Abstract
Clostridium acetobutylicum naturally produces acetone as well as butanol and ethanol. Since acetone cannot be used as a biofuel, its production needs to be minimized or suppressed by cell or bioreactor engineering. Thus, there have been attempts to disrupt or inactivate the acetone formation pathway. Here we present another approach, namely, converting acetone to isopropanol by metabolic engineering. Since isopropanol can be used as a fuel additive, the mixture of isopropanol, butanol, and ethanol (IBE) produced by engineered C. acetobutylicum can be directly used as a biofuel. IBE production is achieved by the expression of a primary/secondary alcohol dehydrogenase gene from Clostridium beijerinckii NRRL B-593 (i.e., adhB-593) in C. acetobutylicum ATCC 824. To increase the total alcohol titer, a synthetic acetone operon (act operon; adc-ctfA-ctfB) was constructed and expressed to increase the flux toward isopropanol formation. When this engineering strategy was applied to the PJC4BK strain lacking in the buk gene (encoding butyrate kinase), a significantly higher titer and yield of IBE could be achieved. The resulting PJC4BK(pIPA3-Cm2) strain produced 20.4 g/liter of total alcohol. Fermentation could be prolonged by in situ removal of solvents by gas stripping, and 35.6 g/liter of the IBE mixture could be produced in 45 h.
INTRODUCTION
Butanol is an important industrial chemical and has been receiving increased attention as a better biofuel than ethanol because of its higher energy density and less hygroscopy. Butanol can be produced by several anaerobic microorganisms that belong to the genus Clostridium. The most extensively studied solventogenic species is Clostridium acetobutylicum, which typically produces butanol, acetone, and ethanol at the mass ratio of 6:3:1 (17, 20, 21). Even though C. acetobutylicum and other solventogenic strains have been used in large-scale butanol production, this so-called acetone-butanol-ethanol (ABE) fermentation process is currently considered less economical than ethanol fermentation using yeasts (32). Since acetone cannot be used as a fuel due to its corrosiveness to engine parts that are composed of rubber or plastic, its coproduction with butanol (and ethanol) is viewed as undesirable because it reduces the butanol yield per unit mass of substrate utilized. Thus, reducing acetone production has been an important objective of clostridial metabolic engineering (28).
Theoretical analysis of organisms' pathway stoichiometry and cellular energetic needs (29, 30) and some experimental results (3) suggest that it is possible to convert most of a sugar substrate into butanol alone. However, attempts to reduce acetone production by metabolic engineering resulted in decreased butanol production and the accumulation of acetic and butyric acids (15, 37, 40, 41). Downregulation of the ctfB gene, encoding the β subunit of coenzyme A transferase (CoAT), using antisense RNA (asRNA) caused a significant decrease of butanol production as well as acetone production. Later, it was found that the expression of the adhE1 gene, encoding the main bifunctional alcohol/aldehyde dehydrogenase (AdhE1), was significantly reduced (41), and the cooverexpression of the adhE1 gene with ctfB asRNA recovered butanol production. Unexpectedly, however, this caused increased production of acetone and ethanol as well (40). Another study aimed to eliminate acetone production by inactivating the gene for acetoacetate decarboxylase (AADC; encoded by the adc gene) using a mobile group II intron (15). This disruption resulted in decreased butanol production and the accumulation of acetic acid unless the electron flow was altered by the addition of methyl viologen.
The inability so far to engineer cells for butanol production without acetone or organic acid production in C. acetobutylicum or other solventogenic clostridia can perhaps be explained based on physiological characteristics during the formation of metabolic products. Solvent production in solventogenic clostridia is widely accepted to be a defensive mechanism against a low culture pH resulting from the production of acetic and butyric acids during exponential cell growth (13, 43). This acidification is remedied by the induction of the solventogenic genes, including adhE1, ctfAB, and adc. As a result, butyric and acetic acids are reassimilated by CoAT, which leads to acetone production, increases culture and cytosolic pH, and also regenerates butyryl-CoA and acetyl-CoA, which are consequently converted to butanol and ethanol, respectively. From this point of view, it appears that acetone formation is essential for cytosolic detoxification from carboxylic acids and protons. Hence, we offer an alternative strategy for avoiding acetone production by solventogenic clostridia.
Isopropanol is the simplest secondary alcohol and is used as an industrial solvent in various applications, such as use as a cleaning agent. It can also be used as a fuel additive for the preparation of high-octane gasoline (33), which is a motivation for this study. Acetone can be converted into isopropanol by a single reduction step. If acetone in ABE fermentation can be converted into isopropanol, C. acetobutylicum can produce mixed alcohols that can be directly used as a biofuel mixture. In this strategy, the acetone production pathway is not disrupted, and thus it is expected that butanol production will not be compromised. Isopropanol is produced in nature by several solventogenic clostridia (4, 7), which produce butanol and ethanol as well. However, studies on isopropanol-producing clostridia have been limited to the characterization of the isopropanol production pathway and enzymes (14) and process optimization (19, 22).
In this study, we report for the first time the development of an isopropanol-butanol-ethanol (IBE)-producing C. acetobutylicum strain by introducing the secondary alcohol dehydrogenase (SADH, encoded by the adhB-593 gene [Fig. 1]) of Clostridium beijerinckii NRRL B-593. Also, it was attempted to further increase total alcohol production by employing a synthetic acetone operon consisting of the adc, ctfA, and ctfB genes. Then, the adhB-593 gene and the synthetic acetone operon were introduced into the butanol-hyperproducing C. acetobutylicum strain PJC4BK, a butyrate kinase-inactivated strain (9). Production of the IBE mixture by the engineered PJC4BK strain was examined by batch fermentation. Also, continuous fermentation with solvent removal by gas stripping was carried out to increase the titer and productivity of the IBE mixture.
Fig 1.
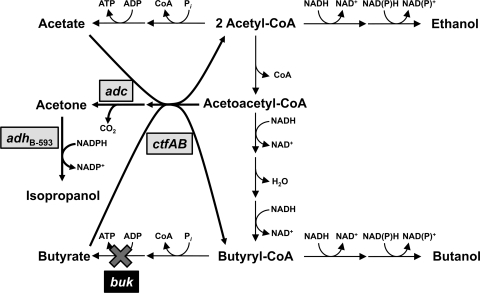
Schematic diagram of the metabolic pathways of engineered C. acetobutylicum strains. Genes that were overexpressed in C. acetobutylicum are shown in gray boxes, and the amplified reactions are shown as thick arrows. The use of a buk-inactivated strain resulted in enhanced IBE production. The genes are adc, the acetoacetate decarboxylase gene, ctfAB, the coenzyme A transferase genes, and adhB-593, the primary/secondary alcohol dehydrogenase gene from C. beijerinckii NRRL B-593.
MATERIALS AND METHODS
Strains, electrotransformation, and flask culture conditions.
