Abstract
Our understanding of microbial ecology has been significantly furthered in recent years by advances in sequencing techniques, but comprehensive surveys of the phenotypic characteristics of environmental bacteria remain rare. Such phenotypic data are crucial for understanding the microbial strategies for growth and the diversity of microbial ecosystems. Here, we describe a high-throughput measurement of the growth of thousands of bacterial colonies using an array of flat-bed scanners coupled with automated image analysis. We used this system to investigate the growth properties of members of a microbial community from untreated soil. The system provides high-quality measurements of the number of CFU, colony growth rates, and appearance times, allowing us to directly study the distribution of these properties in mixed environmental samples. We find that soil bacteria display a wide range of growth strategies which can be grouped into several clusters that cannot be reduced to any of the classical dichotomous divisions of soil bacteria, e.g., into copiotophs and oligotrophs. We also find that, at early times, cells are most likely to form colonies when other, nearby colonies are present but not too dense. This maximization of culturability at intermediate plating densities suggests that the previously observed tendency for high density to lead to fewer colonies is partly offset by the induction of colony formation caused by interactions between microbes. These results suggest new types of growth classification of soil bacteria and potential effects of species interactions on colony growth.
INTRODUCTION
The microbial diversity in natural environments is truly astonishing, as evidenced by the thousands of distinct species inhabiting a single gram of soil (4, 17, 25). Underlying this microbial cornucopia is an impressive range of ecological strategies for survival and proliferation. The traditional species-by-species approach to understanding the ecology of bacteria is clearly impracticable when it comes to capturing community-wide features. Metagenomic approaches based on analyzing microbial DNA from the environment have enabled important discoveries (10, 52, 62, 63) but cannot provide the full picture of ecological function. Both methods thus leave gaps in our understanding of basic ecological questions about the growth and survival of bacteria in the environment. In particular, how diverse are the growth strategies adopted by individual bacterial strains, and can we discern trade-offs or distinct classes of strains? In what ways does the presence of neighbors influence the formation and growth of colonies at the community level?
The classical approach to studying the growth strategies of environmental bacteria, stretching back almost a century (68), divides soil bacteria into two broad classes based on their adaption to specific environmental circumstance, and a number of partly overlapping divisions have been suggested (60). Winogradsky's original division into zymogous and autochthonous species is based on a succession of degradation processes in soil (34, 60), while the most commonly used division today, copiotrophs versus oligotrophs, is based on the preferred nutrient density (32). A third possible classification is the classical ecological distinction between r-selected and K-selected organisms (1, 14, 46). Yet despite this richness of broad ecological concepts to test, comparative empirical studies of bacterial growth have been rare (9, 14, 23).
Any study of growth phenotypes must contend with the issue of unculturability, the disparity between the number of prokaryotic species present in an environmental sample and the much smaller number that will typically grow in the laboratory (“the great plate count anomaly”) (57, 61). This imposes some limitations but simultaneously underscores the importance of studying growth and of understanding the factors that decide which bacteria will grow in the laboratory. Recent studies have made it clear that the failure to grow is a complex ecological response that depends crucially on the environment of the cell (18, 28): changing the environment can increase the fraction of growing cells to 20 to 35% in some samples (6, 41), far more than the 1% typically considered cultivable (61). Methods for improving culturability include using low-nutrient media (29, 51), culturing in the natural environment or on media embedded therein (3, 31, 41, 54, 71), culturing at different concentrations of O2 and CO2 (58), and using long incubation times (7, 29). Two specific cultivation-boosting measures—plating at a low cell density (6) and adding chemical growth stimulants (11, 42)—are particularly intriguing because they suggest that intermicrobial interactions play a role. A comprehensive understanding of the growth and survival of bacteria in the environment must therefore encompass both the growth strategies of individual bacteria (8, 23) and the way growth is modulated by other species in the community (5, 11, 64, 70).
In this study, we used a new methodology to address these questions. Using an automated system of scanners and image analysis capable of simultaneously monitoring thousands of bacterial colonies for several weeks, we performed a large-scale study of the diversity of growth patterns of bacteria from a natural soil community. This method, previously applied to single species (21, 35, 38, 49, 64), significantly increases the number of colonies that can be followed compared to manual tracking (8, 23) or tracking using microscopy (12, 40), allowing a broader view of the microbial community. To capitalize on the strengths of this approach, we focus on exploring how it can shed light on microbial diversity directly at the community level without working explicitly with individual strains.
MATERIALS AND METHODS
Soil samples and preparation of soil stock.
Soil samples were collected from a garden in Somerville, MA, 3 July 2008 (42°23′30.58″N, 71°6′33.23″W). The uppermost 6 cm of the topsoil was removed and soil plugs sampled using sterile 50-ml tubes. In the laboratory, stones and roots were removed, and 100 g of soil was suspended in 400 ml distilled water with 50 mM tetrasodium pyrophosphate (TTSP) and 0.05% Tween 80 added to dislodge and disperse the bacterial cells (2). The soil suspension was then blended vigorously three times for 30 s in a commercial blender (with 60-s pauses on ice) to improve the recovery of cells (37). The stock solution was then filtered through a coarse, sterilized filter (VWR grade 417; 40-μm particle retention) to remove large soil particles. Finally, glycerol was added (20% final concentration) and aliquots stored at −80°C. This protocol was shown by preliminary experiments to give a high fraction of growing colonies while maintaining a highly uniform colony distribution on plates. The improved recovery outweighed the stressful nature of the TTSP-Tween mixture for some bacteria (the pH of 9 to 9.5 corresponds to strongly alkaline soil), though the overall colony count was likely lowered due to the filtering step. Cells in multicell aggregates (29) may occur but are unlikely to have a major effect on the overall set of macroscopic colonies (see further discussion below).
