCel48; lane 3, CtCel9; lane 4, BsCel5′.
Expression and purification of cellulases and mini-CipA in E. coli.
Cellulases used for the assembly of the trifunctional minicellulosome were (i) a noncellulosomal B. subtilis family 5 endoglucanase (BsCel5), (ii) a cellulosomal C. thermocellum family 9 processive endoglucanase (CtCel9), and (iii) a noncellulosomal C. phytofermentans ISDg family 48 cellobiohydrolase (CpCel48) (Fig. 1). BsCel5 contains a catalytic module, a dockerin module from C. thermocellum, and a C-terminal His tag; CtCel9 contains a catalytic module, CBM3c, a dockerin module, and a C-terminal His tag; and CpCel48 contains a catalytic module, CBM3b, a dockerin module, and a C-terminal His tag. Mini-CipA, a truncated miniscaffoldin from CipA of C. thermocellum, contains three cohesins and one CBM3b. Mini-CipA and three cellulase components expressed in E. coli BL21 were purified to a homogeneous protein (Fig. 2B).
The cellulases used in this study were the same as those in our previous work, except for CpCel9 (19). Since the activities of CBM-free BsCel5 and CBM-containing CpCel48 were higher than those of CBM-containing BsCel5 and CBM-free CpCel48, respectively (data not shown), CBM-free BsCel5 and CBM-containing CpCel48 were used. In addition, it was found that CBM-containing CpCel48 was expressed at a much higher level than its CBM-free counterpart in E. coli and B. subtilis (data not shown). In this study, a family 9 cellulase (CtCel9F) from C. thermocellum was used instead of CpCel9 due to the facts that (i) CtCel9F was expressed at higher levels than CpCel9, (ii) these two enzymes exhibited comparable activities at the temperatures tested, and (iii) CtCel9F contained its own dockerin. BsCel5 had one dockerin module from one of the C. thermocellum dockerin-containing cellulases in its C terminus. CpCel48 had another dockerin module in its C terminus. All dockerin modules used had slightly different amino acid sequences because these three cellulases used in this study will be coexpressed in developing consolidated bioprocessing B. subtilis strains (19).
Ex vivo assembly of minicellulosomes on the B. subtilis cell surface.
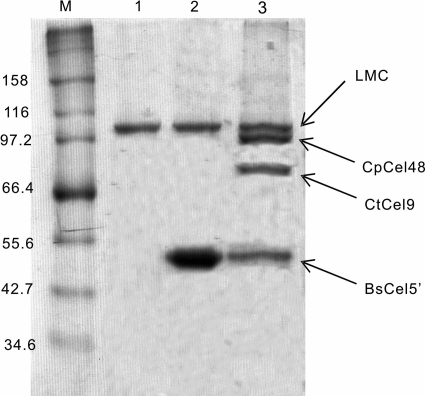
The LMC-displaying B. subtilis cells were mixed with excess Cel5′ or a three-enzyme cellulase mixture containing equimolar Cel5′, Cel48, and Cel9. After LiCl elution followed by RAC-specific adsorption, LMC-Cel5 exhibited only two bands responsible for LMC and Cel5 at an approximate molar ratio of 1:3, as examined by SDS-PAGE (Fig. 3, lane 2), indicating that one LMC molecule can bind with about three Cel5 molecules. When the cells were mixed with the three-cellulase mixture, LMC bound with the three cellulase components nearly equally (Fig. 3, lane 3), indicating that each dockerin-containing cellulase component was nonselectively bound with three cohesins of LMC. Negative-control LysM-displaying B. subtilis cells did not bind any dockerin-containing cellulase (data not shown).
Fig 3.
SDS-PAGE of RAC affinity pulldown for cell wall proteins eluted from B. subtilis strains. Lane 1, cell-bound LMC from B. subtilis(pNWP43N-LMC) cells; lane 2, cell-bound unifunctional minicellulosome from B. subtilis(pNWP43N-LMC) cells premixed with BsCel5′; lane 3, cell-bound trifunctional minicellulosome from B. subtilis(pNWP43N-LMC) cells premixed with BsCel5′, CtCel9, and CpCel48.