The strains used in this study are shown in Table 1. Wild-type C. acetobutylicum ATCC 824 (WT 824) was maintained as spores, as previously reported (26), except that the spores were suspended in clostridial growth medium (CGM) containing (per liter) 0.75 g KH2PO4, 0.75 g K2HPO4, 1.0 g NaCl, 0.017 g MnSO4 · 5H2O, 0.70 g MgSO4 · 7H2O, 0.01 g FeSO4 · 7H2O, 2.0 g l-asparagine, 5.0 g yeast extract, 2.0 g (NH4)2SO4, and 80 g glucose (34) supplemented with 15% (vol/vol) glycerol. Spores were germinated by heating them at 70 to 80°C for 10 min after inoculation. For growth on solid medium, C. acetobutylicum was grown anaerobically at 37°C on 2× YTG (pH 5.8; 16 g Bacto tryptone, 10 g yeast extract, 4 g NaCl, and 5 g glucose per liter) agar plates. For recombinant strains, erythromycin was added to liquid and solid media at the final concentrations of 80 and 40 μg/ml, respectively, and thiamphenicol was added at the final concentration of 5 μg/ml. Electrotransformation of C. acetobutylicum was performed by following the protocol published in a previous report (25). Frozen stocks of recombinant strains were prepared by mixing 1 ml of a mid-exponential-phase CGM culture (optical density at 600 nm [OD600] of ca. 1.5) with 0.5 ml of 50% (vol/vol) glycerol and stored at −80°C. For flask cultures, a single colony was inoculated into a test tube containing 10 ml of CGM and anaerobically cultured at 37°C until the OD600 reached 2.0. Capped flasks containing 150 ml CGM supplemented with 60 g/liter glucose were inoculated with 5 ml of this culture and grown at 37°C. At 48 h after inoculation, the samples were taken from the flasks and used in further analysis.
Table 1.
Strains and plasmids used in this study
| Strain, derivative, or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| C. acetobutylicum ATCC 824 | Wild type | KBRCb |
| C. acetobutylicum PJC4BK | buk::erm | 9 |
| E. coli TOP10 | Invitrogen | |
| Derivatives and plasmids | ||
| pAN1 | Cmr, ϕ3T I gene, p15A origin | 24 |
| pIMP1 | Apr MLSrrepL, ColE1 origin | 25 |
| pIMP2 | pIMP1 derivative with a substituted multiple cloning site | This study |
| pUC18 | Apr, ColE1 origin | 27 |
| pUCadcT | pUC18 derivative with adc terminator insertion | This study |
| pUCadcPT | pUCadcT derivative with adc promoter insertion | This study |
| pSADH1 | pUCadcPT derivative with adhB-593 (adc promoter) | This study |
| pACT | pIMP2 derivative with adc ctfAB (adc promoter) insertion | This study |
| pIPA1 | pIMP2 derivative containing adhB-593 (adc promoter) | This study |
| pIPA3 | pACT derivative containing adhB-593 (adc promoter) | This study |
| pIMP30 | pIMP2 derivative, AprrepL, ColE1 origin | This study |
| pIMP3 | pIMP30 derivative, Cmr | This study |
| pACT-Cm | pIMP3 derivative with adc ctfAB (adc promoter) insertion | This study |
| pIPA3-Cm2 | pACT-Cm derivative containing adhB-593 (adc promoter) | This study |
Cmr, chloramphenicol/thiamphenicol resistance; MLSr, erythromycin resistance; adc, acetoacetate decarboxylase gene from C. acetobutylicum; ctfAB, CoA transferase genes from C. acetobutylicum; adhB-593, a primary/secondary alcohol dehydrogenase gene from C. beijerinckii NRRL B-593; Apr, ampicillin resistance; repL, pIM13g-positive origin of replication.
KBRC, Korea Research Institute of Bioscience and Biotechnology Biological Resource Center, Daejeon, Republic of Korea.
Plasmid construction.
The plasmids used in this study are shown in Table 1. Primers used are listed in Table 2. All restriction enzymes, calf intestine alkaline phosphatase, and Antarctic phosphatase used in this study were purchased from New England BioLabs (Ipswich, MA). T4 DNA ligase (Roche, Basel, Switzerland) was used for ligation, and a proofreading Pfu DNA polymerase (Solgent, Daejeon, Republic of Korea) was used for PCR. Prior to the construction of pACT, the multiple cloning site (MCS) of pIMP1 was modified. Primers P01 and P02 were mixed and annealed to make a synthetic MCS. This product was ligated with the EcoRI-BamHI doubly digested plasmid pIMP1 to obtain pIMP2. To construct pACT, the fragment containing the adc promoter and open reading frame were amplified from C. acetobutylicum genomic DNA using primers P03 and P04. The fragment of the ctfAB genes was amplified using primers P05 and P06. These fragments were used for recombinant PCR using primers P03 and P06, producing a synthetic acetone operon, adc-ctfA-ctfB, under the control of the adc promoter. This product and pIMP2 were digested with BamHI-MluI and ligated to obtain pACT.
Table 2.
Primers used in this study
| Primer | Sequence |
|---|---|
| P01 | 5′-GATCCACTCTAGAGTACGCGTACGCATGCAACCATGGTTCCCGGGATG-3′ |
| P02 | 5′-AATTCATCCCGGGAACCATGGTTGCATGCGTACGCGTACTCTAGAGTG-3′ |
| P03 | 5′-ATATGGATCCAAGTGTACTTTTATTTTCGAAAGC-3′ |
| P04 | 5′-AATCCCTCCTTTCCATTTAAGGTAACTCTTATTTTTA-3′ |
| P05 | 5′-GTTACCTTAAATGGAAAGGAGGGATTAAAATGAACTCT-3′ |
| P06 | 5′-ATATACGCGTCTAAACAGCCATGGGTCTAAGTT-3′ |
| P07 | 5′-GATCCACTACGGCCGTAAAAATAAGAGTTACCTTAAATGGTAACTCTTATTTTTTTAATGC-3′ |
| P08 | 5′-AATTGCATTAAAAAAATAAGAGTTACCATTTAAGGTAACTCTTATTTTTACGGCCGTAGTG-3′ |
| P09 | 5′-ATATCTGCAGAAGTGTACTTTTATTTTCGAAAGC-3′ |
| P10 | 5′-ATATGGATCCTAATAATGTTTAGCTTTTCTAACAT-3′ |
| P11 | 5′-ATATGGATCCTAAGGAGGAACATATTTTATGAAAG-3′ |
| P12 | 5′-ATATCGGCCGTTATAATATAACTACTGCTTTAATTA-3′ |
| P13 | 5′-CGGGCCTCTTCGCTATTACG-3′ |
| P14 | 5′-ATATCCCGGGGGAATTGTGAGCGGATAACA-3′ |
| P15 | 5′-GAGGCAAATGAAATAGATTGACCTC-3′ |
| P16 | 5′-ATATAGATCTGGCGTAATCATGGTCATAGCTG-3′ |
| P17 | 5′-AATTAGATCTATGCAGGAATTGACGATTTAAA-3′ |
| P18 | 5′-GCAAGGCGATTAAGTTGGGT-3′ |
| P19 | 5′-ATATGGATCCTGCATCTAGAAGAATAGCAGATG-3′ |
| P20 | 5′-TTCAAATACCAT GTTTGACCTCCTAAAATTTTATAG-3′ |
| P21 | 5′-AGGAGGTCAAACATGGTATTTGAAAAAATTGATAAAAA-3′ |
| P22 | 5′-ATATCTGCAGTATGAGTCGACATTAAAAAAATAAG-3′ |
Prior to the construction of pSADH1, the adc terminator and promoter were cloned into pUC18. The fragment containing an EagI site and the adc terminator region was obtained by annealing primers P07 and P08. This fragment was ligated with pUC18 doubly digested with BamHI-EcoRI to obtain pUCadcT. The adc promoter region was amplified from the C. acetobutylicum genomic DNA using primers P09 and P10. The product and pUCadcT were digested with PstI-BamHI and ligated to obtain pUCadcPT. The adhB-593 gene was amplified from C. beijerinckii NRRL B-593 genomic DNA using primers P11 and P12. The product was digested with BamHI-EagI and ligated with pUCadcPT doubly digested with BamHI-EagI to obtain pSADH1. Using primers P13 and P14, the adc promoter-adhB-593-adc terminator fragment (F1) was amplified from pSADH1. F1 and pIMP2 were digested with SphI-XmaI and ligated to obtain pIPA1. Plasmid pIPA3 was obtained by ligating pACT and F1 doubly digested with SphI and XmaI.
