Abstract
To address the need for a universal system to assess health status, we previously described a method termed “immunosignaturing” which splays the entire humoral antibody repertoire across a peptide microarray. Two important issues relative to the potential broad use of immunosignatures are sample preparation and stability. In the present study, we compared the immunosignatures developed from serum, plasma, saliva, and antibodies eluted from blood dried onto filter paper. We found that serum and plasma provide identical immunosignatures. Immunosignatures derived from dried blood also correlated well with those from nondried serum from the same individual. Immunosignatures derived from dried blood were capable of distinguishing naïve mice from those infected with influenza virus. Saliva was applied to the arrays, and the IgA immunosignature correlated strongly with that from dried blood. Finally, we demonstrate that dried blood retains immunosignature information even when exposed to high temperature. This work expands the potential diagnostic uses for immunosignatures. These features suggest that different forms of archival samples can be used for diagnosis development and that in prospective studies samples can be easily procured.
INTRODUCTION
The transition of medicine from symptomatic to presymptomatic diagnosis will require a simple, comprehensive diagnostic system. With this goal in mind, we recently introduced a microarray-based system termed “immunosignaturing” (16). This platform consists of a 10,000-feature random peptide array upon which the serum antibody repertoire is displayed in machine-readable form. Random peptides rather than actual epitopes were chosen so that any disease can be detected in an unbiased fashion (16). The resulting pattern of antibody binding, the immunosignature, is unique to the disease, is reproducible, is consistent within a disease (i.e., not personal), quite different across diseases, and has the power to classify individuals on the basis of their health status (16, 20, 21, 27; B. A. Chase, S. A. Johnston, and J. B. Legutki, unpublished data). These random-feature microarrays have been informative in influenza (16), Alzheimer's disease (20, 21), and lupus (S. Williams et al., unpublished data). The usefulness of this technique would be extended if archived and prospectively collected samples had stable immunosignatures. In the present study, we explore the suitability of antibody samples prepared from serum, plasma, saliva, and dried blood for the immunosignature assay.
For the immunosignature array to be effective as a broadly applicable diagnostic, it must be robust and unaffected by variances in sample collection. Such variations in sample collection have impacted the mass spectrometry-based search for biomarkers (reviewed in references1 and 17). In the first reports of the immunosignature platform, we probed an array containing 10,000 spotted peptides 20 amino acids long with the sequences randomly generated (CIM10K array) with antibodies recovered from blood using serum separator tubes. This protocol has generated quality reproducible data in both mice and humans (16, 21; Williams et al., unpublished). However, the ability to compare samples prepared by different preparation protocols is desirable for several reasons. First, archival blood samples exist from prior studies and epidemics. Many of these samples were not collected with microarrays in mind, so nucleic acids are often wholly degraded, but if appropriately frozen and stored, blood, serum, or plasma samples are ideal for immunosignaturing. The different methods used to store these archival samples over the last several decades would likely create differences in assay performance.
Second, it would be ideal for broad use of immunosignaturing, particularly in developing countries, if samples, blood or saliva, could be routinely mailed into a central processing center. Such a capability could facilitate regular health monitoring. Others have previously demonstrated successful collection and retention of serum antibodies from whole-blood samples spotted on absorbent filter paper. In these studies, serum was evaluated by enzyme-linked immunosorbent assay (ELISA) for antibodies to HIV (2, 4, 23), measles virus (6, 11–13, 24, 29), the malaria parasite, hepatitis C virus (3, 5, 15), and other pathogens (7, 22, 28). These studies demonstrated a good correlation between fresh plasma and dried-blood samples using standard ELISA techniques, leading to the expectation that antibodies stored in dried-blood spots should be amenable to immunosignaturing. An ELISA typically evaluates the titer of an antibody against a single antigen, while the immunosignature is based on antibody reactivity to 10,000 array features. The difference in scale between the two assays renders the immunosignature potentially more sensitive to loss of antibody reactivities than ELISA. Therefore, it is critical to evaluate the utility of dried-blood spots on an immunosignaturing diagnostic platform.
In the present study, four sample variables important for using immunosignaturing were investigated. The goal was to determine if samples provided as dried-blood spots, plasma, serum, or saliva would provide comparable information content in our assay. First, we compared serum versus plasma. Second, we determined if antibodies stored as dried-blood spots were stable. Third, relative to mailing in of samples, we examined the effect of temperature. Finally, the suitability of using saliva for immunosignatures was assessed.
MATERIALS AND METHODS
Preparation of peptides and peptide conjugates for injection.
The peptides FT03 (KANWFDFKTFNQMTQVWGSC), FT04 (MMIFRNDFEWLKIHKTRGSC), and FT05 (KTFKSEPAYNIESNSSTGSC) were synthesized in-house on a Symphony 12-channel synthesizer (Protein Technologies, Inc., Tucson, AZ) and purified to greater than 90% purity using a high-pressure liquid chromatograph fitted with a Phenomenex C18 reverse-phase column. Quality control of prepared peptides was conducted by matrix-assisted laser desorption ionization–time of flight mass spectrometry on a Bruker-Daltonics MicroFlex apparatus prior to use. Peptides were conjugated via the C-terminal cysteine to maleimide-activated bovine serum albumin (BSA) and keyhole limpet hemocyanin (KLH) (Pierce, Rockford, IL) following the manufacturer's instructions. Excess peptide was removed using a 10,000-molecular-weight cutoff Slide-A-Lyzer dialysis cassette (Pierce) against four changes of phosphate-buffered saline (PBS). Glycerol was added to a final concentration of 10% and quantified by bicinchoninic acid reaction. Aliquoted conjugates were stored at −20°C until needed.