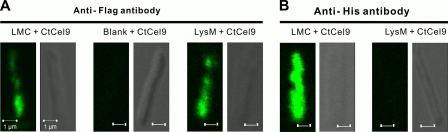
The ex vivo assembly of minicellulosomes on the B. subtilis cell surface was also examined by confocal immunofluorescence microscopy. When the primary anti-Flag antibody against the C-terminal Flag tag in LMC- or LysM-displaying cells was used, green fluorescence signals were observed on the surface of the cells displaying LMC and LysM but not on a negative control (B. subtilis WB600/pNWP43N) (Fig. 4A). These results indicated that LMC and LysM were displayed on the B. subtilis cell surface. LMC- and LysM-displaying B. subtilis cells were mixed with excess CtCel9, followed by the primary anti-His antibody that can bind with the His tag of CtCel9. LMC-displaying B. subtilis cells with CtCel9 exhibited a strong green fluorescence signal (Fig. 4B), suggesting the ex vivo formation of an LMC-CtCel9 complex. In contrast, LysM-displaying B. subtilis cells, as a negative control, did not present a detectable fluorescence signal (Fig. 4B). It was noted that the fluorescence signal for LMC-CtCel9 seen in Fig. 4B was much stronger than that seen in Fig. 4A because three anti-His antibodies can bind with three CtCel9 molecules linked by one LMC molecule, while one anti-Flag antibody can bind with one LMC molecule.
Fig 4.
Confocal fluorescence microscopy images of LMC, a negative control (blank plasmid), and LysM on the surfaces of B. subtilis cells (A) as well as the cell-bound minicellulosome on the surfaces of B. subtilis cells relative to a negative control (B). (A) Cells displaying Flag tag LMC and LysM were probed with an anti-Flag antibody followed by a rabbit anti-mouse IgG conjugated with FITC. (B) The minicellulosome containing LMC and CtCel9 was probed with an anti-His6 antibody followed by a rabbit anti-mouse IgG conjugated with FITC, where CtCel9 contains a His6 tag.
Comparative hydrolysis experiments.
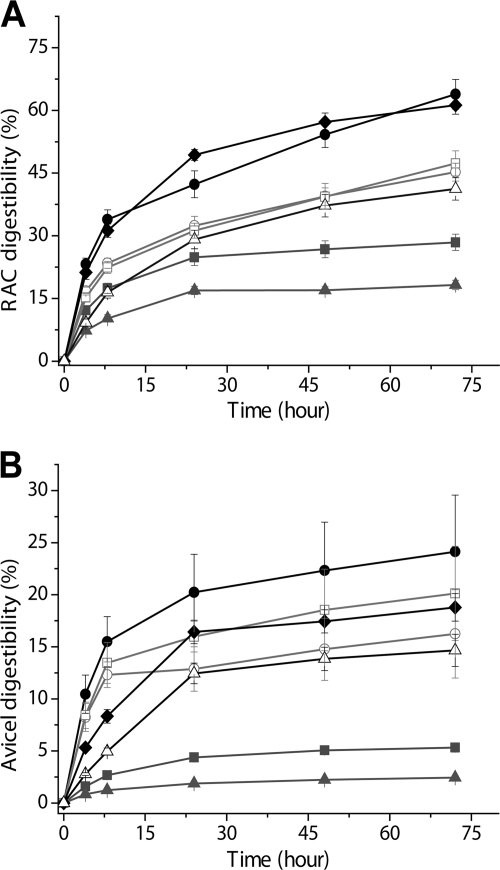
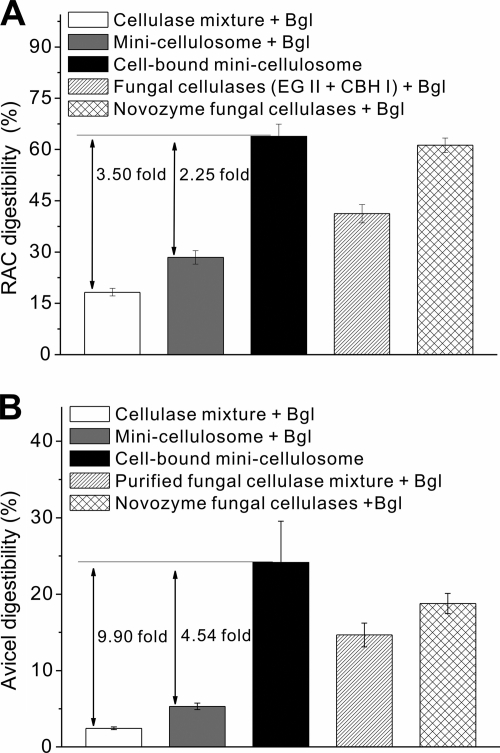
Cellulose hydrolysis activities in the presence of the same mass concentrations of cellulase were compared for the living cell-bound minicellulosome, the cell-free minicellulosome, and a (bacterial) three-cellulase mixture with a BsCel5′/CtCel9/CpCel48 molar ratio of 1:1:1 on two model cellulosic materials, RAC and Avicel. Since the minicellulosome can tightly bind on cellulose, a cellulose-enzyme-microbe (CEM) complex was formed. The cell-bound minicellulosome hydrolyzed RAC more rapidly than the cell-free minicellulosome and the three-enzyme mixture (Fig. 5A). At 