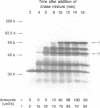
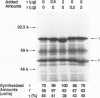
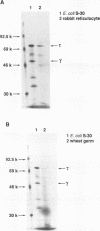
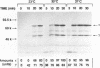
Abstract
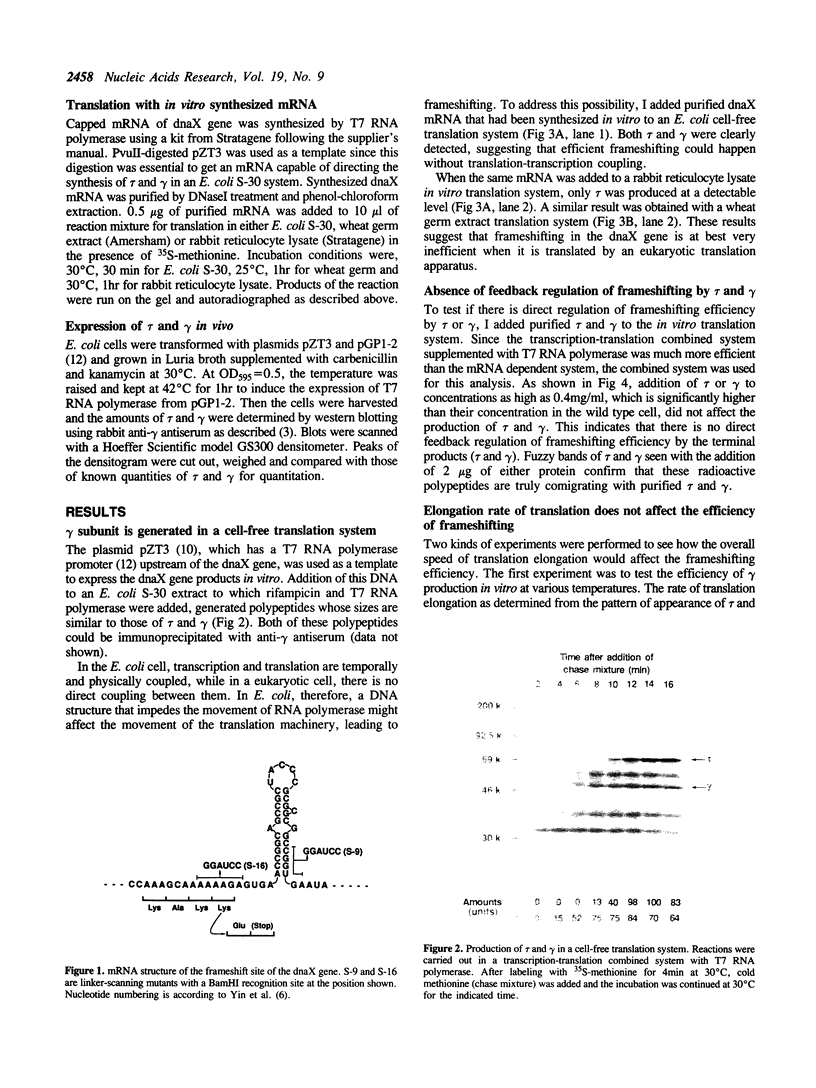
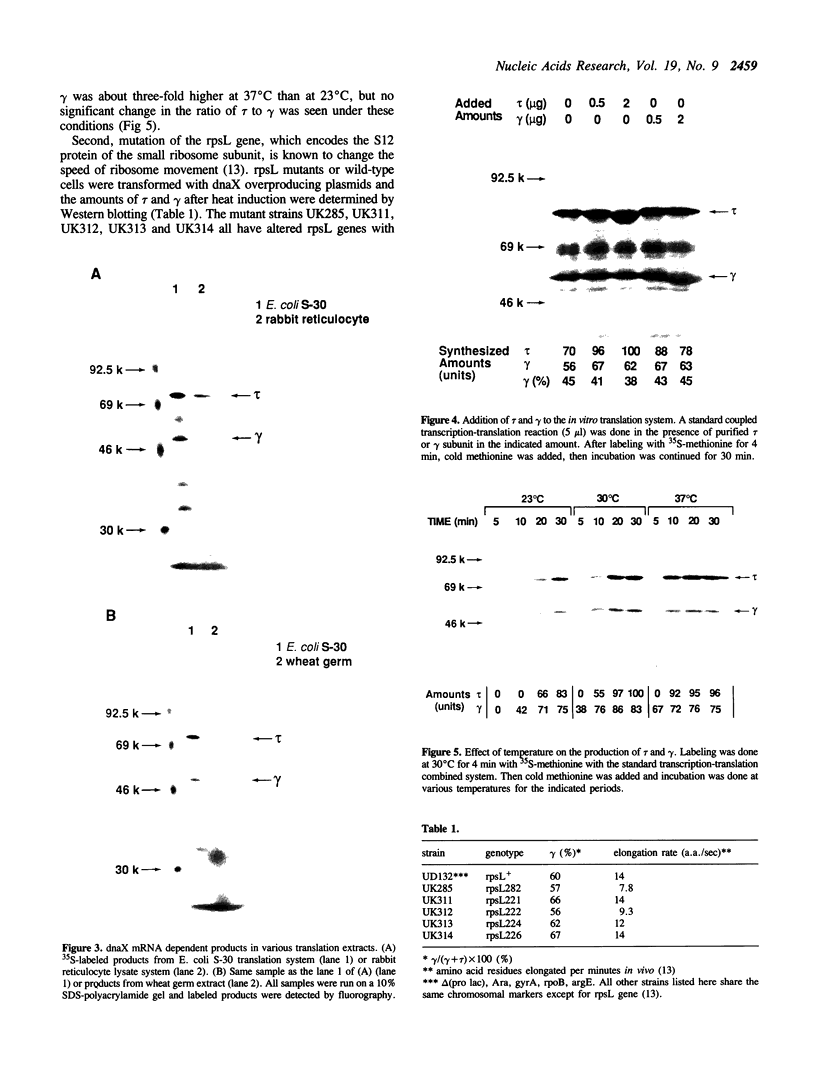
Production of the gamma subunit of Escherichia coli DNA polymerase III holoenzyme is dependent on a very efficient translational frameshif in the dnaX gene. I used an E. coli in vitro translation system to analyze the mechanism of this frameshifting event. In this system, gamma was produced almost to the same extent as the inframe translation product, tau, suggesting that efficient frameshifting was reproduced in vitro. Coupling with transcription was not necessary for frameshifting. Addition of purified tau or gamma had no effect on the frameshifting process suggesting the absence of direct feedback regulation. By use of mutant genes, a strong pausing site was identified at or very close to the frameshift site. This pausing was apparently caused by a potential stem-loop structure which was previously shown to enhance frameshifting. Thus, enhancement of frameshifting by this putative stem-loop seems to be mediated by the translation pausing at the frameshift site. Despite the apparent structural similarity of the dnaX frameshift site to that of the eukaryotic retroviral genes, dnaX mRNA synthesized in vitro failed to direct the production of gamma in eukaryotic translation systems. This suggests that frameshifting in the dnaX gene depends on components specific to the E. coli translation system.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Atkins J. F., Weiss R. B., Gesteland R. F. Ribosome gymnastics--degree of difficulty 9.5, style 10.0. Cell. 1990 Aug 10;62(3):413–423. doi: 10.1016/0092-8674(90)90007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belcourt M. F., Farabaugh P. J. Ribosomal frameshifting in the yeast retrotransposon Ty: tRNAs induce slippage on a 7 nucleotide minimal site. Cell. 1990 Jul 27;62(2):339–352. doi: 10.1016/0092-8674(90)90371-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blinkowa A. L., Walker J. R. Programmed ribosomal frameshifting generates the Escherichia coli DNA polymerase III gamma subunit from within the tau subunit reading frame. Nucleic Acids Res. 1990 Apr 11;18(7):1725–1729. doi: 10.1093/nar/18.7.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohman K., Ruusala T., Jelenc P. C., Kurland C. G. Kinetic impairment of restrictive streptomycin-resistant ribosomes. Mol Gen Genet. 1984;198(2):90–99. doi: 10.1007/BF00328706. [DOI] [PubMed] [Google Scholar]
