Abstract
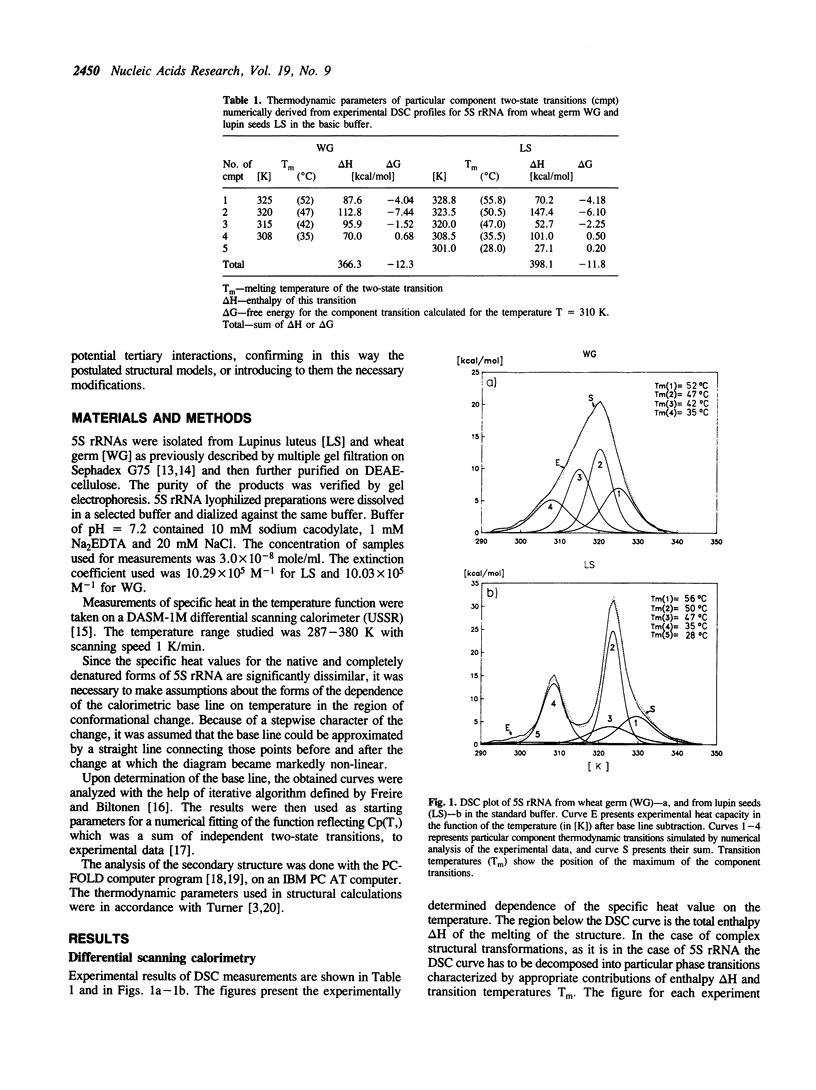
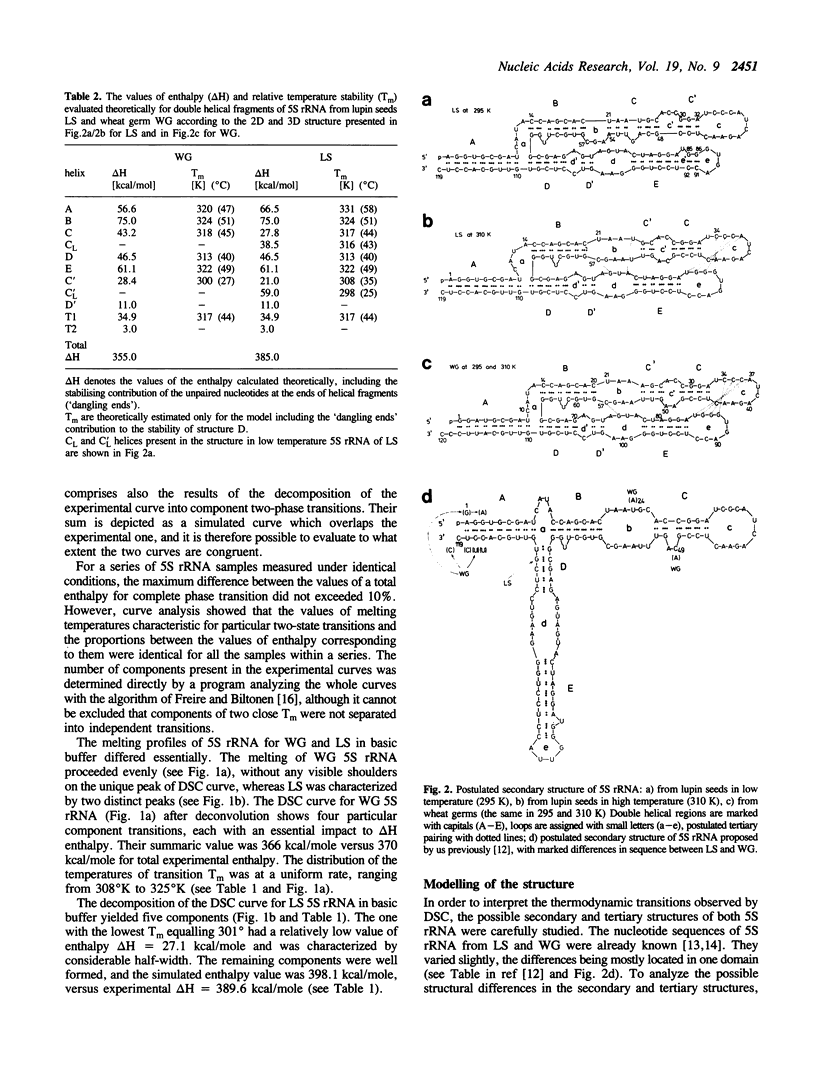
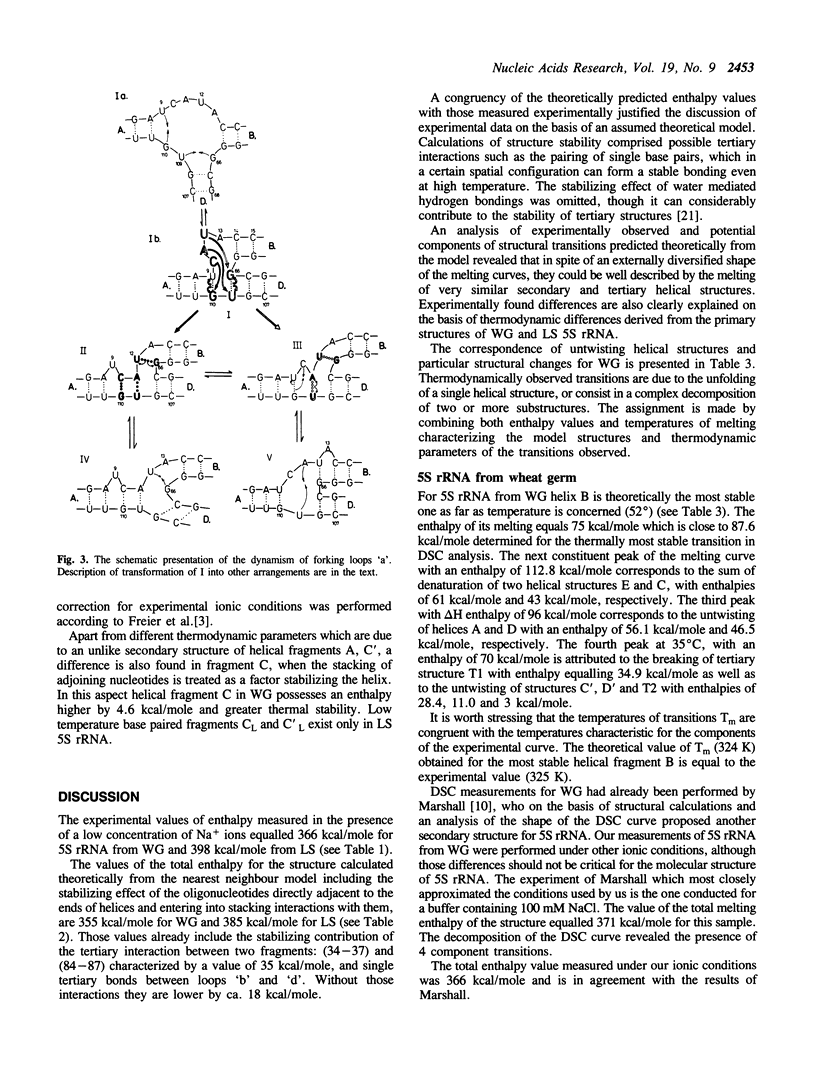
Thermal unfolding of 5S rRNA from wheat germ (WG) and lupin seeds (LS) was studied in solution. Experimental curves of differential scanning calorimetry (DSC) were resolved into particular components according to the thermodynamic model of two-state transitions. The DSC temperature profiles for WG and LS differ significantly in spite of very high similarities in the sequence of both molecules. Those results are interpreted according to a model of the secondary and tertiary molecular structure of 5S rRNA. A comparison of the 'nearest neighbour' model of interaction with the experimental thermodynamic results enables a complete interpretation of the process of the melting of its structures. In light of our observations, the crucial differences between both DSC melting profiles are mainly an outcome of different thermodynamic properties of the first helical fragment 'A' made up of 9 complementary base pairs. It contains 6 differences in the nucleotide sequence of both types of molecules, which still retain 9-meric double helixes. The temperature stability of his helix in WG is much lower than of the LS one. Moreover, the results supply evidence for a strong specific tertiary interaction between the two hairpin loops 'c' and 'e' in both 5S rRNA molecules, modulated by small differences in the thermodynamic properties of both 5S rRNA.
