Abstract
A bacterial community may be resistant to environmental disturbances if some of its species show metabolic flexibility and physiological tolerance to the changing conditions. Alternatively, disturbances can change the composition of the community and thereby potentially affect ecosystem processes. The impact of disturbance on the composition of bacterioplankton communities was examined in continuous seawater cultures. Bacterial assemblages from geographically closely connected areas, the Baltic Sea (salinity 7 and high dissolved organic carbon [DOC]) and Skagerrak (salinity 28 and low DOC), were exposed to gradual opposing changes in salinity and DOC over a 3-week period such that the Baltic community was exposed to Skagerrak salinity and DOC and vice versa. Denaturing gradient gel electrophoresis and clone libraries of PCR-amplified 16S rRNA genes showed that the composition of the transplanted communities differed significantly from those held at constant salinity. Despite this, the growth yields (number of cells ml−1) were similar, which suggests similar levels of substrate utilization. Deep 454 pyrosequencing of 16S rRNA genes showed that the composition of the disturbed communities had changed due to the recruitment of phylotypes present in the rare biosphere of the original community. The study shows that members of the rare biosphere can become abundant in a bacterioplankton community after disturbance and that those bacteria can have important roles in maintaining ecosystem processes.
INTRODUCTION
Recent molecular studies using sequences of small-subunit rRNA genes have shown that the diversity of microbial communities is composed of two components: first, a set of abundant and actively growing taxa and, second, a seed bank of many rare taxa (51, 57). Deep sequencing has shown that this rare biosphere accounts for most of the observed phylogenetic diversity of microbial communities (64). Accordingly, these have strongly skewed rank abundance curves, with a dominance of the most common taxa followed by a long, flattened tail of rare ones.
The uneven distribution of taxa may influence how microbial communities respond to environmental disturbances and to what extent changes in diversity will affect the rates of ecosystem processes provided by the community. Numerous studies have shown that microbial communities are sensitive to environmental changes. Thus, the composition of microbial communities is often altered by disturbances and does not recover over some time (reviewed in reference 3). If the population sizes of the metabolically active part of the community drastically decrease, the processes performed by the community can be significantly altered. Alternatively, the original community might contain functionally redundant taxa such that ecosystem process rates are not affected by community composition changes (3). It has been proposed that the rare biosphere may serve as a reservoir from which new dominant populations can emerge following environmental disturbances (64). This idea is supported by early work on cultivable bacteria (22) and by recent molecular field data (5). However, other studies do not support this (38); the large diversity of the rare biosphere may partly stem from artifacts of the 454 pyrosequencing methodology (35, 58), and it has even been proposed that the rare biosphere is merely dissolved DNA or dead cells (67).
The aim of the present study was to examine to what extent and by which mechanisms aquatic microbial assemblages respond to new environmental conditions. Among the numerous environmental factors thought to regulate bacterioplankton community composition, dissolved organic carbon (DOC) and salinity appear to be of main importance (52). Bacterial growth is often limited by the availability of DOC (11, 13, 39), and the fraction of the DOC pool available to bacteria varies between environments (65). Little information, however, is available about the linkage between the capacity of taxa to utilize particular substrates (14, 44, 60) and community-level exploitation of the DOC pool. For instance, Covert and Moran (15) found that different bacterial groups utilized low- and high-molecular-weight DOC, while other studies suggest that bacterioplankton communities are dominated by generalist bacteria capable of metabolizing a wide variety of organic carbon compounds (48). In salinity gradients, however, the performance of bacterial communities may emerge solely as a response to changed salinity regardless of differences in DOC composition (40), likely due to pronounced compositional changes caused by requirement for or sensitivity to salt (8, 10, 16, 28, 39, 40). However, the results are not consistent; some studies show a homogeneous bacterioplankton community, despite large gradients in salinity (37), whereas in other areas, even moderate changes in salinity may affect bacterial community composition (33, 40), which in turn can lead to altered growth characteristics (40). Consequently, salinity may be a key driver of bacterioplankton community function.
Specifically, we examined effects of gradual changes in salinity on bacterial community composition in continuous cultures using water and inocula from the Baltic Sea (salinity 7) and Skagerrak (salinity 28), the strait between Norway, Sweden, and Denmark connected with the North Sea. These two connected systems also differ in DOC, with concentrations in the Baltic Sea being roughly double those in the Skagerrak (70), as well as in bacterial community composition (55). The Baltic Sea has its outflow of brackish water through the Danish straits, resulting in a stable salinity gradient with increasing salinity from the Baltic toward the Skagerrak. Nevertheless, exchange of water masses does occur occasionally during storm-driven events when high-salinity water from the Skagerrak enters the Baltic Sea. Thereby, the indigenous bacterial assemblages in the respective environments are exposed to pronounced fluctuations in salinity as well as in DOC, and it may be assumed that the bacterial communities are able to respond to these abrupt environmental changes.
Our experimental approach demonstrates that the bacterial communities responded to an environmental disturbance and changed salinity and DOC by compositional succession, partly accommodated by recruitment of members of the rare biosphere. Nevertheless, community growth remained stable.
MATERIALS AND METHODS
Experimental setup.
Natural bacterial communities were exposed to a gradual transformation from a low- to a high-salinity environment and vice versa in continuous seawater cultures. Two experiments were performed: one in June 2007, referred to as experiment 1, and one in August 2007, referred to as experiment 2. In experiment 2, in addition to cultures exposed to changed salinity, control incubations with constant high salinity (salinity 28) or low salinity (salinity 7) were included in the experimental setup. Most data presented are from experiment 2, but some data from experiment 1 are included to illustrate that observed patterns of bacterial dynamics were similar in the two experiments.
Water collection.
Seawater from 2 m depth was collected on two occasions in 2007 in the Baltic Sea (57°27′N, 17°05′E) and the Skagerrak (58°15′N, 11°22′E) using a Niskin bottle: experiment 1, Baltic Sea and Skagerrak on 19 and 20 June, respectively; experiment 2, Baltic Sea on 20 August and Skagerrak on 21 August. Sampling for experiment 1 was performed after the spring bloom, which otherwise would have affected the community composition (54), and sampling for experiment 2 was before the autumn storms, which transfer high-salinity water into the Baltic Sea. Samples were kept in 20-liter polycarbonate bottles and processed within 1.5 h.
Preparation of medium and inoculum.
Baltic and Skagerrak seawater was used as medium and was prepared by consecutive filtration through 3.0-μm-pore-size polycarbonate filters (Whatman) and 0.22-μm-pore-size Sterivex filters (Millipore) using a peristaltic pump. The filtered seawater, one full 20-liter container per continuous culture, was autoclaved before assembly of the continuous culture system. The Sterivex filters were frozen at −80°C until DNA extraction. The seawater inoculum was gravity filtered (once through 3.0-μm-pore-size polycarbonate filters and twice through 0.6-μm-pore-size polycarbonate filters [Whatman], to ensure that no protozoa were present in the inoculum) and stored at 4°C for ∼24 h before inoculation of the continuous seawater cultures.