- Brierley I., Digard P., Inglis S. C. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989 May 19;57(4):537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clare J. J., Belcourt M., Farabaugh P. J. Efficient translational frameshifting occurs within a conserved sequence of the overlap between the two genes of a yeast Ty1 transposon. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6816–6820. doi: 10.1073/pnas.85.18.6816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigen W. J., Caskey C. T. Expression of peptide chain release factor 2 requires high-efficiency frameshift. Nature. 1986 Jul 17;322(6076):273–275. doi: 10.1038/322273a0. [DOI] [PubMed] [Google Scholar]
- Flower A. M., McHenry C. S. The gamma subunit of DNA polymerase III holoenzyme of Escherichia coli is produced by ribosomal frameshifting. Proc Natl Acad Sci U S A. 1990 May;87(10):3713–3717. doi: 10.1073/pnas.87.10.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield D., Feng Y. X., Lee B. J., Rein A., Levin J. G., Oroszlan S. Chromatographic analysis of the aminoacyl-tRNAs which are required for translation of codons at and around the ribosomal frameshift sites of HIV, HTLV-1, and BLV. Virology. 1989 Dec;173(2):736–742. doi: 10.1016/0042-6822(89)90589-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T., Madhani H. D., Masiarz F. R., Varmus H. E. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell. 1988 Nov 4;55(3):447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodaira M., Biswas S. B., Kornberg A. The dnaX gene encodes the DNA polymerase III holoenzyme tau subunit, precursor of the gamma subunit, the dnaZ gene product. Mol Gen Genet. 1983;192(1-2):80–86. doi: 10.1007/BF00327650. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lustig F., Elias P., Axberg T., Samuelsson T., Tittawella I., Lagerkvist U. Codon reading and translational error. Reading of the glutamine and lysine codons during protein synthesis in vitro. J Biol Chem. 1981 Mar 25;256(6):2635–2643. [PubMed] [Google Scholar]
- Mullin D. A., Woldringh C. L., Henson J. M., Walker J. R. Cloning of the Escherichia coli dnaZX region and identification of its products. Mol Gen Genet. 1983;192(1-2):73–79. doi: 10.1007/BF00327649. [DOI] [PubMed] [Google Scholar]