Animal immunization and phlebotomy.
Female BALB/c mice 4 to 5 weeks old (Charles River Laboratories) were housed in barrier isolation housing with standard rodent feed and water provided ad libitum. Mice were acclimated for 1 week prior to immunization. Mice were immunized with 100 μg of the conjugate via intraperitoneal (i.p.) injection three times 2 weeks apart. Blood samples were collected via submandibular venipuncture using a 5.0-mm lancet (MEDIpoint, Inc., Mineola, NY). Fresh blood samples were collected at 211 and 236 days postimmunization. All animal experiments were conducted following an animal use protocol which was reviewed and approved by the Arizona State University Institutional Animal Care and Use Committee (IACUC no. 10-1099R).
Mouse model of influenza virus infection.
Mice were infected as described previously (16). Briefly, sucrose gradient-purified influenza virus A/PR/8/34 was obtained as a frozen stock from Advanced Biotechnologies Inc. (Columbia, MD). A frozen aliquot of virus was thawed and diluted in tissue-culture-grade PBS (Invitrogen, Carlsbad, CA) and kept on ice until needed. Mice were anesthetized prior to injection using a ketamine (42.0 mg/kg of body weight), xylazine (4.8 mg/kg), and acepromazine (0.6 mg/kg) cocktail injected i.p. For the infections, a sublethal dose of 1 × 104 PFU in 30.0 μl per mouse was administered intranasally. Weight was monitored daily postinfection. Mice were bled on day 230 postimmunization.
Human samples and phlebotomy.
Finger sticks for preparation of the dried-blood spots were collected using an Accu-Chek lancet (Roche), followed by blotting the finger onto a Whatman 903 protein saver card. Venous blood draws were collected into serum separator or plasma separator tubes (BD). Saliva samples were collected using sponges or eye sponges (Salvimetrics, State College, PA). Saliva samples were frozen to break apart mucus and centrifuged for 15 min at 3,000 rpm in an Allegra 25R centrifuge. All human specimens were deidentified and handled in accordance with a protocol approved by the Arizona State University Institutional Review Board (IRB no. 09-05004024).
Sample preparation.
Dried samples were prepared by spotting either 10 μl of whole blood, 5 μl of serum, or 20 μl of whole blood mixed 1:1 with Halt protease inhibitor cocktail onto Whatman 903 protein saver cards. Cards with dried-blood spots were stored at either 25°C or 37.8°C until needed. Dried-blood spots were isolated using a 10-mm paper punch and eluted in a 1.5-ml microcentrifuge tube containing 500 μl PBS with agitation on a Vortex Genie 2 tabletop vortex mixer on setting 5 for 30 min. Volumes were selected to give approximately a 1:100 dilution of serum. Following elution, the filter papers were removed and samples were stored at 4°C until needed.
Fresh whole-blood and serum samples were collected at the same time as the dried-blood spots. Fresh serum was isolated by centrifugation at 1,090 relative centrifugal force for 5 min and diluted 1:100 in PBS. A separate set of aliquots was prepared by dilution in PBS containing a 4.29-mg/ml concentration of Halt protease inhibitor cocktail (Thermo Scientific, Waltham, MA). Samples were stored at 4°C until needed.
Application of samples to the CIM10K array.
The peptide arrays were printed as described previously and stored at 4°C with desiccation in an argon atmosphere (16, 19). Prior to use, excess peptide that was not conjugated to the array surface was removed by washing for 5 min in 33% isopropanol, 7.33% acetonitrile, and 0.55% trifluoroacetic acid, followed by a 15-s wash in Tris-buffered saline (19.98 mM Tris, 136 mM NaCl, pH 7.4) with 0.05% Tween 20 (TBST) and a 15-s wash in double-distilled H2O. Arrays were processed on a Tecan HS4800 Pro hybridization station with a protocol adapted to antibody binding. General settings were wash duration of 30 s at 11.0 ml/min with sample agitation set to high. Arrays were blocked for 1 h at 23°C with 200 μl blocking buffer (1× PBS, 3% BSA, 0.05% Tween 20, 0.014% mercaptohexanol) at room temperature in a humidity chamber. The slides were then washed with TBST. A 200-μl volume of diluted sample in incubation buffer (1× PBS, 3.0% BSA, 0.05% Tween 20) was applied to the peptide arrays and incubated for 1 h at 37°C. Dried-blood spots, plasma, and sera were applied at 1:500, and saliva was applied in equal parts clarified saliva and 2× incubation buffer. Slides were washed between each step with TBST. A 200-μl volume of 1.0 nM biotinylated goat anti-mouse IgG (H+L; Bethyl Laboratories, Montgomery, TX) in incubation buffer was applied to each slide, followed by a 1-h incubation at 37°C. After washing, bound secondary antibody was detected by applying to each slide a 200-μl volume of either Alexa Fluor 555-labeled streptavidin or Alexa Fluor 647-labeled streptavidin at 5.0 nM in incubation buffer, followed by 1 h incubation at 37°C. Final washes in TBST and distilled water were done to remove residual salt. Slides were dried by onboard nitrogen flow for 5 min. This was followed by scanning of the arrays using an Agilent C scanner at 552 nm or 635 nm at 100% gain (photo multiplier tube setting). Human samples were similarly processed, except that a biotinylated polyclonal anti-human IgG (H+L; Novus Biologicals, Littleton, CO) or a biotinylated anti-human IgA (alpha chain specific; Vector Laboratories, Burlingame, CA) secondary antibody was used.