Growth medium.
All strains were grown on dilute nutrient broth (DNB) agar (0.08 g nutrient broth powder [Difco] and 15 g of highly purified agar [Fluka impurity-free agar] per liter). This low-nutrient medium allows cultivation of a wide range of soil bacteria (both hard-to-culture and well-characterized groups) (29) but also supports the growth of typically copiotrophic groups such as Gammaproteobacteria (29), in particular Escherichia coli (as we confirmed). Finally, 3 ml black India ink (black India waterproof ink; Sanford) and 50 mg cycloheximide (Sigma-Aldrich) were added to each liter of medium to improve the optical contrast and suppress fungi, respectively.
Preparation and plating of samples.
Two main experiments were performed. For the first experiment, 30 replica petri dishes (25 ml agar per dish) were inoculated with a 1:128 dilution (in phosphate-buffered saline) of the thawed stock; this yielded approximately 15 colonies per plate, allowing precise growth tracking and minimizing potential colony interactions. For the second experiment, dilution stocks were prepared by 11 steps of dilution by a factor of 2, giving 12 solutions ranging from pure thawed stock to a 1:2,048 dilution. For each solution, 5 replica dishes were prepared. We used low-profile petri dishes (VWR Space Saver petri dishes [height, 10 mm; diameter, 100 mm]) to bring the agar surface closer to the scanner. In both experiments, 100 μl of the appropriate solution was inoculated on each dish and spread with 10 to 15 sterile glass beads using vigorous shaking (this gives a more even distribution of colonies than spreading with bent glass or plastic sticks).
Scanner system and scanning protocol.
The colonies were imaged with a modified version of the scanner array described previously (38, 64) (Fig. 1). The system consists of an array of standard flat-bed scanners controlled by an automated script program (AutoIt scripting system) running on a connected PC. The array is housed in a climate-controlled room maintained at 30°C and 70% humidity. The experiments made use of 15 scanners (Epson model 3170) with the glass window lowered to put the agar surface in focus. The color scan images had a resolution of 1,200 dpi (10,200 by 14,040 pixels). Computer-controlled power switches kept all scanners turned off when not in use, and fans circulated air throughout to minimize the effects of generated heat. Since the intensity and frequency of the resulting temperature changes were the same for all scanners, they do not introduce any systematic bias, and we found no correlation between the properties of a colony and the scanner it grew on. At the start of each experiment, all scanners were cleaned with a 70% ethanol solution and loaded with six petri dishes placed upside down in a paper holder to prevent motion and block background light. Replica plates were spread among scanners to avoid systematic effects. Each dish was imaged roughly once every 3.5 h. Each data scan was preceded by several fast (low-resolution) scans to remove any condensation by slightly heating the glass.
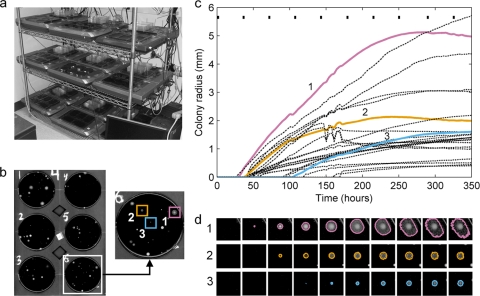
Fig 1.
The scanner system and image analysis process. (a) Scanner array kept in a climate-controlled room. (b) Single scanner image showing six petri dishes with black DNB agar and a zoom of a single dish with three example colonies marked. (c) Graph of the growth of all identified colonies on the example plate from panel b (radius versus time, assuming circular colonies). The three highlighted colonies are shown with solid, colored lines; the rest are shown with black dot-dashed lines. (d) Growth of the three highlighted colonies imaged at 10 different time points (indicated by small black bars at the top of the graph in panel c) with the detected colony outlines shown. For the sake of visual clarity, the contrast has been enhanced in the scan images in panels b and d.
Image analysis.
Images were analyzed with a suite of in-house image analysis programs written in MATLAB (MathWorks) and optimized to ensure consistent tracking of a wide range of bacterial colonies. For each image, possible colonies were automatically identified by conversion to gray scale, background subtraction, and thresholding. A fixed intensity threshold was used (instead of adaptive methods such as Otsu's [19]) to ensure consistent thresholding across all scans. Outlines of a given colony on consecutive scans were automatically matched, and growing colonies were traced backwards to their first occurrence. Hence, even very small colonies could be distinguished from agar defects, down to a limit of just a few pixels set by the slight blur introduced by smoothing. Finally, a semiautomatic program was used to remove all fungal growth and remaining artifacts (e.g., new disconnected parts of existing colonies). The final data consisted of the outline and position of each growing colony on every scan where it appeared. All computer codes are available upon request.
Extracting growth characteristics.
Details of the colony growth were extracted from the 30 dishes in the fixed-dilution experiment. For each colony outline, the colony radius was found (assuming circular colonies, an excellent approximation for early growth in almost all cases) and converted to millimeters using the known diameter of the plate as a reference. The first 15 nonzero radius values were then fitted to the empirical radius formula R(t) = A(t − t0)H for t > t0 and R(t) = 0 for t ≤ t0 (Fig. 2). This formula includes the two standard growth modes as special cases (47): linear radius growth (H0 = 1) and linear area growth [H0 = 1/2 so that area = πR(t)2 = constant × (t − t0)]. Note that t0 is not the time of first detection of the colony but rather an estimate of the actual lag time of the founding cell. Each fit was manually inspected, and cases of inconsistent growth were discarded. To remove colonies that did not arise from cells in the original sample (e.g., colonies formed by dispersed spores from colonies of Streptomyces [15]), colonies appearing later than 200 h into the experiment were also discarded. The final data set consisted of 472 high-quality growth curves. The colony growth rate was measured by the quantity g, the rate of increase in the colony radius at a radius of 10 pixels (0.24 mm) evaluated using the fitted curve (Fig. 2b).