72 h, a digestibility of 28.4% was achieved by the minicellulosome, which was about 1.57-fold higher than that of the three-cellulase mixture. This phenomenon was attributed to the enzyme proximity effect (25, 32, 41). More notable, the cell-bound minicellulosome hydrolyzed RAC at a 2.25-fold-higher digestibility than the minicellulosome (Fig. 5A). A similar hydrolysis trend of an increasing order of the cellulase mixture, cellulosome, and cell-bound minicellulosome was observed on Avicel (Fig. 5B). The cell-bound minicellulosome exhibited 4.54-fold higher digestibility in Avicel than did the minicellulosome (Fig. 5B). A comparison of the CEM synergy (Fig. 6) indicated that the cellulosome-microbe complex increased the cellulose hydrolysis rate more significantly on recalcitrant Avicel than on RAC.
Fig 5.
Hydrolysis of RAC (A) and Avicel (B) by enzyme mixtures supplemented with excess β-glucosidase: the bacterial cellulase mixture (▴), the minicellulosome (■), the Novozymes fungal cellulase mixture (♦), the two-enzyme Trichoderma fungal mixture (EG II and CBH I) (△), the cell-bound minicellulosome (●), the minicellulosome in the presence of LysM-displaying B. subtilis cells (□), and the cell-bound minicellulosome in the presence of 1 g/liter NaN3 (○).
Fig 6.
Comparison of digestibilities of cellulose by the bacterial cellulase mixture, the cell-free minicellulosome, the cell-bound minicellulosome, the commercial fungal cellulase mixture, and the cocktail of two fungal enzymes on RAC (A) and Avicel (B) at 72 h. The error bars represent the standard deviations from triplicate samples.
To understand why the CEM complex hydrolyzed cellulose more rapidly than the minicellulosome, two control experiments were conducted: in the first, minicellulosome-displaying B. subtilis cells were made nonactive by the addition of 1 g/liter NaN3 to inhibit the sugar uptake ability of the cells; in the second, the minicellulosome with active LysM-displaying B. subtilis cells was able to assimilate all soluble sugars in the bulk phase. The nonactive cells associated with the cell-bound cellulosome did not hydrolyze cellulose as rapidly as active cellulosome-bound cells (Fig. 5) possibly due to accumulated sugars in the supernatant, which inhibited minicellulosome activity. The minicellulosome plus active LysM-displaying B. subtilis cells, where no significant soluble sugars were accumulated in the supernatant (data not shown), exhibited less hydrolysis ability than the active cellulosome-bound cells (Fig. 5).