Full text
PDF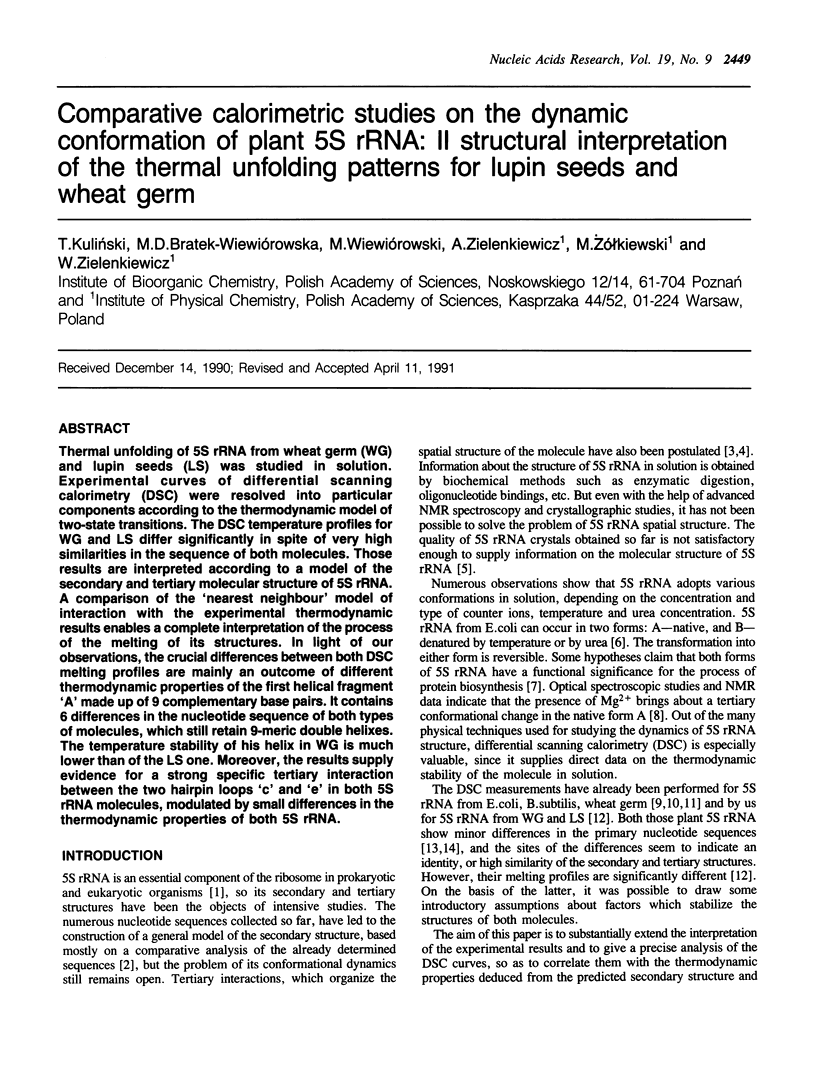



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aubert M., Scott J. F., Reynier M., Monier R. Rearrangement of the conformation of Escherichia coli 5S RNA. Proc Natl Acad Sci U S A. 1968 Sep;61(1):292–299. doi: 10.1073/pnas.61.1.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barciszewska M. Z., Lorenz S., Erdmann V. A., Barciszewski J. Structural analysis of plant ribosomal 5S RNAs. Visualisation of novel tertiary interactions by cleavage of lupin and wheat 5SrRNAs with ribonuclease H. Biochim Biophys Acta. 1990 Sep 10;1087(1):68–72. doi: 10.1016/0167-4781(90)90122-i. [DOI] [PubMed] [Google Scholar]
- Barciszewski J., Bratek-Wiewiórowska M. D., Górnicki P., Naskret-Barciszewska M., Wiewiórowski M., Zielenkiewicz A., Zielenkiewicz W. Comparative calorimetric studies on the dynamic conformation of plant 5S rRNA. I. Thermal unfolding pattern of lupin seeds and wheat germ 5S rRNAs, also in the presence of magnesium and sperminium cations. Nucleic Acids Res. 1988 Jan 25;16(2):685–701. doi: 10.1093/nar/16.2.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L. H., Li S. J., Ricca T. L., Marshall A. G. Theoretical and experimental on-line analysis of multistate melting of polymers by differential scanning microcalorimetry. Anal Chem. 1984 Jul;56(8):1502–1507. doi: 10.1021/ac00272a066. [DOI] [PubMed] [Google Scholar]
- Chang L. H., Marshall A. G. Solution conformations of B. subtilis ribosomal 5S RNA: a calorimetric study. Biopolymers. 1986 Jul;25(7):1299–1313. doi: 10.1002/bip.360250710. [DOI] [PubMed] [Google Scholar]
- Connor J., Schroit A. J. Aminophospholipid translocation in erythrocytes: evidence for the involvement of a specific transporter and an endofacial protein. Biochemistry. 1990 Jan 9;29(1):37–43. doi: 10.1021/bi00453a005. [DOI] [PubMed] [Google Scholar]
- Erdmann V. A. Structure and function of 5S and 5.8 S RNA. Prog Nucleic Acid Res Mol Biol. 1976;18:45–90. [PubMed] [Google Scholar]