Analysis of scanned arrays.
Statistical analysis of microarray data was done with GeneSpring (version 7.3.1) software (Agilent Inc., Palo Alto, CA) by importing image-processed data from GenePix Pro (version 6.0) software (Axon Instruments, Union City, CA). Calculations based on the GenePix-prepared gpr text files were done on the median signal intensity per spot without background subtraction (overall background was considered extremely low and even). Poor-quality spots were excluded from analysis by flagging them as “bad” upon visual inspection; GenePix also autoflags spots on the basis of low threshold circularity. Prior to analysis, each array was normalized to the 50th percentile to eliminate array-to-array variation, and signal intensities of less than 0.01 were set to 0.01. Values from triplicate arrays were averaged and used in the analysis. Array-to-array coefficients of variance averaged <30%; power is 1.3-fold the minimum detectable signal at the 95th percentile per 2 technical replicates (26).
Determination of total protein recovery from dried specimens on collection paper.
Four 5-μl pooled serum samples were spotted on 903 cards. After drying, the samples were eluted as described above for 30 min, 1 h, 2 h, or 3 h. After elution, the total protein concentrations of the reconstituted serum samples were determined with a NanoDrop spectrophotometer.
ELISA detection of antigen-specific IgG reactivity in reconstituted whole-blood samples.
Separate 96-well Nunc Maxisorp microtiter plates were coated with the relevant antigen at a concentration of 100 ng/well in sodium carbonate/bicarbonate buffer (15 mM Na2CO2, 35 mM NaHCO2), followed by overnight incubation at 4°C. The initial recovery experiments were conducted using the FT04 peptide conjugated to BSA or the KLH immunization carrier without peptide. The infected and naïve mice were evaluated against whole influenza virus. After incubation, the plates were washed 3 times with TBST, followed by blocking of the wells with 5% nonfat milk for 1 h at 37°C. Following another three TBST washes, samples were serially diluted across the plate starting at 1:400 and incubated for 1 h at 37°C. Microtiter plates were washed, and bound antibody was detected using 100 μl of 2.5 nM anti-mouse IgG conjugated to horseradish peroxidase (Bethyl Laboratories, Montgomery, TX). Plates were washed and developed using 100 μl 2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid) (ABTS) per well. Development was stopped using 50 μl 1.0% SDS per well. Absorbance was read using a microplate reader. The endpoint titer is defined as the reciprocal of the last dilution giving an absorbance above the limit of detection or twice the background of an empty well.
RESULTS
Immunosignature process and samples used.
The immunosignature assay was developed to splay the serum antibody repertoire across a microarray, the CIM10K array, composed of 10,000 random-sequence peptides (16, 19). Probing the CIM10K with a dilution containing less than 1.0 μl of serum allows the distinction of patients with influenza virus infections from both healthy individuals (16) and patients with noninfectious diseases such as Alzheimer's disease (21) and lupus (Williams et al., unpublished). The present study uses both the classic CIM10K employed in our prior studies and our newest version of the array, CIM10K version 2 (CIM10Kv2). The concept is identical on both arrays; however, the library of peptides is different. Murine samples from peptide-immunized mice are utilized on the arrays to observe any possible change in rank of the immunizing peptide among the features present on the array. Assays are repeated using samples from human donors to ensure clinical relevance.
Plasma and serum samples provide equivalent immunosignatures.
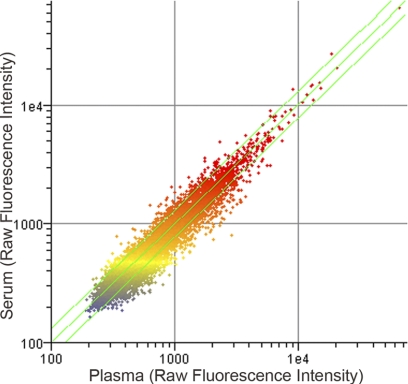
The current sample preparation method for immunosignatures has been to isolate serum from whole blood using serum separator tubes. This format has worked well for immunosignatures; however, not all samples, particularly historical samples, have been collected in the same way. We evaluated whether serum and plasma provide the same immunosignature. Two sequential blood draws were collected from a human donor into either a serum separator tube or a plasma separator tube to discern the influence of clotting factors on the immunosignature. The scatter plot shown in Fig. 1 and reproducibility metrics indicate that there is no sample-associated signature that exceeded the intrinsic noise in the system. Information on the array is driven by the rank order of the peptides and measured by the Spearman nonparametric rank-order test. In this comparison, the Spearman correlation is 0.95, indicating an excellent comparison between plasma and serum. This indicates that prepared plasma may be used in conjunction with prepared serum samples.
Fig 1.