Setup of continuous seawater cultures.
An outline of the experimental setup is shown in Fig. 1. The systems were assembled and autoclaved before inoculation with 0.5 liter of Baltic or Skagerrak bacterial community. Medium was fed dropwise through a glass tube to prevent back growth (31). Air was passed through 0.2-μm-pore-size polytetrafluoroethylene Acrodisc CR filters (Pall Corporation) and used to force the inflow to the cultures. The airflow formed small bubbles, which served as a stirrer for the culture (73).
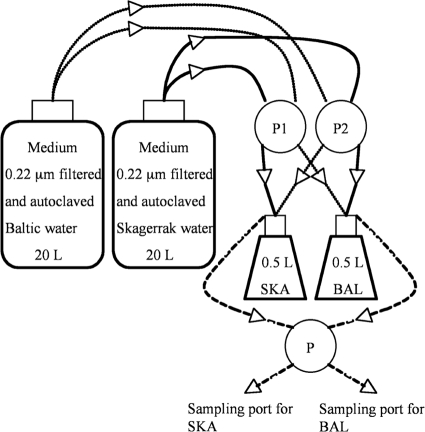
Fig 1.
Setup for continuous seawater cultures. Filtered and autoclaved Baltic and Skagerrak seawater was mixed in the inflow to produce a salinity gradient. Starting with 100% of the inoculum seawater, the gradient was produced by changing the relative speed of the pumps controlling the inflow of media (P1 and P2). The bacterial communities from the Baltic and Skagerrak inocula are referred to as BAL and SKA, respectively.
The experiments were run for 3 to 4 weeks at 20°C, which is the average summer temperature in the Baltic Sea (63), at a constant dilution rate (∼1 day−1), which is close to the median growth rate of marine bacteria ranging from polar to temperate regions (47), and in a 12-h light and 12-h dark cycle. After 6 days of adjustment at constant salinities, the salinities were gradually increased (Baltic sample) or decreased (Skagerrak sample) over the course of 11 days. Seawater medium from each location was mixed in the inflow to the two cultures. By changing the relative speed of the pumps controlling the inflow, while maintaining a constant dilution rate, the supply of Baltic water to the Skagerrak culture and the supply of Skagerrak water to the Baltic culture were successively increased (Fig. 1 and 2A). The cultures were then run for at least an additional 5 days with a constant supply of high-salinity (28, Skagerrak water) or low-salinity (7, Baltic water) water to ensure that all of the water in the cultivation bottles was exchanged and Skagerrak and Baltic salinities, respectively, were reached. In experiment 2, cultures in which the salinity was kept constant at 7 for the Baltic community and 28 for the Skagerrak community were run in parallel to the transplant experiments.
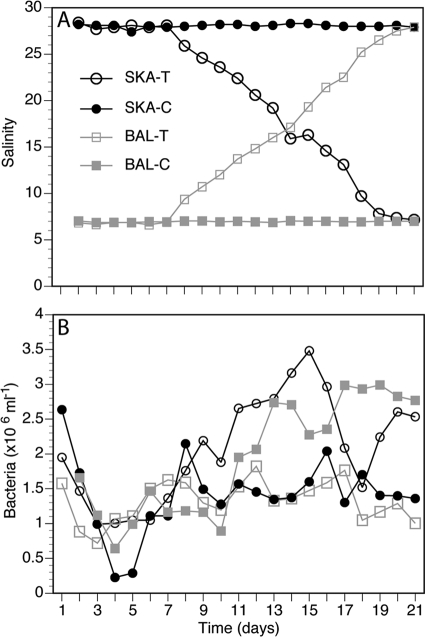
Fig 2.
Effects of seawater exchange on salinity (A) and bacterial abundance (B) in the experimental system. Data from experiment 2. The communities transplanted from low to high salinity are called Baltic transplant (BAL-T), and the ones transplanted from high to low salinity are called Skagerrak transplant (SKA-T). Experiment 2 also included two control cultures with constant salinity, which are referred to as Baltic constant (BAL-C) and Skagerrak constant (SKA-C). The patterns of changes in salinity and bacterial counts for the BAL-T and SKA-T communities were similar in experiment 1.
The communities transplanted from low to high salinity are called Baltic transplant (BAL-T), and the ones transplanted from high to low salinity are called Skagerrak transplant (SKA-T). In experiment 2, control cultures with constant salinity are referred to as Baltic constant (BAL-C) and Skagerrak constant (SKA-C). The bacterial communities from the original Baltic and Skagerrak inocula are referred to as BAL and SKA, respectively. The experimental setup resulted in four different cultures in experiment 2 (BAL-T, SKA-T, BAL-C, and SKA-C) and two in experiment 1 (BAL-T and SKA-T).
Sampling the cultures.
Samples for bacterial enumeration were collected in sterile polypropylene tubes from the outflow of the continuous cultures, fixed for ∼24 h in formaldehyde (final concentration, 3%), and filtered onto 0.2-μm-pore-size black polycarbonate filters (GE Water & Process Technologies). Cells were stained with SYBR gold (final concentration, 1×; Molecular Probes), and at least 200 cells or 30 fields filter−1 were counted at ×1,250 magnification using epifluorescence microscopy (Zeiss Axioplan). The filters were also examined for protozoa, but none were observed. For DNA extraction, 100 ml of sample was filtered onto 0.2-μm-pore-size Supor filters (25 mm; Pall Corporation), which were then frozen. Samples for salinity measurements were stored frozen until measurement of conductivity. Flow rate was recorded daily by weighing the bottles collecting the outflow.
DOC and inorganic nutrients.
Samples were taken from media and cultures at the end of experiment 2. DOC samples were filtered (0.2-μm-pore-size Supor filters; Pall Corporation), acidified, and stored frozen until analysis on a Shimadzu TOC-5000 high-temperature catalytic oxidation instrument. Calculation of carbon concentrations was made with potassium hydrogen phthalate as the standard substance. Samples for inorganic nutrients were filtered through glass fiber filters (GF/F) and stored frozen until analysis on a Bran & Luebbe TRAACS 800 autoanalyzer using standard seawater methods (29).
DNA extraction.
DNA was extracted from Sterivex filters (5 liters Baltic and Skagerrak inocula) and Supor filters (community DNA from the continuous cultures) using an enzyme/phenol-chloroform protocol (25, 62) but with a 30-min lysozyme digestion (final concentrations, 5 mg ml−1 for Sterivex filters and 1 mg ml−1 for Supor filters) at 37°C and an overnight proteinase K digestion (final concentration, 100 μg ml−1) at 55°C (9). DNA was quantified using the PicoGreen assay (Molecular Probes).
PCR, DGGE, clone libraries, and 454 pyrosequencing.
For experiment 2, time course changes in bacterial community compositions were followed by PCR-denaturing gradient gel electrophoresis (DGGE) as described by Riemann et al. (62). Bacterial 16S rRNA genes were PCR amplified using puReTaq Ready-To-Go PCR beads (GE Healthcare), 0.04 ng DNA μl−1, and primers GC341F (50) and 907R (49). Based on the presence/absence of DGGE bands, a dendrogram was constructed by the software Quantity One, version 4.6.2 (Bio-Rad), using the Dice coefficient and cluster analysis by the unweighted-pair group method using arithmetic average (UPGMA).