- Erdmann V. A., Wolters J. Collection of published 5S, 5.8S and 4.5S ribosomal RNA sequences. Nucleic Acids Res. 1986;14 (Suppl):r1–59. doi: 10.1093/nar/14.suppl.r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freier S. M., Petersheim M., Hickey D. R., Turner D. H. Thermodynamic studies of RNA stability. J Biomol Struct Dyn. 1984 Mar;1(5):1229–1242. doi: 10.1080/07391102.1984.10507514. [DOI] [PubMed] [Google Scholar]
- Joachimiak A., Nalaskowska M., Barciszewska M., Barciszewski J., Mashkova T. Higher plant 5S rRNAs share common secondary and tertiary structure. A new three domains model. Int J Biol Macromol. 1990 Oct;12(5):321–327. doi: 10.1016/0141-8130(90)90022-3. [DOI] [PubMed] [Google Scholar]
- Kim K., Jhon M. S. Theoretical study of hydration of RNA. Biochim Biophys Acta. 1979 Nov 22;565(1):131–147. doi: 10.1016/0005-2787(79)90089-3. [DOI] [PubMed] [Google Scholar]
- Kime M. J., Moore P. B. NMR evidence for the existence of two native conformations of 5S RNA. Nucleic Acids Res. 1982 Aug 25;10(16):4973–4983. doi: 10.1093/nar/10.16.4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafalski J. A., Wiewiorowski M., Söll D. Organization and nucleotide sequence of nuclear 5S rRNA genes in yellow lupin (Lupinus luteus). Nucleic Acids Res. 1982 Dec 11;10(23):7635–7642. doi: 10.1093/nar/10.23.7635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studnicka G. M., Eiserling F. A., Lake J. A. A unique secondary folding pattern for 5S RNA corresponds to the lowest energy homologous secondary structure in 17 different prokaryotes. Nucleic Acids Res. 1981 Apr 24;9(8):1885–1904. doi: 10.1093/nar/9.8.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner D. H., Sugimoto N., Freier S. M. RNA structure prediction. Annu Rev Biophys Biophys Chem. 1988;17:167–192. doi: 10.1146/annurev.bb.17.060188.001123. [DOI] [PubMed] [Google Scholar]
- Westhof E. Water: an integral part of nucleic acid structure. Annu Rev Biophys Biophys Chem. 1988;17:125–144. doi: 10.1146/annurev.bb.17.060188.001013. [DOI] [PubMed] [Google Scholar]
- Woese C. R., Pribula C. D., Fox G. E., Zablen L. B. The nucleotide sequence of the 5S ribosomal RNA from a photobacterium. J Mol Evol. 1975 Jun 9;5(1):35–46. doi: 10.1007/BF01732012. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]



