Abstract
Travelers can introduce viruses from disease-endemic to non–disease-endemic areas. Serologic and virologic tests confirmed dengue virus infections in 3 travelers returning to Japan: 2 from Tanzania and 1 from Côte d’Ivoire. Phylogenetic analysis of the envelope gene showed that 2 genetically related virus isolates belonged to dengue virus type 3 genotype III.
Keywords: Dengue, Tanzania, Côte d’Ivoire, viruses, vector-borne infections, dispatch
Dengue virus (DENV) is an arthropod-borne virus that infects ≈100 million persons each year in Southeast Asia, Central and South America, and Africa. Infection with any 1 of the 4 DENV serotypes causes a wide range of disease, from dengue fever to the more severe dengue hemorrhagic fever. Epidemics of dengue-like illness have occurred in Africa, but information about etiology is limited (1). Transmission of another arthropod-borne virus, chikungunya virus, which causes disease similar to dengue, has been documented in Tanzania. Although Tanzania is on the list of countries at risk for DENV transmission (2), to our knowledge, no DENV isolates have yet been identified and studied there.
In Japan as of June 16, 2010, a total of 50 cases of imported dengue had been reported. Among these cases, 2 were dengue fever that had developed in 2 travelers after they returned from Tanzania. We report the molecular characterization of 2 DENV type 3 (DENV-3) isolates from 1 of the travelers who had visited Tanzania in 2010 and from a traveler who had visited Côte d’Ivoire in 2008.
The Study
In 2010, a 55-year-old man (patient 1) and a 23-year-old woman (patient 2) returned to Japan from Tanzania; high fever and thrombocytopenia developed in each on days 1 and 3 days after return, respectively. In 2008, a 65-year-old man (patient 3) returned to Japan from Côte d’Ivoire and subsequently experienced high fever. Serum samples from each of the 3 patients were sent to the National Institute of Infectious Diseases, Japan, for laboratory examination. DENV serotypes were determined by serotype-specific reverse transcription–PCR (RT-PCR) (3). DENV-specific immunoglobulin (Ig) M was detected by Dengue Fever Virus IgM Capture ELISA (Focus Diagnostics, Inc., Cypress, CA, USA) used according to the manufacturer’s instructions. Dengue IgG Indirect ELISA (Panbio Ltd, Sinnamon Park, Queensland, Australia) was used to detect anti-DENV IgG according to the manufacturer’s instructions. Serum from patient 1 was negative for anti-DENV IgM and anti-DENV IgG; serum from patients 2 and 3 was positive for anti-DENV IgM and IgG. All 3 serum samples had positive DENV nonstructural protein (NS) 1 antigen results according to NS1 capture ELISA (Platelia Dengue NS1 Antigen assay; Bio-Rad Laboratories, Marnes-la-Coquette, France) and negative chikungunya viral RNA results by RT-PCR.
DENV-3 (D3/Hu/Tanzania/NIID08/2010) was isolated from patient 1, and DENV-3 (D3/Hu/Côte d’Ivoire/NIID48/2008) was isolated from patient 3 by using the Aedes albopictus mosquito cell line C6/36 and FcγR-expressing baby hamster kidney cells (4). DENV-3 RNA was detected in serum from patient 2 by RT-PCR, but the virus was not isolated. The viral RNA was extracted by using a High Pure Viral RNA Extraction kit (Roche Diagnostics, Mannheim, Germany), transcribed to cDNA, amplified by PCR, and sequenced as described (3).