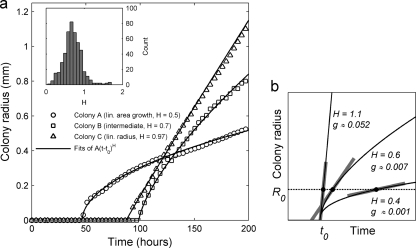
Fig 2.
Examples of different colony growth kinetics. (a) The early radial growth of three colonies from the same petri dish (dilution of 1:128). The curves are fits to the three-parameter formula R(t) = A(t − t0)H for t > t0 and R(t) = 0 for t ≤ t0; to focus on the early stage of growth, only the first 15 nonzero radius values were used for fitting. The three colonies show examples of area growing nearly linearly with time (H = 0.5), radius growing linearly with time (H ≈ 1), and an in-between case (H = 0.7). The inset shows the distribution of fitted values of H for all high-quality growth curves (n = 472 colonies). (b) Schematic illustration of the kinetic parameters (t0, H, and g) extracted from the fits. The three solid curves illustrate different values of the exponent H for the same lag time, t0, and the corresponding values of the colony growth rate g (the value of the parameter A is the same in all cases). The colony growth rate g (in mm/h) is given by the slope of the curve at a fixed colony radius R0 (set to 0.24 mm in our case), as indicated by the thick gray line segments.
Assessing phenotypic variation among clonal colonies.
Colony material was sampled from randomly chosen, well-separated colonies grown from soil stock on DNB agar. Each individual sample was suspended in water with 20% glycerol, aliquoted, and stored at −80°C. Samples were then thawed, plated separately at densities comparable to those used in the growth characteristics experiment (estimated using preliminary plate counts), and monitored by the scanner array. Each dish was analyzed to extract growth parameters for colonies, as described above.
Microscopic counts of cells in soil stock.
An 800-μl volume of thawed soil stock was centrifuged at 3,000 × g for 10 min at 4°C, 700 μl of the supernatant was removed, and the pellet was resuspended in the remaining 100 μl. The sample was stained with SYTO 9 dye (Invitrogen) for 20 min (0.025 mM final concentration) and cells counted on a microscope (20 independent fields of view) at a magnification of ×40 in a Petroff-Hausser counting chamber (Electron Microscopy Sciences) using appropriate fluorescence filters.
RESULTS
We performed two separate main experiments using the scanner array (Fig. 1), one focused on understanding the growth of individual colonies and one focused on the effect of cell density on the formation of colonies.
Growth kinetics of individual colonies.
For the experiment concentrating on individual colonies, we used the early parts of the growth curves to extract three descriptive parameters for each colony: the estimated lag time t0 (time when the first cell started multiplying), the radial growth rate g (measured at the point where the colony radius is 0.24 mm), and the exponent H, which indicates whether the radial growth rate is increasing (H > 1) or decreasing (H < 1) with time (Fig. 2) (see Materials and Methods).
The distribution of H shows a single peak centered at 0.5 (corresponding to the colony area growing linearly with time), but many colonies fall well below or above this value, including a number with H values of above 1 (accelerating radial growth) (Fig. 2a). The growth exponent H is uncorrelated with the fixed-radius colony growth rate g but is negatively correlated with lag time (P = 0.47 and P < 10−5, respectively, by Student's t test).
Distribution of colony growth rates and lag times.
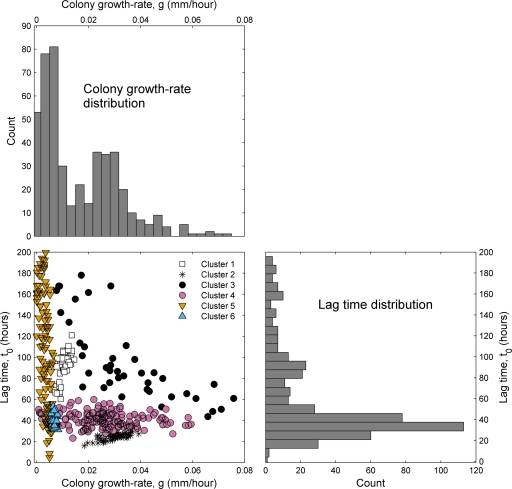
The data for the radial growth rate (g) and lag times (t0) are summarized in Fig. 3. Colony growth rate and lag time are negatively correlated (P = 2 × 10−6 by Student's t test); i.e., fast-growing colonies tend to initiate growth quickly, as previously observed (see, e.g., reference 26). The separate distributions of colony growth rate and lag time (bar plots in Fig. 3) both appear to be bimodal, with a dominant peak and a smaller secondary peak at higher values. While this may suggest the presence of two dominant groups (early/fast and late/slow), the joint scatter plot of these two variables reveals a much more complex pattern, with colonies appearing to fall into a range of different groups (Fig. 3, lower left panel). Using a flexible clustering method (Gaussian mixture model), we clustered the data into groups and found that the optimal division in this framework gives six distinct clusters of growth styles, indicated in the scatter plot; an alternative clustering method also indicated the presence of at least five distinct clusters (see Appendix for details).
Fig 3.
Distribution of colony growth rates and lag times. The bar plots show the measured distribution of g (colony growth rate at a radius of 0.24 mm) and t0 (lag time) at a dilution of 1:128 (n = 472 colonies). Both histograms are bimodal, suggesting the presence of several distinct groups of strains. The lower left panel shows a scatter plot of the same data, with axes matching those of the histograms (eight points with colony growth rates of above 0.08 mm/h are not visible). Clustering the data points using a Gaussian mixtures model yields an optimal fit with six clusters, as indicated by the differently colored markers (see Appendix for details).