Plasmids constructed as described above can be used in the wild-type C. acetobutylicum strain but not in C. acetobutylicum PJC4BK, which is erythromycin resistant (9). Another antibiotic that can be used in C. acetobutylicum is thiamphenicol (Th). To construct a shuttle vector that confers thiamphenicol resistance, the pUC9 portion of pIMP2 was amplified using primers P15 and P16, and the portion containing the repL replicon but not the gene cross-resistant to macrolides, lincosamides, and streptogramins (MLSr) (for erythromycin resistance) (25) was amplified using primers P17 and P18. These two products were digested with HindIII-BglII and ligated to obtain pIMP30. The putative promoter region of the C. beijerinckii NCIMB 8052 thiolase gene (Cbei_0411; nucleotides 498921 to 499120, GenBank accession number NC_009617.1) was amplified from C. beijerinckii NCIMB 8052 genomic DNA using primers P19 and P20, and the cat gene was amplified from pSOS95-Cm using primers P21 and P22. These fragments were used for recombinant PCR with primers P19 and P22, producing a recombinant Th marker. This fragment and pIMP30 were digested with BglII-ClaI and ligated to obtain pIMP3. Using this plasmid, pACT-Cm and pIPA3-Cm2 were obtained by the same procedure used for the construction of pACT and pIPA3, respectively.
Batch fermentation.
Batch fermentation was carried out in a LiFlus GX bioreactor (Biotron, Gyeonggi-Do, Republic of Korea) containing 1.8 liters of CGM supplemented with 80 g/liter glucose. The spore suspension (20 μl) in the case of the wild-type strain and a single colony of the recombinant strain less than 5 days old was inoculated into a capped tube containing 10 ml of CGM, heat shocked, and grown vigorously. This preculture was inoculated into a 500-ml flask containing 200 ml of CGM. When the cell density in the flask reached an OD600 of ∼1.0 to 2.0, the bioreactor was inoculated with the flask culture. The pH was maintained above 5.0 by using ammonia solution.
Enzyme assay.
In order to measure enzyme activities, cells cultured in flasks were harvested at the OD600 of ca. 1.0 (acidogenic-phase sample). Cells were further cultured for an additional 18 h and were harvested again (solventogenic-phase sample). Preparation of crude cell extract was carried out according to the work of Wiesenborn et al. (42), except that cells were anaerobically lysed using a Bioruptor sonicator (Diagenode, Belgium). All procedures were carried out in an anaerobic chamber. Total protein concentration was measured by the Bradford method using bovine serum albumin as a standard. Consumption of the substrate was observed with a GeneQuant 1300 spectrophotometer (GE Healthcare, United Kingdom). Cell extracts (0.1 ml) of the wild-type strain and recombinant strains were used in the enzyme assay. The CoAT activity was measured by following a previous protocol (42). One unit of CoAT activity was defined as the amount of enzyme necessary for the consumption of 1 μmol of acetoacetyl-CoA per min. The assay mixture for SADH contained, in a final volume of 1.0 ml, 50 mM Tris · Cl (pH 7.5), 1 mM dithiothreitol, 0.2 mM NADPH or NADH, and 6.7 mM acetone (14). One unit of SADH activity was defined as 1 μmol of NADPH or NADH oxidized per min. AADC activity was determined as described previously (8), and 1 unit of AADC activity was expressed by production of 1 μl of CO2 per min.
Fed-batch fermentation with gas stripping.
The gas-stripping method used in this study is similar to that described previously (6), except that mechanical agitation at 200 rpm and a pH control were applied. Briefly, the 7-liter bioreactor containing 1.8 liters CGM was used in the fed-batch fermentation to control foam without adding antifoam agent. The inoculum used was 200 ml, thus giving a headspace volume of 5 liters. After 12 h of cultivation, gas stripping was performed by recycling the gas in the headspace of the reactor at 6 liters/min using a vacuum pump. The stripped solvents were condensed using a Dimroth condenser (60 by 800 mm) at −5°C. Prior to the gas stripping, the remaining air in the condenser was discharged by oxygen-free nitrogen gas. Whenever the glucose concentration in the fermentation broth became less than 10 g/liter, 100 ml of a 500-g/liter glucose solution was added to the reactor. The working volume of the reactor was maintained at 2 liters by adding sterilized distilled water to compensate for the loss of water due to the gas stripping.
Analytical procedure.
The concentration of glucose was analyzed with a YSI 2700 Select biochemistry analyzer (YSI Life Sciences, OH). The concentrations of solvents and acids were determined by gas chromatography (model 7890; Agilent, CA) equipped with an 80/120 Carbopack B AW packed glass column (Supelco, PA) and a flame ionization detector (FID). Cell growth was monitored by measuring the OD600 using an Ultrospec 3100 Pro spectrophotometer (Amersham Biosciences, Uppsala, Sweden).
RESULTS
Expression of the secondary alcohol dehydrogenase of C. beijerinckii in C. acetobutylicum.