A total of 10 libraries of 96 clones were generated from experiments 1 and 2. Bacterial 16S rRNA genes were amplified using primers 27F and 1492R (27), and products were purified using a gel extraction kit (E.Z.N.A.), followed by a Cycle-Pure kit (E.Z.N.A.). Products were cloned (TOPO TA cloning kit; Invitrogen), plasmid DNA was extracted (R.E.A.L. Prep 96 plasmid kit; Qiagen), and the inserts (∼750 bp) were sequenced by the dideoxynucleotide termination method with primer 27F (commercially by Macrogen, South Korea).
Partial bacterial 16S rRNA genes (including the variable V4 to V6 regions) were amplified for pyrosequencing from the Baltic inoculum and the transplanted Baltic community from experiment 2 using a primer cocktail containing the degenerate primers 530F (5′-GTGCCAGCMGCNGCGGTA-3′ [19]), but with TA added at the 3 prime end to increase specificity, and 1061R (5′-CRRCACGAGCTGACGAC-3′ [4]). Pyrosequencing was performed at the University of Copenhagen on a 454 GS FLX system (Roche Applied Science) according to the manufacturer's instructions.
Sequence analysis.
Base calling on the Sanger data was performed using the PHRED software (21) and resulted in 960 sequences. For the 454 data (16,793 sequences with an average length of 224 bp), primer and adaptor sequences were trimmed using the SeqClean software (http://compbio.dfci.harvard.edu/tgi/software/) with UniVec_Core as the database (http://www.ncbi.nlm.nih.gov/VecScreen/UniVec.html). Reads <150 bp long and reads with undetermined nucleotides were removed from the 454 data set (34). The Greengenes online tool (http://greengenes.lbl.gov) was used to align sequences and exclude non-16S rRNA genes. Aligned sequences retrieved from Greengenes are truncated if the query sequences do not properly align to any one of the nonchimeric template sequences that Greengenes uses internally for aligning sequences. This is a way to prevent possible chimeras from being included in the Greengenes output. In total, 848 aligned sequences were retrieved from Greengenes for the Sanger data set and 16,760 were retrieved for the 454 data set. An additional, more thorough screening for chimeric sequences was made by use of the ChimeraSlayer utility (30) using default settings, detecting 21 additional chimeras in the Sanger data set and 247 in the 454 data set.
The Greengenes classification tool was used to assign a taxon for each sequence (17). Greengenes currently supports three different taxonomies: RDP, National Center for Biotechnology Information (NCBI), and Hugenholtz (17). If two of these showed the same taxonomic lineage on the phylum/class level, the sequence was kept for further analysis, resulting in 821 sequences for the Sanger data set and 16,497 for the 454 data set. Sequences shorter than 750 nucleotides were removed from the Sanger data set, leaving a final number of 727 sequences.
Maximum likelihood trees were inferred with the aid of RaxML software (version 7.0.4), using the GTRMIX model with default settings (66). The phylum trees were used as input to the RAMI tool to cluster similar sequences (56). RAMI defines clusters on the basis of the patristic distance (branch length) between external nodes in a phylogenetic tree. The patristic distance threshold was set to 0.06, which corresponds approximately to a 97% similarity cutoff (56). The produced clusters are used here as operational taxonomic units (OTUs). OTUs were defined as abundant if they accounted for >1% of the 454 reads (51) and rare if they accounted for <0.1% of the reads (24, 51).
Community distance analysis of the clone libraries was performed using the UniFrac software. To assess whether the UniFrac distance metrics between communities were significant, P value significance tests were done as implemented in the UniFrac program (42). The combinations were constructed so that all pairwise library combinations were tested against each other. The relative differences between all community phylogenies were examined by Jackknife clustering, which performs hierarchical clustering based on the pairwise UniFrac distance metrics. The robustness of the Jackknife analysis for sampling size and evenness is assessed by quantifying how often the cluster nodes are recovered for a random sample from part of the original data. This resampling was permutated 1,000 times using 75% of the smallest library (26 sequences) as the minimum number of sequences kept for each sample in the resampling.
Nucleotide sequence accession numbers.
Sanger sequences have been deposited in the EMBL database under accession numbers FR647485 to FR648326, while pyrotag sequences have been deposited in the NCBI Sequence Read Archive under accession number SRA012481.2.
RESULTS
Salinity, DOC, and inorganic nutrients.
The salinities of the Baltic and Skagerrak samples were ∼7 and ∼28, respectively. In experiment 2, an increase in salinity was recorded on day 7 in the BAL-T community, and the salinity reached the Skagerrak level, salinity 28, on day 21 (Fig. 2A). In contrast, the salinity in the SKA-T community started to decrease on day 7 and reached the Baltic level, salinity 7, on day 21. The DOC concentrations in the Baltic and Skagerrak media were 450 μM and 267 μM, respectively, and were 258 μM and 483 μM in the transplanted communities, BAL-T and SKA-T, respectively, on day 21. Concentrations of inorganic nitrogen and phosphate in the cultures were 1.6 to 2.5 μM and 0.04 to 0.13 μM, respectively. Thus, we do not consider that the growth of the bacteria was limited by nitrogen and phosphate.
Total count and growth yield.
In experiment 2, the bacterial abundance of the transplanted communities, BAL-T and SKA-T, reached the same levels as in the respective controls with the same salinity (SKA-C and BAL-C; Table 1; Fig. 2B). The bacterial abundance in the BAL-T community decreased between days 15 and 18 but then reached the same level as in SKA-C. In experiment 1, a similar time course of development of bacterial abundance was observed, with slightly higher levels in SKA-T than in BAL-T (Table 1). The number of cells per ml is used in this study as a measurement of growth yield.
Table 1.
Bacterial abundance in situ and in the cultures in the beginning and at the end of experiments 1 and 2 and flow rate in the cultures in experiment 2a
| Expt no. (mo) and sampleb | No. of cells (106) ml−1 |
Flow rate (ml day−1) | |||
|---|---|---|---|---|---|
| Days 1–7 | Days 15–21 | Days 22–28 | In situ | ||
| Expt 2 (August) | |||||
| BAL-T | 1.2 ± 0.4 | 1.3 ± 0.2 | 2.6 | 477 ± 71 | |
| BAL-C | 1.2 ± 0.4 | 2.7 ± 0.3 | 456 ± 71 | ||
| SKA-T | 1.3 ± 0.4 | 2.5 ± 0.6 | 0.8 | 483 ± 83 | |
| SKA-C | 1.2 ± 0.4 | 1.5 ± 0.3 | 449 ± 56 | ||
| Expt 1 (June) | |||||
| BAL-T | 1.3 ± 0.2 | 1.7 ± 0.2 | 1.8 | NA | |
| SKA-T | 1.0 ± 0.4 | 2.2 ± 0.2 | 2.3 | NA | |
Values are means ± standard deviations. NA, not available.