Even for a pure strain, there is likely to be some phenotypic variability among clonal colonies due to intrinsic variability or external factors such as the presence of other, nearby colonies (as we discuss further below), and it is even possible that single colonies could be aggregates of several different strains. To investigate the latter possibility, we restreaked randomly chosen colonies grown from our stock on DNB agar and looked for distinct colony types. Only ∼10% (2 of 19) of the restreak plates showed signs of multiple phenotypes, and one of these was due to spontaneous (nonheritable) variation in the phenotype of a single strain. Mixed colonies are thus quite rare. Even for mixed colonies, the measured parameters still describe the growth of the multispecies aggregate, which may in fact be ecologically more relevant than separate measurements for each strain if these are genuine cases of symbiosis.
To estimate the variation between different colonies of a single strain, we measured the growth parameters of multiple clonal colonies from randomly chosen strains (see Materials and Methods). Analysis of five strains showed that the measured phenotypes of clonal colonies exhibited as much variation as the clusters of different strains in Fig. 3 (this includes both natural variation and measurement uncertainty) and showed that there was no statistically significant difference between the coefficients of variation for growth rate and lag time (see the supplemental material).
Effect of dilution on growth.
In the dilution experiment, only the number of colonies detected at a given time for each inoculum dilution level was used (see Materials and Methods). The number of observed colonies is plotted as a function of time for two dilutions in Fig. 4 and as a function of dilution in Fig. 5. To remove the trivial part of the relationship between the number of colonies and the inoculum density, the y axes in Fig. 4 and 5 give the observed number of CFU multiplied by the dilution factor. This normalized CFU value corresponds to the number of colonies formed by a full 100 μl of the undiluted stock after being diluted by the appropriate factor. From the microscopic count, we found a density of (3.15 ± 0.27) × 106 cells/ml in our soil stock (the uncertainty is standard error). Hence, if all cells lead to visible colonies, we would find 3.15 × 105 normalized CFU/plate; our highest value of roughly 4,000 (Fig. 3) corresponds to about 1.3% culturability.
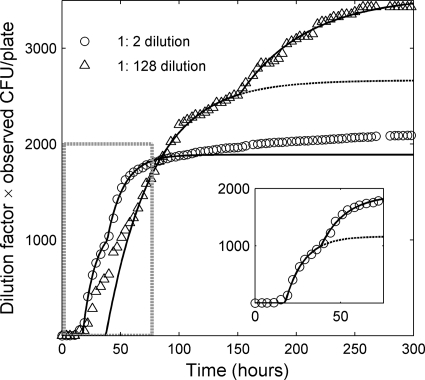
Fig 4.
Average number of observed CFU per plate versus time for two different dilutions. The CFU counts have been multiplied by the dilution factor so that the y coordinate is CFU per 100 μl of undiluted stock (proportional to the fraction of cells in the inoculum that has formed colonies). The inset shows an enlarged version of the 1:2 dilution curve in the area indicated by the dashed gray box in the main plot. Colonies appear faster on the plates with the more concentrated inoculum, but fewer cells give rise to colonies on these plates overall. The curves are lines to guide the eye based on the first-order reaction (FOR) model (26, 27) for communities with a single typical lag time (dashed line) and with two distinct typical lag times (solid line) (see Appendix for details). For clarity, the dashed curve for the 1:2 dilution is shown only in the inset. Each data point is an average over 5 replica petri dishes.
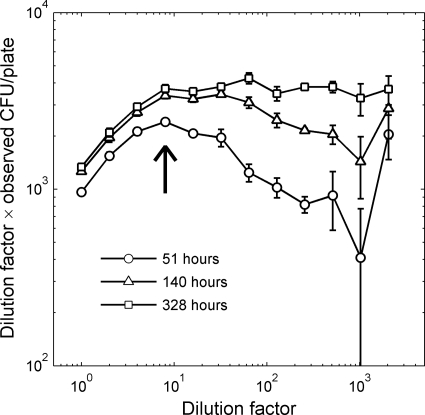
Fig 5.
Number of CFU as a function of dilution factor, measured at three different times. As in Fig. 4, the unit on the y axis is rescaled colony number (proportional to the fraction of colony-forming cells). At early times, the colony-forming fraction peaks at an intermediate dilution (indicated by the arrow). Each point is the average of 5 replicas, and the error bars indicate the standard errors across replicas.
The time of appearance of a given colony depends on both its lag time and its growth rate, but since we can detect even very small colonies, the time that a colony is first observed will be almost entirely determined by its lag time. Hence, for the purpose of understanding how the number of detected colonies changes with time, we can restrict our attention to the distribution of lag times (rightmost histogram in Fig. 3). If the distribution is in fact dominated by the two peaks at 35 and 85 h, we would expect a similar two-phase pattern in the colony appearance over time, and that is indeed the case (Fig. 4). It is particularly clear if we compare the data to the curve shape predicted by the first-order reaction (FOR) model of colony growth (26, 27): assuming two dominant lag time groups rather than one gives a much better fit with a marked “jump” (solid and dashed curves in Fig. 4).
The fact that the two sets of data points in Fig. 4 do not overlap implies that the fraction of inoculated cells that forms colonies varies with the cell density, indicating that density changes lead to global shifts in colony growth that affect many strains. For these two dilutions, we find that (i) on high-density plates, a smaller total fraction of the cells in the inoculum forms visible colonies after several weeks, and (ii) colonies appear faster on the high-density (low-dilution) plates than on the low-density ones. The first observation is an example of what is known as the crowding effect, i.e., the tendency of colonies to compete and inhibit each other (6). The second observation, i.e., the acceleration in colony formation, suggests an opposite phenomenon, namely, that the presence of nearby colonies increases the chance of that a cell will form a visible colony.