To examine the effect of the secondary alcohol dehydrogenase on the conversion of acetone into isopropanol, plasmid pIPA1 was constructed by cloning the secondary alcohol dehydrogenase gene (adhB-593) of C. beijerinckii NRRL B-593 under the control of the C. acetobutylicum adc promoter into pIMP2 (see Materials and Methods), so that this secondary alcohol dehydrogenase gene would be expressed along with the genes needed for acetone production. This plasmid was introduced into wild-type C. acetobutylicum ATCC 824 (WT 824) after methylation, which prevents the digestion by an endogenous restriction endonuclease, Cac824I (23). The WT 824 strain and that transformed with pIPA1, ATCC 824(pIPA1) [824(pIPA1)], were cultured at 37°C in CGM supplemented with 60 g/liter of glucose.
At 48 h after inoculation, WT 824 produced 3.8, 8.2, and 0.8 g/liter of acetone, butanol, and ethanol, respectively, from 40 g/liter of glucose (Table 3). Residual concentrations of acetic and butyric acids were 1.3 and 0.6 g/liter, respectively. In the case of 824(pIPA1), the amount of glucose utilized was slightly lower. Consequently, the concentration of total solvent was also slightly lower than that obtained with WT 824. Additional incubation did not increase the glucose uptake in both the WT 824 and 824(pIPA1) strains. Conversion of acetone into isopropanol was successfully achieved by the expression of the adhB-593 gene; acetone and isopropanol were produced at 0.1 and 3.1 g/liter. The butanol titer obtained with 824(pIPA1) was 7.3 g/liter, which was slightly lower than that obtained with WT 824. The isopropanol/butanol ratio was 0.52 mol/mol, which was slightly lower than the acetone/butanol ratio obtained with WT 824 (0.59 mol/mol).
Table 3.
Flask cultures of the engineered and control strains
| Strain | Titer for characteristic or product (g/liter)a |
||||||
|---|---|---|---|---|---|---|---|
| Glucose utilized | Acetone | Isopropanol | Butanol | Ethanol | Acetic acid | Butyric acid | |
| Wild-type | 40.0 ± 1.0 | 3.8 ± 0.2 | ND | 8.2 ± 0.3 | 0.8 ± 0.0 | 1.3 ± 0.1 | 0.6 ± 0.1 |
| 824(pIPA1) | 38.8 ± 1.4 | 0.1 ± 0.0 | 3.1 ± 0.2 | 7.4 ± 0.2 | 0.8 ± 0.1 | 1.4 ± 0.2 | 0.9 ± 0.1 |
| 824(pACT) | 47.7 ± 2.0 | 5.1 ± 0.2 | ND | 8.9 ± 0.2 | 0.9 ± 0.1 | 0.5 ± 0.1 | 0.5 ± 0.0 |
| 824(pIPA3) | 43.0 ± 1.3 | 0.1 ± 0.0 | 5.1 ± 0.1 | 8.0 ± 0.1 | 0.8 ± 0.1 | 1.0 ± 0.1 | 0.5 ± 0.0 |
All results shown are means ± standard deviations from triplicate experiments. ND, not determined.
Effects of overexpressing a synthetic acetone operon on IBE production.
Overexpression of three key genes of the acetone pathway, adc, ctfA, and ctfB, has been shown to increase total solvent production as well as butanol production (24). Thus, it was thought that greater total alcohol production would be possible by combining enhanced acetone flux with adhB-593 expression. Thus, a synthetic acetone operon comprising the adc, ctfA, and ctfB genes under the control of the adc promoter (act operon) was cloned into pIMP2 to construct pACT. Then, plasmid pIPA3 was constructed by cloning the adhB-593 gene fused to the adc promoter and the adc terminator into pACT (see Materials and Methods for details). These plasmids were introduced into C. acetobutylicum, and flask cultures of the resulting recombinant strains, 824(pACT) and 824(pIPA3), were performed.
Consistent with a previous report (24), 824(pACT) produced more acetone and butanol (Table 3) than the wild-type strain (32% and 8% more, respectively). Also, 824(pACT) consumed more glucose, and the acetone/butanol ratio increased by 22%. The isopropanol titer obtained with 824(pIPA3) was 5.1 g/liter, which is 64% greater than that obtained with 824(pIPA1). The butanol concentration obtained was 8.0 g/liter, which was comparable to that obtained with the wild type (8.2 g/liter) but less than that (8.9 g/liter) obtained with 824(pACT).
IBE production in pH-controlled batch fermentation.
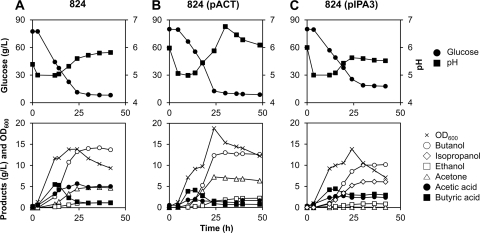
As the results from flask cultures were promising, the pH-controlled batch fermentations were carried out to examine the performance of 824(pIPA3). The results were then compared with those obtained with two control strains, the WT 824 and 824(pACT) strains. The time profiles of duplicate batch fermentations for the WT 824, 824(pACT), and 824(pIPA3) strains are shown in Fig. 2. In the case of the wild-type strain, the pH rose after about 40 g/liter of glucose was consumed. In the case of 824(pACT), acid reassimilation began earlier, and the pH began to rise when less than 25 g/liter of glucose was consumed. The pH decreased after the peak, and the concentrations of acetic and butyric acids slightly increased (Fig. 2B). At the pH of 6.76, the titers of acetic and butyric acids were 1.3 and 0.7 g/liter, and the acetic acid concentration increased to 1.6 g/liter afterwards, with a slight decrease of pH. The concentrations of glucose consumed by the WT 824 and 824(pACT) strains were not significantly different at ∼69.2 to 69.6 g/liter. At the point of the highest butanol titer in 824(pACT), the acetone titer was 7.0 g/liter, which was 46% higher than that (4.8 g/liter) obtained with WT 824. The residual concentrations of acetic and butyric acids were 68 and 42%, respectively, lower for 824(pACT) than for WT 824. Despite the lower residual concentrations of acids, the maximum butanol titer obtained with 824(pACT) was 13.0 g/liter, which is 8% lower than that (14.2 g/liter) obtained with WT 824.
Fig 2.
Time profiles of fermentations of C. acetobutylicum ATCC 824 (A), 824(pACT) (B), and 824(pIPA3) (C) performed at a pH of ≥5.0. The upper panels show the glucose concentrations and pHs, and the lower panels show the optical densities and product concentrations. Fermentations of each strain were conducted in duplicate using independently grown cultures, and average values are presented.
Cooverexpression of the act operon and the adhB-593 gene in the WT 824 strain showed results different from those of the flask cultivation. The 824(pIPA3) strain produced 17.1 g/liter total alcohol, with a yield of 0.28 g/g glucose; individual amounts were 6.1, 10.2, and 0.8 g/liter of isopropanol, butanol, and ethanol, respectively (Fig. 2C). The acetone titer was less than 0.1 g/liter. Even though the molar isopropanol yield of 824(pIPA3) was higher than the molar acetone yield of WT 824, the butanol titer and yield (0.16 versus 0.20 g/g glucose WT 824) were lower than those obtained with WT 824. Unexpectedly, glucose utilization in 824(pIPA3) was less than in the other strains. The peak pH value after solvent production was 5.63, much lower than that of 824(pACT), although 824(pIPA3) showed earlier pH elevation, as in 824(pACT). The residual concentrations of acetic and butyric acids were 2.4 and 2.9 g/liter, respectively, which were higher than those observed with 824(pACT).