BAL-T, Baltic transplant, community transplanted from low to high salinity; BAL-C, Baltic constant, culture with constant low salinity; SKA-T, Skagerrak transplant, community transplanted from high to low salinity; SKA-C, Skagerrak constant, culture with constant high salinity.
Bacterial community dynamics.
DGGE of 16S rRNA genes from experiment 2 showed that the composition changed considerably in the SKA-T treatment as the salinity decreased, with bands disappearing and appearing over time (see lanes 1 to 8 in Fig. S1A in the supplemental material). The SKA inoculum showed 17 discernible bands, which after 3 weeks were reduced to 6 in the SKA-T community, compared to 7 bands in the SKA-C community. In the SKA-C culture (see lanes 9 to 14 in Fig. S1A in the supplemental material), the composition of the bacterial communities also changed over time, but banding patterns were different relative to SKA-T. This was evident from a dendrogram constructed from the DGGE banding patterns (Fig. 3A), where the SKA-T and SKA-C communities formed separate clusters, with the exception that the SKA-C sample from day 4 clustered with the SKA-T samples. The original SKA inoculum formed a discrete branch.
Fig 3.
Changes in bacterial community composition during the exposure of a Skagerrak community to a low-salinity environment (A) and the exposure of a Baltic community to a high-salinity environment (B). The dendrograms were constructed from DGGE profiles (see Fig. S1 in the supplemental material) using average linkage (UPGMA). Data from experiment 2. SKA, Skagerrak inoculum; BAL, Baltic inoculum. Other abbreviations are explained in the legend to Fig. 2.
Analysis of the BAL-T and BAL-C communities from experiment 2 also showed a changed bacterial community composition over time, resulting in different DGGE banding patterns in BAL-T and BAL-C at the end of the experiment (Fig. 3B; see Fig. S1B in the supplemental material).
Community composition determined by clone libraries.
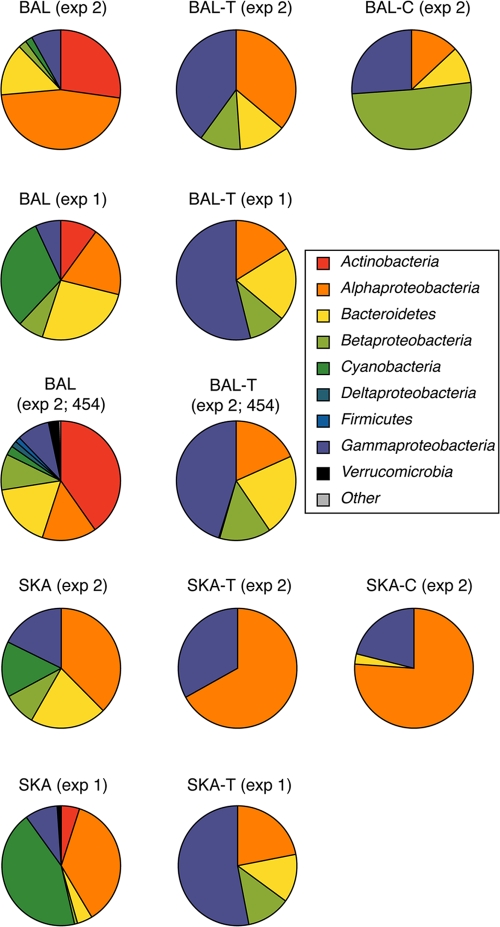
Clone libraries were generated from the Baltic and Skagerrak inocula (BAL and SKA), the last day in BAL-T and SKA-T in experiments 1 and 2, and from BAL-C and SKA-C in experiment 2, resulting in a total of 10 libraries of 96 clones. In all transplant experiments, the phylogenetic richness of the communities decreased; i.e., the inocula contained OTUs representing 5 to 7 phylogenetic groups, while the transplanted communities contained 2 to 4 phylogenetic groups (Fig. 4; see Table S1 in the supplemental material). Clones representing Actinobacteria and Cyanobacteria disappeared as salinity changed in both the BAL and SKA transplanted communities. The clone library analysis also showed that the phylogenetic composition of the inocula from Skagerrak and the Baltic Sea differed significantly (Fig. 4; see Tables S1 and S2 in the supplemental material). Furthermore, there were major differences in the composition between the communities collected in June (experiment 1) and August (experiment 2). In experiment 1, one OTU was shared between all libraries. In experiment 2, none of the clone-based OTUs detected at the end of the water-transplant experiments (in BAL-T or SKA-T) or in the constant cultures (BAL-C or SKA-C) were detected in the original inocula.
Fig 4.
Distribution of major taxonomic groups based on number of sequences determined by clone libraries and 454 sequencing of samples from experiment (exp) 1 (June) and experiment 2 (August). BAL 454 and BAL-T 454 denote that the community composition was determined by 454 sequencing. SKA, Skagerrak inoculum; BAL, Baltic inoculum. Other abbreviations are explained in the legend to Fig. 2. Detailed data are shown in Table S1 in the supplemental material.
Community distance analysis was performed using the UniFrac software, which measures the distance between community phylogenies as the percentage of branch length that is unique to one or the other community in a phylogenetic tree (42). Two identical communities will have all nodes and branch lengths in common, and the UniFrac distance metric will be 0 (0% difference). In contrast, if two samples already differ in the first node of the phylogenetic tree, no common nodes or branches exist and the UniFrac metric will be 1 (100% difference). The UniFrac analysis confirmed that the transplanted and constant communities were significantly different from the inocula (P ≤ 0.05, UniFrac significance test/P test; Fig. 5; see Table S2 in the supplemental material) and that the structures of the communities transplanted from low to high salinity and vice versa were generally significantly different from those held under constant conditions (P ≤ 0.03).
Fig 5.
Relative phylogenetic distances between the communities in the full Sanger data set (experiments 1 and 2). The dendrogram is based on hierarchical clustering analysis of the localities based on the pairwise UniFrac distance metric. Nodes in the tree are colored according to the fraction of the random samples that the nodes were recovered in. SKA, Skagerrak inoculum; BAL, Baltic inoculum. Other abbreviations are explained in the legend to Fig. 2.
454 pyrosequencing of BAL and BAL-T communities.
In experiment 2, a total of 7,775 and 8,722 sequences were retrieved from the transplanted community (BAL-T) and the inoculum (BAL), respectively (Fig. 4; see Table S1 in the supplemental material). The numbers of OTUs identified in BAL and BAL-T were 432 and 116, respectively. The Baltic community was dominated by Actinobacteria, Alphaproteobacteria, and Bacteroidetes, which were also the dominant groups in the clone libraries. The BAL-T community was dominated by Gammaproteobacteria, compared to Gamma- and Alphaproteobacteria in the clone library. The percentages of 454 reads found in the Sanger libraries were 24% and 53% for the BAL and BAL-T communities, respectively, when sequences were compared at a 100% similarity level. It should be noted that the overlapping region was only 150 bp.