- Sekine Y., Ohtsubo E. Frameshifting is required for production of the transposase encoded by insertion sequence 1. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4609–4613. doi: 10.1073/pnas.86.12.4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. Selective inactivation of the exonuclease activity of bacteriophage T7 DNA polymerase by in vitro mutagenesis. J Biol Chem. 1989 Apr 15;264(11):6447–6458. [PubMed] [Google Scholar]
- Tsuchihashi Z., Kornberg A. ATP interactions of the tau and gamma subunits of DNA polymerase III holoenzyme of Escherichia coli. J Biol Chem. 1989 Oct 25;264(30):17790–17795. [PubMed] [Google Scholar]
- Tsuchihashi Z., Kornberg A. Translational frameshifting generates the gamma subunit of DNA polymerase III holoenzyme. Proc Natl Acad Sci U S A. 1990 Apr;87(7):2516–2520. doi: 10.1073/pnas.87.7.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss R. B., Dunn D. M., Atkins J. F., Gesteland R. F. Slippery runs, shifty stops, backward steps, and forward hops: -2, -1, +1, +2, +5, and +6 ribosomal frameshifting. Cold Spring Harb Symp Quant Biol. 1987;52:687–693. doi: 10.1101/sqb.1987.052.01.078. [DOI] [PubMed] [Google Scholar]
- Weiss R. B., Dunn D. M., Shuh M., Atkins J. F., Gesteland R. F. E. coli ribosomes re-phase on retroviral frameshift signals at rates ranging from 2 to 50 percent. New Biol. 1989 Nov;1(2):159–169. [PubMed] [Google Scholar]
- Wilson W., Braddock M., Adams S. E., Rathjen P. D., Kingsman S. M., Kingsman A. J. HIV expression strategies: ribosomal frameshifting is directed by a short sequence in both mammalian and yeast systems. Cell. 1988 Dec 23;55(6):1159–1169. doi: 10.1016/0092-8674(88)90260-7. [DOI] [PubMed] [Google Scholar]
- Yin K. C., Blinkowa A., Walker J. R. Nucleotide sequence of the Escherichia coli replication gene dnaZX. Nucleic Acids Res. 1986 Aug 26;14(16):6541–6549. doi: 10.1093/nar/14.16.6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S., Watanabe T., Murao K., Ishikura H., Yamaizumi Z., Nishimura S., Miyazawa T. Molecular mechanism of codon recognition by tRNA species with modified uridine in the first position of the anticodon. Proc Natl Acad Sci U S A. 1985 Aug;82(15):4905–4909. doi: 10.1073/pnas.82.15.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]