IBE production in a buk-inactivated strain.
Although 824(pIPA3) resulted in increased total alcohol production, butanol production declined. To enhance the butanol production, the C. acetobutylicum PJC4BK strain, which is a buk-inactivated strain by Campbell-like integration and is known to be a better butanol producer than WT 824 (9, 12), was employed. PJC4BK is erythromycin resistant (9), and thus plasmid pIPA3-Cm2 harboring a chloramphenicol/thiamphenicol resistance marker was constructed and used (see Materials and Methods for details). After transformation, the recombinant PJC4BK strain harboring pIPA3-Cm2 was cultured at 37°C in a bioreactor containing 2 liter CGM supplemented with 80 g/liter glucose, and pH was controlled at above 5.0.
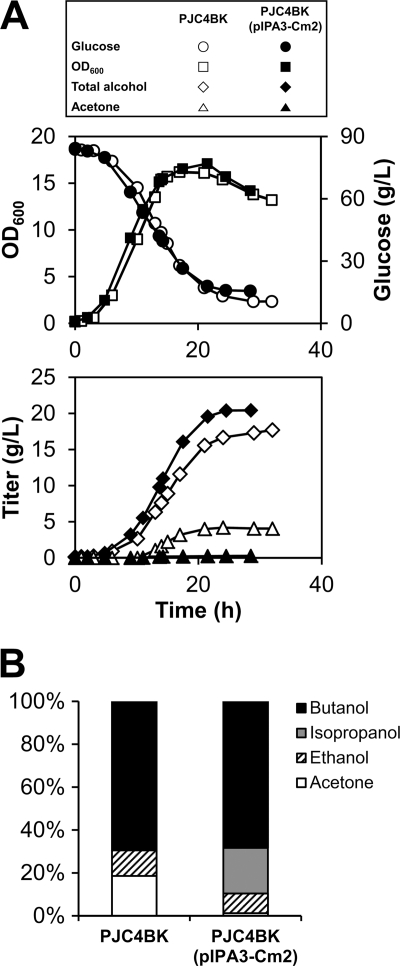
The PJC4BK(pIPA3-Cm2) strain produced 20.4 g/liter and 0.30 g/g glucose of total alcohol, which is 19% and 8% higher than levels produced by 824(pIPA3), respectively. The titers of isopropanol, butanol, and ethanol were 4.4, 14.1, and 1.9 g/liter, respectively. As expected, PJC4BK(pIPA3-Cm2) produced less butyric acid (1.1 versus 2.9 g/liter) but exhibited a higher peak concentration (6.0 versus 2.7 g/liter) and final concentration (3.9 versus 2.4 g/liter) of acetic acid than 824(pIPA3). Acetone was successfully converted to isopropanol by the expression of the adhB-593 gene in PJC4BK(pIPA3-Cm2) (Fig. 3A), but the effect of overexpression of the act operon was not greater in PJC4BK(pIPA3-Cm2) than that in 824(pIPA3); the ratio of isopropanol to butanol in PJC4BK(pIPA3-Cm2) was slightly higher than the ratio of acetone to butanol in PJC4BK (0.38 mol/mol versus 0.34 mol/mol) (Fig. 3B), but the difference was lower than those obtained with WT 824 and 824(pIPA3).
Fig 3.
Effects of buk inactivation on total alcohol production. Fermentations of each strain were conducted in duplicate using independently grown cultures, and average values are presented. (A) Time course of growth, concentration of glucose, total alcohol production, and acetone production during the fermentations of C. acetobutylicum PJC4BK and PJC4BK(pIPA3-Cm2) performed at pHs of ≥5.0. (B) Compositions of solvents produced by C. acetobutylicum PJC4BK and PJC4BK(pIPA3-Cm2).
Enzyme activities.
The activities of the three overexpressed enzymes, CoAT, AADC, and SADH, in two recombinant strains along with the control strain were measured using crude extracts (Table 4). It has been reported that CoAT might be a rate-limiting enzyme for acetone production (10, 41). 824(pIPA3) showed higher CoAT activities both in the acidogenic and in the solventogenic phase than WT 824. The CoAT activity in PJC4BK(pIPA3-Cm2) was lower than that in 824(pIPA3) both in the acidogenic phase (25 versus 40.1 mU/mg protein) and in the solventogenic phase (109 versus 458 mU/mg protein), although CoAT was overexpressed. A similar pattern was observed in the case of NADPH-dependent SADH activity. Interestingly, the CoAT and AADC activities in PJC4BK(pIPA3-Cm2) in the solventogenic phase were similar and slightly higher, respectively, than those in WT 824 (Table 4).
Table 4.
Enzyme activities in the recombinant strains
| Enzyme | Sp acta (mU/mg protein) |
|
|---|---|---|
| Acidogenic | Solventogenic | |
| CoAT in: | ||
| WT 824 | 1.7 ± 1.2 | 105 ± 19.0 |
| 824(pIPA3) | 40.1 ± 4.2 | 458 ± 48.7 |
| PJC4BK(pIPA3-Cm2) | 25 ± 5.8 | 109 ± 21.7 |
| AADC in: | ||
| WT 824 | 28.7 ± 15.3 | 105.6 ± 34.7 |
| 824(pIPA3) | 56.7 ± 20.8 | 243.3 ± 15.3 |
| PJC4BK(pIPA3-Cm2) | 39.6 ± 13.0 | 120.0 ± 20.0 |
| SADH (NADPH dependent)b in: | ||
| WT 824 | ND | ND |
| 824(pIPA3) | 1,910 ± 190 | 7,470 ± 690 |
| PJC4BK(pIPA3-Cm2) | 421 ± 51 | 1,320 ± 190 |
All results shown are means ± standard deviations from triplicate experiments. AADC activities are expressed in U/mg protein. ND, not detected.
NADH-dependent SADH activity was not detected in all strains.
Fed-batch fermentation with in situ removal of the solvents.