The hydrolysis performances of the bacterial cellulase systems were compared to those of a commercial fungal cellulase mixture and a two-enzyme cocktail made of purified Trichoderma CBH I and EG II at the same protein mass concentration. The cocktail of two fungal enzymes hydrolyzed cellulosic materials more efficiently than the cocktail of three bacterial cellulases and the trifunctional minicellulosome at 72 h, although each bacterial cellulase component exhibited a much higher specific activity during short reaction time frames (e.g., 10 min to 1 h) (data not shown). The commercial fungal mixture worked better than the mixture of two fungal cellulases, possibly due to its optimized enzyme ratio. Although the noncomplexed mixture of three bacterial cellulases or the bacterial minicellulosome exhibited less ability to hydrolyze solid cellulosic materials than the commercial fungal cellulase, the cell-bound cellulosome showed equal hydrolytic ability on RAC and approximately 30% higher hydrolytic ability on Avicel (Fig. 6).
DISCUSSION
We assembled an ex vivo trifunctional minicellulosome on the surface of the Gram-positive B. subtilis through high-affinity interaction between the dockerin modules of cellulase components and the three cohesin modules of mini-CipA. This enabled the comparison of the rates of cellulose hydrolysis caused by the cellulose-enzyme-microbe (CEM) complex and by the noncomplexed cellulase mixture or cellulosome (Fig. 5). The CEM synergy was not primarily due to removal of hydrolysis products from the bulk fermentation broth, as suggested by control experiments (Fig. 5). For enzymatic hydrolysis occurring on the surface of a solid cellulosic substrate, the concentration of hydrolysis products in the boundary layer was thought to be much higher than that in bulk phase, according to the boundary layer theory (11). Such high-concentration hydrolysis products, especially for long-chain cellodextrins, in the boundary layer were expected to inhibit cellulase activity more strongly than glucose and cellobiose in the bulk phase because beta-glucosidase that does not have a CBM usually works in the bulk phase. Because the distance between the cell and minicellulosome through an LMC (i.e., 20 to 50 nm) is much shorter than the thickness of the boundary layer on the solid substrate cellulose for cellulolytic microorganisms (e.g., 10 to 100 μm) (33), the adjacent cells located in the boundary layer can assimilate long-chain hydrolysis products before their diffusion to the bulk phase so as to effectively eliminate product inhibition to cellulases and cellulosomes (41). This explanation was partially supported by the observance of some polycellulosomal protuberance between cellulose and C. thermocellum cell under a transmission electron microscope (27) and by the fast assimilation of long-chain cellodextrins by adjacent cellulolytic cells rather than further hydrolysis to cellobiose and glucose by cellulases in the bulk phase (41, 42).
The CEM synergy was more significant on the recalcitrant Avicel than on the highly reactive amorphous cellulose (Fig. 6). This difference may be explained by stronger boundary layer product inhibition on crystalline cellulose than on amorphous cellulose. Because cellobiohydrolase is more sensitive to product inhibition than endoglucanase (i.e., KI,CBH ≪ KI,EG) and endoglucanase exhibits more hydrolysis ability on amorphous cellulose than on Avicel (19), the aggregated cellulosome exhibited less product inhibition on amorphous cellulose than on recalcitrant Avicel (17). Displaying the cellulosome on the surface of a microorganism would be effective in enhancing the cellulolytic host's ability to effectively hydrolyze a recalcitrant cellulosic fragment of pretreated heterologous biomass.