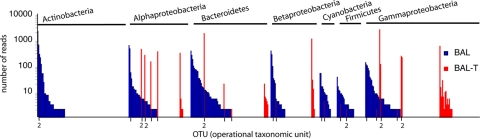
To identify OTUs shared between BAL and BAL-T, a phylogenetic tree was constructed with the 454 sequences from both libraries (see Fig. S2 in the supplemental material). In theory, all OTUs in the BAL-T sample should have been present in the BAL sample. Constrained by the depth of sequencing, however, only 27 OTUs in the BAL-T community were also detected in the BAL sample (Fig. 6; see Table S3 in the supplemental material). The relative abundances of 11 of these 27 OTUs were higher in BAL-T than in BAL. Identical sequences (100.0% nucleotide similarity) in BAL and BAL-T were found for 5 of the OTUs with higher abundance in BAL-T than in BAL, and in total, 14 of the 27 OTUs shared between BAL and BAL-T contained identical sequences (see Table S3 in the supplemental material). For example, within the alphaproteobacterial OTU 1, 1 single read in the BAL sample resulted in 247 identical reads recruited in BAL-T. Similarly, 2 identical reads in BAL within the gammaproteobacterial OTU 1 resulted in 1,136 identical reads in the BAL-T community.
Fig 6.
Results from 454 sequencing in experiment 2 showing the 27 OTUs in common between the Baltic inoculum (BAL) and the transplanted community (BAL-T). OTUs are sorted by phylum and abundance in BAL. Shared OTUs between BAL and BAL-T are illustrated by vertical lines on the x axis. Numbers of OTUs are shown when a line represents more than one OTU. Only phyla/classes with shared OTUs are included. Further details on the OTUs present in both BAL and BAL-T are given in Table 2 and Table S3 in the supplemental material.
Recruitment of initially rare taxa (<0.1% of the 454 reads) in the BAL community in response to the water transplant was observed among Alphaproteobacteria, Bacteroidetes, and Gammaproteobacteria (Fig. 6; see Table S3 in the supplemental material). In the BAL-T community, 12 OTUs accounting for >1% of the 454 reads were found, and 9 of these OTUs were also detected in the BAL community (Table 2). The taxonomy of these 9 OTUs was determined at the family level and at the genus level, if the classification from the three different taxonomies (RDP, NCBI, and Hugenholtz) showed the same taxonomic lineage (Table 2). Two dominant gammaproteobacterial OTUs in the BAL-T community belonged to the family Pseudomonadaceae, genus Pseudomonas (30.5% and 2.3% of the reads), which accounted for only 0.06% and 0.01% of the reads in the inoculum, respectively. Other dominant gammaproteobacterial OTUs belonged to the families Chromatiaceae and Idiomarinaceae (genus Idiomarina). The most prevalent Bacteroidetes OTU in the BAL-T community belonged to the family Flammeovirgaceae and accounted for 22.2% of the reads, but accounted for 0.15% of the reads in the inoculum. Two of the dominant alphaproteobacterial OTUs were members of the family Hyphomonadaceae and the genera Maricaulis and Hyphomonas, respectively. Other dominant OTUs belonged to the families Hyphomicrobiaceae and Caulobacteraceae (genus Phenylobacterium).
Table 2.
Phylogenetic affiliations of the main OTUs recruited from the inoculum (BAL) upon exposure to Skagerrak water, as revealed by 454 pyrosequencinga
| Phylum/class | Family | Genus | % of reads in: |
|
|---|---|---|---|---|
| BAL | BAL-T | |||
| Gammaproteobacteria OTU 1* | Pseudomonadaceae | Pseudomonas | 0.06 | 30.5 |
| Bacteroidetes OTU2* | Flammeovirgaceae | Unclassified | 0.15 | 22.2 |
| Alphaproteobacteria OTU 1* | Hyphomonadaceae | Maricaulis | 0.06 | 5.2 |
| Alphaproteobacteria OTU 6 | Hyphomicrobiaceae | Unclassified | 0.01 | 4.1 |
| Alphaproteobacteria OTU 7* | Caulobacteraceae | Phenylobacterium | 0.04 | 3.0 |
| Gammaproteobacteria OTU 5 | Chromatiaceae | Unclassified | 0.01 | 2.9 |
| Gammaproteobacteria OTU 12* | Pseudomonadaceae | Pseudomonas | 0.01 | 2.3 |
| Alphaproteobacteria OTU 5 | Hyphomonadaceae | Hyphomonas | 0.02 | 1.7 |
| Gammaproteobacteria OTU 6 | Idiomarinaceae | Idiomarina | 0.06 | 1.4 |
*, OTUs that at least in one case contain identical sequences (100% sequence similarity) in BAL and BAL-T. For all OTUs shared between BAL and BAL-T, see Table S3 in the supplemental material. BAL, Baltic inoculum; BAL-T, Baltic transplant.
DISCUSSION
We used continuous seawater cultures to investigate how and to what extent bacterial assemblages respond to changes in salinity and DOC. Using DGGE and clone library analyses, we found that unique multispecies communities with nearly no OTUs in common with the original inocula were recruited. However, the increased depth of sequencing provided by 454 pyrosequencing demonstrated that some species from the rare biosphere in an inoculum became dominant in its transplanted community in response to environmental change. Despite major differences in composition between the disturbed and constant communities, the growth yields (number of cells ml−1) were similar, findings which suggest similar levels of substrate utilization.
The bacterial community composition in the Baltic Sea and in Skagerrak was dominated by the phylogenetic groups commonly found in the areas (Fig. 4) (5, 55, 61). Community composition differed between the inocula, presumably reflecting seasonal variations in the bacterial communities in the Baltic Sea and in Skagerrak (54). As anticipated, the prevalence of phylogenetic groups that are common in freshwaters, like Actinobacteria and Verrucomicrobia (28, 72), decreased in the communities exposed to increased salinity. However, the parallel decrease of Actinobacteria in the Baltic community held at a low and stable salinity level suggests that the decrease may also be related to the factors leading to difficulty of cultivation of members of this phylum from planktonic systems. Gammaproteobacteria increased in abundance in all cultures, which is probably due to a high growth capacity and ability to exploit nutrients when available (20, 23, 53). Betaproteobacteria are common in freshwater (28, 72) but are also present in coastal waters (59). Interestingly, in our cultures Betaproteobacteria showed a slight increase with increasing salinity and were, thus, actively growing at salinity 28. Hence, if advected from the Baltic Sea and into the Skagerrak, some Betaproteobacteria may thrive under the local high-saline conditions. This is consistent with the observation of Betaproteobacteria in the SKA community in June.
Bacterial abundance in the continuous cultures was higher in water from the Baltic Sea than that from Skagerrak. This was consistent with the DOC concentration in the Baltic Sea medium or cultures being almost twice as high as in Skagerrak water. The higher DOC concentration in the Baltic Sea (32, 70) is due to a large load of riverine DOC, which constitutes an important nutrient and energy source for bacterioplankton (74). In accordance with our results, experiments from Swedish lakes showed that DOC concentration was positively correlated with bacterial abundance and that the origin of the medium affected bacterial activity and DOC utilization (41). Interestingly, Tranvik and Höfle (69) found that DOC from humic and clear-water lakes was utilized to the same extent regardless of the origin of the inoculum, indicating that bacterial communities can adapt quickly to new conditions, at least in terms of utilization of carbon sources. Hence, in our experimental system without grazers and with available inorganic nutrients, the level of bioavailable DOC was presumably the main determinant of bacterial growth yield.