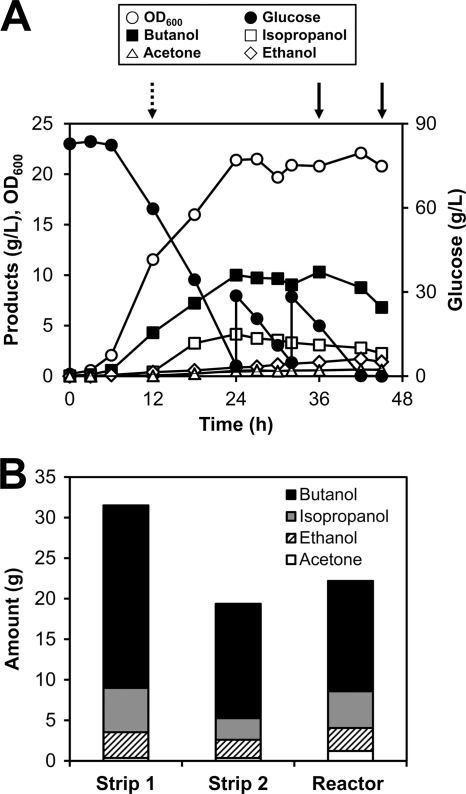
It is well known that the fermentation of solventogenic clostridia can be extended with in situ removal of solvents (6). To date, however, this experiment has not been conducted using metabolically engineered clostridial strains. Hence, the potential of strain PJC4BK(pIPA3-Cm2) for increased IBE production using a gas-stripping system was examined. The PJC4BK(pIPA3-Cm2) strain was cultured at 37°C in a bioreactor containing 2 liters CGM supplemented with 80 g/liter glucose, and pH was maintained above 5.0. The gas stripping was initiated at the 12th h of fermentation by circulating the headspace gas through the condenser at the flow rate of 6 liters/min. The pH increased from 14 h, reaching 5.68 at the 24th h of the fermentation and then decreased again. At this point, the pH was recontrolled at pH 5.6 with ammonia solution. Addition of ammonia solution to maintain the pH was crucial for maintaining fermentation (see Discussion for details). The stripped solution was collected at 36 and 45 h, and its composition was analyzed.
With in situ gas stripping, the PJC4BK(pIPA3-Cm2) strain completely consumed 132.9 g/liter of glucose, producing 35.6 g/liter of the IBE mixture (Fig. 4). The IBE yield was 0.27 g/g glucose, which is slightly lower than that obtained with the batch fermentation. In this experiment, the butanol titer was maintained at around 10 g/liter, and cell death was not observed until 42 h, at which time glucose was depleted (Fig. 4A). The maximum OD600 of 22.1 reached during gas-stripping fermentation was higher than that obtained from the batch fermentation (OD600, 17.1). This is due to the fact that the lower butanol concentration in the broth positively affected cell growth. Interestingly, the titers of acetic and butyric acids did not decrease even though solventogenesis was extended by gas stripping; they were maintained at about 3.0 and 1.8 g/liter, respectively. The amounts of acetone relative to the amounts of total solvents in stripped solutions 1 and 2 were 0.01 and 0.02 g/g total solvents, respectively (Fig. 4B), while that in the bioreactor was about 0.06 g/g total solvents.
Fig 4.
Gas-stripping fermentation of the PJC4BK(pIPA3-Cm2) strain. (A) Time profiles of the fed-batch fermentation of PJC4BK(pIPA3-Cm2). The dotted arrow indicates the starting point of gas stripping, while the solid arrows indicate the sampling points of stripped solution. (B) Compositions of solvents present in each stripped solution and the broth in the bioreactor after the fermentation.
DISCUSSION
C. acetobutylicum is a model organism for ABE fermentation. Several studies to produce butanol in more friendly organisms, such as Escherichia coli, at a level comparable to that of native producers have been conducted recently, but C. acetobutylicum is still an attractive platform due to its versatility of carbon source utilization (18) and its higher productivity than that of E. coli. Since 1-butanol is a promising biofuel, while acetone is not considered suitable as a fuel alternative, studies have been conducted to reduce acetone production. As already discussed earlier, this approach has not been successful to further increase butanol titers so far, indicating that the metabolic regulation of C. acetobutylicum is more complicated than is currently known. Here, we sought an alternative and simpler approach, namely, adding the conversion step from acetone to isopropanol, which can be used as a fuel additive, into C. acetobutylicum.
Our study thus aims to develop engineered strains suitable for IBE production by introducing a primary/secondary alcohol dehydrogenase from C. beijerinckii NRRL B-593. Flask cultures showed that the overexpression of the adc and ctfAB genes enhances total alcohol production by increasing the assimilation of organic acids (Table 3). Also, a higher yield (0.30 g/g glucose) and titer (20.4 g/liter) of the IBE mixture could be obtained by applying our strategy in the buk-deficient PJC4BK strain (Fig. 3). By combining in situ gas stripping with the fed-batch fermentation, very efficient IBE production was possible (35.4 g/liter within 45 h) (Fig. 3). This performance of PJC4BK(pIPA3-Cm2) exceeds that of natural IBE producers. In a previous study, several clostridial strains, including C. beijerinckii NRRL B-593, were shown to produce isopropanol without acetone production, but the range of titers is 3.2 to 5.1 g/liter from 20 g/liter of initial glucose (7). According to Survase et al. (38), C. beijerinckii NRRL B-593 produced 2.2 and 3.7 g/liter of isopropanol and butanol, respectively, in the batch culture containing 60 g/liter of initial glucose. Shaheen et al. (35) demonstrated that C. beijerinckii NRRL B-592 produced about 16 g/liter of total solvents, including isopropanol, from 80 g/liter of maize mash, but the efficiency of acetone conversion in this strain is not known; the NRRL B-592 strain did not produce isopropanol in another study (7).
Although a high yield and titer of butanol could be achieved using an engineered E. coli strain anaerobically (36), the productivity was still lower than what can be achieved with solventogenic clostridia (30 g/liter with gas stripping for ca. 180 h). The butanol productivity as well as the IBE productivity of our engineered strain exceeds that obtained using engineered E. coli.
In previous studies on the isopropanol production of E. coli, the conversion of acetone into isopropanol was incomplete. Hanai et al. (11) reported the production of ca. 2.4 g/liter of isopropanol with ca. 0.4 g/liter of acetone, while Jojima et al. (16) reported the production of 13.6 g/liter of isopropanol with ca. 3 g/liter of acetone. Considering that the secondary alcohol dehydrogenase used in these studies is NADPH dependent (14), the insufficient conversion of acetone into isopropanol in engineered E. coli strains seems to be due to the cofactor imbalance. In the present study, all adhB-593-expressing clostridial strains efficiently converted acetone into isopropanol and only trace amounts of acetone remained in the culture broth (Table 3 and Fig. 2 and 3). Unlike many bacteria that use the oxidative pentose phosphate (PP) pathway, C. acetobutylicum does not have the oxidative PP pathway (2, 5). The major route for NADPH generation in C. acetobutylicum consists of pyruvate-ferredoxin oxidoreductase, which converts pyruvate into acetyl-CoA and carbon dioxide, producing reduced ferredoxin. This reduced ferredoxin can be used for the production of either NADH, NADPH, or molecular hydrogen according to its cellular state (31).