The dependence of culturability on cell density and growth time becomes evident when plotting the normalized colony number as a function of the inoculum dilution factor (Fig. 5). Strikingly, the early formation of colonies does not simply increase with density (as the crowding effect would suggest) but is instead most marked at the 8-fold dilution of the stock, leading to a local peak in the fraction of colony-forming cells in the early phase of the experiment (arrow in Fig. 5). As time progresses and colonies continue to appear, the colony counts at low cell densities catch up, and after 2 weeks the peak has disappeared. Comparable growth experiments extending over several months indicate that the tail of the curve would continue to rise so that it is eventually uniformly increasing with increasing dilution (6).
DISCUSSION
Summary.
The use of a scanner array coupled with image analysis allowed us to survey the growth phenotypes of thousands of colony-forming soil bacteria, measuring growth kinetic parameters and overall colony formation with high precision. We found that grouping strains by colony growth rate and lag time yields not the traditional two but rather several more distinct groups (six by our clustering method and at least five with an alternative method). Our study of the effect of inoculum density on colony formation shows that colonies form most rapidly at an intermediate cell density.
These results indicate a potentially important role for broad phenotypic studies of environmental microbes alongside targeted cultivation and culture-independent molecular approaches. Such methods could be particularly useful in detecting global patterns that would be invisible in experiments limited to a few strains or based solely on DNA. Conversely, understanding the physiological basis of the growth characteristics that we observe will require complementary approaches, e.g., work on exemplar strains from each cluster to allow the experimental fine-tuning that must be relinquished when working with a broad range of strains.
Growth strategies of individual strains.
The parameter H introduced here measures how fast radial growth is decelerating and is hence a rough measure of how quickly a colony becomes growth limited: if a large fraction of cells in the colony are still able to divide around the time of detection, H will be large (44, 47). While this characteristic is likely primarily determined by the nutrient utilization of each strain, it will also reflect, e.g., the ratio of vertical to horizontal growth, investment in sporulation, or an intrinsic tendency to form small colonies (7, 31, 65). The overall negative correlation between lag time on one hand and colony growth rate and H on the other indicates a coordination of small lag and fast growth across the clusters we observe, in contrast with the trade-offs observed between, e.g., growth rate and yield or substrate affinity (16, 20, 30, 45).
The diversity of growth modes is more directly reflected by the clustering of strains in the space of colony growth rate and lag time (Fig. 3). In contrast to the dichotomous classification of strains (1, 14, 32, 46, 60, 68), the more numerous groups that we observe indicate that a different and more fine-grained classification based on growth strategies is possible. Our clustering should be seen as complementary to the classical schemes, as it does not subsume them. Thus, while both colony growth rate and lag time relate to the K versus r division, fast growth in our experiment does not directly imply a general r-strategy, since the low-nutrient medium could give an overall advantage to otherwise poor growers (34).
It would be natural to hypothesize that each cluster corresponds to specific adaptations, such as a preference for certain nutrient types (e.g., easily digestible versus recalcitrant). Future targeted studies of individual strains would be needed to uncover the physiological traits and ecological circumstances that may characterize the members of each cluster. Additional single-strain studies would also further illuminate the potential role of within-strain variation, which may complicate classifications based on growth type. Thus, it has been suggested that environmental bacteria may use a stochastic growth strategy in which most cells remain dormant but occasional “scout cells” randomly resume growth and found a new population (colony) if conditions are favorable (13). If so, even single-species samples could show a spread in the observed lag times and conceivably even separated clusters. We found some lag time spread but no signs of multiple clusters in five strains for which we measured growth parameters for several clonal colonies (see Fig. S6 in the supplemental material); further studies would be needed for a more comprehensive overview.
We have chosen to state our results entirely in terms of the (radial) growth rate of visible colonies (g), a well-defined and directly measurable quantitative phenotype. The radial growth rate can often serve as a qualitative proxy for the underlying biomass growth rate: the two are directly proportional for a fixed-thickness colony that grows only at the edge (44, 47). Clearly, other physiological parameters can play a role (33, 67), and colony morphology may also be shaped by swarming and chemotaxis, by nutrient availability (56), and by the spatial structure of soil (22). However, measuring growth in liquid is not a feasible general alternative, since liquid supports the growth of fewer species than solid media (53) and many soil bacteria form aggregates when growing in liquid media (e.g., Streptomyces [24]). The colony growth rate therefore offers a good compromise between experimental accessibility and ecological information.
Community-wide growth effects and induction of colony formation.
The fact that the early rate of colony formation peaks at an intermediate dilution, i.e., in the presence of some neighboring colonies, indicates that both negative and positive interactions between colonies play a role in colony formation. Since we observe this effect in mixed communities, the underlying interactions would have to be common in the community (66). Indeed, these observations are consistent with previous studies indicating the widespread presence of quorum sensing (39, 55, 59, 66) and with the wide range of chemical compounds, many produced by microbes, known to influence (or even be essential to) bacterial growth (5, 11, 31, 36, 42, 64, 69, 70). The observations of an intermediate colony density which best promotes growth can have practical implications for maximizing culturability. While one must be careful in extrapolating from laboratory result to conditions in natural soil, the fact that bacteria in soil tend to grow in localized clumps (43) suggests that neighbor effects like ones we observe could also potentially play a role in situ.