Comparison of serum and plasma from a single donor. Sequential blood draws were collected from a single donor and used to probe the array. Raw fluorescence intensities are plotted. To illustrate the relation of individual data points between the plasma and serum, the individual data points are colored on the basis of the plasma (vertical axis). The color used to plot data points represents the relation of the data point to the median intensity (yellow spots). Red represents features three times above the median value, and blue represents features three times below the median. Data plotted are from a single donor and are representative of two independent donor samples (data for the second sample are not shown).
Evaluation of serum antibody recovery from dried-blood spots.
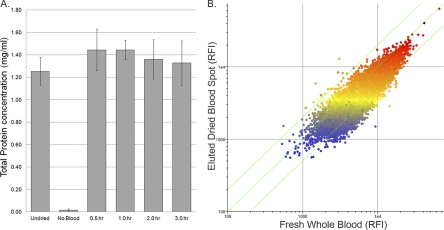
In-home collection is an attractive alternative to clinical visits. Patients self-collect small drops of blood onto a Whatman 903 protein saver card for mailing to the institute conducting the study. Such cards have been used by others to evaluate titers of antibodies to specific antigens (4, 7, 11, 12). Since the immunosignature is a measure of total relative antibody reactivity, the utility of collecting dried-blood spots for immunosignature-based diagnostics is highly dependent on the ability to achieve near complete recovery of the antibody repertoire. The percent recovery of protein from the collection paper was determined. Figure 2A shows maximum recovery of a dried human serum sample after 30 min of agitation. Rates of recovery in separate elution experiments have ranged from 78 to 100% protein recovery for both mouse and human samples. No protein was eluted from blank 903 cards in any test.
Fig 2.
Analysis of serum protein recovery and immunoglobulin reactivity observed in dried whole-blood specimens. (A) Total protein recovery from Whatman 903 protein saver cards. The total protein content of the 1:100-diluted human serum samples was measured with the NanoDrop spectrophotometer. Data presented are the average of four separate dried-blood spots from the same donor and are representative of recoveries from different donors (data not shown). (B) Scatter plot showing the correlation between the immunosignatures of the fresh mouse whole-blood sample (x axis) and dried mouse whole-blood sample on collection paper (y axis) (R2 = 0.832) represented by raw fluorescence intensities (RFIs). The black dot indicates the immunizing peptide. The color used to plot data points represents the relation of the data point to the median intensity of the eluted dried-blood spot (yellow spots). Red represents features three times above the median value, and blue represents features three times below the median. Diagonal green lines represent the values that are 1.3-fold different between the two samples. Data presented are representative of three separate peptide-KLH conjugates.
Recovery of functional antigen-specific antibodies from the 903 cards was evaluated using sera collected from peptide-KLH-immunized mice. In an endpoint ELISA, both fresh serum- and dried-blood-spot-eluted antibodies had a titer of 1:51,200 against KLH and a titer of 1:204,800 against the peptide-BSA, indicating that the amount of reactive antibody is not changed by the drying process. These results have been replicated using samples from other peptide-KLH-immunized mice. Our elution conditions and sample recovery agree with those previously reported (7). These results demonstrate effective recovery of serum proteins, including functional antibodies, from dried blood.
Dried-blood samples provide immunosignatures comparable to those for fresh serum samples.
Since identical endpoint titers were obtained in an ELISA for both the fresh serum and dried-blood spots, we asked whether the fine specificity of the antibody repertoire was retained in the immunosignature. Fresh serum and reconstituted dried-blood spots from peptide-KLH-immunized mice were used to probe the CIM10K. Previously, we demonstrated that the serum antibodies from peptide-immunized mice generate on the CIM10K unique immunosignatures which include a specific rank of the immunizing peptide (10). We reasoned that similarly immunized mice would provide an ideal means to assess sample preparation effects on the immunosignature. The immunosignatures from three separate peptide-KLH immunogens were evaluated for both broad assay performance and specific information content. In each case, the fresh serum samples had higher relative fluorescence intensities at key metrics than did dried-blood spots (Table 1); however, the higher fluorescence was uniform across all features of the array and did not affect the rank order of the features (Table 2). A Spearman nonparametric rank-order test value of 0.87 to 0.9 indicates that the order of peptide binding is not affected in dried blood versus fresh serum, although the intensity may change. In each case, the rank of the immunizing peptide was identical or within the variance of technical replicates. Uniform changes in intensity are accounted for by median normalization in analysis of microarray data, as illustrated in the fold change data presented in Table 2. The median Spearman correlation for each fresh serum and dried-blood spot pair was within the range found for technical replicates of the same peptide. Taken together, the immunosignature recovered from dried blood is comparable to that of fresh serum.
Table 1.
Summary statistics for three peptide-KLH conjugatesa
| Statistic | FT03 |
FT04 |
FT05 |
|||
|---|---|---|---|---|---|---|
| Fresh WB | DBS | Fresh WB | DBS | Fresh WB | DBS | |
| 95th percentile | 5,262 | 14,664 | 14,480 | 10,697 | 26,676 | 18,981 |
| Median intensity | 1,066 | 1,078 | 5,226 | 3,513 | 6,250 | 3,289 |
| Blank featureb | 264 | 611 | 1,889 | 1,279 | 2,369 | 1,369 |
| Dynamic rangec | 19.93 | 24.00 | 7.67 | 8.36 | 11.26 | 9.68 |
| Immunogen rankd | 26 (100) | 10 (100) | 2 (100) | 2 (100) | 41 (100) | 38 (100) |
Microarray summary statistics were calculated using GeneSpring (version 7.3.1) and Microsoft Office Excel 2003 software. Raw fluorescence intensities from the average of triplicate arrays were used. BALB/c mice were immunized with peptide conjugates of peptides also present as features on the array.