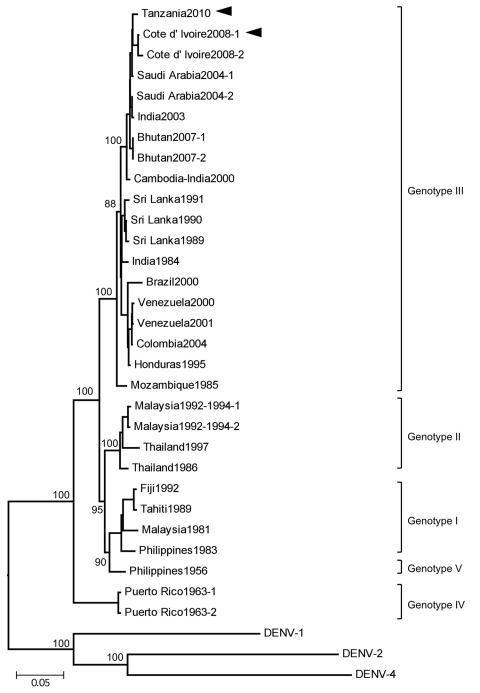
Nucleotide sequences of the isolates were compared with selected sequences of DENV-3 (Table). Sequence alignment and phylogenetic analysis was performed by the Genetyx analysis program (Genetyx Corp., Tokyo, Japan). The phylogenetic tree was constructed by using the neighbor-joining method. The selected DENV-3 strains were grouped into 5 genotypes (5). Confidence values for virus groupings were assessed by bootstrap assembling analysis of 1,000 replicates. The 2 DENV-3 isolates belonged to DENV-3 genotype III (Figure). The envelope (E)-protein sequence showed that the DENV-3 (D3/Hu/Tanzania/NIID08/2010 strain) isolated from patient 1 had a sequence homology of 98% to the DENV-3 D3/Hu/Côte d’Ivoire/NIID48/2008 strain and 99% to a DENV-3 6805 strain isolated in Saudi Arabia in 2004 (GenBank accession no. AM746229) (6).
Table. Comparison of dengue virus type 3 sequences from travelers returning from Tanzania and Côte d’Ivoire with selected dengue virus type 3 sequences*.
| Year isolated | Name | Strain | Isolate origin | GenBank accession no. |
|---|---|---|---|---|
| 2010 | Tanzania2010 | D3/Hu/Tanzania/NIID08/2010 | Tanzania | AB549332 |
| 2008 | Côte d’Ivoire2008–1 | D3/Hu/Côte d'Ivoire/NIID48/2008 | Côte d’Ivoire | AB447989 |
| 2008 | Côte d’Ivoire2008–2 | 2008/00510 | Côte d’Ivoire | FM213456 |
| 2007 | Bhutan2007–1 | SV0786_07 | Bhutan | FJ606712 |
| 2007 | Bhutan2007–2 | SV0837_07 | Bhutan | FJ606708 |
| 2004 | Saudi Arabia2004–1 | 6805 | Saudi Arabia | AM746229 |
| 2004 | Saudi Arabia2004–2 | 6475 | Saudi Arabia | AM746232 |
| 2004 | Colombia2004 | 22379_MEDELLIN/04 | Colombia | FJ389910 |
| 2003 | India2003 | GWL-25 | India | AY770511 |
| 2001 | Venezuela2001 | LARD6667 | Venezuela | AY146773 |
| 2000 | Brazil2000 | 68784 | Brazil | AY038605 |
| 2000 | Venezuela2000 | LARD6315 | Venezuela | AY146767 |
| 2000 | Cambodia–India2000† | 00–28–1HuNIID | Cambodia/India | AB111081 |
| 1997 | Thailand1997 | D97–0106 | Thailand | AY145728 |
| 1995 | Honduras1995 | HN179 | Honduras | FJ189469 |
| 1992–1994 | Malaysia1992–1994–1 | LN1746 | Malaysia | AF147458 |
| 1992–1994 | Malaysia1992–1994–2 | LN2632 | Malaysia | AF147459 |
| 1992 | Fiji1992 | 29472 | Fiji | L11422 |
| 1991 | Sri Lanka1991 | 2783 | Sri Lanka | L11438 |
| 1990 | Sri Lanka1990 | SK698 | Sri Lanka | FJ189449 |
| 1989 | Tahiti1989 | 2167 | Tahiti | L11619 |
| 1989 | Sri Lanka1989 | 260698 | Sri Lanka | L11437 |
| 1986 | Thailand1986 | D86–007 | Thailand | L11441 |
| 1985 | Mozambique1985 | 1559 | Mozambique | L11430 |
| 1984 | India1984 | 1416 | India | L11424 |
| 1983 | Philippines1983 | 168.AP-2 | Philippines | L11432 |
| 1981 | Malaysia1981 | 29586 | Malaysia | L11427 |
| 1963 | Puerto Rico1963–1 | PR6 | Puerto Rico | L11433 |
| 1963 | Puerto Rico1963–2 | BS-PRico63 | Puerto Rico | AY146762 |
| 1956 | Philippines1956 | H87 | Philippines | L11423 |
| 2001 | DENV-1 | 01–44–1HuNIID | Tahiti | AB111070 |
| 2005 | DENV-2 | D2/Hu/OPD030NIID/2005 | East Timor | AB219135 |
| 2001 | DENV-4 | MY01–22713 | Malaysia | AJ428556 |
*DENV, dengue virus. †Strain 00–28–1HuNIID was isolated from a traveler who returned to Japan after visiting Cambodia and India.