Both B. subtilis and S. cerevisiae are important industrial microorganisms. As a potential CBP host, B. subtilis could be better than S. cerevisiae due to (i) a natural ability to take up long-chain cellodextrins, (ii) a natural ability to coutilize C5 and C6 sugars, (iii) an inherent ability to secrete a large amount of proteins, and (iv) a small cell (0.7 by 2 μm) versus a large cell size for yeast (2.5 to 10 by 4.5 to 21 μm) (i.e., a better mass transfer for a smaller cell). The first two features have been introduced into recombinant yeasts (12, 28). In spite of intensive efforts, recombinant cellulose-utilizing yeasts that can produce ample cellulase and hydrolyze cellulose to support cell growth and cellulase synthesis without the help of other soluble organic nutrients are not yet available (18). In contrast, a recombinant cellulose-utilizing B. subtilis strain has been created to produce lactate from cellulose without the addition of exogenous cellulase or any water-soluble organic nutrients (37). Since anaerobic cellulolytic microorganisms must produce more secretory cellulase than do their aerobic counterparts based on the weight ratio of cellulase to cellular protein for supporting their growth on cellulose (23), cellulase synthesis always represents a significant bioenergetic burden for anaerobic microorganisms (42). The bacterium C. thermocellum, for example, produces ∼10 to 20% (wt/wt) cellulase relative to cellular proteins for fast cellulose hydrolysis, with nearly all of the cellulosome displayed on its cell surface (40). It appears that cellulolytic, anaerobic bacteria evolved cell-bound cellulosomes so to increase specific cellulase activity and to decrease their bioenergetic burden (42). However, this cellulase evolution mechanism is speculated not to occur in fungi and yeasts because (i) the ATP supply is much more plentiful and (ii) relatively large cellulolytic fungi and yeasts may not have enough cell surface to display 10% to 20% (wt/wt) cellulase relative to cellular protein due to low surface/volume ratios, where the surface/volume ratio is inversely proportion to the radius of a cell. Therefore, it is hypothesized that in nature cellulolytic fungi evolved to secrete a large amount of cellulases.
For high-yield biofuel production from cellulosic material, it is vital to increase the carbohydrate allocation of the desired biofuels by decreasing the carbohydrate allocation to synthesis of cellulase and cell mass under anaerobic conditions (14, 22). This study showed that displaying a cellulosome on the surface of a microbe can enhance the microbial cellulose hydrolysis rate by severalfold without increasing the cellulase synthesis burden. Since fungal cellulases exhibited higher hydrolysis ability over a long time range (19), the coexpression of dockerin-containing fungal cellulases by recombinant cellulolytic B. subtilis strains may be another worthy direction for study. Another potential direction would be in vitro assembly of dockerin-containing fungal cellulases produced by Trichoderma spp. and a recombinant yeast or bacterium that can produce a cell surface-displayed scaffoldin.
ACKNOWLEDGMENTS
This work was supported mainly by the DOE BioEnergy Science Center. The BioEnergy Science Center is a U.S. Department of Energy Bioenergy Research Center supported by the Office of Biological and Environmental Research in the DOE Office of Science. This work was also partially supported by the CALS Biodesign and Bioprocessing Research Center and the Integrated Internal Competitive Grants Program at Virginia Tech.
Footnotes
Published ahead of print 30 December 2011
REFERENCES
- 1. Anderson TD, et al. 2011. Assembly of minicellulosomes on the surface of Bacillus subtilis. Appl. Environ. Microbiol. 77:4849–4858 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 2. Arai T, et al. 2007. Synthesis of Clostridium cellulovorans minicellulosomes by intercellular complementation. Proc. Natl. Acad. Sci. U. S. A. 104:1456–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bayer EA, Belaich JP, Shoham Y, Lamed R. 2004. The cellulosomes: multienzyme machines for degradation of plant cell wall polysaccharides. Annu. Rev. Microbiol. 58:521–554 [DOI] [PubMed] [Google Scholar]