Blank features are features printed containing only spotting buffer.
The dynamic range is calculated as the ratio of the 95th percentile over the blank feature intensity.
Immunogen rank is the feature corresponding to the peptide immunogen, with 1 being the feature with the highest RFI. Data in parentheses indicate the percentile of the feature corresponding to the peptide immunogen.
Table 2.
Comparison of fresh whole blood and antibodies eluted from dried-blood spotsa
| Statistic | FT03 | FT04 | FT05 |
|---|---|---|---|
| Spearman correlationb | 0.92 | 0.87 | 0.90 |
| Fold changec | |||
| Raw | 1.00 ± 0.34 | 1.52 ± 0.46 | 1.99 ± 0.66 |
| Normalizedd | 0.99 ± 0.30 | 1.05 ± 0.35 | 1.04 ± 0.42 |
Microarray summary statistics were calculated using GeneSpring (version 7.3.1) and Microsoft Office Excel 2003 software. Raw fluorescence intensities from the average of triplicate arrays were used. BALB/c mice were immunized with peptide conjugates of peptides also present as features on the array.
Spearman correlation considers only the relative rank of two arrays.
The fold change is the ratio of individual features on the fresh sample array to the same feature on the dried-blood spot arrays.
Microarrays were normalized to the per chip median.
Dried-blood spots retain functional antibodies after storage in a simulated mailing environment.
It would be of utility for both large-scale epidemiology studies and health monitoring via immunosignaturing if samples could be shipped by mail for analysis. To test this, we examined the heat stability of the immunosignature. The stability of a dried whole-blood sample over time was assessed by comparing the correlation of the immunosignature obtained from dried-blood spots to that obtained from fresh samples. Immunosignature assays from dried-blood spots stored at either 25°C or 37.8°C were run at selected time points. The immunosignature remained relatively stable over time at 25°C, maintaining a Spearman correlation of 0.91 after 2 weeks (Table 3). At 37.8°C, the sample was relatively stable overnight (Spearman correlation = 0.95) but declined significantly after 2 weeks at the high temperature (Table 4).
Table 3.
Correlations between fresh blood and dried-blood spot immunosignatures across storage conditionsa
| Storage time | 25°C |
37.8°C |
||
|---|---|---|---|---|
| Correlationb | Immunogen rankc | Correlation | Immunogen rank | |
| Fresh serum | NAd | 2 | NA | 2 |
| Overnight | 0.96 | 2 | 0.95 | 2 |
| 1 wk | 0.95 | 4 | 0.89 | 2 |
| 2 wk | 0.91 | 3 | 0.85 | 5 |
Microarray summary statistics were calculated using GeneSpring (version 7.3.1) and Microsoft Office Excel 2003 software. Raw fluorescence intensities from the average of triplicate arrays were used. BALB/c mice were immunized with the FT04-peptide conjugates.
Correlation is the Spearman correlation, which considers only the relative rank of array features.
Immunogen rank is the feature corresponding to the peptide immunogen, with 1 being the feature with the highest raw fluorescence intensity. The top-ranked peptide in this experiment is highly reactive to the detection reagents.
NA, not applicable.
Table 4.
Correlations between protease inhibitor-treated fresh blood and dried-blood-spot immunosignatures at 25.0°C and 37.8°Ca
Microarray summary statistics were calculated using GeneSpring (version 7.3.1) and Microsoft Office Excel 2003 software. Raw fluorescence intensities from the average of triplicate arrays were used. Replicate dried-blood spots were collected from a human donor. These samples were run on the CIM10Kv2.0 library.
The correlation between the dried-blood spot and the fresh serum is the Spearman correlation, which considers only the relative rank of array features.
NA, not applicable.
Treatment of blood samples with protease inhibitor prior to deposition on collection paper improves the stability of the immunosignature.
Immunoglobulins are sensitive to proteases secreted by commensal and environmental bacteria (14, 18). These bacteria may be introduced into the sample during collection or transportation. To address this, whole blood was treated with a protease inhibitor cocktail prior to sample deposition. The protease inhibitor cocktail was previously tested in an inhibition assay and had no impact on antibody binding (data not shown). Human samples stored as dried-blood spots were assayed on the CIM10K version 2 array. After 1 week storage at 25°C or 37°C, the samples remained stable with a Spearman correlation of 0.85 compared to the fresh serum. Similarly treated murine samples from peptide-immunized mice run on the classic CIM10K array did not show a change in rank of the immunizing peptide within the immunosignature (data not shown). Taken together, these results suggest that protease inhibitor can protect the information quality of the dried-blood sample but that degradation is not particularly biased against any subpopulation of antibody. To facilitate self-collection by patients, the protease inhibitor may be impregnated in the collection paper.
A dried-whole-blood immunosignature can distinguish previously infected from naïve mice.