Figure.
Phylogenetic tree based on the envelope genome sequence of selected dengue virus type 3 (DENV-3) strains. The tree was rooted to DENV-1, DENV-2 and DENV-4. Multiple sequence alignments were performed, and the tree was constructed by using the neighbor-joining method. The percentage of successful bootstrap replication is indicated at the nodes. DENV-3 genotypes are indicated on the right. The isolated DENV-3 strains, D3/Hu/Tanzania/NIID08/2010 strain (Tanzania2010) and D3/Hu/Côte d'Ivoire/NIID48/2008 strain (Côte d’Ivoire2008), are indicated with arrowheads. Scale bar indicates nucleotide substitutions per site.
Conclusions
DENV transmission has occurred in western Africa and some parts of eastern Africa (7–10). We isolated DENV-3 from 2 patients in Japan in whom dengue fever developed after they returned from Côte d’Ivoire (western Africa) and Tanzania (eastern Africa). Detection of DENV-3 in patients 1 and 2 suggests local DENV-3 transmission in Tanzania. As of April 2010, at least 17 suspected cases of dengue have been reported among residents of Dar es Salaam, Tanzania, but the molecular epidemiology of DENV in Tanzania has not been analyzed (11). In the absence of such analyses, data on molecular epidemiology of DENV isolated from returning travelers offers timely information to countries where dengue surveillance is not routinely performed. Data and reports of the presence of a competent DENV vector, Ae. aegypti mosquitoes, suggest the need for further studies on local DENV transmission in Tanzania (12).
It is assumed that travelers increase the risk for introduction of DENV serotypes or strains from disease-endemic to non–disease-endemic areas where competent vectors such as Ae. aegypti or Ae. albopictus mosquitoes are present. The sequence homology among the DENV-3 strain isolated from the traveler to Tanzania (patient 1), the DENV-3 strain isolated from the traveler from Côte d’Ivoire in 2008 (patient 3), and a DENV-3 strain isolated in Saudi Arabia in 2004 ranged from 98% to 99% (GenBank accession nos. AB549332, AB447989, AM746229, respectively). Because the E-protein gene of the isolate from Tanzania was highly similar to those in viruses circulating regionally and the Middle East, the disease could have been introduced or reintroduced into the country from neighboring areas. These data suggest the need for further studies of the route of disease dissemination and surveillance of dengue in Africa. Genetic differences among subtypes may result in differences in virus virulence and epidemic potential (13). DENV-3 genotype III, previously isolated from several parts of Africa, Latin America, and the Indian subcontinent, has been associated with higher incidence of major epidemics of dengue and dengue hemorrhagic fever (14). The DENV isolates from Tanzania and Côte d’ Ivoire were closely related to a DENV-3 genotype III strain isolated from a major DENV outbreak in northern India in 2003–2004; the E-protein gene homology was 98% (15).
Emergence of DENV-3 genotype III in geographically diverse areas may thus result from higher epidemic potential of the virus, although further studies are needed to understand the clinical and epidemiologic implications of emergence or reemergence of the virus in Tanzania and Côte d’ Ivoire. Whereas other vector-borne diseases such as malaria and yellow fever have been well studied in Africa, dengue needs more attention with regard to identification of epidemics, clinical implications, and disease management.
Acknowledgments
This work was supported by grants for Research on Emerging and Re-emerging Infectious Diseases (H21-shinkou-ippan-005 and H20-shinkou-ippan-015) from the Ministry of Health, Labour and Welfare, Japan.
Biography
Dr Moi is a researcher at the National Institute of Infectious Diseases, Tokyo, Japan. Her research interest is the immunopathogenesis of flavivirus infection.