If intermicrobial interactions play a role in the growth of environmental bacteria, one should be cautious in applying rigid ecological categories based on measurements of strains in isolation. Understanding if and how phenotypic groups change with density presents an interesting direction for future extensions of the present study. Likewise, it is known that some soil bacteria form colonies only on time scales longer than the ones addressed here (6, 7). Extending the maximum growth time and testing other growth conditions may thus reveal further phenotypic diversity. Nevertheless, the results presented here offer concrete examples of broad, cross-species phenotypic patterns and their complex dependency on species interactions.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Kalin Vetsigian for extensive discussions and help with the scanner system. We also thank Michael Baym, Tobias Bollenbach, Pedro Bordalo, Remy Chait, Rishi Jajoo, Eric Kelsic, Jean-Baptiste Michel, Tenzin Phulchung, Elizabeth A. Shank, Nicholas Stoustrop, Erdal Toprak, and Zhizhong Yao for discussions and practical assistance. The automated image analysis described in this paper was performed on the Orchestra computing cluster supported by the Harvard Medical School Research Information Technology Group.
This investigation was aided by a postdoctoral fellowship from the Jane Coffin Childs Memorial Fund for Medical Research to M.E. R.K. acknowledges the support of a James S. McDonnell Foundation 21st Century Science Initiative in Studying Complex Systems Research Award and from National Institutes of Health grants 5P50 GM068763-07 (Murray) SUB0015 and R01 GM081617.
Appendix
Cluster analysis of the data in Fig. 3.
The clustering of the data in the scatter plot in Fig. 3 was done using a Gaussian mixtures model (48). This method allows clusters to have an arbitrary elliptic shape and orientation and so, in particular, renders the clustering insensitive to the choice of units for the two axes. In contrast, the widely used k-means clustering forces clusters to be spherical, and standard hierarchical clustering methods also allow less freedom in cluster shape (48). Briefly, with a Gaussian mixtures model one describes the data as being drawn from a set of some number, K, of Gaussian distributions (two-dimensional in our case) and then searches for the distribution parameters that maximize the likelihood of the actual data. Points are clustered by assigning each one to the individual Gaussian distributions from which it was most likely drawn. We performed the clustering using the “fit” and “cluster” functions in the @gmdistribution class from the MATLAB Statistical Toolbox (MathWorks). The eight colonies not shown in Fig. 3 were excluded from the analysis. The clustering was done 2,5000 times with random starting parameters for each K from 2 to 11, and the clustering with the best likelihood score was then selected for each K. The optimal K value was then determined using the Bayesian information criterion (BIC), which combines the likelihood of the data (given the fit) with a penalty for each extra fitted parameter (50). The best fits were for K = 6 and K = 7, with hardly any difference in the BIC scores (less than 0.15% difference); in the interest of parsimony, we therefore chose to cluster with K = 6.
To further ascertain the presence of clustering and evaluate the reasonability of a division into six clusters, we performed two further analyses of our data: (i) comparing the Gaussian mixtures clustering in the actual data to that in randomized version of it and (ii) clustering using a different, hierarchical method and evaluating the result by an alternative method. The detailed results are presented in the supplemental material. In summary, we found that (i) our data do indeed show significant clustering and (ii) using the hierarchical method, clustering fits are poor if we assume fewer than five clusters, while six clusters yield a good-quality fit (the exact clusters differ somewhat from the ones found above due to the stronger constraints in the hierarchical method).
The FOR model of colony formation.
The FOR model of colony formation predicts that for a single species, the number of visible colonies, N(t), at time t should be given by the following equations (27): N(t) = 0 for t < tr and N(t) = N∞{1 − exp[−λ(t − tr)]} for t ≥ tr. Here, N∞ is the number of colonies after an infinitely long incubation, tr is the retardation time (time before colony formation starts), and λ is the rate at which colonies form. If two groups of species with different lag times are present, the total number of colonies at any given time will be given by the sum of two terms of the form above, with different parameter values (26).
Since we have a wide range of variation in the colony formation kinetics, a two-group FOR model cannot fully describe our observation, and a formal fitting of the complete data to the model yields very poor fits. Instead, the curves in Fig. 4 are meant only as guides to the eye. They take the form predicted by the FOR model for a single type of bacteria (dashed line) and two types of bacteria (solid line), but parameters were found by fitting only sections of the data to achieve curves which trace the initial increase in the colony number and, for the two-type model, the “jumps” in the data (indicating the presence of two typical lag times).
Footnotes
Published ahead of print 22 December 2011
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Andrews JH, Harris RF. 1986. R-selection and K-selection and microbial ecology. Adv. Microb. Ecol. 9:99–147 [Google Scholar]
- 2. Burmølle M, Hansen LH, Oregaard G, Sørensen SJ. 2003. Presence of N-acyl homoserine lactones in soil detected by a whole-cell biosensor and flow cytometry. Microb. Ecol. 45:226–236 [DOI] [PubMed] [Google Scholar]