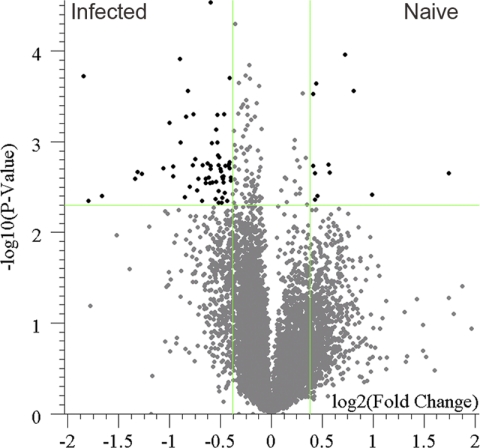
We have demonstrated that the immunosignature recovered from dried whole-blood samples is comparable to that recovered from fresh whole blood in terms of signal strength and correlation. To verify that the information content recovered is as informative as that recovered from fresh samples, dried-blood spots were prepared from mice that were previously infected with influenza virus A/PR/8/34 and age-matched naïve mice. In an endpoint ELISA against whole virus, the infected mice had an IgG titer of 1:102,400 and the naïve mice had an IgG titer of 1:800. Serum antibodies were eluted from the filter paper and run on the CIM10K array, and the immunosignatures were compared. To select for peptides whose recognition was significantly influenced by the infection, peptides either increasing or decreasing more than the minimal detectable fold change were compared. Testing the array features with a Student's t test resulted in 165 peptides having a P value of less than 0.005. The list was then filtered using the calculated minimal detectable fold change of 1.3, resulting in 75 peptides passing both restrictions. These peptides and their relationship to the remainder of the array are presented in Fig. 3. The ability to separate two samples on the basis of the resulting immunosignature and to identify peptides characteristic of an influenza virus infection demonstrates the utility of dried-blood spots as a sampling method for immunosignature-based diagnostics.
Fig 3.
Comparison of influenza virus-infected and naïve murine immunosignatures recovered from dried-blood spots. Whole blood from convalescent-phase mice that were either infected with influenza virus A/PR/8/34 or mock infected with PBS were collected on 903 protein saver cards. The resulting dried-blood spots were than processed in the immunosignature assay. The resulting immunosignatures are plotted in a volcano plot to illustrate the fold change difference between samples and the significance of the difference. Each array feature is plotted using its fold change between conditions (x axis, log2 scale) against its P value significance (y axis, −log10 scale). The vertical lines represent the minimal detectable fold change cutoff (1.3-fold), and the horizontal line represents the P value cutoff of 0.005. Black features meet both cutoffs, and the gray features do not meet the cutoffs. Data presented represent convalescent-phase sera from one infection experiment using a pool of blood from three mice under each condition.
Saliva contains sufficient immunoglobulin to derive an immunosignature.
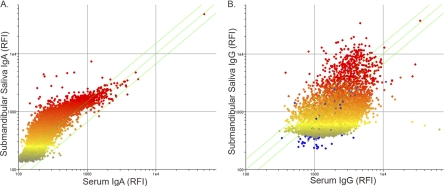
Saliva has been demonstrated by others to contain antigen-specific immunoglobulin (5, 13, 24, 28, 29). To determine whether the immunoglobulins in saliva were sufficient to generate an informative immunosignature, we collected submandibular and parotid saliva from people to probe the array. Both saliva sources contained IgG giving a median fluorescence score of 670 for parotid saliva and 669 for submandibular saliva, compared to a 1,843 median spot intensity for dried blood from the same donor. The dynamic range of the signal was less in the saliva samples, 3.62 for parotid saliva and 3.48 for submandibular saliva, than in dried blood, 8.2. The submandibular saliva contained more IgG and had a low rank-order Spearman correlation of 0.53 compared to the dried-blood spot. The same samples assayed for IgA content had a high Spearman correlation of 0.88 between submandibular saliva and dried blood. Comparison of IgG and IgA immunosignatures in the submandibular saliva samples to those in dried blood is shown in Fig. 4. Taken together these findings suggest that saliva may be used as a sample source for an IgA immunosignature but may need to be concentrated if used for IgG immunosignatures.
Fig 4.
Comparison of submandibular saliva to dried-blood spots for IgA (A) and IgG (B). Saliva samples were compared to dried-blood spots collected at the same time from a single human donor. Raw intensities are plotted, and color represents the median intensity, where yellow dots represent features at the median for the saliva samples. Saliva is plotted on the vertical axis, and the dried-blood spot is plotted on the x axis. Data plotted are from a single human donor and are representative of four individual donors. RFI, raw fluorescence intensity.
DISCUSSION
We have tested practical aspects of assaying immunosignatures from various samples. We first demonstrated that immunosignatures from serum and plasma from the same human donor correlate well. Second, we determined that the immunosignature derived from dried blood correlated with that derived from serum. Third, we demonstrated that storage of dried blood at high temperatures did not inhibit the sample for use in immunosignaturing assays and that correlations improved when the sample was dried in the presence of protease inhibitors. Fourth, we demonstrated that the information from the dried-blood immunosignature could distinguish influenza virus-infected from naïve mice. Finally, we demonstrated that the IgA immunosignature derived from saliva was comparable to that derived from serum. We previously demonstrated that immunosignatures are inexpensive and informative assays that discern health status (16, 21). In the present study, we built upon the utility of the assay by demonstrating that antibodies collected in different ways are generally acceptable for use in immunosignature-based diagnostics.