The recruited low-salinity community (SKA-T) obtained the same growth yield as the low-salinity control community (BAL-C). Similarly, the bacterial abundance in the BAL-T community reached the same level as in the SKA-C community. Hence, despite the fact that the communities differed significantly in composition, similar growth yields were obtained, indicating that the recruited communities exploited the resource (DOC) to the same extent. This may suggest that the different communities were functionally redundant; i.e., the rare taxa favored by the environmental disturbance fulfilled similar ecological roles as the abundant species being replaced (12). Alternatively, the rare taxa may fill new functional niches and become abundant as a result of lower predation pressure. Moreover, a portfolio effect (3), where positive responses of some taxa are averaged with negative responses of other taxa (18), may have contributed to the stable growth yield.
The occasional mixing of Baltic and Skagerrak waters can be assumed to maintain low-abundance populations of Baltic species in the Skagerrak and vice versa. Hence, assuming that “everything is everywhere, but the environment selects” (7), it should be possible to recruit a Skagerrak community from a Baltic inoculum and vice versa. However, analysis of the clone libraries showed that the recruited communities differed significantly from the native communities found in the Baltic Sea and Skagerrak (Fig. 5). There may be several explanations for the limited overlap between the compositions of the recruited and native communities: (i) everything is not everywhere, as suggested from the noncosmopolitan distribution of the rare biosphere in the Arctic Ocean (26); (ii) if present in the inocula, some rare species may not be actively growing (dead or dormant) and therefore risk being flushed out of the container before optimal growth conditions are met; (iii) bottle effects were incompatible with the required growth conditions for the rare species; and (iv) removal of grazers allowed specific organisms to proliferate. These may have been suppressed by grazing under natural conditions.
Exposure of the Baltic inoculum to Skagerrak water led to a recruitment of taxa. While several factors, e.g., changed salt and DOC levels, lack of grazing, and confinement, could have caused this recruitment, it is noteworthy that several of the main OTUs, accounting for 0.01 to 0.15% of the reads from the inoculum but 1.4 to 30.5% of the reads from the transplanted community (Table 2), showed a high salt tolerance. For instance, the genera of the recruited gammaproteobacterial OTUs, Pseudomonas and Idiomarina, prefer salinities of 30 to 50 (43) and 10 to 100 (68), respectively. Similarly, the alphaproteobacterial genus Maricaulis contains marine, halophilic caulobacteria with optimal growth at salinities of 20 to 60 (1, 2). Other recruited alphaproteobacterial genera, Phenylobacterium and Hyphomonas, also contain species with a wide salt tolerance (43, 46). It should be noted, however, that characteristics other than salt tolerance may have contributed to the recruitment of certain taxa. For instance, some members of the Maricaulis and Hyphomonas genera possess a prostheca used to attach to surfaces (2, 46, 71), which may be advantageous during confinement. Nevertheless, upon storm-driven intrusions of high-saline water from the Skagerrak, these indigenous Baltic taxa would conceivably be able to respond to the abrupt environmental change and proliferate.
DGGE analysis, clone libraries, and 454 sequencing all showed a decreased diversity in the transplanted community compared to the inoculum, when the numbers of bands, phyla, and OTUs, respectively, were compared. Similar to our findings, other studies have shown that bacterial communities in stressed environments may have lower diversity relative to undisturbed communities (6, 45). Such stressed communities may show enhanced physiological tolerance and substrate utilization, suggesting proliferation and dominance of generalist bacterial populations as a consequence of disturbance (6, 45). Therefore, it is possible that the dominant OTUs in the transplanted communities, which were recruited from the rare biosphere in the inoculum, are generalists. These may be unable to compete for a narrow niche, but since they are able to withstand a wide range of conditions, they can persist at low levels in an environment and proliferate upon disturbance (45). However, an alternative explanation may be that the dominant OTUs in the transplanted communities are specialists that were rare in the inocula because of intense predation control or increased in abundance because of confinement.
The experimental data presented here suggest that disturbance (salinity and DOC change) elicits pronounced changes in bacterial community composition, partly accommodated by recruitment of members of the rare biosphere. This rebuts speculation that the rare biosphere exclusively consists of dead cells or free DNA (67). Consistent with early observations of recruitment of cultivable bacteria during experimental confinement (22), it has been suggested that rare bacterial taxa are active and have the potential to increase in abundance (36). Concordantly, the present experimental evidence documents that rare species detected by deep sequencing indeed can become abundant in response to environmental change, hence enabling a bacterial community to rapidly respond to and recover from shifts in environmental conditions, while sustaining the growth yield. Growth yield is a coarse measure of community function, and patterns could have looked different under different conditions or if a different measure for community function was used. Nevertheless, the finding that highly divergent bacterial communities can obtain similar growth yields adds uncertainty to the value of information about composition from an ecosystem modeling perspective. This highlights, as previously emphasized (3), that more empirical work is needed to understand the functional consequences of changes in microbial composition in the face of environmental disturbance.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grant 621-20085385 from the Swedish Research Council to Å.H. and by grant 217-2006476 from the Swedish Research Council for Environmental Agricultural Science and Spatial Planning (FORMAS) to P.L., Å.H., A.T., and L.R.
We are grateful for the use of laboratory and sampling facilities at Kristineberg Marine Research Station. We thank the three anonymous reviewers for valuable comments on the manuscript.