Expression of the adhB-593 gene in C. acetobutylicum unexpectedly resulted in increased titers of residual acids. In order to reduce the amounts of residual acids, C. acetobutylicum PJC4BK, a buk-inactivated strain (9), was employed. The engineered PJC4BK strain produced 38% more butanol but 28% less isopropanol than the 824(pIPA3) strain. The lower activities of CoAT, AADC, and SADH in PJC4BK(pIPA3-Cm2) than in 824(pIPA3) seem to be one of the reasons for lower isopropanol production in the former strain (Table 4). The reason for this phenomenon is not clear.
Also, previous metabolic flux analysis of the PJC4BK strain suggested that the flux through acetoacetyl-CoA to butyryl-CoA in PJC4BK was enhanced compared to that of the wild type (12), suggesting that the activities of the corresponding enzymes, such as 3-hydroxybutyryl-CoA dehydrogenase, are upregulated in PJC4BK. In addition, it was suggested that formation of acetoacetyl-CoA from two acetyl-CoA molecules might be a rate-limiting step of butanol formation (44). Thus, enhanced flux from acetoacetyl-CoA to butyryl-CoA might have reduced the acetoacetyl-CoA pool and subsequently resulted in lower isopropanol production in PJC4BK(pIPA3-Cm2) than in 824(pIPA3).
The metabolic activity of C. acetobutylicum greatly decreases with the age of culture (28), due to metabolite inhibition (1) and/or sporulation. Thus, it was reasoned that alleviating solvent toxicity using gas stripping (6) would allow prolonged solvent production, resulting in the production of more butanol as well as isopropanol and ethanol. Indeed, by coupling the gas stripping with fermentation, the amount of glucose consumption in PJC4BK(pIPA3-Cm2) was dramatically increased (Fig. 4A) (132.9 g/liter). The total titer of IBE was 35.6 g/liter, including 25.1 g/liter of butanol at 45 h.
In conclusion, metabolically engineered C. acetobutylicum strains were developed for the efficient production of a biofuel mixture consisting of isopropanol, butanol, and ethanol. Also, the complete consumption of glucose in our gas-stripping fermentation without strain degeneration suggests that the engineered strain developed here can be used for long-term fermentation. Recently, the population dynamics of C. acetobutylicum was successfully investigated using flow cytometry (39), which might be useful for our future studies toward better understanding clostridial physiology and solventogenesis during gas-stripping-coupled fermentation.
ACKNOWLEDGMENTS
This work was supported by the Advanced Biomass R&D Center of Korea (ABC-2010-0029799) through the Global Frontier Research Program of the Ministry of Education, Science and Technology (MEST). Further support by GS Caltex, BioFuelChem, the EEWS program of KAIST, and the World Class University program (R32-2008-000-10142-0) of the MEST is appreciated.
Footnotes
Published ahead of print 30 December 2011
REFERENCES
- 1. Alsaker KV, Paredes C, Papoutsakis ET. 2010. Metabolite stress and tolerance in the production of biofuels and chemicals: gene-expression-based systems analysis of butanol, butyrate, and acetate stresses in the anaerobe Clostridium acetobutylicum. Biotechnol. Bioeng. 105:1131–1147 [DOI] [PubMed] [Google Scholar]
- 2. Amador-Noguez D, et al. 2010. Systems-level metabolic flux profiling elucidates a complete, bifurcated tricarboxylic acid cycle in Clostridium acetobutylicum. J. Bacteriol. 192:4452–4461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bahl H, et al. 1986. Nutritional factors affecting the ratio of solvents produced by Clostridium acetobutylicum. Appl. Environ. Microbiol. 52:169–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen J-S, Hiu SF. 1986. Acetone-butanol-isopropanol production by Clostridium beijerinckii (synonym, Clostridium butylicum). Biotechnol. Lett. 8:371–376 [Google Scholar]
- 5. Crown SB, et al. 2011. Resolving the TCA cycle and pentose-phosphate pathway of Clostridium acetobutylicum ATCC 824: isotopomer analysis, in vitro activities and expression analysis. Biotechnol. J. 6:300–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ezeji TC, Qureshi N, Blaschek HP. 2004. Acetone butanol ethanol (ABE) production from concentrated substrate: reduction in substrate inhibition by fed-batch technique and product inhibition by gas stripping. Appl. Microbiol. Biotechnol. 63:653–658 [DOI] [PubMed] [Google Scholar]
- 7. George HA, Johnson JL, Moore WE, Holdeman LV, Chen JS. 1983. Acetone, isopropanol, and butanol production by Clostridium beijerinckii (syn. Clostridium butylicum) and Clostridium aurantibutyricum. Appl. Environ. Microbiol. 45:1160–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gerischer U, Durre P. 1990. Cloning, sequencing, and molecular analysis of the acetoacetate decarboxylase gene region from Clostridium acetobutylicum. J. Bacteriol. 172:6907–6918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Green EM, et al. 1996. Genetic manipulation of acid formation pathways by gene inactivation in Clostridium acetobutylicum ATCC 824. Microbiology 142(Part 8):2079–2086 [DOI] [PubMed] [Google Scholar]
- 10. Han B, Gopalan V, Ezeji T. 2011. Acetone production in solventogenic Clostridium species: new insights from non-enzymatic decarboxylation of acetoacetate. Appl. Microbiol. Biotechnol. 91:565–576 [DOI] [PubMed] [Google Scholar]
- 11. Hanai T, Atsumi S, Liao JC. 2007. Engineered synthetic pathway for isopropanol production in Escherichia coli. Appl. Environ. Microbiol. 73:7814–7818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Harris LM, Desai RP, Welker NE, Papoutsakis ET. 2000. Characterization of recombinant strains of the Clostridium acetobutylicum butyrate kinase inactivation mutant: need for new phenomenological models for solventogenesis and butanol inhibition? Biotechnol. Bioeng. 67:1–11 [PubMed] [Google Scholar]
- 13. Husemann MHW, Papoutsakis ET. 1988. Solventogenesis in Clostridium acetobutylicum fermentations related to carboxylic acid and proton concentrations. Biotechnol. Bioeng. 32:843–852 [DOI] [PubMed] [Google Scholar]
- 14. Ismaiel AA, Zhu CX, Colby GD, Chen JS. 1993. Purification and characterization of a primary-secondary alcohol dehydrogenase from two strains of Clostridium beijerinckii. J. Bacteriol. 175:5097–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jiang Y, et al. 2009. Disruption of the acetoacetate decarboxylase gene in solvent-producing Clostridium acetobutylicum increases the butanol ratio. Metab. Eng. 11:284–291 [DOI] [PubMed] [Google Scholar]