Serum and plasma differ largely by the presence of clotting factors. It has been shown that proteins can bind to the CIM10K peptide arrays (8), raising the concern that clotting factors could influence the immunosignature through competition. The high correlation between serum and plasma collected from the same human donor indicates that any clotting factor protein in plasma samples binding to the array has a negligible effect. The interchangeability of serum and plasma is in agreement with the high correlation seen in ELISA titers of antibodies to mycobacterial antigens by others (25). This indicates that historical samples stored as either serum or plasma can be compared for their immunosignatures.
A second practical concern is whether samples could be sent through the mail for analysis. Application of antibodies collected as dried-blood spots on a 903 card for obtaining titers of antibody against a single antigen has been demonstrated by others (2, 3, 4–7, 11–13, 15, 22–24, 28, 29). The immunosignature assay is instead based on the ensemble binding properties of the repertoire of antibodies against 10,000 peptides. We have demonstrated using peptide-immunized mice that the immunosignatures obtained from both fresh samples and dried-blood spots are highly correlative. In each case, the immunizing peptide was recognized at a comparable rank in both the fresh and dried samples. Serum from mice immunized with one of the peptide conjugates demonstrated that dried-blood spots would yield a usable immunosignature when subjected to a simulated mailing environment. We have demonstrated that collection of blood samples on a 903 card stored at high temperatures then eluted yields similar to those for fresh blood. At both room temperature and an elevated temperature, the immunizing peptide was recognized at a comparable rank. The remainder of the immunosignature did lose some correlation to that for fresh samples over time but did not decrease below the variance in technical replicates. These observations are in line with previous reports of the use of dried blood to collect blood antibodies for ELISA in tropical and subtropical regions (2, 6, 9, 22). Addition of a protease inhibitor to the spotted sample reduced the degree of degradation, keeping the correlation at 2 weeks within the range of acceptability for technical replicates. Furthermore, influenza virus-infected mice were distinguished from naïve mice using the immunosignature from dried-blood spots. The ability to elute antibodies from the dried-blood spots was not limited to murine samples; the dried-blood spots from healthy human donors generated signatures which correlated to those for fresh samples and provided a range of reactivities on the array. Given the previously reported difference in the immunosignature between diseases, these results imply several applications. It may be possible to perform epidemiology studies with participants mailing blood samples.
Noninvasive collection of antibodies is the most attractive sample collection format to ensure compliance of self-sample collection for health monitoring. Saliva has been reported to contain both IgA and IgG. For human donors, the correlation of IgG between serum and saliva was low. However, a strong correlation of IgA in saliva and serum was seen. IgA works as well as IgG for immunosignatures and is the critical immunoglobulin for some diseases (15). Saliva thus has utility for IgA-based immunosignaturing.
This study expands the practical utility of the immunosignature assay. Two immediate benefits result from the ability to use multiple sample formats. First, a wealth of well-characterized archival samples which can be used to develop a database of disease immunosignatures exists. Second, the self-collection of blood or saliva will enhance prospective trials using immunosignaturing technology. Both features indicate that flexibility in sample preparation for immunosignature diagnostics could have advantages relative to other diagnostic technologies.
ACKNOWLEDGMENTS
We acknowledge John Lainson for printing the peptide microarrays. Min Hahn and Michael Ewing synthesized and purified the peptides. Phillip Stafford provided helpful advice on the analysis and provided critical reviews of the manuscript. Pattie Madjidi, Penny Gwynne, and Phillip Stafford assisted with collection of samples. The CIM10Kv2 arrays were run by the Center for Innovations in Medicine Peptide Array Core.
This work was supported by an Innovator Award to S.A.J. from the U.S. Department of Defense.
Stephen Albert Johnston declares ownership in HealthTell LLC, a diagnostic chip manufacturing company. Joseph Barten Legutki and Brian Andrew Chase declare no competing interests.
Footnotes
Published ahead of print 11 January 2012
REFERENCES
- 1. Ahmed FE. 2009. Sample preparation and fractionation for proteome analysis and cancer biomarker discovery by mass spectrometry. J. Separation Sci. 32: 771–798 [DOI] [PubMed] [Google Scholar]
- 2. Boillot F, Peeters M, Kosia A, Delaporte E. 1997. Prevalence of the human immunodeficiency virus among patients with tuberculosis in Sierra Leone, established from dried blood spots on filter paper. Int. J. Tuberc. Lung Dis. 1: 493–497 [PubMed] [Google Scholar]
- 3. Bradley JS, Graham S, Picchio GR, Vugia DJ, Kharrazi M. 2011. Prevalence of hepatitis C virus antibody in newborn infants in Southern California in 2003. Pediatr. Infect. Dis. J. 30: 618–620 [DOI] [PubMed] [Google Scholar]