Footnotes
Published ahead of print 22 December 2011
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Abraham WR, et al. 2002. Phylogeny of Maricaulis Abraham et al. 1999 and proposal of Maricaulis virginensis sp. nov., M. parjimensis sp. nov., M. washingtonensis sp. nov. and M. salignorans sp. nov. Int. J. Syst. Evol. Microbiol. 52:2191–2201 [DOI] [PubMed] [Google Scholar]
- 2. Abraham WR, et al. 1999. Phylogeny and polyphasic taxonomy of Caulobacter species. Proposal of Maricaulis gen. nov. with Maricaulis maris (Poindexter) comb. nov. as the type species, and emended description of the genera Brevundimonas and Caulobacter. Int. J. Syst. Bacteriol. 49:1053–1073 [DOI] [PubMed] [Google Scholar]
- 3. Allison SD, Martiny JB. 2008. Colloquium paper: resistance, resilience, and redundancy in microbial communities. Proc. Natl. Acad. Sci. U. S. A. 105:11512–11519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Andersson AF, et al. 2008. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS One 3:e2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Andersson AF, Riemann L, Bertilsson S. 2010. Pyrosequencing reveals contrasting seasonal dynamics of taxa within Baltic Sea bacterioplankton communities. ISME J. 4:171–181 [DOI] [PubMed] [Google Scholar]
- 6. Atlas RM, Horowitz A, Krichevsky M, Bej AK. 1991. Response of microbial populations to environmental disturbance. Microb. Ecol. 22:249–256 [DOI] [PubMed] [Google Scholar]
- 7. Baas Becking LGM 1934. Geobiologie of inleiding tot de milieukunde.W. P. Van Stockum & Zoon, The Hague, The Netherlands [Google Scholar]
- 8. Barberán A, Casamayor E. 2010. Global phylogenetic community structure and β-diversity patterns in surface bacterioplankton metacommunities. Aquat. Microb. Ecol. 59:1–10 [Google Scholar]
- 9. Boström HK, Simu K, Hagström Å, Riemann L. 2004. Optimization of DNA extraction for quantitative marine bacterioplankton community analysis. Limnol. Oceanogr. Methods 2:365–373 [Google Scholar]
- 10. Bouvier TC, del Giorgio PA. 2002. Compositional changes in free-living bacterial communities along a salinity gradient in two temperate estuaries. Limnol. Oceanogr. 47:453–470 [Google Scholar]
- 11. Carlson CA, Ducklow HW. 1996. Growth of bacterioplankton and consumption of dissolved organic carbon in the Sargasso Sea. Aquat. Microb. Ecol. 10:69–85 [Google Scholar]
- 12. Caron DA, Countway PD. 2009. Hypotheses on the role of the protistian rare biosphere in a changing world. Aquat. Microb. Ecol. 57:227–238 [Google Scholar]
- 13. Chin-Leo G, Benner R. 1992. Enhanced bacterioplankton production and respiration at intermediate salinities in the Mississippi River plume. Mar. Ecol. Prog. Ser. 87:87–103 [Google Scholar]
- 14. Cottrell MT, Kirchman DL. 2000. Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl. Environ. Microbiol. 66:1692–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Covert JS, Moran MA. 2001. Molecular characterization of estuarine bacterial communities that use high- and low-molecular weight fractions of dissolved organic carbon. Aquat. Microb. Ecol. 25:127–139 [Google Scholar]
- 16. Crump BC, Hopkinson CS, Sogin ML, Hobbie JE. 2004. Microbial biogeography along an estuarine salinity gradient: combined influences of bacterial growth and residence time. Appl. Environ. Microbiol. 70:1494–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. DeSantis TZ, et al. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Doak DF, et al. 1998. The statistical inevitability of stability-diversity relationships in community ecology. Am. Nat. 151:264–276 [DOI] [PubMed] [Google Scholar]
- 19. Dowd SE, Sun Y, Wolcott RD, Domingo A, Carroll JA. 2008. Bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) for microbiome studies: bacterial diversity in the ileum of newly weaned Salmonella-infected pigs. Foodborne Pathog. Dis. 5:459–472 [DOI] [PubMed] [Google Scholar]
- 20. Eilers H, Pernthaler J, Amann R. 2000. Succession of pelagic marine bacteria during enrichment: a close look at cultivation-induced shifts. Appl. Environ. Microbiol. 66:4634–4640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ewing B, Green P. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186–194 [PubMed] [Google Scholar]
- 22. Ferguson R, Buckley E, Palumbo A. 1984. Response of marine bacterioplankton to differential filtration and confinement. Appl. Environ. Microbiol. 47:49–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fuchs BM, Zubkov MV, Sahm K, Burkill PH, Amann R. 2000. Changes in community composition during dilution cultures of marine bacterioplankton as assessed by flow cytometric and molecular biological techniques. Environ. Microbiol. 2:191–201 [DOI] [PubMed] [Google Scholar]
- 24. Fuhrman JA. 2009. Microbial community structure and its functional implications. Nature 459:193–199 [DOI] [PubMed] [Google Scholar]
- 25. Fuhrman JA, Comeau DE, Hagström Å, Chan AM. 1988. Extraction from natural planktonic microorganisms of DNA suitable for molecular biological studies. Appl. Environ. Microbiol. 54:1426–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Galand PE, Casamayor EO, Kirchman DL, Lovejoy C. 2009. Ecology of the rare microbial biosphere of the Arctic Ocean. Proc. Natl. Acad. Sci. U. S. A. 106:22427–22432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Giovannoni SJ. 1991. The polymerase chain reaction, p 177–201 In Stackebrandt E, Goodfelow M. (ed), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Inc., New York, NY [Google Scholar]
- 28. Glöckner FO, et al. 2000. Comparative 16S rRNA analysis of lake bacterioplankton reveals globally distributed phylogenetic clusters including an abundant group of actinobacteria. Appl. Environ. Microbiol. 66:5053–5065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Grasshoff K, Ehrhardt M, Kremling K. 1983. Methods of seawater analysis.Verlag Chemie, GmbH, Weinheim, Germany [Google Scholar]
- 30. Haas B, et al. 2011. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequencing PCR amplicons. Genome Res. 21:494–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hagström Å, Ammerman JW, Henrichs S, Azam F. 1984. Bacterioplankton growth in seawater. II. Organic matter utilization during steady-state growth in seawater cultures. Mar. Ecol. Prog. Ser. 18:41–48 [Google Scholar]
- 32. Hagström Å, Azam F, Kuparinen J, Zweifel UL. 2001. Pelagic plankton growth and resource limitations in the Baltic Sea, p 177–210 In Wulff F, Rahm L, Larsson P. (ed), Ecological studies, a system analysis of the Baltic Sea. Springer, Berlin, Germany [Google Scholar]
- 33. Holmfeldt K, et al. 2009. Diversity and abundance of freshwater Actinobacteria along environmental gradients in the brackish northern Baltic Sea. Environ. Microbiol. 11:2042–2054 [DOI] [PubMed] [Google Scholar]
- 34. Huse SM, Huber JA, Morrison HG, Sogin ML, Welch DM. 2007. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 8:R143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Huse SM, Welch DM, Morrison HG, Sogin ML. 2010. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ. Microbiol. 12:1889–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jones SE, Lennon JT. 2010. Dormancy contributes to the maintenance of microbial diversity. Proc. Natl. Acad. Sci. U. S. A. 107:5881–5886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kan J, Suzuki MT, Wang K, Evans SE, Chen F. 2007. High temporal but low spatial heterogeneity of bacterioplankton in the Chesapeake Bay. Appl. Environ. Microbiol. 73:6776–6789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kirchman DL, Cottrell MT, Lovejoy C. 2010. The structure of bacterial communities in the western Arctic Ocean as revealed by pyrosequencing of 16S rRNA genes. Environ. Microbiol. 12:1132–1143 [DOI] [PubMed] [Google Scholar]