- 16. Jojima T, Inui M, Yukawa H. 2008. Production of isopropanol by metabolically engineered Escherichia coli. Appl. Microbiol. Biotechnol. 77:1219–1224 [DOI] [PubMed] [Google Scholar]
- 17. Jones DT, Woods DR. 1986. Acetone-butanol fermentation revisited. Microbiol. Rev. 50:484–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Keis S, Shaheen R, Jones DT. 2001. Emended descriptions of Clostridium acetobutylicum and Clostridium beijerinckii, and descriptions of Clostridium saccharoperbutylacetonicum sp. nov. and Clostridium saccharobutylicum sp. nov. Int. J. Syst. Evol. Microbiol. 51:2095–2103 [DOI] [PubMed] [Google Scholar]
- 19. Krouwel PG, Groot WJ, Kossen NWF, van der Laan WFM. 1983. Continuous isopropanol-butanol-ethanol fermentation by immobilized Clostridium beijerinckii cells in a packed bed fermenter. Enzyme Microb. Technol. 5:46–54 [Google Scholar]
- 20. Lee JY, Jang YS, Lee J, Papoutsakis ET, Lee SY. 2009. Metabolic engineering of Clostridium acetobutylicum M5 for highly selective butanol production. Biotechnol. J. 4:1432–1440 [DOI] [PubMed] [Google Scholar]
- 21. Lee SY, et al. 2008. Fermentative butanol production by Clostridia. Biotechnol. Bioeng. 101:209–228 [DOI] [PubMed] [Google Scholar]
- 22. Matsumura M, Takehara S, Kataoka H. 1992. Continuous butanol/isopropanol fermentation in down-flow column reactor coupled with pervaporation using supported liquid membrane. Biotechnol. Bioeng. 39:148–156 [DOI] [PubMed] [Google Scholar]
- 23. Mermelstein LD, Papoutsakis ET. 1993. In vivo methylation in Escherichia coli by the Bacillus subtilis phage ϕ3T I methyltransferase to protect plasmids from restriction upon transformation of Clostridium acetobutylicum ATCC 824. Appl. Environ. Microbiol. 59:1077–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mermelstein LD, Papoutsakis ET, Petersen DJ, Bennett GN. 1993. Metabolic engineering of Clostridium acetobutylicum ATCC 824 for increased solvent production by enhancement of acetone formation enzyme activities using a synthetic acetone operon. Biotechnol. Bioeng. 42:1053–1060 [DOI] [PubMed] [Google Scholar]
- 25. Mermelstein LD, Welker NE, Bennett GN, Papoutsakis ET. 1992. Expression of cloned homologous fermentative genes in Clostridium acetobutylicum ATCC 824. Biotechnology 10:190–195 [DOI] [PubMed] [Google Scholar]
- 26. Nakotte S, Schaffer S, Bohringer M, Durre P. 1998. Electroporation of, plasmid isolation from and plasmid conservation in Clostridium acetobutylicum DSM 792. Appl. Microbiol. Biotechnol. 50:564–567 [DOI] [PubMed] [Google Scholar]
- 27. Norrander J, Kempe T, Messing J. 1983. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene 26:101–106 [DOI] [PubMed] [Google Scholar]
- 28. Papoutsakis ET. 2008. Engineering solventogenic clostridia. Curr. Opin. Biotechnol. 19:420–429 [DOI] [PubMed] [Google Scholar]
- 29. Papoutsakis ET. 1984. Equations and calculations for fermentations of butyric acid bacteria. Biotechnol. Bioeng. 26:174–187 [DOI] [PubMed] [Google Scholar]
- 30. Papoutsakis ET, Meyer CL. 1985. Fermentation equations for propionic-acid bacteria and production of assorted oxychemicals from various sugars. Biotechnol. Bioeng. 27:67–80 [DOI] [PubMed] [Google Scholar]
- 31. Petitdemange H, Cherrier C, Bengone JM, Gay R. 1977. Study of the NADH and NADPH-ferredoxin oxidoreductase activities in Clostridium acetobutylicum. Can. J. Microbiol. 23:152–160 [PubMed] [Google Scholar]
- 32. Pfromm PH, Amanor-Boadu V, Nelson R, Vadlani P, Madl R. 2010. Bio-butanol bio-ethanol: a technical and economic assessment for corn and switchgrass fermented by yeast or Clostridium acetobutylicum. Biomass Bioenergy 34:515–524 [Google Scholar]
- 33. Rassadin V, Shlygin O, Likhterova N, Slavin V, Zharov A. 2006. Problems in production of high-octane, unleaded automotive gasolines. Chem. Technol. Fuels Oil 42:235–242 [Google Scholar]
- 34. Roos JW, McLaughlin JK, Papoutsakis ET. 1985. The effect of pH on nitrogen supply, cell lysis, and solvent production in fermentations of Clostridium acetobutylicum. Biotechnol. Bioeng. 27:681–694 [DOI] [PubMed] [Google Scholar]
- 35. Shaheen R, Shirley M, Jones DT. 2000. Comparative fermentation studies of industrial strains belonging to four species of solvent-producing clostridia. J. Mol. Microbiol. Biotechnol. 2:115–124 [PubMed] [Google Scholar]
- 36. Shen CR, et al. 2011. Driving forces enable high-titer anaerobic 1-butanol synthesis in Escherichia coli. Appl. Environ. Microbiol. 77:2905–2915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sillers R, Al-Hinai MA, Papoutsakis ET. 2009. Aldehyde-alcohol dehydrogenase and/or thiolase overexpression coupled with CoA transferase downregulation lead to higher alcohol titers and selectivity in Clostridium acetobutylicum fermentations. Biotechnol. Bioeng. 102:38–49 [DOI] [PubMed] [Google Scholar]
- 38. Survase SA, Jurgens G, van Heiningen A, Granstrom T. 2011. Continuous production of isopropanol and butanol using Clostridium beijerinckii DSM 6423. Appl. Microbiol. Biotechnol. 91:1305–1313 [DOI] [PubMed] [Google Scholar]
- 39. Tracy BP, Gaida SM, Papoutsakis ET. 2008. Development and application of flow-cytometric techniques for analyzing and sorting endospore-forming clostridia. Appl. Environ. Microbiol. 74:7497–7506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tummala SB, Junne SG, Papoutsakis ET. 2003. Antisense RNA downregulation of coenzyme A transferase combined with alcohol-aldehyde dehydrogenase overexpression leads to predominantly alcohologenic Clostridium acetobutylicum fermentations. J. Bacteriol. 185:3644–3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tummala SB, Welker NE, Papoutsakis ET. 2003. Design of antisense RNA constructs for downregulation of the acetone formation pathway of Clostridium acetobutylicum. J. Bacteriol. 185:1923–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wiesenborn DP, Rudolph FB, Papoutsakis ET. 1989. Coenzyme A transferase from Clostridium acetobutylicum ATCC 824 and its role in the uptake of acids. Appl. Environ. Microbiol. 55:323–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhao Y, Tomas CA, Rudolph FB, Papoutsakis ET, Bennett GN. 2005. Intracellular butyryl phosphate and acetyl phosphate concentrations in Clostridium acetobutylicum and their implications for solvent formation. Appl. Environ. Microbiol. 71:530–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zheng YN, et al. 2009. Problems with the microbial production of butanol. J. Ind. Microbiol. Biotechnol. 36:1127–1138 [DOI] [PubMed] [Google Scholar]