- 4. Castro AC, Borges LG, Souza Rda S, Grudzinski M, D'Azevedo PA. 2008. Evaluation of the human immunodeficiency virus type 1 and 2 antibodies detection in dried whole blood spots (DBS) samples. Rev. Inst. Med. Trop. Sao Paulo 50: 151–156 [DOI] [PubMed] [Google Scholar]
- 5. De Cock L, et al. 2004. Detection of HCV antibodies in oral fluid. J. Virol. Methods 122: 179–183 [DOI] [PubMed] [Google Scholar]
- 6. El Mubarak HS, et al. 2004. Surveillance of measles in the Sudan using filter paper blood samples. J. Med. Virol. 73: 624–630 [DOI] [PubMed] [Google Scholar]
- 7. Fachiroh J, et al. 2008. Dried-blood sampling for Epstein-Barr virus immunoglobulin G (IgG) and IgA serology in nasopharyngeal carcinoma screening. J. Clin. Microbiol. 46: 1374–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Greving MP, et al. 2010. Thermodynamic additivity of sequence variations: an algorithm for creating high affinity peptides without large libraries or structural information. PLoS One 5: e15432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Habluetzel A, Esposito F, Lombardi S. 1989. Immunotechniques for epidemiology of malaria: appropriate tools for integration of primary health care with malaria research and control. Trans. R. Soc. Trop. Med. Hyg. 83 Suppl: 15–19 [DOI] [PubMed] [Google Scholar]
- 10. Halperin RF, Stafford P, Johnston SA. 2011. Exploring antibody recognition of sequence space through random-sequence peptide microarrays. Mol. Cell. Proteomics 10: M110.000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hardelid P, et al. 2008. Agreement of rubella IgG antibody measured in serum and dried blood spots using two commercial enzyme-linked immunosorbent assays. J. Med. Virol. 80: 360–364 [DOI] [PubMed] [Google Scholar]
- 12. Hardelid P, et al. 2008. Analysis of rubella antibody distribution from newborn dried blood spots using finite mixture models. Epidemiol. Infect. 136: 1698–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hutse V, et al. 2010. Oral fluid for the serological and molecular diagnosis of measles. Int. J. Infect. Dis. 14: e991–e997 [DOI] [PubMed] [Google Scholar]
- 14. Jansen HJ, Grenier D, Van der Hoeven JS. 1995. Characterization of immunoglobulin G-degrading proteases of Prevotella intermedia and Prevotella nigrescens. Oral Microbiol. Immunol. 10: 138–145 [DOI] [PubMed] [Google Scholar]
- 15. Judd A, et al. 2003. Evaluation of a modified commercial assay in detecting antibody to hepatitis C virus in oral fluids and dried blood spots. J. Med. Virol. 71: 49–55 [DOI] [PubMed] [Google Scholar]
- 16. Legutki JB, Magee DM, Stafford P, Johnston SA. 2010. A general method for characterization of humoral immunity induced by a vaccine or infection. Vaccine 28: 4529–4537 [DOI] [PubMed] [Google Scholar]
- 17. Luque-Garcia JL, Neubert TA. 2007. Sample preparation for serum/plasma profiling and biomarker identification by mass spectrometry. J. Chromatogr. A 1153: 259–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Molla A, Kagimoto T, Maeda H. 1988. Cleavage of immunoglobulin G (IgG) and IgA around the hinge region by proteases from Serratia marcescens. Infect. Immun. 56: 916–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Morales Betanzos C, et al. 2009. Bacterial glycoprofiling by using random sequence peptide microarrays. ChemBioChem 10: 877–888 [DOI] [PubMed] [Google Scholar]
- 20. Reddy MM, et al. 2011. Identification of candidate IgG biomarkers for Alzheimer's disease via combinatorial library screening. Cell 144: 132–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Restrepo L, Stafford P, Magee DM, Johnston SA. 2011. Application of immunosignatures to the assessment of Alzheimer's disease. Ann. Neurol. 70: 286–295 [DOI] [PubMed] [Google Scholar]
- 22. Rodriguez-Perez MA, Danis-Lozano R, Rodriguez MH, Bradley JE. 1999. Application of an enzyme-linked immunosorbent assay to detect antibodies to Onchocerca volvulus on filter-paper blood spots: effect of storage and temperature on antibody decay. Trans. R. Soc. Trop. Med. Hyg. 93: 523–524 [DOI] [PubMed] [Google Scholar]
- 23. Rollins N, Mzolo S, Moodley T, Esterhuizen T, van Rooyen H. 2009. Universal HIV testing of infants at immunization clinics: an acceptable and feasible approach for early infant diagnosis in high HIV prevalence settings. AIDS 23: 1851–1857 [DOI] [PubMed] [Google Scholar]
- 24. Samuel D, et al. 2003. Development of a measles specific IgM ELISA for use with serum and oral fluid samples using recombinant measles nucleoprotein produced in Saccharomyces cerevisiae. J. Clin. Virol. 28: 121–129 [DOI] [PubMed] [Google Scholar]
- 25. Siev M, et al. 2011. Correlation between serum and plasma antibody titers to mycobacterial antigens. Clin. Vaccine Immunol. 18: 173–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stafford P, Brun M. 2007. Three methods for optimization of cross-laboratory and cross-platform microarray expression data. Nucleic Acids Res. 35: e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stafford P, Johnston S. 2011. Microarray technology displays the complexities of the humoral immune response. Expert Rev. Mol. Diagn. 11: 5–8 [DOI] [PubMed] [Google Scholar]
- 28. Talukder Y, et al. 2005. Development and evaluation of varicella zoster virus ELISA for oral fluid suitable for epidemiological studies. J. Virol. Methods 128: 162–167 [DOI] [PubMed] [Google Scholar]
- 29. Vainio K, et al. 2008. Detection of measles- and mumps-specific IgG antibodies in paired serum and oral fluid samples from Norwegian conscripts. Eur. J. Clin. Microbiol. Infect. Dis. 27: 461–465 [DOI] [PubMed] [Google Scholar]