- 3. Button DK, Schut F, Quang P, Martin R, Robertson BR. 1993. Viability and isolation of marine bacteria by dilution culture: theory, procedures, and initial results. Appl. Environ. Microbiol. 59:881–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Curtis TP, et al. 2006. What is the extent of prokaryotic diversity? Philos. Trans. R. Soc. B Biol. Sci. 361:2023–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Davelos AL, Xiao K, Flor JM, Kinkel LL. 2004. Genetic and phenotypic traits of streptomycetes used to characterize antibiotic activities of field-collected microbes. Can. J. Microbiol. 50:79–89 [DOI] [PubMed] [Google Scholar]
- 6. Davis KER, Joseph SJ, Janssen PH. 2005. Effects of growth medium, inoculum size, and incubation time on culturability and isolation of soil bacteria. Appl. Environ. Microbiol. 71:826–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Davis KER, Sangwan P, Janssen PH. 2011. Acidobacteria, Rubrobacteridae and Chloroflexi are abundant among very slow-growing and mini-colony-forming soil bacteria. Environ. Microbiol. 13:798–805 [DOI] [PubMed] [Google Scholar]
- 8. De Leij FAAM, Whipps JM, Lynch JM. 1994. The use of colony development for the characterization of bacterial communities in soil and on roots. Microb. Ecol. 27:81–97 [DOI] [PubMed] [Google Scholar]
- 9. Di Mattia E, Grego S, Cacciari I. 2002. Eco-physiological characterization of soil bacterial populations in different states of growth. Microb. Ecol. 43:34–43 [DOI] [PubMed] [Google Scholar]
- 10. Dinsdale EA, et al. 2008. Functional metagenomic profiling of nine biomes. Nature 452:629–632 [DOI] [PubMed] [Google Scholar]
- 11. D'Onofrio A, et al. 2010. Siderophores from neighboring organisms promote the growth of uncultured bacteria. Chem. Biol. 17:254–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Elfwing A, LeMarc Y, Baranyi J, Ballagi A. 2004. Observing growth and division of large numbers of individual bacteria by image analysis. Appl. Environ. Microbiol. 70:675–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Epstein SS. 2009. Microbial awakenings. Nature 457:1083. [DOI] [PubMed] [Google Scholar]
- 14. Fierer N, Bradford MA, Jackson RB. 2007. Toward an ecological classification of soil bacteria. Ecology 88:1354–1364 [DOI] [PubMed] [Google Scholar]
- 15. Flärdh K, Buttner MJ. 2009. Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat. Rev. Microbiol. 7:36–49 [DOI] [PubMed] [Google Scholar]
- 16. Frank SA. 2010. The trade-off between rate and yield in the design of microbial metabolism. J. Evol. Biol. 23:609–613 [DOI] [PubMed] [Google Scholar]
- 17. Gans J, Wolinsky M, Dunbar J. 2005. Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science 309:1387–1390 [DOI] [PubMed] [Google Scholar]
- 18. Giovannoni SJ, Foster RA, Rappe MS, Epstein S. 2007. New cultivation strategies bring more microbial plankton species into the laboratory. Oceanography 20:62–69 [Google Scholar]
- 19. Gonzales Rafael C, Woods RE. 2008. Digital image processing, 3rd ed. Pearson Prentice Hall, Upper Saddle River, NJ [Google Scholar]
- 20. Gudelj I, Beardmore RE, Arkin SS, Maclean RC. 2007. Constraints on microbial metabolism drive evolutionary diversification in homogeneous environments. J. Evol. Biol. 20:1882–1889 [DOI] [PubMed] [Google Scholar]
- 21. Guillier L, Pardon P, Augustin JC. 2006. Automated image analysis of bacterial colony growth as a tool to study individual lag time distributions of immobilized cells. J. Microbiol. Methods 65:324–334 [DOI] [PubMed] [Google Scholar]
- 22. Hattori T, Hattori R, McLaren AD. 1976. The physical environment in soil microbiology: an attempt to extend principles of microbiology to soil microorganisms. Crit. Rev. Microbiol. 4:423–461 [DOI] [PubMed] [Google Scholar]
- 23. Hattori T, et al. 1997. Advances in soil microbial ecology and the biodiversity. Antonie Van Leeuwenhoek 72:21–28 [DOI] [PubMed] [Google Scholar]
- 24. Hobbs G, Frazer CM, Gardner DCJ, Cullum JA, Oliver SG. 1989. Dispersed growth of Streptomyces in liquid culture. Appl. Microbiol. Biotechnol. 31:272–277 [Google Scholar]
- 25. Hong SH, Bunge J, Jeon SO, Epstein SS. 2006. Predicting microbial species richness. Proc. Natl. Acad. Sci. U. S. A. 103:117–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ishikuri S, Hattori T. 1987. Analysis of colony-forming curves of soil bacteria. Soil Sci. Plant Nutr. 33:355–362 [Google Scholar]
- 27. Ishikuri S, Hattori T. 1985. Formation of bacterial colonies in successive time intervals. Appl. Environ. Microbiol. 49:870–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Janssen PH. 2008. New cultivation strategies for terrestrial microorganisms, p 173–192 In Zengler K. (ed), Accessing uncultivated microorganisms. ASM Press, Washington, DC [Google Scholar]
- 29. Janssen PH, Yates PS, Grinton BE, Taylor PM, Sait M. 2002. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl. Environ. Microbiol. 68:2391–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jessup CM, Bohannan BJM. 2008. The shape of an ecological trade-off varies with environment. Ecol. Lett. 11:947–959 [DOI] [PubMed] [Google Scholar]
- 31. Kaeberlein T, Lewis K, Epstein SS. 2002. Isolating “uncultivable” microorganisms in pure culture in a simulated natural environment. Science 296:1127–1129 [DOI] [PubMed] [Google Scholar]
- 32. Koch AL. 2001. Oligotrophs versus copiotrophs. Bioessays 23:657–661 [DOI] [PubMed] [Google Scholar]
- 33. Kreft J-U, Booth G, Wimpenny JWT. 1998. BacSim, a simulator for individual-based modelling of bacterial colony growth. Microbiology 144:3275–3287 [DOI] [PubMed] [Google Scholar]
- 34. Kuznetsov SI, Dubinina GA, Lapteva NA. 1979. Biology of oligotrophic bacteria. Annu. Rev. Microbiol. 33:377–387 [DOI] [PubMed] [Google Scholar]
- 35. Levin-Reisman I, et al. 2010. Automated imaging with ScanLag reveals previously undetectable bacterial growth phenotypes. Nat. Methods 7:737–739 [DOI] [PubMed] [Google Scholar]