Footnotes
Suggested citation for this article: Moi ML, Takasaki T, Kotaki A, Tajima S, Lim C-K, Sakamoto M, et al. Importation of dengue virus type 3 to Japan from Tanzania and Côte d’Ivoire. Emerg Infect Dis [serial on the Internet]. 2010 Nov [date cited]. http://dx.doi.org/10.3201/eid1611.101061
References
- 1.Gubler DJ, Sather GE, Kuno G, Cabral JR. Dengue 3 virus transmission in Africa. Am J Trop Med Hyg. 1986;35:1280–4. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. Dengue in the Western Pacific Region. 2010. [cited 2010 Jun 10]. http://www.wpro.who.int/health_topics/dengue/
- 3.Ito M, Takasaki T, Yamada K, Nerome R, Tajima S, Kurane I. Development and evaluation of fluorogenic TaqMan reverse transcriptase PCR assays for detection of dengue virus types 1 to 4. J Clin Microbiol. 2004;42:5935–7. 10.1128/JCM.42.12.5935-5937.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moi ML, Lim CK, Kotaki A, Takasaki T, Kurane I. Development of an antibody-dependent enhancement assay for dengue virus using stable BHK-21 cell lines expressing FcgammaRIIA. J Virol Methods. 2010;163:205–9. 10.1016/j.jviromet.2009.09.018 [DOI] [PubMed] [Google Scholar]
- 5.Wittke V, Robb TE, Thu HM, Nisalak A, Nimmannitya S, Kalanrooj S, et al. Extinction and rapid emergence of strains of dengue 3 virus during an interepidemic period. Virology. 2002;301:148–56. 10.1006/viro.2002.1549 [DOI] [PubMed] [Google Scholar]
- 6.Zaki A, Perera D, Jahan SS, Cardosa MJ. Phylogeny of dengue viruses circulating in Jeddah, Saudi Arabia: 1994 to 2006. Trop Med Int Health. 2008;13:584–92. 10.1111/j.1365-3156.2008.02037.x [DOI] [PubMed] [Google Scholar]
- 7.Takasaki T. Dengue/DHF update (15). ProMed. 2010. Mar 23 [cited 2010 Jun 10]. http://www.promedmail.org, archive no. 20100323.0922.
- 8.Takasaki T. Dengue/DHF update (35). ProMed. 2008. Aug 18 [cited 2010 Jun 10]. http://www.promedmail.org, archive no. 20080818.2573.
- 9.Gautret P, Simon F, Hervius Askling H, Bouchaud O, Leparc-Goffart I, Ninove L, et al. EuroTravNet. Dengue type 3 virus infectious in European travelers returning from Comoros and Zanzibar, February–April 2010. Euro Surveill. 2010;15:19451. [PubMed] [Google Scholar]
- 10.Ninove L, Parola P, Baronti C, De Lamballerie X, Gautret P, Doudier B, et al. Dengue virus type 3 infection in traveler returning from west Africa. Emerg Infect Dis. 2009;15:1871–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klaassen B. Dengue/DHF update (23). ProMed. 2010 May 17 [cited 2010 Jun 10]. http://www.promedmail.org, archive no. 20100517.1620.
- 12.Trpis M. Seasonal changes in the larval populations of Aedes aegypti in two biotopes in Dar es Salaam, Tanzania. Bull World Health Organ. 1972;47:245–55. [PMC free article] [PubMed] [Google Scholar]
- 13.Messer WB, Gubler DJ, Harris E, Sivananthan K, Silva AM. Emergence and global spread of a dengue serotype 3, subtype III virus. Emerg Infect Dis. 2003;9:800–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lanciotti RS, Lewis JG, Gubler DJ, Trent DW. Molecular evolution and epidemiology of dengue-3 viruses. J Gen Virol. 1994;75:65–75. 10.1099/0022-1317-75-1-65 [DOI] [PubMed] [Google Scholar]
- 15.Dash PK, Parida MM, Saxena P, Abhyankar A, Singh CP, Tewari KN, et al. Reemergence of dengue virus type-3 (subtype-III) in India: implications for increased incidence of DHF & DSS. Virol J. 2006;3:55. 10.1186/1743-422X-3-55 [DOI] [PMC free article] [PubMed] [Google Scholar]