- 4. Chen C-L, et al. 2008. Development of a LytE-based high-density surface display system in Bacillus subtilis. Microb. Biotechnol. 1:177–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cho H-Y, Yukawa H, Inui M, Doi RH, Wong S-L. 2004. Production of minicellulosomes from Clostridium cellulovorans in Bacillus subtilis WB800. Appl. Environ. Microbiol. 70:5704–5707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Demain AL, Newcomb M, Wu JHD. 2005. Cellulase, clostridia, and ethanol. Microbiol. Mol. Biol. Rev. 69:124–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ding SY, et al. 2008. A biophysical perspective on the cellulosome: new opportunities for biomass conversion. Curr. Opin. Biotechnol. 19:218–227 [DOI] [PubMed] [Google Scholar]
- 8. Doi RH. 2008. Cellulases of mesophilic microorganisms: cellulosome and noncellulosome producers. Ann. N. Y. Acad. Sci. 1125:267–279 [DOI] [PubMed] [Google Scholar]
- 9. Doi RH, Goldstein M, Hashida S, Park JS, Takagi M. 1994. The Clostridium cellulovorans cellulosome. Crit. Rev. Microbiol. 20:87–93 [DOI] [PubMed] [Google Scholar]
- 10. Doi RH, Kosugi A. 2004. Cellulosomes: plant-cell-wall-degrading enzyme complexes. Nat. Rev. Microbiol. 2:541–551 [DOI] [PubMed] [Google Scholar]
- 11. Fogler HS. 1999. Elements of chemical reaction engineering. Prentice Hall PTR, Upper Saddle River, NJ [Google Scholar]
- 12. Galazka JM, et al. 2010. Cellodextrin transport in yeast for improved biofuel production. Science 330:84–86 [DOI] [PubMed] [Google Scholar]
- 13. Gerngross UT, Romaniec MP, Kobayashi T, Huskisson NS, Demain AL. 1993. Sequencing of a Clostridium thermocellum gene (cipA) encoding the cellulosomal SL-protein reveals an unusual degree of internal homology. Mol. Microbiol. 8:325–334 [DOI] [PubMed] [Google Scholar]
- 14. Huang WD, Zhang Y-HP. 2011. Analysis of biofuels production from sugar based on three criteria: thermodynamics, bioenergetics, and product separation. Energy Environ. Sci. 4:784–792 [Google Scholar]
- 15. Ingram LO, et al. 1998. Metabolic engineering of bacteria for ethanol production. Biotechnol. Bioeng. 58:204–214 [DOI] [PubMed] [Google Scholar]
- 16. Ito J, et al. 2009. Regulation of the display ratio of enzymes on the Saccharomyces cerevisiae cell surface by the immunoglobulin G and cellulosomal enzyme binding domains. Appl. Environ. Microbiol. 75:4149–4154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Johnson EA, Reese ET, Demain AL. 1982. Inhibition of Clostridium thermocellum cellulase by end products of cellulolysis. J. Appl. Biochem. 4:64–71 [Google Scholar]
- 18. la Grange D, den Haan R, van Zyl W. 2010. Engineering cellulolytic ability into bioprocessing organisms. Appl. Microbiol. Biotechnol. 87:1195–1208 [DOI] [PubMed] [Google Scholar]
- 19. Liao HH, Zhang XZ, Rollin JA, Zhang Y-HP. 2011. A minimal set of bacterial cellulases for consolidated bioprocessing of lignocellulose. Biotechnol. J. 6:1409–1418 [DOI] [PubMed] [Google Scholar]
- 20. Linding R, et al. 2003. Protein disorder prediction: implications for structural proteomics. Structure 11:1453–1459 [DOI] [PubMed] [Google Scholar]
- 21. Lu Y, Zhang Y-HP, Lynd LR. 2006. Enzyme-microbe synergy during cellulose hydrolysis by Clostridium thermocellum. Proc. Natl. Acad. Sci. U. S. A. 103:16165–16169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lynd LR, et al. 2008. How biotech can transform biofuels. Nat. Biotechnol. 26:169–172 [DOI] [PubMed] [Google Scholar]
- 23. Lynd LR, van Zyl WH, McBride JE, Laser M. 2005. Consolidated bioprocessing of cellulosic biomass: an update. Curr. Opin. Biotechnol. 16:577–583 [DOI] [PubMed] [Google Scholar]
- 24. Lynd LR, Weimer PJ, van Zyl WH, Pretorius IS. 2002. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 66:506–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Moraïs S, et al. 2010. Cellulase-xylanase synergy in designer cellulosomes for enhanced degradation of a complex cellulosic substrate. mBio 1:e00285–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Romero-Garcia S, Hernandez-Bustos C, Merino E, Gosset G, Martinez A. 2009. Homolactic fermentation from glucose and cellobiose using Bacillus subtilis. Microb. Cell Fact. 8:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shoham Y, Lamed R, Bayer EA. 1999. The cellulosome concept as an efficient microbial strategy for the degradation of insoluble polysaccharides. Trends Microbiol. 7:275–281 [DOI] [PubMed] [Google Scholar]