- 36. Linares JF, Gustafsson I, Baquero F, Martinez JL. 2006. Antibiotics as intermicrobial signaling agents instead of weapons. Proc. Natl. Acad. Sci. U. S. A. 103:19484–19489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lindahl V, Bakken LR. 1995. Evaluation of methods for extraction of bacteria from soil. FEMS Microbiol. Ecol. 16:135–142 [Google Scholar]
- 38. Michel JB, Yeh PJ, Chait R, Moellering RC, Kishony R. 2008. Drug interactions modulate the potential for evolution of resistance. Proc. Natl. Acad. Sci. U. S. A. 105:14918–14923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Miller MB, Bassler BL. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55:165–199 [DOI] [PubMed] [Google Scholar]
- 40. Møller S, Kristensen CS, Poulsen LK, Carstensen JM, Molin S. 1995. Bacterial growth on surfaces: automated image analysis for quantification of growth rate-related parameters. Appl. Environ. Microbiol. 61:741–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nichols D, et al. 2010. Use of Ichip for high-throughput in situ cultivation of “uncultivable” microbial species. Appl. Environ. Microbiol. 76:2445–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nichols D, et al. 2008. Short peptide induces an “uncultivable” microorganism to grow in vitro. Appl. Environ. Microbiol. 74:4889–4897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nunan N, Wu K, Young IM, Crawford JW, Ritz K. 2002. In situ spatial patterns of soil bacterial populations, mapped at multiple scales, in an arable soil. Microb. Ecol. 44:296–305 [DOI] [PubMed] [Google Scholar]
- 44. Panikov NS. 1995. Microbial growth dynamics. Chapman & Hall, London, United Kingdom [Google Scholar]
- 45. Pernthaler A, Pernthaler J, Eilers H, Amann R. 2001. Growth patterns of two marine isolates: adaptations to substrate patchiness? Appl. Environ. Microbiol. 67:4077–4083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pianka ER. 1970. R-selection and K-selection. Am. Nat. 104:592–597 [Google Scholar]
- 47. Pirt SJ. 1975. Principles of microbe and cell cultivation. John Wiley & Sons, New York, NY [Google Scholar]
- 48. Press WH, Teukolsky SA, Vetterling WT, Flannery BP. 2007. Numerical recipes, 3rd ed Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 49. Puchkov EO. 2010. Computer image analysis of microbial colonies. Microbiology 79:141–146 [Google Scholar]
- 50. Quinn GP, Keough MJ. 2002. Experimental design and data analysis for biologists. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 51. Rappe MS, Connon SA, Vergin KL, Giovannoni SJ. 2002. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418:630–633 [DOI] [PubMed] [Google Scholar]
- 52. Roesch LF, et al. 2007. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 1:283–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schoenborn L, Yates PS, Grinton BE, Hugenholtz PH. 2004. Liquid serial dilution is inferior to solid media for isolation of cultures representative of the phylum-level diversity of soil bacteria. Appl. Environ. Microbiol. 70:4363–4366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schut F, et al. 1993. Isolation of typical marine bacteria by dilution culture: growth, maintenance, and characteristics of isolates under laboratory conditions. Appl. Environ. Microbiol. 59:2150–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shank EA, Kolter R. 2009. New developments in microbial interspecies signaling. Curr. Opin. Microbiol. 12:205–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shapiro JA. 1995. The significances of bacterial colony patterns. Bioessays 17:597–607 [DOI] [PubMed] [Google Scholar]
- 57. Staley JT, Konopka A. 1985. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu. Rev. Microbiol. 39:321–346 [DOI] [PubMed] [Google Scholar]
- 58. Stevenson BS, Eichorst SA, Wertz JT, Schmidt TM, Breznak JA. 2004. New strategies for cultivation and detection of previously uncultured microbes. Appl. Environ. Microbiol. 70:4748–4755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Straight PD, Kolter R. 2009. Interspecies chemical communication in bacterial development. Annu. Rev. Microbiol. 63:99–118 [DOI] [PubMed] [Google Scholar]
- 60. Sylvia DM, Fuhrmann JJ, Hartel PG, Zuberer DA. (ed). 1998. Principles and applications of soil microbiology. Prentice-Hall, Upper Saddle River, NJ [Google Scholar]
- 61. Torsvik V, Øvreås L. 2002. Microbial diversity and function in soil: from genes to ecosystems. Curr. Opin. Microbiol. 5:240–245 [DOI] [PubMed] [Google Scholar]
- 62. Tringe SG, et al. 2005. Comparative metagenomics of microbial communities. Science 308:554–557 [DOI] [PubMed] [Google Scholar]
- 63. Venter JC, et al. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66–74 [DOI] [PubMed] [Google Scholar]
- 64. Vetsigian K, Jajoo R, Kishony R. 2011. Structure and evolution of Streptomyces interaction networks in soil and in silico. PLoS Biol. 9:e1001184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Watve M, et al. 2000. The ‘K’ selected oligophilic bacteria: a key to uncultured diversity? Curr. Sci. (India) 78:1535–1542 [Google Scholar]
- 66. Williams P, Winzer K, Chan WC, Camara M. 2007. Look who's talking: communication and quorum sensing in the bacterial world. Philos. Trans. R. Soc. B Biol. Sci. 362:1119–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wimpenny JWT. 1979. The growth and form of bacterial colonies. J. Gen. Microbiol. 114:483–486 [DOI] [PubMed] [Google Scholar]
- 68. Winogradsky WS. 1924. Sur la microflora autochtone de la terre arable. C. R. Hebd. Seances Acad. Sci. 124:1236–1239 [Google Scholar]
- 69. Yim G, Wang HHM, Davies J. 2006. The truth about antibiotics. Int. J. Med. Microbiol. 296:163–170 [DOI] [PubMed] [Google Scholar]
- 70. Yim G, Wang HMH, Davies J. 2007. Antibiotics as signalling molecules. Philos. Trans. R. Soc. B Biol. Sci. 362:1195–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zengler K, et al. 2002. Cultivating the uncultured. Proc. Natl. Acad. Sci. U. S. A. 99:15681–15686 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.