- 39. Kirchman DL, Rich JH. 1996. Regulation of bacterial growth rates by dissolved organic carbon and temperature in the equatorial Pacific Ocean. Microb. Ecol. 33:11. [DOI] [PubMed] [Google Scholar]
- 40. Langenheder S, Kisand V, Wikner J, Tranvik LJ. 2003. Salinity as a structuring factor for the composition and performance of bacterioplankton degrading riverine DOC. FEMS Microbiol. Ecol. 45:189–202 [DOI] [PubMed] [Google Scholar]
- 41. Langenheder S, Lindström ES, Tranvik LJ. 2005. Weak coupling between community composition and functioning of aquatic bacteria. Limnol. Oceanogr. 50:957–967 [Google Scholar]
- 42. Lozupone C, Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71:8228–8235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lysenko O. 1961. Pseudomonas—an attempt at a general classification. J. Gen. Microbiol. 25:379–408 [DOI] [PubMed] [Google Scholar]
- 44. Martinez J, Smith DC, Steward GF, Azam F. 1996. Variability in ectohydrolytic enzyme activities of pelagic marine bacteria and its significance for substrate processing in the sea. Aquat. Microb. Ecol. 10:223–230 [Google Scholar]
- 45. Mills AL, Mallory LM. 1987. The community structure of sessile heterotrophic bacteria stressed by acid mine drainage. Microb. Ecol. 14:219–232 [DOI] [PubMed] [Google Scholar]
- 46. Moore RL, Weiner RM, Gebers R. 1984. Genus Hypomonas Pongratz 1957 nom. rev. emend., Hypomonas polymorpha Pongratz 1957 nom. rev. emend., and Hyphomonas neptunium (Leifson 1964) comb. nov. emend. (Hyphomicrobium neptunium). Int. J. Syst. Bacteriol. 34:71–73 [Google Scholar]
- 47. Moriarty DJW. 1986. Measurement of bacterial-growth rates in aquatic systems using rates of nucleic-acid synthesis. Adv. Microb. Ecol. 9:245–292 [Google Scholar]
- 48. Mou X, Sun S, Edwards RA, Hodson RE, Moran MA. 2008. Bacterial carbon processing by generalist species in the coastal ocean. Nature 451:708–711 [DOI] [PubMed] [Google Scholar]
- 49. Muyzer G, et al. 1998. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology, p 1–27 In Akkermans ADL, van Elsas JD, de Bruin FJ. (ed), Molecular microbial ecology manual. Kluwer Academic Publishers, London, United Kingdom [Google Scholar]
- 50. Muyzer G, de Waal EC, Uitterlinden AG. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pedrós-Alió C. 2006. Marine microbial diversity: can it be determined? Trends Microbiol. 14:257–263 [DOI] [PubMed] [Google Scholar]
- 52. Pernthaler J, Amann R. 2005. Fate of heterotrophic microbes in pelagic habitats: focus on populations. Microbiol. Mol. Biol. Rev. 69:440–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pinhassi J, Berman T. 2003. Differential growth response of colony-forming α- and γ-proteobacteria in dilution culture and nutrient addition experiments from Lake Kinneret (Israel), the eastern Mediterranean Sea, and the Gulf of Eilat. Appl. Environ. Microbiol. 69:199–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pinhassi J, Hagström Å. 2000. Seasonal succession in marine bacterioplankton. Aquat. Microb. Ecol. 21:245–256 [Google Scholar]
- 55. Pinhassi J, et al. 2003. Spatial variability in bacterioplankton community composition at the Skagerrak-Kattegat Front. Mar. Ecol. Prog. Ser. 255:1–13 [Google Scholar]
- 56. Pommier T, Canbäck B, Lundberg P, Hagström Å, Tunlid A. 2009. RAMI: a tool for identification and characterization of phylogenetic clusters in microbial communities. Bioinformatics 25:736–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pommier T, et al. 2007. Global patterns of diversity and community structure in marine bacterioplankton. Mol. Ecol. 16:867–880 [DOI] [PubMed] [Google Scholar]
- 58. Quince C, et al. 2009. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat. Methods 6:639–641 [DOI] [PubMed] [Google Scholar]
- 59. Rappé MS, Vergin K, Giovannoni SJ. 2000. Phylogenetic comparisons of a coastal bacterioplankton community with its counterparts in open ocean and freshwater systems. FEMS Microbiol. Ecol. 33:219–232 [DOI] [PubMed] [Google Scholar]
- 60. Riemann L, Azam F. 2002. Widespread N-acetyl-d-glucosamine uptake among pelagic marine bacteria and its ecological implications. Appl. Environ. Microbiol. 68:5554–5562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Riemann L, et al. 2008. The native bacterioplankton community in the central Baltic Sea is influenced by freshwater bacterial species. Appl. Environ. Microbiol. 74:503–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Riemann L, Steward GF, Azam F. 2000. Dynamics of bacterial community composition and activity during a mesocosm diatom bloom. Appl. Environ. Microbiol. 66:578–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Siegel H, Gerth M, Tschersich G. 2006. Sea surface temperature development of the Baltic Sea in the period 1990-2004. Oceanologia 48:119–131 [Google Scholar]
- 64. Sogin ML, et al. 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere.” Proc. Natl. Acad. Sci. U. S. A. 103:12115–12120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Søndergaard M, Middelboe M. 1995. A cross-system analysis of labile organic carbon. Mar. Ecol. Prog. Ser. 118:283–294 [Google Scholar]
- 66. Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690 [DOI] [PubMed] [Google Scholar]
- 67. Stoeck T, Epstein S. 2009. Crystal ball-2009: protist and the rare biosphere. Environ. Microbiol. Rep. 1:20–22 [Google Scholar]
- 68. Taborda M, et al. 2009. Description of Idiomarina insulisalsae sp. nov., isolated from the soil of a sea salt evaporation pond, proposal to transfer the species of the genus Pseudidiomarina to the genus Idiomarina and emended description of the genus Idiomarina. Syst. Appl. Microbiol. 32:371–378 [DOI] [PubMed] [Google Scholar]
- 69. Tranvik LJ, Höfle MG. 1987. Bacterial growth in mixed cultures on dissolved organic carbon from humic and clear waters. Appl. Environ. Microbiol. 53:482–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wedborg M, Skoog A, Fogelqvist E. 1994. Organic carbon and humic substances in the Baltic Sea, Kattegat and Skagerrak, p 917–942 In Sense N, Miano TM. (ed), Humic substances in the global environment and implication on human health. Elsevier, Amsterdam, The Netherlands [Google Scholar]
- 71. Weiner RM, Devine RA, Powell DM, Dagasan L, Moore RL. 1985. Hyphomonas oceanitis sp. nov., Hyphomonas hirschiana sp. nov., and Hyphomonas jannaschiana sp. nov. Int. J. Syst. Bacteriol. 35:237–243 [Google Scholar]
- 72. Zwart G, et al. 2003. Rapid screening for freshwater bacterial groups by using reverse line blot hybridization. Appl. Environ. Microbiol. 69:5875–5883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zweifel UL, Blackburn N, Hagström Å. 1996. Cycling of marine dissolved organic matter. I. An experimental system. Aquat. Microb. Ecol. 11:65–77 [Google Scholar]
- 74. Zweifel UL, Wikner J, Hagström Å, Lundberg E, Norrmann B. 1995. Dynamics of dissolved organic carbon in a coastal ecosystem. Limnol. Oceanogr. 40:299–305 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.