- 28. Stevis PE, Ho NWY. 1989. Construction of yeast xylulokinase mutant by recombinant DNA techniques. Appl. Biochem. Biotechnol. 20:327–334 [Google Scholar]
- 29. Tamaru Y, et al. 2011. Comparison of the mesophilic cellulosome-producing Clostridium cellulovorans genome with other cellulosome-related clostridial genomes. Microb. Biotechnol. 4:64–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tolonen AC, et al. 2011. Proteome-wide systems analysis of a cellulosic biofuel-producing microbe. Mol. Syst. Biol. 7:461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tsai SL, Oh J, Singh S, Chen R, Chen W. 2009. Functional assembly of minicellulosomes on the Saccharomyces cerevisiae cell surface for cellulose hydrolysis and ethanol production. Appl. Environ. Microbiol. 75:6087–6093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vazana Y, Morais S, Barak Y, Lamed R, Bayer EA. 2010. Interplay between Clostridium thermocellum family 48 and family 9 cellulases in cellulosomal versus noncellulosomal states. Appl. Environ. Microbiol. 76:3236–3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang Z-W, Hamilton-Brehm SD, Lochner A, Elkins JG, Morrell-Falvey JL. 2011. Mathematical modeling of hydrolysate diffusion and utilization in cellulolytic biofilms of the extreme thermophile Caldicellulosiruptor obsidiansis. Biores. Technol. 102:3155–3162 [DOI] [PubMed] [Google Scholar]
- 34. Wen F, Sun J, Zhao H. 2010. Yeast surface display of trifunctional minicellulosomes for simultaneous saccharification and fermentation of cellulose to ethanol. Appl. Environ. Microbiol. 76:1251–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wu XC, Lee W, Tran L, Wong SL. 1991. Engineering a Bacillus subtilis expression-secretion system with a strain deficient in six extracellular proteases. J. Bacteriol. 173:4952–4958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yamamoto H, S-i, Kurosawa Sekiguchi J. 2003. Localization of the vegetative cell wall hydrolases LytC, LytE, and LytF on the Bacillus subtilis cell surface and stability of these enzymes to cell wall-bound or extracellular proteases. J. Bacteriol. 185:6666–6677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang X-Z, Sathitsuksanoh N, Zhu Z, Zhang Y-HP. 2011. One-step production of lactate from cellulose as sole carbon source without any other organic nutrient by recombinant cellulolytic Bacillus subtilis. Metab. Eng. 13:364–372 [DOI] [PubMed] [Google Scholar]
- 38. Zhang X-Z, Zhang Y-HP. 2011. Simple, fast and high-efficiency transformation system for directed evolution of cellulase in Bacillus subtilis. Microb. Biotechnol. 4:98–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang XZ, et al. 2010. The noncellulosomal family 48 cellobiohydrolase from Clostridium phytofermentans ISDg: heterologous expression, characterization, and processivity. Appl. Microbiol. Biotechnol. 86:525–533 [DOI] [PubMed] [Google Scholar]
- 40. Zhang Y-H, Lynd LR. 2003. Quantification of cell and cellulase mass concentrations during anaerobic cellulose fermentation: development of an ELISA-based method with application to Clostridium thermocellum batch cultures. Anal. Chem. 75:219–227 [DOI] [PubMed] [Google Scholar]
- 41. Zhang Y-HP. 2011. Substrate channeling and enzyme complexes for biotechnological applications. Biotechnol. Adv. 29:715–725 [DOI] [PubMed] [Google Scholar]
- 42. Zhang Y-HP, Lynd LR. 2005. Cellulose utilization by Clostridium thermocellum: bioenergetics and hydrolysis product assimilation. Proc. Natl. Acad. Sci. U. S. A. 102:7321–7325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zverlov VV, Klupp M, Krauss J, Schwarz WH. 2008. Mutations in the scaffoldin gene, cipA, of Clostridium thermocellum with impaired cellulosome formation and cellulose hydrolysis: insertions of a new transposable element, IS1447, and implications for cellulase synergism on crystalline cellulose. J. Bacteriol. 190:4321–4327 [DOI] [PMC free article] [PubMed] [Google Scholar]